Background: New regulators of the ubiquitin-proteasome system (UPS) were sought in yeast.
Results: Cuz1 (Cdc48-associated ubiquitin-like/zinc finger protein-1) interacts with the Cdc48/p97 ATPase and promotes endoplasmic reticulum-associated degradation.
Conclusion: Cuz1 is a highly conserved Cdc48 cofactor that also binds proteasomes, especially in cells exposed to arsenite.
Significance: This first characterization of the Cuz1 protein family links it to specific Cdc48/p97 complexes.
Keywords: ATPases, ER-associated Degradation, Proteasome, Ubiquitin, Yeast, Zinc Finger, Cdc48, Cuz1
Abstract
Regulated protein degradation mediated by the ubiquitin-proteasome system (UPS) is critical to eukaryotic protein homeostasis. Often vital to degradation of protein substrates is their disassembly, unfolding, or extraction from membranes. These processes are catalyzed by the conserved AAA-ATPase Cdc48 (also known as p97). Here we characterize the Cuz1 protein (Cdc48-associated UBL/zinc finger protein-1), encoded by a previously uncharacterized arsenite-inducible gene in budding yeast. Cuz1, like its human ortholog ZFAND1, has both an AN1-type zinc finger (Zf_AN1) and a divergent ubiquitin-like domain (UBL). We show that Cuz1 modulates Cdc48 function in the UPS. The two proteins directly interact, and the Cuz1 UBL, but not Zf_AN1, is necessary for binding to the Cdc48 N-terminal domain. Cuz1 also associates, albeit more weakly, with the proteasome, and the UBL is dispensable for this interaction. Cuz1-proteasome interaction is strongly enhanced by exposure of cells to the environmental toxin arsenite, and in a proteasome mutant, loss of Cuz1 enhances arsenite sensitivity. Whereas loss of Cuz1 alone causes only minor UPS degradation defects, its combination with mutations in the Cdc48Npl4-Ufd1 complex leads to much greater impairment. Cuz1 helps limit the accumulation of ubiquitin conjugates on both the proteasome and Cdc48, suggesting a possible role in the transfer of ubiquitylated substrates from Cdc48 to the proteasome or in their release from these complexes.
Introduction
The ubiquitin-proteasome system (UPS)6 is an elaborate network of enzymes and proteins that ensures the specific and timely degradation of proteins in eukaryotic cells (1). Substrate proteins include both regulatory proteins that must be inactivated for proper cell and organismal function and quality control substrates such as misfolded or misassembled proteins (1). Attachment of ubiquitin polymers to substrates is generally required for their efficient recognition by the 26 S proteasome, a 2.6-MDa complex that ultimately degrades the substrate into short peptides and recycles ubiquitin (2).
The 26 S proteasome is composed of two major subcomplexes: the 20 S proteasome core particle (CP) and the 19 S regulatory particle (RP) (3, 4). The RP recognizes polyubiquitylated substrates, and uses a heterohexameric ring of AAA+ ATPases to unfold substrates and translocate them into the central chamber of the CP where the proteolytic sites are located. Additional subunits in the RP function either as intrinsic receptors for polyubiquitin via their ubiquitin-binding domains or as binding sites for more mobile extrinsic ubiquitin receptors or shuttle factors such as Rad23 (5).
Much of the machinery of the UPS has been identified, but key parts of the system remain poorly understood. Prominent among the open questions is the mechanism by which polyubiquitylated proteins are released by E3 ligases and transferred to ubiquitin receptors and the proteasome for degradation. The highly conserved, multifunctional Cdc48 protein, also called valosin-containing protein or p97 in mammals (6), is an AAA-ATPase that forms a homohexameric ring (7). It has been shown to be required for UPS-mediated protein degradation of several classes of substrates, including those degraded by the endoplasmic reticulum-associated degradation (ERAD) and ubiquitin-fusion degradation (UFD) branches of the UPS (8–11). The general mechanistic function of Cdc48 appears to be that of a “segregase,” a protein that uses the energy of ATP hydrolysis to unfold or disassemble protein complexes or to extract proteins from membranes (12, 13).
Cdc48 consists of a globular N-terminal domain; two type II AAA (ATPases associated with diverse cellular activities) ATPase domains called D1 and D2, which share 40% sequence identity; and a disordered C-terminal tail (14, 15). Cdc48 interacts with a plethora of cofactors, and these proteins mediate the wide range of functions in which Cdc48 has been implicated. Members of the largest family of Cdc48 cofactors are related by the UBX (ubiquitin regulatory X) domain; there are seven UBX proteins in Saccharomyces cerevisiae. Structural determination of the UBX domain, which binds specifically to the Cdc48 N-terminal domain (16), revealed a β-grasp fold similar to that of ubiquitin (17).
The first characterized UBX domain-containing Cdc48 cofactor was p47 (called Ubx1 or Shp1 in yeast), which is required for homotypic membrane fusion in the nuclear envelope, Golgi, and ER (18, 19). Ubx2 is a transmembrane protein of the ER that helps recruit Cdc48 to ubiquitin-ligase complexes in the ER membrane (20), whereas Ubx5 appears to recruit Cdc48 complexes to chromatin sites of DNA repair (21). Precise functions for the other UBX proteins have not yet been as clearly defined (see “Discussion”).
In the ERAD and UFD pathways, a key Cdc48 cofactor is the Npl4-Ufd1 heterodimer (22, 23). One Cdc48 hexamer interacts with one Npl4-Ufd1 heterodimer via a short binding site (BS1) in the C-terminal region of Ufd1 and a region in Npl4 with a similar fold to UBX and ubiquitin (the ubiquitin-D or UBX-related domain) (24). The Cdc48Npl4-Ufd1 complex is required for the extraction of ERAD substrates from the ER membrane (25). Another Cdc48 cofactor, Vms1, has recently been shown to function in both ERAD and mitochondrial protein degradation (26, 27); in mitochondrial degradation, Vms1 is necessary to recruit Cdc48 to the mitochondrial membrane.
The physical and functional coordination between Cdc48 and the proteasome is still poorly understood. Here we describe a previously uncharacterized yeast protein, YNL155w, which associates with Cdc48 and contains both a conserved AN1-type zinc finger (Zf_AN1) domain and an extremely diverged C-terminal ubiquitin-like domain (UBL). Based on these features, we have named the protein Cdc48-associated UBL/zinc finger protein-1 (Cuz1). Cuz1 is highly conserved, with the most similar human protein being the uncharacterized ZFAND1 polypeptide. Cuz1 binds directly to Cdc48 and also to the proteasome; the latter interaction is strongly augmented in cells exposed to arsenite (As2O3). Deletion of CUZ1 causes minor UPS degradation defects; however, when cuz1Δ is combined with mutations in the Cdc48Npl4-Ufd1 complex, the proteolytic deficiency is enhanced. Loss of Cuz1 also increases the accumulation of polyubiquitin conjugates on the proteasome and Cdc48. These data indicate that Cuz1 is a novel Cdc48 cofactor that may promote transfer of ubiquitylated substrates from Cdc48 to the proteasome or facilitate the disassembly of Cdc48-polyubiquitin-substrate complexes on the proteasome.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, and Plasmid Constructions
Yeast rich (YPD) and minimal (SD) media were prepared as described previously, and all yeast manipulations were carried out according to standard procedures (28). Yeast chromosomal gene deletions were made by PCR-mediated marker amplification and gene replacement in diploid cells; the resulting diploid heterozygotes were dissected to verify 2:2 marker segregation and to isolate haploid deletion strains. Complete lists of Saccharomyces cerevisiae strains and plasmids used in this study are presented in supplemental Tables S1 and S2, respectively.
The YNL155W (CUZ1) and YOR052C genes were isolated by PCR amplification from genomic yeast DNA, and inserted into various plasmids. The absence of mutations was verified by DNA sequencing of the entire inserts. Plasmid pRS314-FLAG-Cuz1 was derived from pRS314-Cuz1 using site-directed, ligase-independent mutagenesis (SLIM) (29). DNA sequences encoding His6-Cdc48 or His6-Cdc48(1–220) were PCR amplified from genomic yeast DNA using an oligonucleotide that introduced a His6 tag and then cloned into pET42b using NdeI and XhoI restriction sites, which removed the sequence for the GST tag from the plasmid. DNA sequencing confirmed that the ORF contained no mutations. Plasmid pGEX-KT was used to express full-length Cuz1 and different Cuz1 deletion variants as GST fusions in Escherichia coli. CUZ1 sequences were obtained by amplifying the desired DNA fragments from yeast genomic DNA and inserting them downstream of the GST coding sequence in pGEX-KT. To make pGEX-KT-Cuz1–4S, Cys to Ser codon mutations were introduced into the CUZ1 sequence using two sequential QuikChange (Stratagene) site-directed mutagenesis reactions.
To fuse the endogenous CUZ1 gene to an upstream FLAG epitope sequence and maintain the normal promoter sequences, we used the delitto perfetto methodology (30). After insertion of the CORE-I-SceI cassette from pGSKU, the FLAG coding sequence was amplified from pRS314-FLAG-Cuz1 using primers whose 5′ segments had 40 nucleotides of identity to sequences upstream and downstream, respectively, of the CORE cassette insertion. This PCR product was then transformed into yeast to replace the CORE cassette in CUZ1 by homologous recombination. Correct recombination was verified by DNA sequencing and anti-FLAG immunoblotting.
Identification of Cuz1-binding Proteins by LC-MS/MS
Late log-phase 2-liter cultures of yeast cells were harvested by centrifugation. Cell pellets were washed with ice-cold water, centrifuged, flash frozen in liquid nitrogen, and stored at −80 °C. Cell lysis was achieved by grinding cells to a fine powder in liquid nitrogen (31). The powder was resuspended in a buffer containing 50 mm HEPES, pH 7.5, 200 mm NaCl, 10% glycerol, 0.5% Triton X-100 and Complete Protease Inhibitor tablets (Roche Applied Science). The extract was centrifuged for 25 min at 30,000 × g to remove cell debris. The protein concentration was determined using the BCA assay (Pierce), and 96 mg of protein extract (∼40 ml) were mixed with 0.4 ml of FLAG-M2 antibody resin (50% slurry; Sigma). After 2 h rotating at 4 °C, the beads were washed four times with 10 ml of the resuspension buffer. Beads were resuspended in 0.6 ml of buffer and then transferred to a new tube to which 3× FLAG peptide was added to a final concentration of 0.2 mg/ml. After incubation for 45 min at 4 °C, the batch eluate was concentrated using a Vivaspin 500 concentrator (MWCO 10,000 kDa; GE Healthcare). SDS-PAGE followed by silver staining was used to evaluate 10% of the concentrated eluate. The remainder was frozen with liquid nitrogen and used for liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) analysis.
The mass spectrometry analysis was performed according to an optimized procedure for LC-MS/MS (32). Briefly, the immunoprecipitated proteins were resolved and excised from a Coomassie Blue-stained SDS gel and digested with trypsin. The extracted peptides were loaded on a C18 capillary column (75 μm inner diameter, 10 cm length, 2.7 μm HALO C18 resin, tip size 15 μm; New Objective, MA), and then eluted during a 60-min gradient of 10–40% solvent B (solvent A, 0.1% formic acid; solvent B, 70% acetonitrile, 0.1% formic acid, flow rate of 300 nl/min). The eluted peptides were analyzed on a hybrid LTQ Orbitrap Velos MS (ThermoFisher Scientific) with one MS survey scan and up to 10 data-dependent MS/MS scans. Acquired MS/MS spectra were searched against yeast Uniprot database using the SEQUEST algorithm. Searching parameters included mass tolerance of precursor ions (±20 ppm) and product ion (±0.5 Da), tryptic restriction, dynamic mass shifts for oxidized Met (+15.9949), two maximal modification sites, two maximal missed cleavages, as well as only b and y ions counted. To evaluate the false discovery rate during spectrum-peptide matching, all original protein sequences were reversed to generate a decoy database that was concatenated to the original database (33). Assigned peptides were grouped by charge state and then filtered by matching scores (XCorr and ΔCn) to reduce the protein false discovery rate to 1%.
Co-immunoprecipitation and Immunoblot Analyses
For co-immunoprecipitation (co-IP) experiments, cultures were grown at 30 °C to mid-logarithmic phase (A600 ∼1); where indicated, As2O3 was added to a final concentration of 0.2 mm, and the cultures were then incubated with shaking for 2 h, except where indicated otherwise. Cells were harvested by centrifugation. To test the interaction of Cdc48 with Cuz1, lysates were prepared by resuspending cell pellets in co-IP buffer A (25 mm Tris-HCl, pH 8.0, 200 mm NaCl, 2 mm MgCl2, 5% glycerol, 1% Triton X-100 and protease inhibitors) and, when indicated, 2 mm ATP. The resuspended cells were disrupted using glass beads in an MP Biomedicals FastPrep bead-beater followed by centrifugation at 21,000 × g for 10 min to remove cell debris. After protein quantification, 2.5 mg of protein extract were incubated with 50 μl of FLAG-M2 slurry for 2 h at 4 °C. The beads were washed three times with 1 ml of co-IP buffer and then resuspended in 25 μl of gel loading buffer. Proteins were resolved by SDS-PAGE and analyzed by immunoblotting.
To test for interaction of Cuz1 with proteasomes, the same co-IP methodology was used except for a modified co-IP buffer (co-IP buffer B, 50 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol, 5 mm MgCl2, 0.1% Triton X-100, 5 mm ATP and protease inhibitors). After protein binding, the resin was washed three times with 1 ml of PBS containing 0.2% Tween 20. To test the interaction of polyubiquitylated substrates with Cdc48, cells expressing V5-tagged Cdc48 were transformed with a plasmid expressing an HA-tagged ubiquitin gene under the control of the CUP1 promoter (34). CuSO4 was added to a final concentration of 0.1 mm when the cultures were diluted. Cells were resuspended in co-IP buffer B without ATP, lysed, and after protein quantification, 1.5 mg of protein was incubated with 40 μl of anti-V5-agarose (Sigma). After 2 h, the beads were washed 3 times with 1 ml of PBS containing 0.2% Tween 20. To test the interaction of the proteasome with polyubiquitylated substrates, cells were grown in the presence of CuSO4 to logarithmic phase and exposed to As2O3 for 2 h. Extracts were prepared with co-IP buffer B without ATP; 1 mg of protein was incubated with anti-FLAG resin, which was then washed with PBS containing 0.2% Tween 20. For testing the interaction of Cdc48 with the proteasome, co-IP buffer C was used: 50 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol, 5 mm MgCl2, 0.5% Triton X-100.
Recombinant Protein Purification and in Vitro Binding Assays
Expression of GST and GST fusion proteins was induced in E. coli BL21(DE3) transformants by addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside and overnight growth at 30 °C. The GST fusion proteins were purified with glutathione-agarose (ThermoScientific) and eluted with reduced glutathione according to the manufacturer's instructions. Expression of His6-Cdc48 in E. coli Rosetta2 (DE3) pLysS cells was induced by adding 1 mm isopropyl 1-thio-β-d-galactopyranoside for 4 h at 30 °C. The recombinant protein was purified using HisPur Cobalt Resin (ThermoScientific) and eluted using a buffer containing 150 mm imidazole. All the purified recombinant proteins were dialyzed against 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 5 mm MgCl2, and 10% glycerol.
For testing the interaction of Cdc48 with GST-Cuz1 and Cuz1 deletion derivatives, the purified proteins were used in a 1:1 molar ratio of Cuz1 monomer to Cdc48 monomer in GST pulldown assays. Binding reactions were incubated for 2 h at 4 °C in a final volume of 0.4 ml of co-IP buffer A. To examine binding of GST-Cuz1 and its deletion derivatives to purified 26 S proteasomes, proteasomes were first purified from yeast as in Ref. 35, and binding reactions were done in 19 S co-IP buffer. Finally, for measuring interaction of ubiquitylated substrates with GST-Cuz1 and its derivatives, recombinant proteins (3 μg) were incubated with 800 μg of yeast extract from cells overexpressing HA-tagged ubiquitin. Extract preparation as well as GST pulldowns were performed with co-IP buffer D: 50 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol, 5 mm MgCl2, 1% Triton X-100. After incubation for 2 h at 4 °C, beads were washed with the same buffer and proteins were eluted by boiling in gel loading buffer.
Cycloheximide Chase/Immunoblot and Pulse-Chase Analyses
For analysis of substrate degradation by cycloheximide chase/immunoblot assays, cultures were grown at room temperature (∼23 °C) to logarithmic phase and switched for 1 h to 37 °C. Cycloheximide was added to a final concentration of 0.25 mg/ml, and 2.5 A600 equivalents of cells were harvested at each time point. The chase was performed at 37 °C. Cell pellets were resuspended in 0.1 ml of water plus 0.1 ml of 0.2 m NaOH and incubated for 5 min at room temperature. Cells were pelleted by centrifugation, resuspended in gel loading buffer, and heated to 100 °C for 5 min; the lysates were centrifuged for 2 min at 21,000 × g to remove cell debris.
Pulse-chase analysis was performed essentially as described previously (36). Cultures were grown at 23 °C to exponential phase; after washing, cells (∼10 A600 eq) were incubated for 4 min at 28 °C and labeled with ∼0.2 mCi of Tran35S-label (MP Biomedicals) for 10 min and chased with excess cold methionine and cysteine at 28 °C. Immunoprecipitation was performed using anti-β-galactosidase antibody and protein A-agarose (Repligen). Immunoprecipitated proteins were separated by SDS-PAGE and analyzed by autoradiography using a Storm 860 PhosphorImager system and ImageQuant 5.2 software (Molecular Dynamics).
Antibodies
An anti-Cuz1 polyclonal antiserum was raised in rabbits and subsequently purified. Additional antibodies used in this study were anti-HA (Covance), anti-GST (Abcam), anti-FLAG (Sigma), anti-V5 (Invitrogen), anti-PGK (Invitrogen), anti-β-galactosidase (Millipore), anti-Tetra His (Qiagen), and anti-Cdc48 (a gift from Dr. Thomas Sommer, Berlin).
RESULTS
Yeast Cuz1/YNL155w and YOR052c as Potential UPS Factors
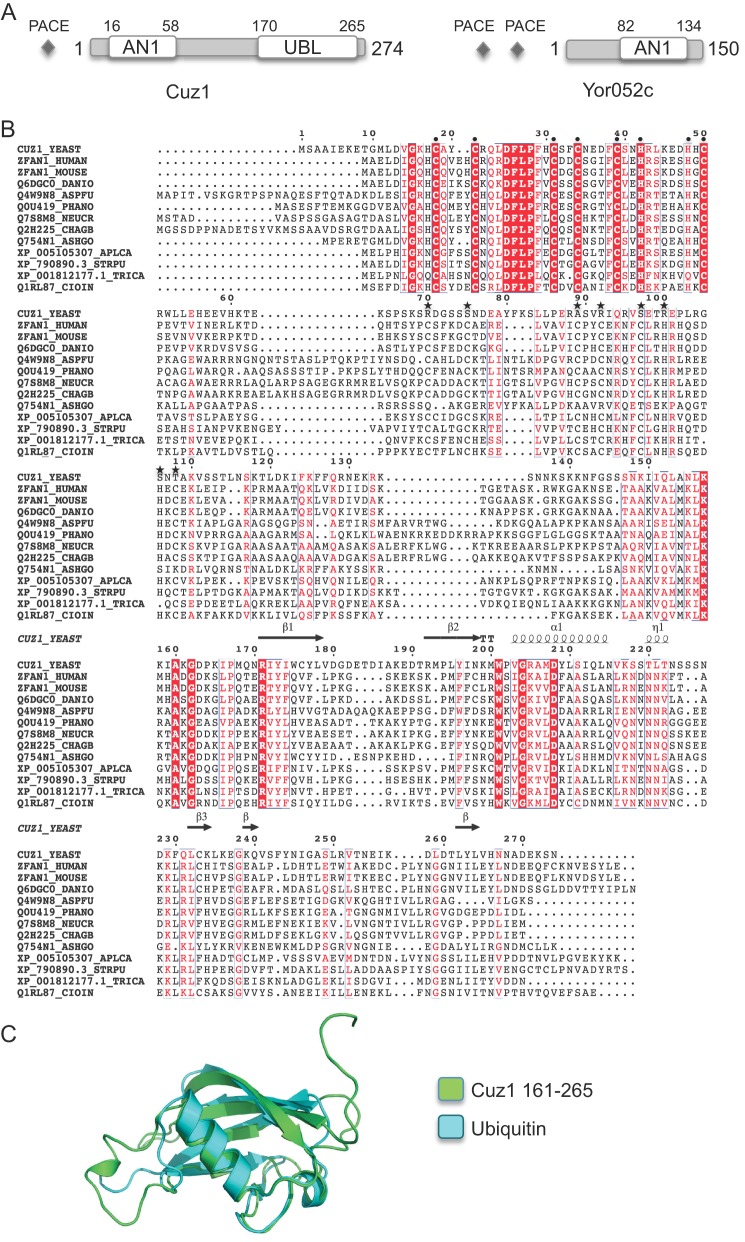
An initial bioinformatic search for uncharacterized S. cerevisiae genes that might function in the UPS led us to two genes, YNL155W and YOR052C (Fig. 1A). These genes are preceded by one or two PACE (proteasome-associated control element) sequences; such nonamer DNA elements (consensus: 5′-GGTGGCAAA-3′) are binding sites for the Rpn4 transcription factor and are found upstream of most proteasome genes as well as other genes involved in the UPS or in other stress response systems (37, 38). Rpn4 is required for normal levels of proteasome gene expression. The presence of upstream proteasome-associated control elements was our original search criterion. Both YNL155W and YOR052C encode predicted proteins with AN1-type zinc finger (Zf_AN1) domains, which coordinate a pair of zinc ions and are part of a widespread structural motif known as the treble-clef domain (39). Treble-clef domains include the RING and IBR domains, both sequence signatures of ubiquitin ligases. Similarity between YNL155w and YOR052c is restricted to the Zf_AN1 domains (46% sequence identity over 32 residues). Two mammalian Zf_AN1 proteins, ZFAND2A/AIRAP (zinc finger-AN1 domain/arsenite-inducible RNA-associated protein) and ZFAND2B/AIRAPL (AIRAP-like), were reported to bind the 26 S proteasome and may modulate its activity (40, 41). The Caenorhabditis elegans ortholog of AIRAPL, AIP-1 has been genetically linked to proteotoxic stress resistance and increased longevity (40).
FIGURE 1.
Sequence features of yeast Cuz1/YNL155w and YOR052c. A, schematic depicting domain organization of Cuz1 and Yor052c. Proteasome-associated control elements (PACE) are found upstream of the corresponding genes. B, Cuz1 is evolutionarily conserved, containing an AN1-type zinc finger (Zf_AN1) domain in its N-terminal region. Aligned proteins were chosen based on the phylogenetic tree of Cuz1 orthologs from the Phylome database. Alignments were performed with ClustalOmega and edited with ESPript. The expected zinc-coordinating residues of the N-terminal Zf_AN1 domain are indicated with a filled circle (●). Except for Cuz1 and Q754N1, all other proteins possess a second Zf_AN1 domain; the putative metal-coordinating residues of this second domain are marked with a filled star (★). Proteins are from the following species: YEAST, S. cerevisiae; HUMAN, Homo sapiens; MOUSE, Mus musculus; DANIO, Danio rerio; ASPFU, Aspergillus fumigatus; PHANO, Phaeosphaeria nodorum SN15; NEUCR, Neurospora crassa; CHAGB, Chaetomium globosum; ASHGO, Ashbya gossypii; APLCA, Aplysia californica; STRPU, Strongylocentrotus purpuratus; TRICA, Tribolium castaneum; CIOIN, Ciona intestinalis. Secondary structure elements were added using ESPript based on the Protein Data Bank file of the obtained model for the ubiquitin-like domain. C, Cuz1 contains a C-terminal UBL. Protein fold and three-dimensional structure predictions were obtained using Phyre2 (43). Model includes fragment 161–265 in green and aligned to ubiquitin (Protein Data Bank 1UBQ) in blue using PyMol. Phyre2 output model was predicted based on human ubiquilin 3 (d1yqba1).
Both YNL155w and YOR052c belong to the set of proteins in Cluster of Orthologous Groups 3582 (COG3582). More detailed sequence comparisons showed that the human ZFAND family member closest to YNL155w is ZFAND1, although the latter protein has an additional Zf_AN1 motif at residues 64–105 (Fig. 1B). Nothing is known about the biochemical function of ZFAND1, but ZFAND1 mutations have been linked to several cancers including ovarian carcinoma (42). YOR052c is a much more divergent protein, with only a low level of similarity to mammalian proteins. Human ZFAND1 has diverged substantially from AIRAP (28% identity over 140 residues) and AIRAPL (29% identity over 130 residues).
In addition to the AN1-type zinc finger, sequence and structural homology searches revealed that both YNL155w and ZFAND1 contain a UBL near the C terminus, which is not true of YOR052c, AIRAP, or AIRAPL. Using the Phyre2 structural modeling program (43), the YNL155w sequence between residues 161 and 265 could be readily fit to the structure of ubiquitin (90% confidence score over 79 residues). The two structures were aligned with a root mean square deviation of 2.15 Å over 64 core residues (Fig. 1C). Ramachandran plot analysis (MolProbity) of the modeled polypeptide showed 88% of the backbone conformations in allowed regions. Similar results were obtained with human ZFAND1 residues 150–250. Because of these structural features and the association of YNL155w with Cdc48 (next section), we named the YNL155w protein Cuz1 for Cdc48-associated UBL/Zn-finger protein-1.
A Proteomic Screen for Cuz1-binding Proteins
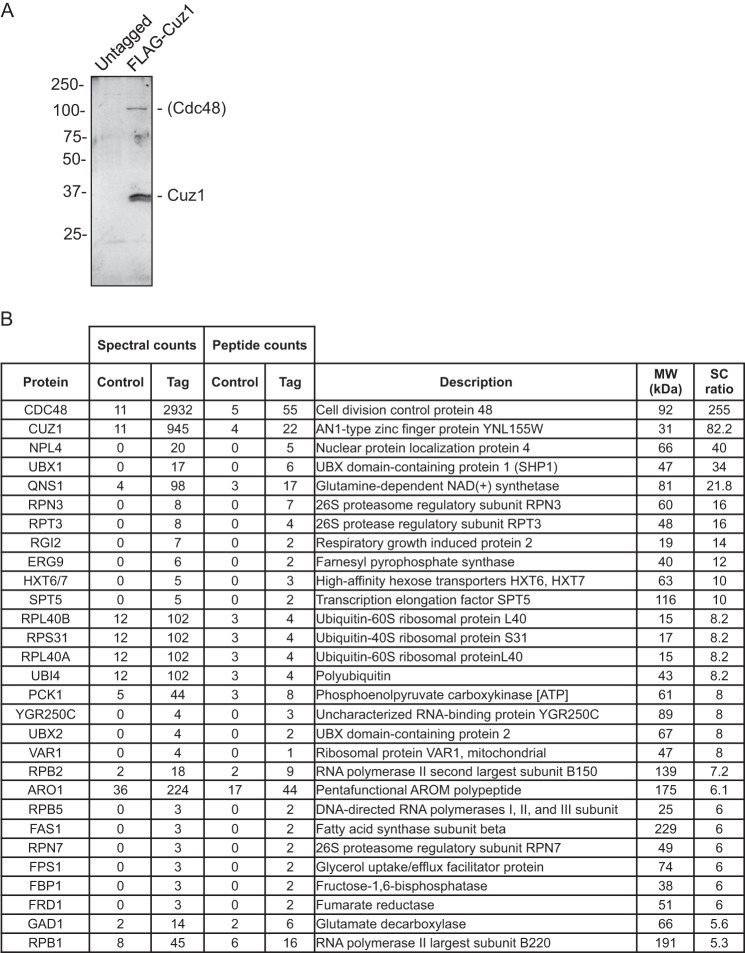
As a first step to determine potential functions for Cuz1 in the UPS, we searched for interactions of Cuz1 with other proteins. Toward this end, the chromosomal CUZ1 locus was modified to encode an N terminally FLAG-tagged Cuz1 protein. FLAG-Cuz1 was purified under nondenaturing conditions on an anti-FLAG affinity resin with elution from the resin by excess 3× FLAG peptide. As a negative control, a parallel purification from yeast cells expressing untagged Cuz1 was used. A fraction of each eluate was first evaluated by SDS-PAGE and silver staining (Fig. 2A). The band corresponding to FLAG-Cuz1 was the most prominent species, and a second protein close to 100 kDa in mass was also seen in the FLAG-Cuz1 eluate and not in the untagged control.
FIGURE 2.
Identification of Cuz1-binding proteins in vivo. A, proteins from a strain expressing FLAG-Cuz1 from the chromosomal CUZ1 locus, or an untagged control strain, were affinity purified on an anti-FLAG resin; 10% of the purified sample was resolved in a 10% SDS-PAGE gel followed by silver staining. B, the remaining sample from the immunopurification was subjected to LC-MS/MS analysis. Using spectral counts as a semi-quantitative index, the majority of proteins showed similar abundance in both samples (untagged versus FLAG-Cuz1). The table shows the proteins from the FLAG-Cuz1 purification that had a spectral count (SC) ratio ≥5-fold above the untagged control.
The remainder of each purified sample was analyzed by LC-MS/MS. Peptides from over 370 different proteins were identified in both the control (untagged Cuz1) and FLAG-Cuz1 preparations. Only those proteins represented by at least five times as many spectral counts in the tagged sample are shown in Fig. 2B. From this analysis, Cdc48 appeared to be the major Cuz1-interacting protein in vivo. Cdc48, with a predicted molecular mass of 92 kDa, is likely to be the protein migrating near the 100-kDa size standard in Fig. 2A. Notably, we also detected several proteins known to interact with Cdc48: Npl4, Ubx1/Shp1, and more weakly, Ubx2. Npl4 is a Cdc48 cofactor that is usually bound to the Cdc48 hexameric ring as part of an Npl4-Ufd1 heterodimer (12, 22). No Ufd1 peptides were detected in this sample but were detected in two subsequent purifications (supplemental Table S3).
Other proteins identified in the FLAG-Cuz1 purification are also likely to be significant. Ubiquitin was represented by more than eight times the number of spectral counts seen in the control purification (Fig. 2B). Ubiquitin-conjugate binding by Cuz1 (likely indirect) was verified by co-immunoprecipitation analysis (see Fig. 5, below). Rpn3, Rpn7, and Rpt3, all subunits of the proteasome RP, were also identified (interaction between Cuz1 and the proteasome was subsequently validated; see below). The remaining proteins in Fig. 2B were not pursued further.
FIGURE 5.
Cuz1 associates with ubiquitin-protein conjugates in vivo. A, recombinant GST-Cuz1 was incubated with extracts from yeast overexpressing HA-tagged ubiquitin, and protein eluted from the glutathione resin was analyzed by anti-HA and anti-GST immunoblotting. B, the Cuz1 UBL domain is required for ubiquitin-conjugate interaction, but the Zf_AN1 domain is neither necessary nor sufficient. The indicated GST-tagged constructs were used in GST pulldown assays performed as in A. C, loss of Cuz1 enhances association of polyubiquitinated conjugates with Cdc48 in vivo. Analysis was done with cultures of CUZ1 or cuz1Δ cells expressing Cdc48-V5 from the endogenous CDC48 locus and expressing HA-tagged ubiquitin from a plasmid.
In summary, the mass spectrometry data suggest that Cuz1 functions primarily with the Cdc48 ATPase in vivo, possibly with multiple distinct Cdc48-cofactor complexes. The apparent association of Cuz1 with proteasomes and ubiquitin, together with its binding to Cdc48, indicates that Cuz1 may indeed act as a component of the UPS, as was originally suggested by the UBL and Zf_AN1 domains in its polypeptide sequence.
Cuz1 Associates Directly with the Cdc48 AAA-ATPase
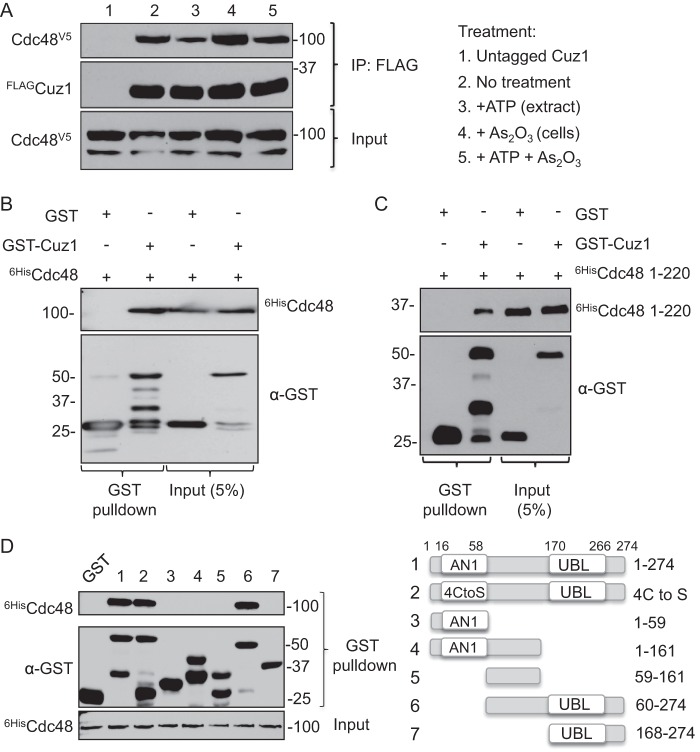
To validate the in vivo interaction of Cuz1 and Cdc48 that was suggested by the LC-MS/MS analysis, we fused the chromosomal copy of CDC48 with a sequence encoding a V5 epitope tag. The tag on the essential Cdc48 protein caused no detectable growth defect. Using a strain that had both the chromosomal CDC48-V5 and FLAG-CUZ1 alleles, we generated whole cell extracts under nondenaturing conditions and immune-precipitated FLAG-Cuz1 and any associated proteins with anti-FLAG antibody beads. As seen in Fig. 3A, FLAG-Cuz1 efficiently co-precipitated the Cdc48-V5 protein. Interestingly, addition of 2 mm ATP to the extraction buffer consistently reduced the amount of co-precipitated Cdc48 protein (Fig. 3A, lanes 3 and 5 versus lanes 2 and 4). This effect of ATP was confirmed by LC-MS/MS analysis; by comparing Cdc48 spectral counts in FLAG-Cuz1 purifications in buffers with or without added ATP, a greater than 2-fold reduction in counts was seen when ATP was added compared with extracts without the added nucleotide (supplemental Table S3).
FIGURE 3.
Cuz1 interacts directly with Cdc48. A, Cuz1 and Cdc48 associate in vivo, and ATP reduces their interaction. Cells expressed FLAG-tagged Cuz1 and Cdc48-V5 or, as a control, untagged Cuz1 and Cdc48-V5. Cultures were treated with 0.2 mm As2O3 for 2 h prior to lysis where indicated. ATP (2 mm) was added to the extracts where indicated. Following immunoprecipitation of FLAG-Cuz1, anti-V5 and anti-FLAG immunoblot analysis was performed. B, Cdc48 interaction with Cuz1 is direct. His6-Cdc48, GST, and GST-Cuz1 were expressed in and purified from E. coli. GST pulldowns were followed by anti-His tag and anti-GST immunoblotting. C, Cuz1 interacts with N-terminal domain of Cdc48. Binding of GST-Cuz1 to His6-Cdc48(1–220) was tested as described for the full-length construct in B. D, the Cuz1 Zf_AN1 domain is neither necessary nor sufficient for Cdc48 interaction, whereas the UBL domain is required. All proteins used in the GST pulldown analysis were purified from E. coli.
To determine whether the association of Cuz1 with Cdc48 was direct or mediated by other proteins, in vitro binding assays were carried out using recombinant GST-Cuz1 and His6-Cdc48 proteins both purified from E. coli. GST or GST-Cuz1 was immobilized on glutathione-agarose beads and incubated with His6-Cdc48. GST-Cuz1, but not GST, was able to pull down His6-Cdc48, indicating that their interaction did not require any other yeast proteins (Fig. 3B). Cdc48 interacts with its cofactors through either the Cdc48 N-terminal domain or its C-terminal tail, although the N-terminal domain is the predominant interaction site (44). Therefore, we asked whether a fragment encompassing the Cdc48 N-terminal domain (residues 1–220) would be sufficient for Cuz1 binding. As shown in Fig. 3C, this was in fact the case.
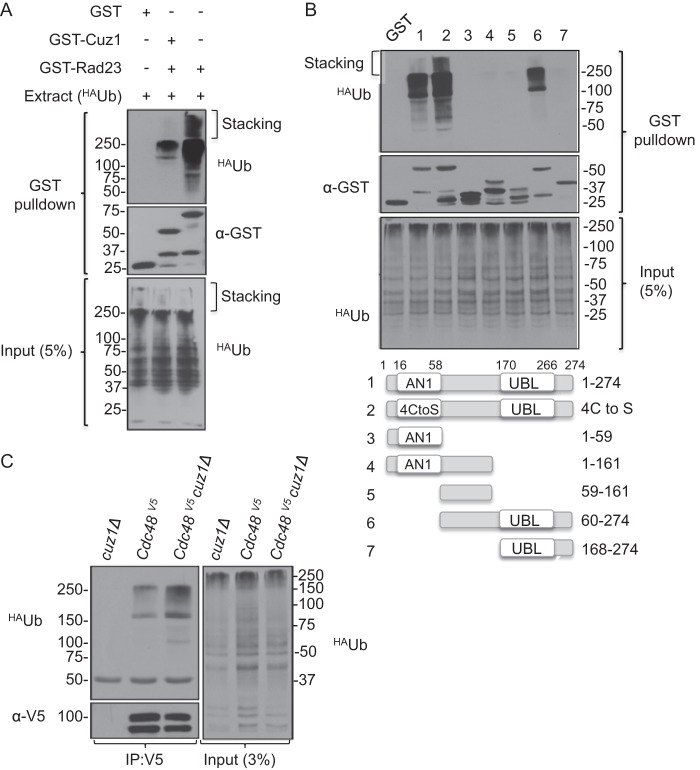
To identify which region(s) of Cuz1 was involved in Cdc48 binding, several GST-Cuz1 deletion variants were created (Fig. 3D). Neither the Cuz1 zinc finger domain by itself (GST-Cuz1(1–59)) nor a fragment consisting of the less conserved central region between the Zf_AN1 and UBL domains (GST-Cuz1(59–161)) showed detectable binding to Cdc48. Moreover, a fragment including both of these regions (GST-Cuz1(1–161)) also failed to pull down Cdc48. Conversely, mutation of four cysteines in Zf_AN1 to serines, which should eliminate zinc binding, did not impair binding to Cdc48 (4-Cys to Ser, construct 2). Together, these data indicate that the Cuz1 zinc finger is neither necessary nor sufficient for Cdc48 interaction.
Most of the known Cdc48 adaptors bind Cdc48 via a UBX domain, which has a ubiquitin-like (β-grasp) fold. The Cdc48 cofactor Npl4 binds to Cdc48 through a UBX-related domain (24). Cuz1 possesses a domain predicted to be a UBL (Fig. 1), and a C-terminal deletion that removed the UBL and little else prevented Cdc48 binding (Fig. 3D, GST-Cuz1(1–161)). This indicates that the Cuz1 UBL is necessary for Cdc48 binding.
We tested two N-terminal Cuz1 truncations to help narrow down the region that is sufficient for Cdc48 interaction. GST-Cuz1(60–274) and GST-Cuz1(168–274) were expressed at similar levels, but only the longer construct was able to bind Cdc48 (Fig. 3D, constructs 6 and 7). This confirmed the lack of a requirement for the Zf_AN1 domain for Cdc48 binding and suggested either that the UBL is not sufficient for binding or that additional sequences N-terminal to residue 168 are necessary for a fully functional UBL. Considered together, the deletion data indicate that the Cuz1 UBL is necessary for Cdc48 binding but may not be sufficient, whereas the zinc finger domain is neither necessary nor sufficient for this interaction.
Cuz1 Functions with Cdc48 in the Ubiquitin-Proteasome System
Cdc48 has a broad array of functions, but one of its best characterized roles is in protein degradation by the UPS (45). We used pulse-chase analyses to determine whether Cuz1 contributes to the degradation of the UFD substrate UbVal-76-β-galactosidase (UbVal-76-β-gal) (46). Deletion of CUZ1 caused a very mild but reproducible slowdown in UbVal-76-β-gal degradation kinetics (Fig. 4A). We then combined the cuz1Δ allele with temperature-sensitive mutations in Cdc48Npl4-Ufd1 components. The defects in UbVal-76-β-gal degradation were already sufficiently severe in these single mutants at the semipermissive temperature of 28 °C that we could not detect any additional defect when the mutations were combined with cuz1Δ (not shown).
FIGURE 4.
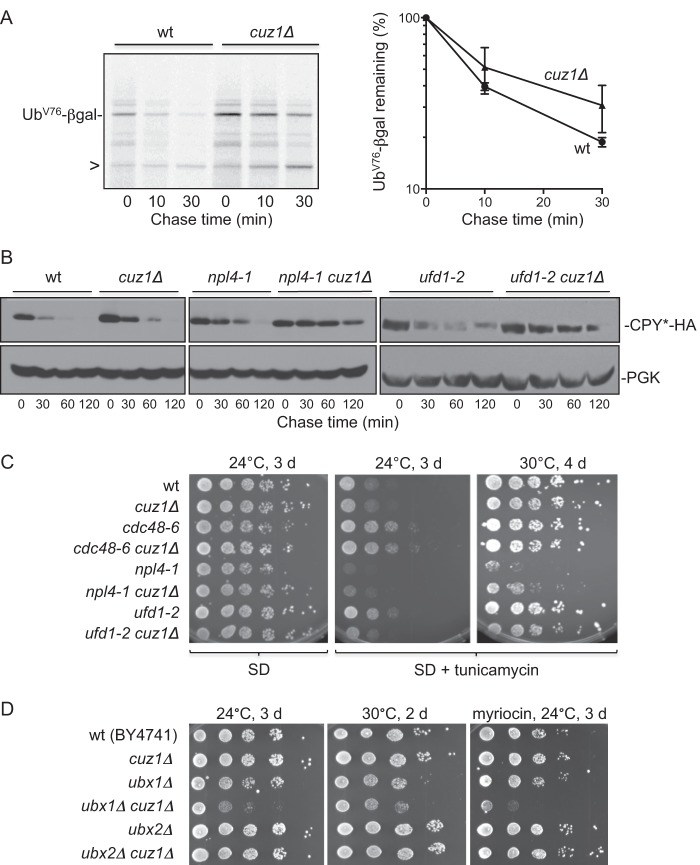
Deletion of Cuz1 causes cellular protein degradation defects. A, pulse-chase analysis of UbVal-76-β-gal in the indicated yeast strains. Representative autoradiograph of a gel is shown at left. Arrowhead indicates a 90-kDa degradation product observed with UbVal-76-β-gal degradation in yeast. Bands above the primary UbVal-76-β-gal are polyubiquitylated species. The graph at right shows the mean degradation rates observed from three independent experiments. Error bars represent S.E. B, degradation of CPY*-HA was analyzed by cycloheximide chase/immunoblot analysis. CPY*-HA was detected by anti-HA immunoblotting. As a loading control, the membrane was subsequently probed with anti-PGK antibodies (bottom panels). C, growth assays reveal genetic interactions of cuz1Δ with mutations in Cdc48Npl4-Ufd1. 6-Fold serial dilutions of cultures were spotted onto plates (SD minimal medium or SD with 0.5 μg/ml of tunicamycin). The apparent growth advantage of cdc48-6 and cdc48-6 cuz1Δ in medium containing tunicamycin could in principle be due to a low constitutive induction of the ER unfolded-protein response in these cells. D, double mutant analysis of cuz1Δ with different UBX gene deletions. Myriocin was used at 0.2 μg/ml. 10-Fold serial dilutions of cultures were spotted onto the plates. Negative genetic interactions between cuz1Δ and ubx1Δ were also observed in a different strain background (not shown).
We also examined degradation of the classical ERAD substrate CPY*, a mutant derivative of the vacuolar carboxypeptidase Y enzyme that is retrotranslocated from the ER lumen for degradation by the cytoplasmic proteasome (47). Degradation of an HA-tagged CPY* protein at 37 °C appeared to be weakly impaired by loss of Cuz1 (Fig. 4B). Notably, when cuz1Δ was combined with mutations in Npl4 or Ufd1, CPY*-HA degradation was further impeded (the cdc48-6 single mutant was already strongly defective; not shown). These results indicate that Cuz1 has an auxiliary or partially redundant role in Cdc48Npl4-Ufd1-dependent protein degradation by the UPS.
When growth of these same strains was examined under various conditions, distinct genetic interactions between cuz1Δ and different Cdc48Npl4-Ufd1 mutations were observed. The ufd1-2 cuz1Δ double mutant grew more slowly than the ufd1-2 single mutant in the presence of tunicamycin, an ER stress inducer (Fig. 4C). We did not see clear differences in growth when cuz1Δ was combined with cdc48-6, and in combination with npl4-1, an apparent increase in growth rate was seen relative to the npl4-1 single mutant. This was seen with cells derived from two different npl4-1 cuz1Δ spores, but when CUZ1 was reintroduced into these cells on a plasmid, no change in growth was seen, suggesting that the double mutants carried a cryptic suppressor of npl4-1 (not shown). Collectively, the growth data suggest an overlap in Cuz1 and Ufd1 function, whereas the results with the cdc48-6 and npl4-1 mutants are not readily interpreted.
Because the Cdc48-binding Ubx1/Shp1 and Ubx2 proteins were found to copurify with Cuz1 based on LC-MS/MS, we tested for genetic interactions between cuz1Δ and deletions of all seven yeast UBX genes as well as the gene for the Cdc48 cofactor Vms1. For ubx2Δ–ubx7Δ and vms1Δ, no obvious differences in growth rate were seen under a variety of conditions when these alleles were combined with cuz1Δ (Fig. 4D and not shown). In contrast, a ubx1Δ cuz1Δ double mutant grew markedly slower than either single mutant at lower temperatures (24 °C), but this effect was attenuated at 30 °C and no longer detected at 36.5 °C (Fig. 4D and not shown). Membrane stressors such tunicamycin or the sphingolipid synthesis inhibitor myriocin, exacerbated the synthetic defect. The ubx1Δ cuz1Δ genetic interaction, together with the enhanced defects seen in the ufd1-2 cuz1Δ mutant, is consistent with our LC-MS/MS results and supports a function for Cuz1 in multiple Cdc48-adaptor complexes.
Cuz1 Interacts with Ubiquitylated Proteins in Vivo
Many Cdc48 cofactors help recruit or process polyubiquitylated protein substrates (48). This prompted us to investigate a possible interaction between Cuz1 and ubiquitylated proteins. GST-Cuz1 was immobilized on glutathione-agarose beads and incubated with whole cell extracts from yeast that expressed HA-tagged ubiquitin. GST and GST-Rad23 were used as negative and positive controls, respectively. GST-Cuz1 bound to high molecular mass ubiquitylated species that were visualized near the top of the resolving gel and in the stacking gel (Fig. 5A). The pattern of ubiquitin conjugates that bound was similar to those bound by the well characterized ubiquitin chain-binding GST-Rad23 protein, although the level of bound conjugates was lower than found with Rad23. No conjugates were detected when GST was used (Fig. 5A).
We tested whether the ubiquitylated protein-Cuz1 interaction was likely to be direct by performing the pulldown assay with purified polyubiquitin chains. Neither Lys48-linked chains (chain lengths of 2 to 7 ubiquitins) nor Lys63-linked tetraubiquitin chains bound detectably to GST-Cuz1, in contrast to GST-Rad23 (data not shown). Although it remains possible that Cuz1 directly interacts with ubiquitin chains of distinct linkages, it is more likely that the interactions seen in yeast lysates were indirect (or only occur in the context of a protein complex, such as one bearing additional ubiquitin-binding sites). Using the same Cuz1 deletion constructs described above for examining Cdc48 interactions, we mapped the region(s) in Cuz1 responsible for interaction with ubiquitylated proteins in yeast extracts (Fig. 5B). We observed the exact same binding behaviors as seen when Cdc48 interaction was evaluated (Fig. 3D). Specifically, binding did not require the Zf_AN1 domain, but association was observed with a C-terminal Cuz1 fragment including the UBL (GST-Cuz1(60–274)) but with no other truncations. A parsimonious explanation of these results is that Cdc48 mediates the interaction of ubiquitylated proteins with Cuz1, presumably in the context of Cdc48 complexes with one or more of its known cofactors.
These results raise the question of whether Cuz1 influences the interaction of Cdc48 with ubiquitylated substrates in vivo. We transformed a plasmid overexpressing HA-tagged ubiquitin into cuz1Δ and CUZ1 strains carrying the chromosomal CDC48-V5 allele. If Cuz1 were involved in the recruitment of substrates to Cdc48, loss of Cuz1 might reduce ubiquitin-conjugate interaction with the ATPase complex. Conversely, if Cuz1 were affecting substrate release from Cdc48, an increase in the level of ubiquitylated substrates on Cdc48 may be observed. In fact, when Cdc48-V5 was immunoprecipitated from extracts derived from cuz1Δ cells, there was a reproducible increase in the levels of high molecular mass ubiquitylated species that were co-precipitated (Fig. 5C). This suggests a potential role for Cuz1 in releasing ubiquitylated substrates from Cdc48 or in transferring them from Cdc48 to the proteasome or proteasome shuttle factors.
Cuz1 Binds to the Proteasome and Is Stimulated by Exposure of Cells to Arsenite
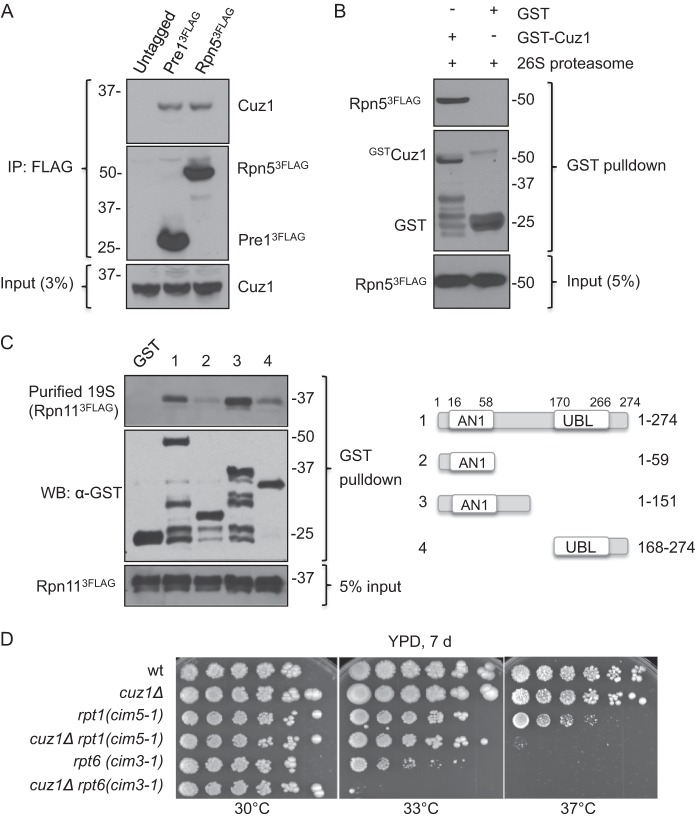
The preceding data raised the possibility that Cuz1 is involved in transferring ubiquitylated substrates from Cdc48 to the proteasome. This idea is supported by our mass spectrometry data, which revealed low levels of proteasome subunits co-purifying with Cuz1 (Fig. 2B). Moreover, previous studies had shown that AIRAP and AIRAPL, mammalian proteins related to Cuz1, can interact with the 26 S proteasome (40, 41). To determine whether Cuz1 interacts with the yeast 26 S proteasome, we affinity purified 26 S proteasomes from yeast cells that expressed proteasomes with a FLAG epitope tag on either the Pre1 (β4) subunit of the CP or the Rpn5 subunit of the RP. Anti-Cuz1 immunoblot analysis of the purified complexes revealed association of Cuz1 in both cases (Fig. 6A). Additionally, GST-tagged Cuz1 was purified from E. coli associated with isolated 26 S proteasome particles, suggesting that the Cuz1-proteasome interaction is direct (Fig. 6B). We noted that Cuz1 is a long-lived protein, so its association with the proteasome is unlikely to be as a substrate (data not shown). A subset of the GST-Cuz1 deletion derivatives described above was used for pulldown assays with purified yeast RP. In contrast to Cdc48 and polyubiquitin (Figs. 3D and 5B), the UBL was not necessary for RP binding (Fig. 6C). A fragment containing the Zf_AN1 domain and the linker region was sufficient for maximal binding (GST-Cuz1(1–151)). The Zf_AN1 domain itself does not appear to be sufficient for proteasome binding, but whether it is necessary remains to be determined.
FIGURE 6.
Genetic and physical interactions between Cuz1 and the proteasome. A, Cuz1 interacts in vivo with the 26 S proteasome. Proteasomes were affinity purified from extracts of yeast expressing either FLAG-tagged Pre1 (CP) or Rpn11 (RP) from the respective endogenous locus. The bound material was analyzed by anti-Cuz1 immunoblotting. B, interaction between Cuz1 and 26 S proteasomes in vitro. Recombinant GST-Cuz1 was incubated with yeast 26 S proteasomes affinity-purified from an Rpn5–3FLAG-expressing strain. C, the UBL domain is not required for Cuz1 interaction with proteasomes. Recombinant GST-Cuz1 constructs were mixed with purified proteasomal 19 S RP (purified from an Rpn11–3FLAG-expressing strain), and the proteins were subjected to GST pulldown analysis. D, deletion of CUZ1 exacerbates the growth defects of cim3-1 and cim5-1 proteasome mutants. Serial dilutions of cultures were done as described in the legend to Fig. 4C.
The composition of the S. cerevisiae proteasome has been thoroughly analyzed, and Cuz1 association was not reported (49, 50). It is likely that the amount of Cuz1 on the proteasome is strongly substoichiometric and its interaction transient. Measurements of epitope-tagged Cuz1 levels in yeast compared with those of the proteasome and Cdc48 suggest that the latter proteins are present at 5–20 times the amount of Cuz1 (51, 52). Nevertheless, in support of the physiological relevance of the Cuz1-proteasome interaction, deletion of CUZ1 enhanced the growth defects of both rpt6 (cim3-1) and rpt1 (cim5-1) temperature-sensitive mutants (Fig. 6D).
Arsenite is a well known inducer of protein misfolding and causes accumulation of polyubiquitylated conjugates in vivo (41, 53). CUZ1 mRNA levels were reported to be elevated by exposure of cells to arsenite (54), and we detected a modest increase in FLAG-Cuz1 protein levels when cells were exposed to 0.2 mm As2O3 for 2 h (Fig. 7A, input lanes). It is noteworthy that both mammalian AIRAP and p97 (Cdc48) display increased binding to the proteasome upon arsenite stress (41, 55). Based on these results, we tested whether arsenite might affect interaction of Cuz1 with proteasomes in yeast. In the presence of arsenite, we observed a marked increase in Cuz1 binding to proteasomes immunoprecipitated from yeast extracts; the increase greatly exceeded the modest overall increase of Cuz1 levels in the treated cells (Fig. 7A). When normalized to the amount of precipitated FLAG-Rpn11, a ∼10-fold increase in Cuz1-proteasome association was seen following arsenite treatment (Fig. 7A, right). The increase in this association was supported by LC-MS/MS analysis (supplemental Table S3). Addition of arsenite to cells before FLAG-Cuz1 purification led to the identification of 15 of the 19 RP subunits (but no CP subunits).
FIGURE 7.
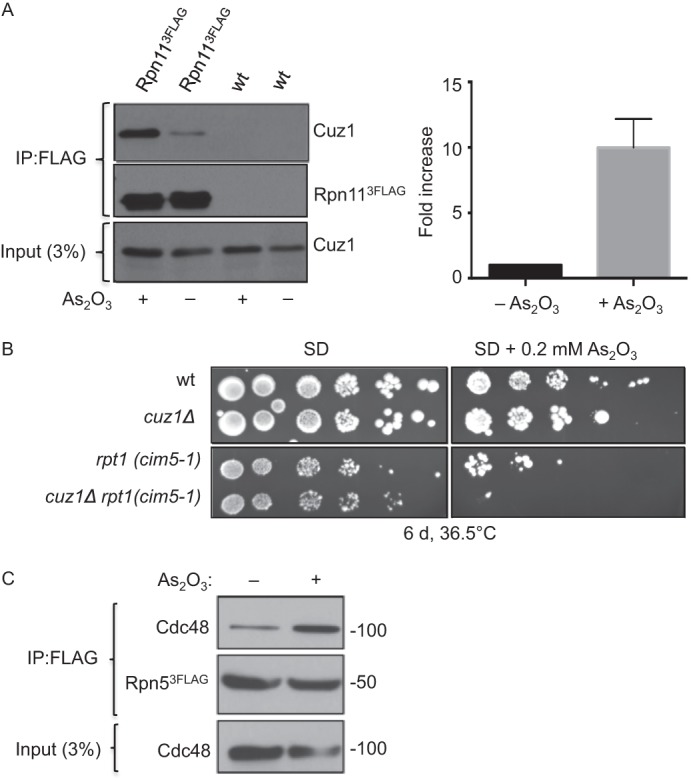
Cell exposure to arsenite enhances proteasome association with Cuz1. A, FLAG-tagged Rpn11 was immunoprecipitated and the amount of co-purifying Cuz1 was quantified using a Syngene G-box. A representative experiment is shown at the left. Cuz1 values were normalized to the levels of precipitated Rpn11–3FLAG. The quantification at the right is derived from three independent experiments and shows the normalized fold-increase of coprecipitated Cuz1; error bars denote S.D. B, in the presence of arsenite, deletion of CUZ1 worsens the growth defect of a temperature-sensitive cim5-1 mutant at high temperature. C, Cdc48 interaction with the proteasome is enhanced by arsenite. Proteasomes tagged with Rpn5–3FLAG were immunoprecipitated with anti-FLAG resin from extracts derived from cells grown in the presence of arsenite for 30 min. Co-purified Cdc48 levels was analyzed by anti-Cdc48 immunoblotting.
Loss of Cuz1 alone caused no reduction in cell growth in the presence of arsenite, but when cuz1Δ cim5-1 cells grown at high temperature were examined, arsenite exacerbated the already slow growth seen in the cim5-1 single mutant (Fig. 7B). This supports the potential biological importance of Cuz1 in promoting proteasome function in arsenite resistance. Finally, because arsenite seemed to increase association of mammalian p97 with the proteasome (55), we checked if this was also true for Cdc48. In fact, Cdc48 co-precipitation with the proteasome increased already after just a 30-min exposure to the metalloid (Fig. 7C). These data suggest that Cdc48 and the proteasome have linked functions in promoting arsenite resistance.
Interestingly, loss of Cuz1 caused a small but reproducible increase in Cdc48 interaction with proteasomes with and without arsenite stress (Fig. 8A). A parallel increase in polyubiquitylated proteins on the proteasome was observed when CUZ1 was deleted (Fig. 8B). These data would be consistent with the participation of Cuz1 in either the transfer of polyubiquitin conjugates from Cdc48 to the proteasome, which would normally end with the release of Cdc48 from the proteasome, or, more directly, in Cdc48-proteasome dissociation.
FIGURE 8.
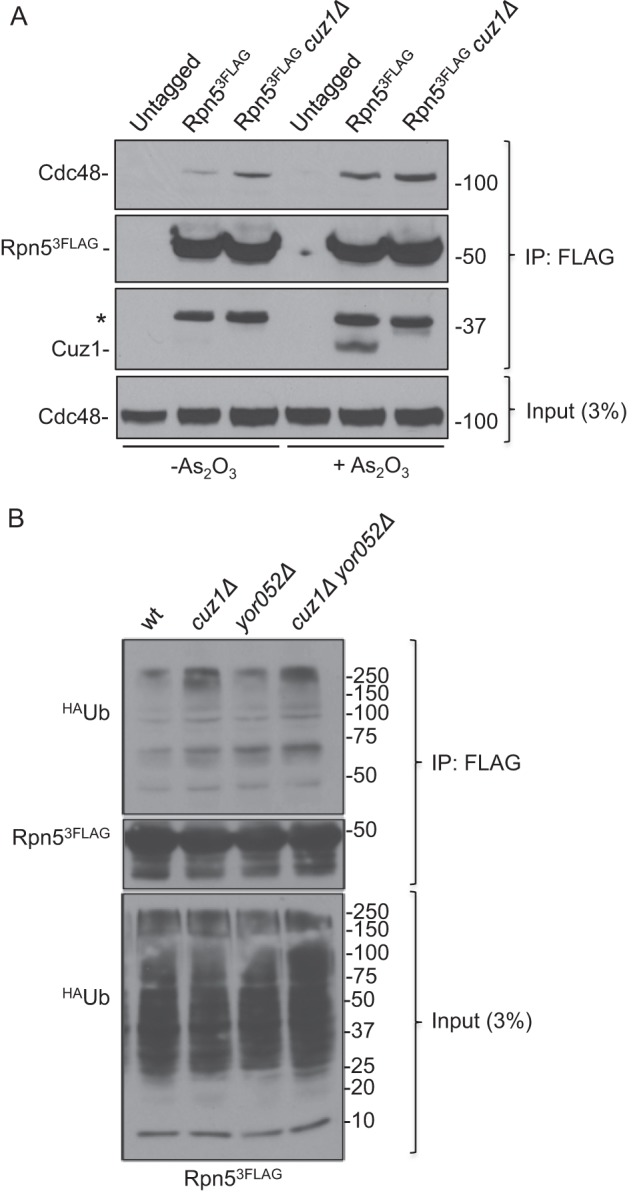
Cuz1 affects the interaction of polyubiquitinated substrates with proteasomes. A, a slight increase in Cdc48 bound to the proteasome is observed in the absence of Cuz1, both in the presence and absence of arsenite. FLAG-tagged proteasomes were immunoprecipitated, and the amount of bound Cdc48 was analyzed by anti-Cdc48 immunoblotting. An unspecific band is indicated with an asterisk. B, an increase in polyubiquitylated proteins on the proteasome is observed when CUZ1 is deleted. After exposing cells for 2 h to 0.2 mm arsenite, cells were lysed and proteasomes were immunoprecipitated. Co-purified polyubiquitinated proteins were analyzed by anti-HA immunoblot analysis.
DISCUSSION
In this study we have described a novel arsenite-inducible yeast protein, Cuz1, which associates under normal growth conditions primarily with Cdc48; however, upon cellular exposure to arsenite, which activates multiple stress-response pathways (40), a ∼10-fold increase in Cuz1-proteasome association occurs (Fig. 7A). Cuz1 provides the first reported functional connection between the extremely widespread AN1-type zinc finger (Zf_AN1) motif and the phylogenetically conserved Cdc48 AAA-ATPase. The Zf_AN1 domain is not required for Cdc48 binding but appears to contribute to proteasome association. Conversely, a highly divergent UBL domain near the C terminus of Cuz1 is required for its binding to Cdc48 but not the proteasome. Cuz1 plays an ancillary or partially redundant role in the degradation of UPS substrates that depend on Cdc48. Protein-protein interaction data and other results suggest possible roles for Cuz1 in promoting polyubiquitylated substrate release from Cdc48 or the proteasome (or both).
The Zf_AN1 domain has an extremely broad distribution in both eukaryotes and archaea, with a few examples in bacteria, presumably due to lateral gene transfer (39). Most remarkably, this domain is part of many highly divergent multidomain proteins, but most, if not all, are linked to membrane-localized proteolytic systems. When we performed sequence searches with the Zf_AN1 domain, it was found in a wide range of euryarchaeota species and several thaumarchaeotes. The domain most commonly found to be part of archaeal Zf_AN1 proteins was a rhomboid-related protease domain. Rhomboid proteases are polytopic membrane proteins bearing a protease active site deep within the lipid bilayer (56). It may be relevant in this context that some eukaryotic rhomboid and pseudorhomboid proteins function in ERAD (57, 58).
In eukaryotes, Zf_AN1 domains are commonly part of proteins that also contain ubiquitin, polyubiquitin, or ubiquitin-like sequences; however, the domain order in the protein is often permuted relative to Cuz1. For example, the fungus Rhizopus oryzae has a protein (GenBankTM EIE85715) with two N-terminal, near-exact ubiquitin repeats, which should be cleavable by deubiquitylating enzymes, and a C-terminal Zf_AN1 domain. The joining of divergent ubiquitin and UBL domains to Zf_AN1 motifs may be an example of convergent evolution based on their phylogenetic distribution and differences in Zf_AN1 and ubiquitin/UBL sequence; this might have been driven by a common role for many Zf_AN1 domain proteins in proteasome binding. Like Cuz1, the human AIRAP/ZFAND2A and AIRAPL/ZFAND2B proteins both bind 26 S proteasomes, as does their C. elegans ortholog, AIP-1 (40, 41). Our data with yeast Cuz1 (Fig. 6) and domain swaps with the AIRAP and AIRAPL proteins (40, 41) both suggest that the Zf_AN1 domain participates in proteasome association. Finally, a subfamily of Zf_AN1 proteins also contain A20 zinc finger domains, which in some cases have been shown to have ubiquitin ligase activity (59). In plants these proteins are associated with stress responses (60), whereas in mammals they are usually involved in the immune system (61, 62). It is likely that Zf_AN1 proteins have widespread and disparate functions in the UPS.
Additional support for links between Cuz1 and both proteasomes and Cdc48 can be found in available genomic and proteomic databases. Cluster analysis of aggregated RNA microarray data shows that CUZ1 transcriptional regulation clusters very closely with that of UBX4, which encodes a UBX protein that has been suggested to promote dissociation of ubiquitinated proteins from Cdc48 based on results similar to those reported here for Cuz1 (63). Another gene with a similar transcription profile is UFD1. Of the 25 genes most closely correlated transcriptionally with CUZ1 (in addition to YOR052C), 19 are proteasome subunit genes. This co-regulation is consistent with the fact that CUZ1, YOR052C, and proteasome genes all have upstream proteasome-associated control element sequences, the target for the Rpn4 transcription factor. Large scale protein interaction studies using yeast two-hybrid or mass spectrometry approaches also identified, among other proteins, Cdc48, Ubx1, and Npl4. Although not pursued further, these earlier genomic and proteomic studies are fully consistent with our findings.
Cdc48 adaptors bind to the ATPase in both mutually exclusive and interdependent fashion. Ubx1 and Npl4-Ufd1 do not bind the same Cdc48 hexamer (18, 22), and conversely, certain UBX proteins only bind to Cdc48 if Npl4-Ufd1 is also present (64, 65). Intriguingly, the mass spectrometry analysis of Cuz1-associated proteins identified Ubx1 as well as Npl4 and Ufd1 (Fig. 2B and supplemental Table S3). Moreover, mutations in either adaptor when combined with loss of Cuz1 cause enhanced growth defects (Fig. 4). This suggests that Cuz1 is part of distinct Cdc48-adaptor complexes. A C-terminal GFP fusion of Cuz1 localizes diffusely throughout the cell, with a slight concentration in the nucleus (66), so Cuz1 would be well placed to function in multiple Cdc48 complexes.
As is true for the majority of Cdc48 cofactors, Cuz1 interacts directly with the N-terminal domain of the ATPase (Fig. 3C). Notably, addition of ATP to Cuz1-Cdc48 binding reactions attenuates their association (Fig. 3A), and this was also apparent in our LC-MS/MS samples with ATP supplementation (supplemental Table S3). ATP has been reported to modulate the recruitment of other cofactors to Cdc48. ATP binding to the D1 domain enhances interaction of the Npl4-Ufd1 heterodimer with the N-terminal domain of p97 (67), and disassembly of Ufd2-Rad23 complexes by Cdc48 depends on Cdc48 binding to Ufd2 and ATP (68). Ubx2 interaction with Cdc48 is strongly decreased in the presence of ATP (69). Cuz1 occupancy of Cdc48 may similarly be tied to nucleotide-dependent conformation changes or specific steps of the ATPase catalytic cycle. It remains to be determined whether this modulation of Cuz1-Cdc48 association is due to ATP binding or hydrolysis. A recent study suggests that Cdc48 may directly control proteolytic activity of the eukaryotic 20 S proteasome (70); Cuz1 might modulate this potential Cdc48 function as well.
Many Cdc48 cofactors interact with polyubiquitin-protein conjugates, some of them directly. For example, Ubx1, Ubx2, and Ubx5 are UBA-UBX proteins in which the UBA domain mediates polyubiquitin binding (71). Cdc48, Npl4, and Ufd1 also all have ubiquitin-binding sites (12, 72, 73). Cuz1 interacts in vivo with polyubiquitylated proteins (Fig. 5A), but this is likely to be indirect; this is not unexpected given that Cuz1 lacks any known ubiquitin-binding motif. The binding behavior of a series of Cuz1 deletion variants is identical for Cdc48 and polyubiquitylated species in cell extracts, consistent with the possibility that interaction between Cuz1 and polyubiquitin occurs in the context of a Cdc48 complex. Although it is known that recruitment of polyubiquitinated substrates to Cdc48 is largely mediated by the Npl4-Ufd1 dimer (73), their release to downstream components is less well understood. Ubx4 has been identified as one possible releasing factor (61) and Vms1 as another (27). As observed upon deletion of these factors in the earlier studies, loss of Cuz1 also leads to an increase in the levels of polyubiquitylated substrates associated with Cdc48 (Fig. 5C). Therefore, Cuz1 might have a related mechanism of action. Moreover, cuz1Δ cells also accumulate higher levels of polyubiquitin conjugates on proteasomes (Fig. 8B).
The exact mechanistic function of Cuz1, like that of most other Cdc48 cofactors, remains to be determined. Based on the genetic interactions we observe between mutations in Cuz1 and Npl4-Ufd1 (Fig. 4) and their co-purification (Fig. 2B and supplemental Table S3), Cuz1 might work with Npl4-Ufd1 on a subset of Cdc48 complexes. Cuz1 function in these complexes remains unclear, but it may stimulate ATP binding or hydrolysis by Cdc48, inducing conformation changes that weaken ubiquitylated substrate binding. This would facilitate substrate transfer to Rad23 or related shuttle factors. Other factors such as Ufd2 bind to both Cdc48 and Rad23 and may recruit Rad23 to Cdc48-bound substrates (68). Following release from Ufd2, Rad23 and related shuttle proteins can bind the proteasome via their UBL domain interaction with Rpn1. In the absence of Cuz1, the Cdc48 complex may tend to remain bound to polyubiquitylated substrates and accompany them to the proteasome. Alternatively, Cuz1 might enhance the transfer of certain ubiquitin conjugates from Cdc48 directly to the proteasome or promote Cdc48-proteasome dissociation. Arsenite may alter Cuz1 itself in a way that impacts these processes, or the metalloid might act more indirectly, such as by modifying the cellular pool of ubiquitin conjugates. These ideas are testable and will be the subject of future studies.
This work was supported, in whole or in part, by National Institutes of Health Grants GM083050 and GM046904 (to M. H.).

This article contains supplemental Tables S1–S3.
- UPS
- ubiquitin-proteasome system
- CP
- core particle
- RP
- regulatory particle
- ERAD
- endoplasmic reticulum-associated degradation
- UFD
- ubiquitin-fusion degradation
- UBL
- ubiquitin-like domain
- Cuz1
- Cdc48-associated UBL/zinc-finger protein-1
- co-IP
- co-immunoprecipitation.
REFERENCES
- 1. Ravid T., Hochstrasser M. (2008) Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peters J. M., Franke W. W., Kleinschmidt J. A. (1994) Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J. Biol. Chem. 269, 7709–7718 [PubMed] [Google Scholar]
- 4. Tomko R. J., Jr., Hochstrasser M. (2013) Molecular architecture and assembly of the eukaryotic proteasome. Annu. Rev. Biochem. 82, 415–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schmidt M., Hanna J., Elsasser S., Finley D. (2005) Proteasome-associated proteins. Regulation of a proteolytic machine. Biol. Chem. 386, 725–737 [DOI] [PubMed] [Google Scholar]
- 6. Fröhlich K. U., Fries H. W., Rüdiger M., Erdmann R., Botstein D., Mecke D. (1991) Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J. Cell Biol. 114, 443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peters J. M., Walsh M. J., Franke W. W. (1990) An abundant and ubiquitous homo-oligomeric ring-shaped ATPase particle related to the putative vesicle fusion proteins Sec18p and NSF. EMBO J. 9, 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ye Y., Meyer H. H., Rapoport T. A. (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414, 652–656 [DOI] [PubMed] [Google Scholar]
- 9. Rabinovich E., Kerem A., Fröhlich K. U., Diamant N., Bar-Nun S. (2002) AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22, 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bays N. W., Wilhovsky S. K., Goradia A., Hodgkiss-Harlow K., Hampton R. Y. (2001) HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell 12, 4114–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ghislain M., Dohmen R. J., Levy F., Varshavsky A. (1996) Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15, 4884–4899 [PMC free article] [PubMed] [Google Scholar]
- 12. Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S. (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107, 667–677 [DOI] [PubMed] [Google Scholar]
- 13. Braun S., Matuschewski K., Rape M., Thoms S., Jentsch S. (2002) Role of the ubiquitin-selective CDC48(UFD1/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 21, 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. DeLaBarre B., Brunger A. T. (2003) Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 10, 856–863 [DOI] [PubMed] [Google Scholar]
- 15. Huyton T., Pye V. E., Briggs L. C., Flynn T. C., Beuron F., Kondo H., Ma J., Zhang X., Freemont P. S. (2003) The crystal structure of murine p97/VCP at 3.6 Å. J. Struct. Biol. 144, 337–348 [DOI] [PubMed] [Google Scholar]
- 16. Dreveny I., Kondo H., Uchiyama K., Shaw A., Zhang X., Freemont P. S. (2004) Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. EMBO J. 23, 1030–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buchberger A., Howard M. J., Proctor M., Bycroft M. (2001) The UBX domain. A widespread ubiquitin-like module. J. Mol. Biol. 307, 17–24 [DOI] [PubMed] [Google Scholar]
- 18. Kondo H., Rabouille C., Newman R., Levine T. P., Pappin D., Freemont P., Warren G. (1997) p47 is a cofactor for p97-mediated membrane fusion. Nature 388, 75–78 [DOI] [PubMed] [Google Scholar]
- 19. Uchiyama K., Jokitalo E., Kano F., Murata M., Zhang X., Canas B., Newman R., Rabouille C., Pappin D., Freemont P., Kondo H. (2002) VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J. Cell Biol. 159, 855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Neuber O., Jarosch E., Volkwein C., Walter J., Sommer T. (2005) Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat. Cell. Biol. 7, 993–998 [DOI] [PubMed] [Google Scholar]
- 21. Verma R., Oania R., Fang R., Smith G. T., Deshaies R. J. (2011) Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol. Cell 41, 82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Meyer H. H., Shorter J. G., Seemann J., Pappin D., Warren G. (2000) A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 19, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hitchcock A. L., Krebber H., Frietze S., Lin A., Latterich M., Silver P. A. (2001) The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol. Biol. Cell 12, 3226–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bruderer R. M., Brasseur C., Meyer H. H. (2004) The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism. J. Biol. Chem. 279, 49609–49616 [DOI] [PubMed] [Google Scholar]
- 25. Jarosch E., Taxis C., Volkwein C., Bordallo J., Finley D., Wolf D. H., Sommer T. (2002) Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4, 134–139 [DOI] [PubMed] [Google Scholar]
- 26. Heo J. M., Livnat-Levanon N., Taylor E. B., Jones K. T., Dephoure N., Ring J., Xie J., Brodsky J. L., Madeo F., Gygi S. P., Ashrafi K., Glickman M. H., Rutter J. (2010) A stress-responsive system for mitochondrial protein degradation. Mol. Cell 40, 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tran J. R., Tomsic L. R., Brodsky J. L. (2011) A Cdc48p-associated factor modulates endoplasmic reticulum-associated degradation, cell stress, and ubiquitinated protein homeostasis. J. Biol. Chem. 286, 5744–5755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guthrie C., Fink G. R. (eds) (1991) Guide to yeast genetics and molecular biology. Methods Enzymol. 194, 1–863 [PubMed] [Google Scholar]
- 29. Chiu J., March P. E., Lee R., Tillett D. (2004) Site-directed, ligase-independent mutagenesis (SLIM). A single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 32, e174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Storici F., Resnick M. A. (2006) The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 409, 329–345 [DOI] [PubMed] [Google Scholar]
- 31. Hochstrasser M., Funakoshi M. (2012) Disulfide engineering to map subunit interactions in the proteasome and other macromolecular complexes. Methods Mol. Biol. 832, 349–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu P., Duong D. M., Peng J. (2009) Systematical optimization of reverse-phase chromatography for shotgun proteomics. J. Proteome Res. 8, 3944–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P. (2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis. The yeast proteome. J. Proteome Res. 2, 43–50 [DOI] [PubMed] [Google Scholar]
- 34. Hochstrasser M., Ellison M. J., Chau V., Varshavsky A. (1991) The short-lived MAT α2 transcriptional regulator is ubiquitinated in vivo. Proc. Natl. Acad. Sci. U.S.A. 88, 4606–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tomko R. J., Jr., Hochstrasser M. (2011) Incorporation of the Rpn12 subunit couples completion of proteasome regulatory particle lid assembly to lid-base joining. Mol. Cell 44, 907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen P., Johnson P., Sommer T., Jentsch S., Hochstrasser M. (1993) Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT α2 repressor. Cell 74, 357–369 [DOI] [PubMed] [Google Scholar]
- 37. Mannhaupt G., Schnall R., Karpov V., Vetter I., Feldmann H. (1999) Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 450, 27–34 [DOI] [PubMed] [Google Scholar]
- 38. Xie Y., Varshavsky A. (2001) RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome. A negative feedback circuit. Proc. Natl. Acad. Sci. U.S.A. 98, 3056–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burroughs A. M., Iyer L. M., Aravind L. (2011) Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Mol. Biosyst. 7, 2261–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yun C., Stanhill A., Yang Y., Zhang Y., Haynes C. M., Xu C. F., Neubert T. A., Mor A., Philips M. R., Ron D. (2008) Proteasomal adaptation to environmental stress links resistance to proteotoxicity with longevity in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 105, 7094–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stanhill A., Haynes C. M., Zhang Y., Min G., Steele M. C., Kalinina J., Martinez E., Pickart C. M., Kong X. P., Ron D. (2006) An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol. Cell 23, 875–885 [DOI] [PubMed] [Google Scholar]
- 42. Pharoah P. D., Tsai Y. Y., Ramus S. J., Phelan C. M., Goode E. L., Lawrenson K., Buckley M., Fridley B. L., Tyrer J. P., Shen H., Weber R., Karevan R., Larson M. C., Song H., Tessier D. C., Bacot F., Vincent D., Cunningham J. M., Dennis J., Dicks E., Australian Cancer Study, Australian Ovarian Cancer Study Group, Aben K. K., Anton-Culver H., Antonenkova N., Armasu S. M., Baglietto L., Bandera E. V., Beckmann M. W., Birrer M. J., Bloom G., Bogdanova N., Brenton J. D., Brinton L. A., Brooks-Wilson A., Brown R., Butzow R., Campbell I., Carney M. E., Carvalho R. S., Chang-Claude J., Chen Y. A., Chen Z., Chow W. H., Cicek M. S., Coetzee G., Cook L. S., Cramer D. W., Cybulski C., Dansonka-Mieszkowska A., Despierre E., et al. (2013) GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 45, 362–370, 370e1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web. A case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 44. Wolf D. H., Stolz A. (2012) The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim. Biophys. Acta 1823, 117–124 [DOI] [PubMed] [Google Scholar]
- 45. Stolz A., Hilt W., Buchberger A., Wolf D. H. (2011) Cdc48. A power machine in protein degradation. Trends Biochem. Sci. 36, 515–523 [DOI] [PubMed] [Google Scholar]
- 46. Johnson E. S., Ma P. C., Ota I. M., Varshavsky A. (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270, 17442–17456 [DOI] [PubMed] [Google Scholar]
- 47. Hiller M. M., Finger A., Schweiger M., Wolf D. H. (1996) ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273, 1725–1728 [DOI] [PubMed] [Google Scholar]
- 48. Buchberger A. (2010) Control of ubiquitin conjugation by cdc48 and its cofactors. Subcell. Biochem. 54, 17–30 [DOI] [PubMed] [Google Scholar]
- 49. Guerrero C., Milenkovic T., Przulj N., Kaiser P., Huang L. (2008) Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc. Natl. Acad. Sci. U.S.A. 105, 13333–13338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Verma R., Chen S., Feldman R., Schieltz D., Yates J., Dohmen J., Deshaies R. J. (2000) Proteasomal proteomics. Identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11, 3425–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Russell S. J., Steger K. A., Johnston S. A. (1999) Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 274, 21943–21952 [DOI] [PubMed] [Google Scholar]
- 52. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 53. Kirkpatrick D. S., Dale K. V., Catania J. M., Gandolfi A. J. (2003) Low-level arsenite causes accumulation of ubiquitinated proteins in rabbit renal cortical slices and HEK293 cells. Toxicol. Appl. Pharmacol. 186, 101–109 [DOI] [PubMed] [Google Scholar]
- 54. Haugen A. C., Kelley R., Collins J. B., Tucker C. J., Deng C., Afshari C. A., Brown J. M., Ideker T., Van Houten B. (2004) Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 5, R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Isakov E., Stanhill A. (2011) Stalled proteasomes are directly relieved by P97 recruitment. J. Biol. Chem. 286, 30274–30283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Freeman M. (2008) Rhomboid proteases and their biological functions. Annu. Rev. Genet. 42, 191–210 [DOI] [PubMed] [Google Scholar]
- 57. Zettl M., Adrain C., Strisovsky K., Lastun V., Freeman M. (2011) Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell 145, 79–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Greenblatt E. J., Olzmann J. A., Kopito R. R. (2011) Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat. Struct. Mol. Biol. 18, 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vij S., Tyagi A. K. (2008) A20/AN1 zinc-finger domain-containing proteins in plants and animals represent common elements in stress response. Funct. Integr. Genomics 8, 301–307 [DOI] [PubMed] [Google Scholar]
- 60. Mukhopadhyay A., Vij S., Tyagi A. K. (2004) Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. U.S.A. 101, 6309–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Heyninck K., Beyaert R. (2005) A20 inhibits NF-κB activation by dual ubiquitin-editing functions. Trends Biochem. Sci. 30, 1–4 [DOI] [PubMed] [Google Scholar]
- 62. Huang J., Teng L., Li L., Liu T., Li L., Chen D., Xu L. G., Zhai Z., Shu H. B. (2004) ZNF216 is an A20-like and IκB kinase γ-interacting inhibitor of NFκB activation. J. Biol. Chem. 279, 16847–16853 [DOI] [PubMed] [Google Scholar]
- 63. Alberts S. M., Sonntag C., Schäfer A., Wolf D. H. (2009) Ubx4 modulates cdc48 activity and influences degradation of misfolded proteins of the endoplasmic reticulum. J. Biol. Chem. 284, 16082–16089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schuberth C., Buchberger A. (2005) Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat. Cell Biol. 7, 999–1006 [DOI] [PubMed] [Google Scholar]
- 65. Rumpf S., Jentsch S. (2006) Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol. Cell 21, 261–269 [DOI] [PubMed] [Google Scholar]
- 66. Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 67. Chia W. S., Chia D. X., Rao F., Bar Nun S., Geifman Shochat S. (2012) ATP binding to p97/VCP D1 domain regulates selective recruitment of adaptors to its proximal N-domain. PLoS One 7, e50490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Baek G. H., Kim I., Rao H. (2011) The Cdc48 ATPase modulates the interaction between two proteolytic factors Ufd2 and Rad23. Proc. Natl. Acad. Sci. U.S.A. 108, 13558–13563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wilson J. D., Liu Y., Bentivoglio C. M., Barlowe C. (2006) Sel1p/Ubx2p participates in a distinct Cdc48p-dependent endoplasmic reticulum-associated degradation pathway. Traffic 7, 1213–1223 [DOI] [PubMed] [Google Scholar]
- 70. Barthelme D., Sauer R. T. (2013) Bipartite determinants mediate an evolutionarily conserved interaction between Cdc48 and the 20S peptidase. Proc. Natl. Acad. Sci. U.S.A. 110, 3327–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schuberth C., Richly H., Rumpf S., Buchberger A. (2004) Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep. 5, 818–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang B., Alam S. L., Meyer H. H., Payne M., Stemmler T. L., Davis D. R., Sundquist W. I. (2003) Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4. J. Biol. Chem. 278, 20225–20234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ye Y., Meyer H. H., Rapoport T. A. (2003) Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol. Dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]