Background: Autophagy inhibition up-regulates p62/sequestosome-1, an ubiquitin-binding and multifunctional protein.
Results: Up-regulation of p62 promotes caspase-8 self-aggregation and activation by ABT-263, resulting in apoptosis that can be disrupted by p62 mutations at its functional domains. Caspase-8 aggregates co-localize with p62 and the autophagosome marker, LC3.
Conclusion: p62 up-regulation can mediate ABT-263-induced apoptosis via caspase-8 activation.
Significance: p62 can mediate apoptosis in the setting of autophagy inhibition.
Keywords: Apoptosis, Autophagy, Bcl-2 Family Proteins, Cancer Therapy, Caspase, BH3 Mimetic, p62/Sequestosome 1
Abstract
Autophagy and apoptosis regulate cancer cell viability in response to cytotoxic stress; however, their functional relationship remains unclear. p62/sequestosome 1 is a multifunctional protein and a signaling hub that shuttles ubiquitinated proteins to the lysosome during autophagy. Autophagy inhibition up-regulates p62, and prior data suggest that p62 may mediate apoptosis. Here, we demonstrate that p62 can regulate a caspase-8-dependent apoptosis in response to the BH3 mimetic agent, ABT-263. Up-regulation of p62 was shown to enhance ABT-263-induced caspase-8 activation that was Bax-dependent and resulted from mitochondrial amplification. Dependence upon caspase-8 was confirmed using caspase-8-deficient cells and by caspase-8 siRNA. Ectopic wild-type p62, but not p62 mutants with loss of ability to promote apoptosis, was shown to co-localize with caspase-8 and to promote its self-aggregation in ABT-263-treated cells, shown using a bimolecular fluorescence complementation assay. Endogenous p62 co-localized with caspase-8 in the presence of ABT-263 plus an autophagy inhibitor. Caspase-8 was shown to interact and co-localize with the autophagosome marker, LC3II. Knockdown of p62 attenuated binding between caspase-8 and LC3II, whereas p62 overexpression enhanced the co-localization of caspase-8 aggregates with LC3. LC3 knockdown did not affect interaction between caspase-8 and p62, suggesting that p62 may facilitate caspase-8 translocation to the autophagosomal membrane. A direct activator of caspase-8, i.e., TRAIL, alone or combined with ABT-263, induced caspase-8 aggregation and co-localization with p62 that was associated with a synergistic drug interaction. Together, these results demonstrate that up-regulation of p62 can mediate apoptosis via caspase-8 in the setting of autophagy inhibition.
Introduction
Macroautophagy is a catabolic process whereby cytoplasmic proteins and organelles are sequestered into autophagosomes and transported to lysosomes for degradation (1). During autophagy, the mammalian homolog of yeast Atg8, LC3, undergoes conversion from LC1 to LC3II and serves as a reliable marker of autophagosomes (1). Autophagy can function to maintain the survival of established tumor cells under stressful stimuli or, alternatively, may contribute to cell death depending upon the cellular context (2, 3). Autophagy inhibition has been shown to promote tumor cell apoptosis, thereby establishing autophagy as a therapeutic target (2). Inhibition of autophagy can enhance both the caspase-8-dependent and the mitochondria-mediated apoptotic pathways by a mechanism(s) that has yet to be defined (4, 5). Although the functional relationship between autophagy and apoptosis remains unclear, a molecular link may be p62/sequestosome 1 (herein referred to as p62) that is up-regulated by autophagy deficiency or inhibition (6). p62 is a multidomain protein that serves as a signaling hub through its ability to interact with key components of various signaling mechanisms (6). p62 serves as an ubiquitin-binding protein that chaperones protein aggregates to the lysosome or the proteasome (7) for degradation by autophagy (8–10). In addition to genetic means, autophagy can be disabled by the lysosomal inhibitor chloroquine (11) or by bortezomib, which blocks autophagy via a cathepsin-dependent mechanism resulting in up-regulation of p62 (12). p62 is rich in protein-interacting sequences and contains domains that include a protein-protein interaction module (PB1) that facilitates its oligomerization, a ubiquitin-associated (UBA)3 domain, and an LC3 interaction region that are essential for p62 function (8, 13, 14). In a prior study, we found that p62 overexpression can mediate a caspase-dependent apoptosis (16) by an as yet undefined mechanism.
The BH3 mimetic ABT-263 is a high affinity small molecule inhibitor of Bcl-2 and Bcl-xL that promotes apoptosis and shows anti-tumor activity both in vitro and in vivo (17). Recent evidence indicates that BH3 mimetic agents can induce autophagy in tumor cells mainly because of the release of Beclin 1 (which contains a BH3 domain) from Bcl-2 and Bcl-xL that stimulates a Beclin 1-dependent activation of the pro-autophagic lipid kinase, Vps34 (18–21). Given early phase data showing limited efficacy, strategies are needed to enhance the anti-tumor effects of BH3 mimetic agents, particularly in patients with solid tumors. BH3 mimetics are known to trigger mitochondria-mediated apoptosis; however, ABT-737 was reported to activate caspase-8 via mitochondrial membrane permeabilization (22) and was also shown to up-regulate death receptor 5 expression (23). A direct activator of caspase-8 is the anti-cancer drug TRAIL (TNF-related apoptosis-inducing ligand) that engages death receptors followed by recruitment of Fas-associated death domain (FADD) to form the death-inducing signaling complex (DISC) (24, 25). Because caspase-8 can cleave its substrate BID whose truncated form translocates to and interacts with the mitochondria (26), both apoptotic pathways converge on the mitochondria and can activate downstream effector caspases that execute cell death. Importantly, both apoptotic pathways are negatively regulated at the mitochondria by anti-apoptotic Bcl-2 proteins, which are antagonized by BH3 mimetics.
To test the hypothesis that p62 up-regulation is a molecular link between autophagy and apoptosis, we determined whether p62 can promote caspase-8 aggregation/activation and whether co-localization with LC3 occurs on autophagosomal membranes. Using wild-type and p62 functional mutants, we demonstrate the ability of p62 to regulate ABT-263-induced caspase-8 self-aggregation, activation, and co-localization in human colon carcinoma cell lines. Furthermore, ectopic p62 enhanced caspase-8 aggregation on the autophagosome shown by caspase-8 and LC3 interaction and co-localization. Together, these data indicate that p62 is a mediator of cross-talk between autophagy and apoptosis via caspase-8 activation, and suggest a mechanism by which autophagy inhibition can promote tumor cell death to enhance chemotherapeutic efficacy.
EXPERIMENTAL PROCEDURES
Cell Culture and Drugs
Human colon carcinoma cell lines (HCT116, DLD1), including HCT116 Bax−/− cells (gift from Dr. B. Volgelstein, Johns Hopkins University), were grown as a monolayer in RPMI 1640 (Invitrogen) supplemented with 10% (v/v) FBS and 1% antibiotic-antimycotic (Invitrogen). Caspase-8-deficient (I9.2), FADD-deficient (I2.1), and parental Jurkat cells were provided by Dr. S. H. Kaufmann (Mayo Clinic). The cells were treated with ABT-263, obtained from Dr. C. Yu (Roswell Park Institute, Buffalo, NY), in the presence or absence of chloroquine or Bafilomycin A1 (both from Sigma Co.) or bortezomib (LC labs, Woburn, MA). Cells were also treated with TRAIL (R&D Systems, St. Paul, MN) alone or combined with ABT-263. ABT-263 and Bafilomycin A1 were prepared as 10 mmol/liter and 10 μmol/liter stock solutions, respectively, in Me2SO and stored at −20 °C. A caspase-3 inhibitor, Z-DEVD-FMK (R&D Systems), was utilized.
Plasmid Constructs and Transfection of siRNA
The cDNA sequences of p62 was fused to the C terminus of mCherry, and then cloned into a lentiviral vector pCDH1-MCS-EF1-neomycin (System Biosciences, Mountain View, CA). 3tag-p62 and two mutants with mutations in UBA and PB1 domains were previously described (16). The p62 mutant with defective LC3 interaction region (D335A/D336A/D337A/W338A) (8) was generated by site-directed mutagenesis. Transfection of caspase-8 siRNA (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) into cells was performed using LipofectamineTM RNAiMax reagent (Invitrogen), as previously described (16).
Pseudo-typed Virus Production and Transduction
p62 shRNA, LC3 shRNA (with target sequence GAAGGCGCTTACAGCTCAA), and a nonsilencing shRNA, as well as mCherry constructs, were packaged into pseudo-typed lentivirus. The production of virus and transduction of target cells were performed utilizing a standard procedure that has been previously described (27). Retroviral overexpression of p62 and its mutants were also described previously (16).
Biomolecular Fluorescence Complementation (BiFC) Assay and Confocal Microscopy
HCT116 cells with stable expression of a lentiviral mCherry-p62 were sorted by flow cytometry to isolate cells with a high level of expression of mCherry fusion proteins. Human noncleavable caspase-8 (C360A) was cloned with pBiFC-VN155 (I152L) or pBiFC-VC155 to generate a fluorescent complementation pair (gift from Dr. H. Wang, Pennsylvania State University) that was transfected into HCT116 cells with stable expression of mCherry-p62 using Lipofectamine LTX (Invitrogen). Cells were then plated onto glass-bottomed microwell dishes (MatTek Corporation, Ashland, MA) and treated with drugs. Live images of the fluorescently tagged cells were captured using a confocal microscope with appropriate settings. Similar procedures were utilized for immunofluorescence staining and confocal microscopy of endogenous p62 and caspase-8, as previously described (16).
Apoptosis Assay
Apoptosis was analyzed by annexin V+ staining and then quantified as previously described (28). Briefly, cells were incubated with drugs, and at prespecified time points, the adherent cells were trypsinized and then combined with the floating cells. The cells were pelleted by centrifugation and then washed three times with cold PBS. The cells were incubated with annexin V+ (BD Biosciences, San Jose, CA), and the cell populations were labeled with the fluorescent dyes to enable their quantitation by flow cytometry.
MTS Assay and Calculation of a Combination Index (CI)
The effect of drugs upon tumor cell viability was determined by MTS assay as previously described (27). To test whether a drug combination exerts an additive or synergistic effect, the study drugs were combined using a fixed ratio and then added to cells for performance of the MTS assay. The means of triplicate experiments were utilized as the input to calculate the CI (29). A combination index that is less than 1 indicates synergy.
Caspase-8 Activity Assay
Caspase-8 activity was measured using a Caspase Glo 8 assay kit (Promega, Madison, WI) that utilizes luminogenic caspase-8 substrates, per the manufacturer's instructions. The activity was read by a SYNEGYMx multimode microplate reader (BioTek, Winooski, VT).
Immunoprecipitation and Immunoblotting
Protein samples were prepared in a lysis buffer (5 mmol/liter MgCl2, 137 mmol/liter KCl, 1 mmol/liter EDTA, 1 mmol/liter EGTA, 10 mmol/liter HEPES, pH 7.5; AppliChem, Boca Raton, FL) supplemented with a protease inhibitor mixture (Sigma). Protein samples were normalized using a nanodrop measurement method (Thermo Scientific, Franklin, MA) or a protein assay kit (Bio-Rad). Cell lysates were incubated with primary antibodies overnight at 4 °C. Immunocomplexes were captured with magnetic beads conjugated with protein A/G (Pierce) and washed three times in lysis buffer. Immunoprecipitated proteins were eluted with 2× LDS sample buffer (Invitrogen) and then loaded onto a 14% SDS-PAGE gel for protein separation that was followed by an electrical transfer onto a PVDF membrane (Bio-Rad). Immunoblotting was performed using a standard procedure that was previously described (16). All primary antibodies were obtained from Cell Signaling (Boston, MA), with the exception of antibodies against FLAG (Sigma), p62/SQSTM 1 (MBL, Woburn, MA), and β-tubulin (Sigma).
RESULTS
Autophagy Inhibition Enhances ABT-263-induced Apoptosis That Is Regulated by p62/Sequestosome-1
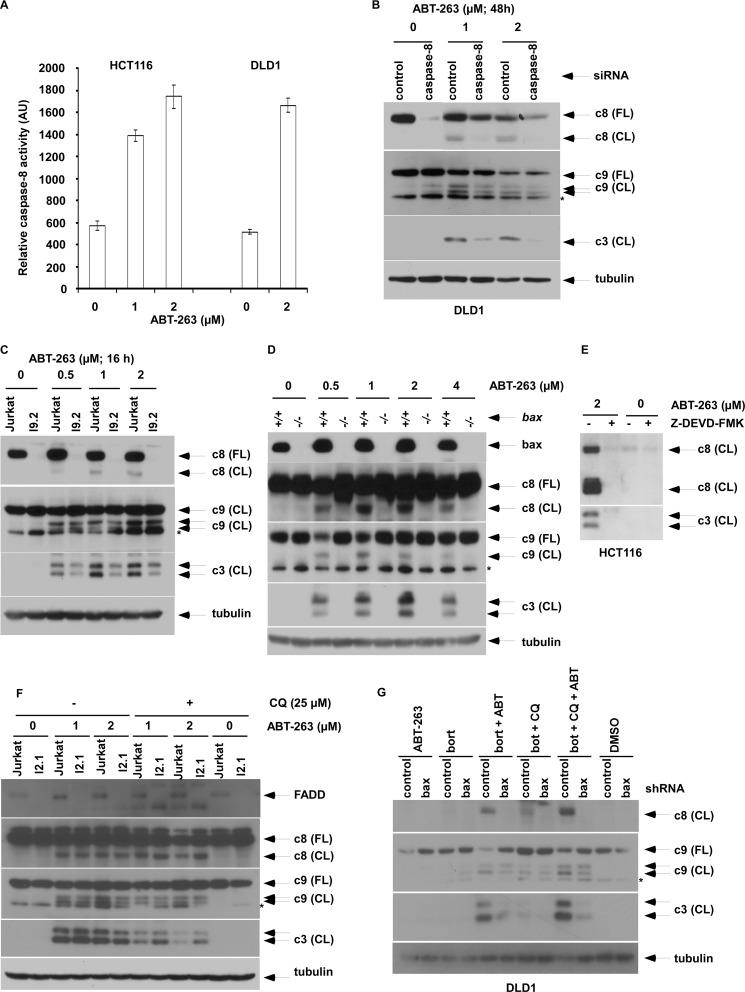
In addition to the ability of ABT-263 to induce apoptosis (30), we demonstrate that ABT-263 can induce autophagy, as shown by translocation of a fusion protein consisting of GFP with LC3 (GFP-LC3) from the cytosol to the autophagosomal membrane indicated by punctate localization using confocal microscopy (Fig. 1A). During autophagy, LC3 undergoes conversion from LC3I to its lipidated form LC3II (3), and therefore, GFP-LC3 serves as a fluorescent marker of autophagosomes (31). To exclude the possibility that punctate GFP-LC3 localization may derive from lysosomal inhibition, autophagic flux was examined. Using the lysosomal inhibitor bafilomycin A1, we observed increased conversion of LC3I to LC3II by ABT-263, confirming that this drug can induce autophagy in colon cancer cell lines.
FIGURE 1.
Modulation of p62/sequestosome 1 expression sensitizes colon cancer cell lines to undergo apoptosis by ABT-263. A, HCT116 cells were stably transfected with a lentiviral GFP-LC3B to fluorescently label autophagosomes. Autophagy induction is indicated by an increase in the number of LC3B puncta compared with a diffuse staining pattern in vehicle-treated (dimethyl sulfoxide, DMSO) cells (top). To confirm autophagy induction, autophagic flux was studied using bafilomycin (baf) A1 with analysis of the conversion of LC3I to its lipidated form, LC3II (bottom). Densitometry was performed to quantify protein bands and normalized to tubulin. B, effect of chloroquine (CQ, 25 μm; 48 h) pretreatment on caspase cleavage (CL) induced by ABT-263 (24 h) in whole cell lysates from HCT116 and DLD1 cell lines. Tubulin serves as control for protein loading. C, effect of autophagy inhibition by bortezomib (bort) alone (12) and combined with chloroquine on p62 expression. D, effect of bortezomib on ABT-263-induced (24 h) caspase cleavage in the presence of chloroquine. E, cells containing stable expression of p62 shRNA or control were treated with ABT-263 (24 h) alone or combined with chloroquine and/or bortezomib. LC3 expression and caspase cleavage products were analyzed by immunoblotting. *, nonspecific band.
In human cancer cells, autophagy can function to maintain cell survival or, in some contexts, may contribute to cell death (2, 3). The lysosomal autophagy inhibitor chloroquine was shown to increase the expression of p62 proteins (Fig. 1B) and to sensitize cells to ABT-263-induced caspase cleavage, with the effect being most evident for caspase-3 cleavage in both cell lines (Fig. 1B). Because p62 is degraded by autophagy, its level can be increased by autophagy inhibition. Specifically, autophagy blockade by chloroquine and/or by cathepsin-dependent inhibition using bortezomib (12) were shown to up-regulate p62 expression (Fig. 1C) and to enhance ABT-263-induced caspase cleavage (Fig. 1D). Dependence upon p62 in mediating apoptosis was shown in cells with suppression of p62 by a lentiviral shRNA that attenuated caspase cleavage induced by ABT-263 combined with autophagy inhibitors compared with control shRNA cells (Fig. 1E). These results indicate that p62 up-regulation can sensitize tumor cells to ABT-263-induced apoptosis.
To further examine the ability of p62 to regulate apoptosis, we utilized stable cells expressing a lentiviral shRNA against p62 and determined whether p62 is involved in chemotherapy-induced apoptosis. In cells treated with ABT-263 (48 h), we found that p62 knockdown can significantly attenuate apoptosis induction by ABT-263 or to a lesser extent by the topoisomerase 1 inhibitor, irinotecan (CPT-11), as shown by annexin V labeling (Fig. 2, A and B). Knockdown of p62 reduced ABT-263-induced caspase-8 and -3 cleavage in both HCT116 and DLD1 cell lines (Fig. 2, C and D). Furthermore, we demonstrate that p62 suppression can attenuate CPT-11-induced caspase-3 cleavage (Fig. 2E).
FIGURE 2.
Suppression of p62 attenuates drug-induced apoptosis. A and B, HCT116 cells with stable expression of lentiviral p62 shRNA or control (ctrl) were incubated with ABT-263 (48 h) (A) or CPT-11 (5 days) (B), and apoptosis was quantified by annexin V+ staining using flow cytometry. The mean values of triplicate experiments are shown; bars represent S.D. C and D, HCT116 (C) or DLD1 (D) cells with suppression of p62 by shRNA versus control were incubated with ABT-263 at the indicated doses for 48 h, and expression of p62 and caspase cleavage were analyzed by immunoblotting. E, effect of p62 suppression by shRNA on ABT-263-induced caspase-3 cleavage.
ABT-263 was shown to cleave caspase-8 (Fig. 2, C and D) and was also found to significantly activate caspase-8 using a luminogenic assay (Fig. 3A). Furthermore, the role of caspase-8 in ABT-263-induced apoptosis was shown using caspase-8 siRNA that attenuated caspase-3 cleavage (Fig. 3B). To confirm the critical role of caspase-8 as a mediator of ABT-263-induced apoptosis, we utilized caspase-8-deficient I9.2 cells where caspase-3 cleavage was attenuated compared with parental Jurkat cells (Fig. 3C). We then determined whether caspase-8 activation occurs upstream or downstream of the mitochondria given that BH3 mimetics promote mitochondrial apoptotic signaling. Using Bax−/− HCT116 cells, we found that Bax deficiency was able to block ABT-263-induced cleavage of caspase-8, -9, and -3 (Fig. 3D). Furthermore, a caspase-3 inhibitor was able to block ABT-263-induced caspase-8 cleavage (Fig. 3E), indicating that caspase-8 activation results from a caspase-3-mediated mitochondrial amplification loop. Caspase-8 has been shown to be activated by self-aggregation in response to death receptor stimulation upon ligand binding that enables its recruitment to the DISC (32). To exclude the possibility that caspase-8 activation is mediated by the death-inducing signaling complex, we utilized FADD-deficient Jurkat I2.1 cells and demonstrated that ABT-263 alone or combined with chloroquine can induce casapse-8, -9, and -3 cleavage that was unaffected by FADD deficiency (Fig. 3F). We also found that Bax deficiency can attenuate ABT-263-induced caspase-8 cleavage in the presence of autophagy inhibitors (Fig. 3G). Together, these data indicate that caspase-8 activation by ABT-263 occurs downstream of the mitochondrion and is mediated by a feedback loop.
FIGURE 3.
Caspase-8 activation contributes to ABT-263-induced apoptosis. A, HCT116 or DLD1 cells were incubated with ABT-263 (24 h) and caspase-8 activity was measured with a luminescent activity assay kit. The mean values are shown from assays performed in triplicate. Error bars, S.D. B and C, ability of ABT-263 to induce caspase cleavage was analyzed in cells with caspase-8 knockdown by siRNA (B) or in caspase-8-deficient I9.2 cells versus parental Jurkat cells (C). D, effect of wild-type (+/+) or Bax knock-out (−/−) status on caspase cleavage was determined in HCT116 cells treated with ABT-263 (48 h). E, effect of a caspase-3 inhibitor, Z-DEVD-FMK (50 μm), on ABT-263-induced caspase-8 and -3 cleavage is shown in HCT116 cells. F, FADD-deficient I2.1 cells versus parental Jurkat cells were treated with ABT-263 + chloroquine (CQ), and caspase cleavage was then examined. G, effect of Bax knockdown by shRNA in DLD-1 cells was determined in cells treated with ABT-263 (2 μm) alone or combined with chloroquine (25 μm) and/or bortezomib (bort, 20 nm).
p62 Interacts with Caspase-8 to Promote Apoptosis
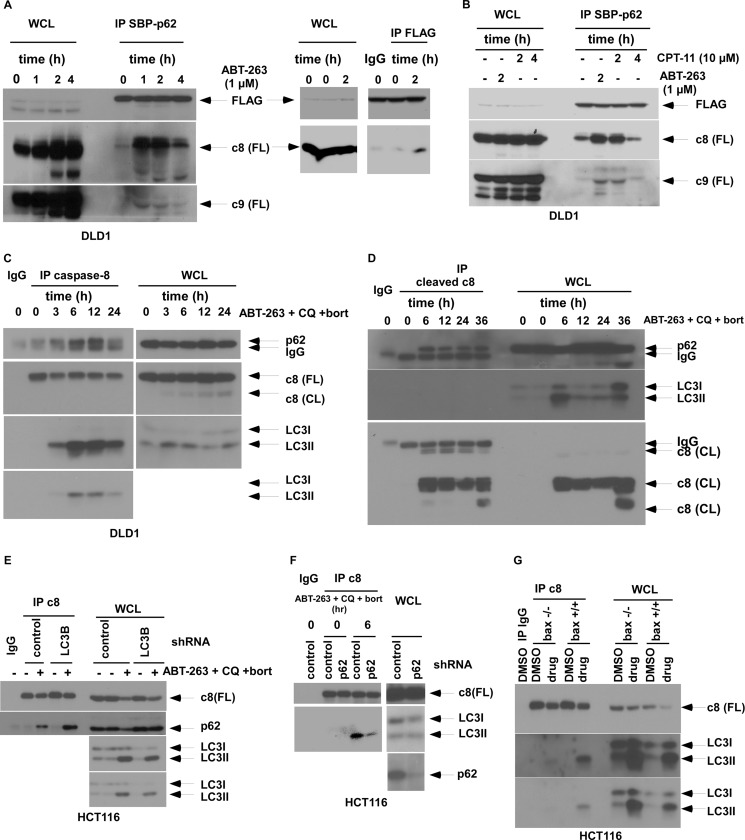
Evidence indicates that p62 can oligomerize important signaling proteins (33). We determined whether the regulation of apoptosis by p62 involves its interaction with proximal caspases that require oligomerization and activation (32, 34). Given that inhibition of autophagy can up-regulate p62 and can enhance chemotherapy-induced apoptosis, we generated cells with ectopic p62 expression to mimic p62 up-regulation. Using immunoprecipitation, we found that ABT-263 treatment promotes binding between ectopic p62 and caspase-8 to a greater extent than with caspase-9 (Fig. 4A). Similarly, irinotecan was shown to enhance p62 and caspase-8 binding (Fig. 4B), indicating that this interaction can be promoted by drugs with diverse mechanisms of action. In cells treated with ABT-263 in the presence of autophagy inhibitors, full-length caspase-8 was shown to interact with endogenous p62 and the autophagosomal membrane marker LC3II in a time-dependent manner (Fig. 4C). Furthermore, cleaved caspase-8 was also shown to interact with p62 in these cells (Fig. 4D), indicating the presence of activated caspase-8 within the p62-containing complex. To explore the relative sequence of association of caspase-8 with p62 and LC3, we utilized stable cells with knockdown of p62 or LC3. Although interaction of caspase-8 with p62 was unaffected by LC3 knockdown (Fig. 4E), suppression of p62 attenuated the association between caspase-8 and LC3II (Fig. 4F), suggesting that p62 may bind to caspase-8 with subsequent translocation of the complex to the autophagosomal membrane (LC3II). Bax deficiency abrogated the binding of caspase-8 to LC3II (Fig. 4G), indicating that this event occurs downstream of mitochondria and is consistent with the observed dependence of caspase-8 activation upon Bax (Fig. 3G).
FIGURE 4.
Regulation of drug-induced binding of caspase-8 to p62 or LC3II. A and B, DLD1 cells with stable expression of a retroviral p62 tagged with three tandem tags (3tag: s-tag, 2×FLAG, and streptavidin-binding protein (SBP)) were incubated with ABT-263 (A) or CPT-11 (B) at the indicated doses and times. SBP-p62 fusion proteins were captured using anti-FLAG antibody (A) or streptavidin-coated magnetic beads (A and B) in whole cell lysates (WCL). Caspase-8 or -9 protein and FLAG-p62 expression was probed by immunoblotting. C, DLD1 cells treated with ABT-263 (μm), chloroquine (CQ; 25 μm), and bortezomib (bort; 20 nm) (versus untreated cells) were subjected to immunoprecipitation by an anti-caspase-8 antibody and then probed for p62 or LC3II proteins by immunoblotting. D, immunoprecipitation of cleaved caspase-8 was performed in DLD1 cells treated with ABT-263 (2 μm), bortezomib (20 nm), and chloroquine (25 μm), and binding of p62 or LC3 was detected by immunoblotting. E, HCT116 cells containing a lentiviral control or LC3B shRNA were treated with ABT-263, chloroquine, and bortezomib (versus untreated cells), and immunoprecipitation of cell lysates for caspase-8 was performed and probed for p62. F, HCT116 cells with stable expression of a lentiviral p62 or control shRNA were treated with drug (ABT-263, chloroquine, and bortezomib) or vehicle. Caspase-8 was then immunoprecipitated and probed for LC3II. G, immunoprecipitation of caspase-8 was performed in wild-type (+/+) or Bax knock-out (−/−) HCT116 cells and probed for LC3II.
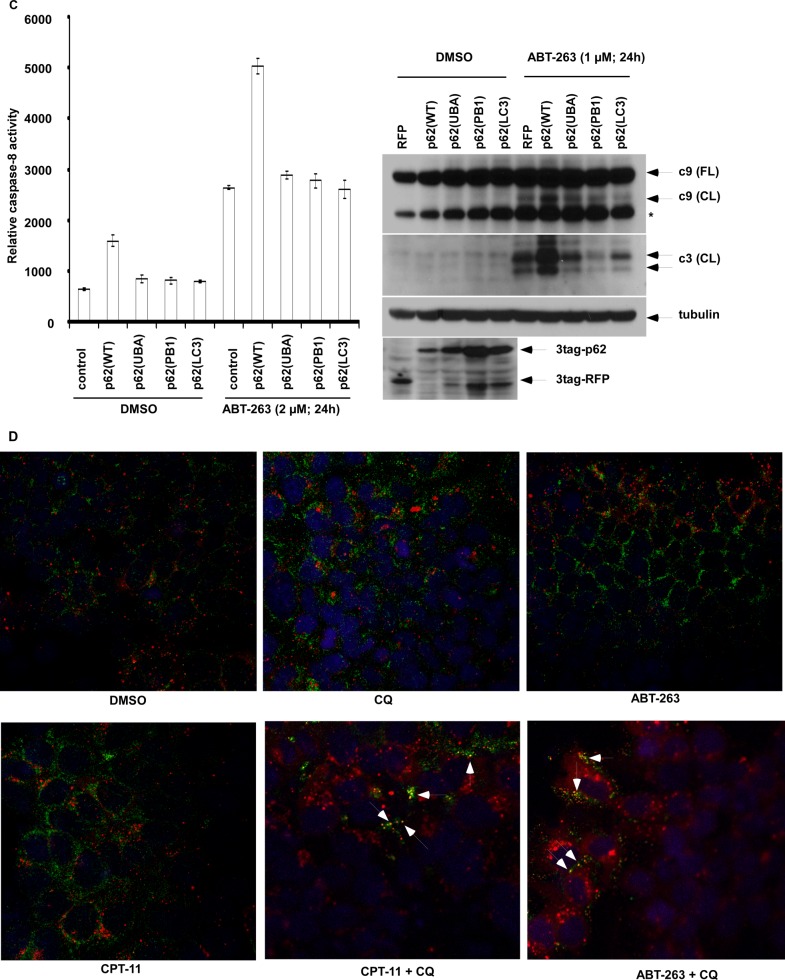
p62 Co-localizes with Caspase-8 Aggregates and Promotes Caspase-8 Activation That Is Disrupted by p62 Mutation
To determine the mechanism by which ectopic p62 can enhance apoptosis, we generated p62 mutants using site-directed mutagenesis to disable p62 function. Given that PB1 and UBA domains are essential for p62 function (13, 14), mutation at UBA:I431A and PB1:D69A results in loss of the ability to form p62 foci, as previously shown by our laboratory (16). We also generated cells with a p62 mutant that displays loss of ability to bind to LC3 (8). Although chloroquine-induced co-localization of wild-type p62 foci with LC3 foci, all the three p62 mutants failed to form a complex with LC3 as shown by fluorescence confocal microscopy (Fig. 5A). We determined whether ABT-263 can induce caspase-8 aggregation and co-localize with p62 using a BiFC assay (35). To avoid cell death induction caused by overexpression of caspase-8, we utilized a catalytically inactive mutant (C360A) of pro-caspase-8 (36) that was fused to the split Venus proteins, Venus N1–173 (VN) and Venus C155–239 (VC). These constructs were transfected into HCT116 cells that had been stably transfected with a lentiviral mCherry-p62 construct. ABT-263 treatment was shown to induce caspase-8 aggregation, indicated by the punctate green fluorescence of Venus (Fig. 5B), which occurs when caspase-8 self-oligomerization pulls the VN and VC together and does not occur in vehicle-treated cells. The caspase-8 aggregates were shown to co-localize with p62 cellular foci (Fig. 5B), suggesting that p62 enables caspase-8 aggregation in ABT-263-treated cells. In contrast, the three p62 mutants failed to co-localize with caspase-8 and to promote its aggregation (Fig. 5B). In cells with ectopic wild-type but not mutant p62, treatment with ABT-263 was shown to enhance caspase-8 activation and cleavage of caspase-9 and -3 (Fig. 5C). To determine whether up-regulation of endogenous p62 can aggregate caspase-8, we treated cells with chloroquine combined with ABT-263 or CPT-11, which were shown to induce co-localization of caspase-8 with p62 (Fig. 5D).
FIGURE 5.
p62/sequestosome 1 co-localizes with caspase-8 aggregates and promotes caspase-8 activation that is disrupted by p62 mutation in ABT-263-treated cells. A, DLD1 cells stably expressing p62 variants (wild-type or mutations in UBA, PB1, and LC3-binding domains) were treated with chloroquine (CQ, 25 μm), and cells were stained with anti-FLAG or anti-LC3 antibodies followed by labeling with fluorescence dye-conjugated secondary antibodies. Red, anti-FLAG; green, anti-LC3; yellow punctae indicate co-localization. The nuclei were counterstained with DAPI (blue). B, HCT116 cells stably expressing a lentiviral mCherry-p62 were transiently transfected with a catalytically inactive procaspase-8 (C360A) fused to Venus N-terminal (VN) or C-terminal (VC). The cells were treated with ABT-263, and localization of mCherry-p62 variants (wild type or mutations in UBA, PB1 domains) and caspase-8-Venus was determined by fluorescence confocal microscopy. Co-localization is shown in merged images (gold color). C, HCT116 cells with stable expression of retroviral p62 variants (wild type or mutations in UBA, PB1 and LC3-binding domains) were treated with ABT-263, and activation of caspase-8 (left) and cleavage of caspase-9 and -3 (right) were determined. Red fluorescence protein (RFP) serves as a control. *, nonspecific band. D, DLD1 cells were treated with ABT-263 (2 μm) or CPT-11 (10 μm) alone or combined with chloroquine (25 μm) for 12 h, and immunofluorescence staining of p62 and caspase-8 was performed. Red, p62; green, caspase-8; nuclei are counterstained with DAPI (blue). Co-localization of caspase-8 with p62 is shown by yellow punctae. DMSO, dimethyl sulfoxide.
p62-mediated Caspase-8 Aggregation Occurs on the Autophagosome
We determined whether co-localization of p62 with caspase-8 aggregates occurs on the autophagosome. To address this issue, we ectopically expressed mCherry-LC3 in p62-overexpressed cells and observed caspase-8 aggregation using the BiFC assay. As expected, ectopic p62 enhanced caspase-8 aggregation in ABT-263-treated cells, as compared with empty vector cells (Fig. 6). Moreover, ectopic p62 expression enhanced the co-localization of caspase-8 aggregates with mCherry-LC3, suggesting that these events occur on autophagosomal membranes (Fig. 6).
FIGURE 6.
Ectopic p62 promotes co-localization of caspase-8 aggregates with LC3 at the autophagosome. HCT116 cells with stable expression of mCherry-LC3 were stably transfected with a retroviral 3tag-p62 (versus empty control (EV)) and transiently transfected with procaspase-8 (C360A)-VN and procaspase-8 (C360A)-VC. The cells were then treated with ABT-263 and imaged to determine the localization of fluorescent caspase-8-Venus and mCherry-LC3 by confocal microscopy. Co-localization is shown in the merged images.
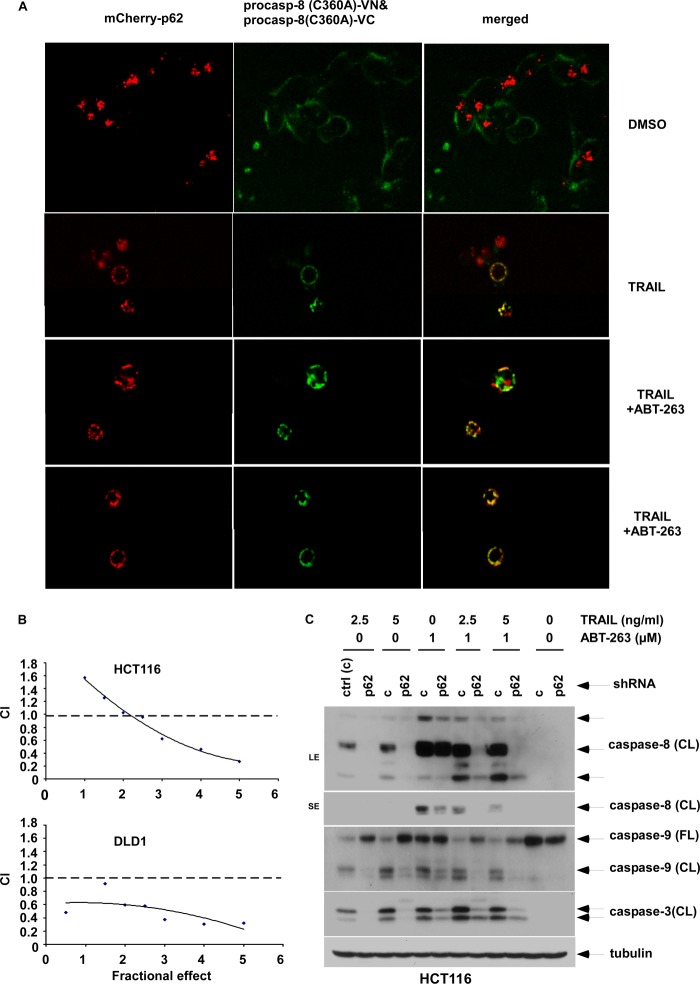
Because ABT-263 was shown to activate caspase-8 indirectly through a feedback loop, we utilized TRAIL as a direct activator of caspase-8 to determine whether this drug can cooperatively enhance p62-mediated caspase-8 activation and apoptosis when combined with ABT-263. TRAIL treatment alone or combined with ABT-263 was shown to promote caspase-8 aggregation and promoted its co-localization with p62 foci compared with vehicle-treated cells (Fig. 7A). Furthermore, the combination of TRAIL and ABT-263 synergistically reduced cell viability in both HCT116 and DLD1 colon cancer cell lines as shown by calculation of the CI using the method of Chou and Talalay (29) (Fig. 7B). We also found that the drug combination was dependent upon p62 in that caspase cleavage was significantly attenuated in p62 knockdown cells (Fig. 7C). These data confirm the important role of p62-regulated caspase-8 aggregation/activation in mediating ABT-263-induced apoptosis.
FIGURE 7.
p62 regulates caspase-8 aggregation and apoptosis by TRAIL alone or combined with ABT-263. A, HCT116 cells containing stable expression of mCherry-p62 were transfected with a VN- or VC-labeled procaspase-8 (C360A) pair (as per Fig. 5) and were treated with TRAIL alone or combined with ABT-263. Localization of mCherry-p62 and caspase-8-Venus were determined by fluorescence confocal microscopy. B, HCT116 and DLD1 cells were treated with TRAIL and ABT-263 for 24 h at a fixed ratio, and cell viability was determined using MTS assay. The CI was calculated, as described under “Experimental Procedures,” and an isobologram was plotted. A CI value <1 indicates a synergistic interaction. C, HCT116 cells with stable expression of p62 versus control (ctrl) shRNA were incubated with TRAIL (10 ng/ml) ±ABT-263 (2 μm) at the indicated doses for 8 h. Expression of caspase cleavage was analyzed by immunoblotting. DMSO, dimethyl sulfoxide.
DISCUSSION
We studied the effect of up-regulation of p62 expression that was hypothesized to mediate cross-talk between autophagy and apoptosis. We demonstrate that ABT-263 can induce both autophagy and apoptosis and observed that the addition of autophagy inhibitors to up-regulate p62 can significantly enhance apoptosis that was attenuated in p62 knockdown cells. Given that p62 can function as a signaling hub by aggregating important molecules (33), we examined its ability to bind and aggregate proximal caspases whose self-aggregation is required for their activation and autocatalytic processing, in contrast to effector caspases that depend strictly on proteolytic cleavage for activation (37). We found that ectopic p62 preferentially binds to caspase-8 versus caspase-9 in cells treated with ABT-263 or CPT-11, and that both ectopic and endogenous p62 co-localize with caspase-8. The importance of caspase-8 activation in p62-mediated apoptosis was shown using caspase-8-deficient or knockdown cells where downstream effector caspase cleavage was markedly attenuated. We demonstrate that up-regulated p62 can physically interact with caspase-8 to promote caspase-8 self-aggregation and activation in response to ABT-263. Furthermore, p62 binds to the full-length and catalytically active forms of caspase-8. Consistent with our findings, p62 has also been shown to mediate caspase-8 oligomerization and activation in response to a pan sphingokinase inhibitor (35). We found that ectopic p62 expression promotes the co-localization of caspase-8 with LC3 and that caspase-8 interacts with LC3II in ABT-263-treated cells where autophagy was inhibited, suggesting that caspase-8 aggregation occurs on the autophagosomal membrane. This process was shown to require p62, because suppression of p62 attenuated the interaction between caspase-8 and LC3II, whereas LC3 knockdown had no effect on the caspase-8 and p62 interaction. We speculate that a potential sequence of events may include the binding of p62 to initiate caspase-8 aggregation followed by an association of p62 with LC3II on the autophagosomal membrane.
p62 is rich in protein-interacting sequences, and its signaling mechanisms are mediated by functional domains. These domains include UBA that binds ubiquitinated proteins, PB1 that mediates dimerization/oligomerization, and an LC3 interaction region (8, 13, 14). In contrast to wild-type p62, mutations at p62 functional domains (UBA and PB1) that are defective in p62 foci formation or in LC3 binding were shown to markedly attenuate caspase-8 aggregation/activation as well as the ability of p62 to promote chemotherapy-induced apoptosis. These data indicate that p62 functional domains (UBA, PB1, and LC3 interaction region) are required for p62 to activate caspase-8.
Activation of caspase-8 by ABT-263 was shown to be dependent upon pro-apoptotic Bax, because Bax deficiency blocked ABT-263-induced caspase-8 cleavage. This finding and the ability of caspase-3 inhibition to block caspase-8 cleavage indicate that ABT-263 induces caspase-8 activation via a mitochondrial feedback amplification loop. Similarly, epirubicin and paclitaxel-induced caspase-8 activation was shown to be independent of CD95/Fas receptor-ligand interaction and to occur downstream of caspase-3 (38). We found that FADD-deficient cells had no effect on ABT-263-induced caspase-8 activation, confirming that the observed events were independent of death receptor-induced signaling. In this regard, caspase-8 activation can occur independently of the death receptors (35, 39) where the critical step is its oligomerization that facilitates autoactivation through self-cleavage (32). A recent study found that ABT-737 can activate caspase-8 independently of the extrinsic apoptosis pathway via mitochondrial membrane permeabilization in human platelets (22). Caspase-8 has been reported to function on mitochondrial membranes through its interaction with cardiolipin (40) that provides an activating platform for caspase-8 translocation to the mitochondrial membrane where it oligomerizes and is further activated to enable an apoptotic response (40). We sought to confirm the observed p62-dependent mechanism of caspase-8 activation using TRAIL, which is a direct activator of caspase-8 via DISC formation (32). As with ABT-263, TRAIL treatment promoted caspase-8 self-aggregation that co-localized with p62 foci by confocal microscopy and was shown to synergistically enhance cell death. The ability of p62 knockdown to attenuate caspase-8 activation induced by TRAIL, ABT-263, or their combination, underscores the critical role played by p62 in the regulation of caspase-8 activation.
Because autophagy inhibition results in the up-regulation of p62 proteins that are normally degraded by autophagy (41), our data provide a mechanism by which autophagy inhibitors, including chloroquine (11, 16, 42–44) or bortezomib, can sensitize cancer cells to chemotherapy-induced apoptosis. In addition to chloroquine, recent data indicate that bortezomib can also inhibit autophagy via a cathepsin-dependent mechanism that results in p62 up-regulation (12). Our data indicate that p62 up-regulation is critical to its ability to aggregate caspase-8 on the autophagosome (LC3II) in ABT-263-treated cells. Given that p62 may be vulnerable to degradation on autophagosomes, we confine our interpretation of its interaction with caspase-8 on autophagosomes to the setting of autophagy inhibition. Strategies to directly up-regulate p62 expression have the potential to chemosensitize tumor cells, and a recent report found that a farnesoid X receptor ligand can transcriptionally up-regulate p62 mRNA and protein expression in vivo (15).
In conclusion, our data demonstrate that up-regulation of p62 in the setting of autophagy inhibition promotes a caspase-8-dependent apoptosis by ABT-263. The mechanism of this effect involves co-localization of p62 with caspase-8 to promote its self-aggregation/activation and apoptosis that can be disrupted by p62 mutations at its functional domains. p62 overexpression promotes the co-localization of caspase-8 with LC3, suggesting that these events occur on the autophagosome. Because autophagy deficiency or inhibition accumulates p62, our findings establish p62 as a mediator of cross-talk between autophagy and apoptosis and provide a mechanism by which autophagy inhibition can promote tumor cell death.
Acknowledgments
We express appreciation to Deborah Frank for very capable secretarial assistance in the preparation of this manuscript and to Dr. Scott H. Kaufmann for software used to calculate the combination index.
This work was supported, in whole or in part, by National Institutes of Health Grants K05CA142885, R01CA132991, and R01CA113681 (to F. A. S.). This work was also supported by Mayo Cancer Center Core Grant CA15083 and Mayo Clinic Center for Cell Signaling in Gastroenterology Grant P30DK084567.
- UBA
- ubiquitin-associated
- FADD
- Fas-associated death domain
- BiFC
- biomolecular fluorescence complementation
- VN
- Venus N1–173
- VC
- Venus C155–239
- CI
- combination index
- DISC
- death-inducing signaling complex.
REFERENCES
- 1. Klionsky D. J., Abdalla F. C., et al. (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. White E., DiPaola R. S. (2009) The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 15, 5308–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Denton D., Nicolson S., Kumar S. (2012) Cell death by autophagy. Facts and apparent artefacts. Cell Death Differ. 19, 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han J., Hou W., Goldstein L. A., Lu C., Stolz D. B., Yin X. M., Rabinowich H. (2008) Involvement of protective autophagy in TRAIL resistance of apoptosis-defective tumor cells. J. Biol. Chem. 283, 19665–19677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang S., Sinicrope F. A. (2010) Celecoxib-induced apoptosis is enhanced by ABT-737 and by inhibition of autophagy in human colorectal cancer cells. Autophagy 6, 256–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Moscat J., Diaz-Meco M. T. (2009) p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137, 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seibenhener M. L., Babu J. R., Geetha T., Wong H. C., Krishna N. R., Wooten M. W. (2004) Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol. Cell Biol. 24, 8055–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pankiv S., Clausen T. H., Lamark T., Brech A., Bruun J. A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 [DOI] [PubMed] [Google Scholar]
- 9. Parkhitko A., Myachina F., Morrison T. A., Hindi K. M., Auricchio N., Karbowniczek M., Wu J. J., Finkel T., Kwiatkowski D. J., Yu J. J., Henske E. P. (2011) Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. Proc. Natl. Acad. Sci. U.S.A. 108, 12455–12460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ni H. M., Boggess N., McGill M. R., Lebofsky M., Borude P., Apte U., Jaeschke H., Ding W. X. (2012) Liver-specific loss of Atg5 causes persistent activation of Nrf2 and protects against acetaminophen-induced liver injury. Toxicol. Sci. 127, 438–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Amaravadi R. K., Lippincott-Schwartz J., Yin X. M., Weiss W. A., Takebe N., Timmer W., DiPaola R. S., Lotze M. T., White E. (2011) Principles and current strategies for targeting autophagy for cancer treatment. Clin. Cancer Res. 17, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Periyasamy-Thandavan S., Jackson W. H., Samaddar J. S., Erickson B., Barrett J. R., Raney L., Gopal E., Ganapathy V., Hill W. D., Bhalla K. N., Schoenlein P. V. (2010) Bortezomib blocks the catabolic process of autophagy via a cathepsin-dependent mechanism, affects endoplasmic reticulum stress and induces caspase-dependent cell death in antiestrogen-sensitive and resistant ER+ breast cancer cells. Autophagy 6, 19–35 [DOI] [PubMed] [Google Scholar]
- 13. Moscat J., Diaz-Meco M. T., Albert A., Campuzano S. (2006) Cell signaling and function organized by PB1 domain interactions. Mol. Cell 23, 631–640 [DOI] [PubMed] [Google Scholar]
- 14. Moscat J., Diaz-Meco M. T., Wooten M. W. (2007) Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci 32, 95–100 [DOI] [PubMed] [Google Scholar]
- 15. Williams J. A., Thomas A. M., Li G., Kong B., Zhan L., Inaba Y., Xie W., Ding W. X., Guo G. L. (2012) Tissue specific induction of p62/Sqstm1 by farnesoid X receptor. PLoS One 7, e43961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang S., Yang Z. J., Yu C., Sinicrope F. A. (2011) Inhibition of mTOR kinase by AZD8055 can antagonize chemotherapy-induced cell death through autophagy induction and down-regulation of p62/sequestosome 1. J. Biol. Chem. 286, 40002–40012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tse C., Shoemaker A. R., Adickes J., Anderson M. G., Chen J., Jin S., Johnson E. F., Marsh K. C., Mitten M. J., Nimmer P., Roberts L., Tahir S. K., Xiao Y., Yang X., Zhang H., Fesik S., Rosenberg S. H., Elmore S. W. (2008) ABT-263. A potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 68, 3421–3428 [DOI] [PubMed] [Google Scholar]
- 18. Sinha S., Levine B. (2008) The autophagy effector Beclin 1. A novel BH3-only protein. Oncogene 27, S137–S148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 20. Malik S. A., Orhon I., Morselli E., Criollo A., Shen S., Mariño G., BenYounes A., Bénit P., Rustin P., Maiuri M. C., Kroemer G. (2011) BH3 mimetics activate multiple pro-autophagic pathways. Oncogene 30, 3918–3929 [DOI] [PubMed] [Google Scholar]
- 21. Maiuri M. C., Criollo A., Tasdemir E., Vicencio J. M., Tajeddine N., Hickman J. A., Geneste O., Kroemer G. (2007) BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-XL. Autophagy 3, 374–376 [DOI] [PubMed] [Google Scholar]
- 22. Mutlu A., Gyulkhandanyan A. V., Freedman J., Leytin V. (2012) Activation of caspases-9, -3 and -8 in human platelets triggered by BH3-only mimetic ABT-737 and calcium ionophore A23187. Caspase-8 is activated via bypass of the death receptors. Br. J. Haematol. 159, 565–571 [DOI] [PubMed] [Google Scholar]
- 23. Song J. H., Kandasamy K., Kraft A. S. (2008) ABT-737 induces expression of the death receptor 5 and sensitizes human cancer cells to TRAIL-induced apoptosis. J. Biol. Chem. 283, 25003–25013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fulda S., Debatin K. M. (2006) Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene 25, 4798–4811 [DOI] [PubMed] [Google Scholar]
- 25. Li J., Yuan J. (2008) Caspases in apoptosis and beyond. Oncogene 27, 6194–6206 [DOI] [PubMed] [Google Scholar]
- 26. Li H., Zhu H., Xu C. J., Yuan J. (1998) Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94, 491–501 [DOI] [PubMed] [Google Scholar]
- 27. Huang S., Sinicrope F. A. (2008) BH3 mimetic ABT-737 potentiates TRAIL-mediated apoptotic signaling by unsequestering Bim and Bak in human pancreatic cancer cells. Cancer Res. 68, 2944–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang S., Okumura K., Sinicrope F. A. (2009) BH3 mimetic obatoclax enhances TRAIL-mediated apoptosis in human pancreatic cancer cells. Clin. Cancer Res. 15, 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chou T. C., Talalay P. (1984) Quantitative analysis of dose-effect relationships. The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul 22, 27–55 [DOI] [PubMed] [Google Scholar]
- 30. Adams J. M., Cory S. (2007) The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene 26, 1324–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jin Z., Li Y., Pitti R., Lawrence D., Pham V. C., Lill J. R., Ashkenazi A. (2009) Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 137, 721–735 [DOI] [PubMed] [Google Scholar]
- 33. Moscat J., Diaz-Meco M. T. (2009) To aggregate or not to aggregate? A new role for p62. EMBO Rep. 10, 804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Riedl S. J., Salvesen G. S. (2007) The apoptosome. Signalling platform of cell death. Nat. Rev. Mol. Cell Biol. 8, 405–413 [DOI] [PubMed] [Google Scholar]
- 35. Young M. M., Takahashi Y., Khan O., Park S., Hori T., Yun J., Sharma A. K., Amin S., Hu C. D., Zhang J., Kester M., Wang H. G. (2012) Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J. Biol. Chem. 287, 12455–12468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boldin M. P., Goncharov T. M., Goltsev Y. V., Wallach D. (1996) Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 85, 803–815 [DOI] [PubMed] [Google Scholar]
- 37. Boatright K. M., Salvesen G. S. (2003) Mechanisms of caspase activation. Curr. Opin. Cell Biol. 15, 725–731 [DOI] [PubMed] [Google Scholar]
- 38. Wieder T., Essmann F., Prokop A., Schmelz K., Schulze-Osthoff K., Beyaert R., Dörken B., Daniel P. T. (2001) Activation of caspase-8 in drug-induced apoptosis of B-lymphoid cells is independent of CD95/Fas receptor-ligand interaction and occurs downstream of caspase-3. Blood 97, 1378–1387 [DOI] [PubMed] [Google Scholar]
- 39. Pan J. A., Ullman E., Dou Z., Zong W. X. (2011) Inhibition of protein degradation induces apoptosis through a microtubule-associated protein 1 light chain 3-mediated activation of caspase-8 at intracellular membranes. Mol. Cell Biol. 31, 3158–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gonzalvez F., Schug Z. T., Houtkooper R. H., MacKenzie E. D., Brooks D. G., Wanders R. J., Petit P. X., Vaz F. M., Gottlieb E. (2008) Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J. Cell Biol. 183, 681–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bjørkøy G., Lamark T., Brech A., Outzen H., Perander M., Overvatn A., Stenmark H., Johansen T. (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bristol M. L., Emery S. M., Maycotte P., Thorburn A., Chakradeo S., Gewirtz D. A. (2013) Autophagy inhibition for chemosensitization and radiosensitization in cancer. Do the preclinical data support this therapeutic strategy? J. Pharmacol. Exp. Ther. 344, 544–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lamoureux F., Thomas C., Crafter C., Kumano M., Zhang F., Davies B. R., Gleave M. E., Zoubeidi A. (2013) Blocked autophagy using lysosomotropic agents sensitizes resistant prostate tumor cells to the novel Akt inhibitor AZD5363. Clin. Cancer Res. 19, 833–844 [DOI] [PubMed] [Google Scholar]
- 44. Bellodi C., Lidonnici M. R., Hamilton A., Helgason G. V., Soliera A. R., Ronchetti M., Galavotti S., Young K. W., Selmi T., Yacobi R., Van Etten R. A., Donato N., Hunter A., Dinsdale D., Tirrò E., Vigneri P., Nicotera P., Dyer M. J., Holyoake T., Salomoni P., Calabretta B. (2009) Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J. Clin. Invest. 119, 1109–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]