Background: Rpl4, Rpl17, and Rpl39 expose patch-like domains to the empty ribosomal polypeptide tunnel.
Results: Rpl4 contacts various nascent chain residues, even far from its patch-like domain. Rpl17 contacts only hydrophobic α-helical segments, other nearby nascent chain segments are excluded.
Conclusion: The tunnel topology of Rpl4 and Rpl17 must be flexible.
Significance: The amino acid sequence of nascent chains affects the topology of Rpl4 and Rpl17.
Keywords: Protein Folding, Protein Synthesis, Ribosomes, Translation, Yeast, Newly Synthesized Polypeptides, Site-specific Cross-linking
Abstract
As translation proceeds, nascent polypeptides pass through an exit tunnel that traverses the large ribosomal subunit. Three ribosomal proteins, termed Rpl4, Rpl17, and Rpl39 expose domains to the interior of the exit tunnel of eukaryotic ribosomes. Here we generated ribosome-bound nascent chains in a homologous yeast translation system to analyze contacts between the tunnel proteins and nascent chains. As model proteins we employed Dap2, which contains a hydrophobic signal anchor (SA) segment, and the chimera Dap2α, in which the SA was replaced with a hydrophilic segment, with the propensity to form an α-helix. Employing a newly developed FLAG exposure assay, we find that the nascent SA segment but not the hydrophilic segment adopted a stable, α-helical structure within the tunnel when the most C-terminal SA residue was separated by 14 residues from the peptidyl transferase center. Using UV cross-linking, antibodies specifically recognizing Rpl17 or Rpl39, and a His6-tagged version of Rpl4, we established that all three tunnel proteins of yeast contact the SA, whereas only Rpl4 and Rpl39 also contact the hydrophilic segment. Consistent with the localization of the tunnel exposed domains of Rpl17 and Rpl39, the SA was in contact with Rpl17 in the middle region and with Rpl39 in the exit region of the tunnel. In contrast, Rpl4 was in contact with nascent chain residues throughout the ribosomal tunnel.
Introduction
Newly synthesized polypeptides exit the ribosome through a tunnel of ∼100 Å. The exit tunnel is conserved and structural details of the ribosomal tunnels from all kingdoms of life are available (1–10). In contrast to other polypeptide-conducting assemblies, for example the translocon in the endoplasmic reticulum membrane, the ribosomal tunnel is not exclusively made of proteins but built of ribosomal RNA with intermingled domains of only three ribosomal proteins (see Fig. 1, A and B). The tunnel originates at the peptidyl transferase center (PTC),2 traverses the body of the large subunit, and exits at a platform on the opposite side. Near the PTC the tunnel wall consists of ribosomal RNA only; at a distance of ∼20 Å, the tunnel bends and its diameter narrows to ∼10 Å. Adjacent to this most narrow constriction, two universally conserved ribosomal proteins contribute to the tunnel wall (see Fig. 1B). Rpl4 (L4 in archaea and eubacteria) exposes a patch-like domain to the lumen (see Fig. 1C), Rpl17 (L22 in archaea and eubacteria) exposes a long β-hairpin with a unique, twisted conformation (see Fig. 1D). Distal to the tip of the β-hairpin, the tunnel continues with more or less constant diameter to widen into the so-called vestibule ∼20 Å before the tunnel exit (11). In archaea and eukaryotes, part of the vestibule wall is formed by Rpl39 (L39 in archaea). Rpl4 and Rpl17 possess surface exposed globular domains (Fig. 1, A, C, and D). The globular domain of Rpl17 is arranged at the exit platform, and the globular domain of Rpl4 is positioned close to the subunit interface near the L1 protuberance. In yeast, Rpl4 and Rpl17 are essential proteins, whereas Rpl39 is dispensable. However, loss of Rpl39 causes severe growth defects similar to aminoglycoside sensitivity and cold sensitivity (12, 13).
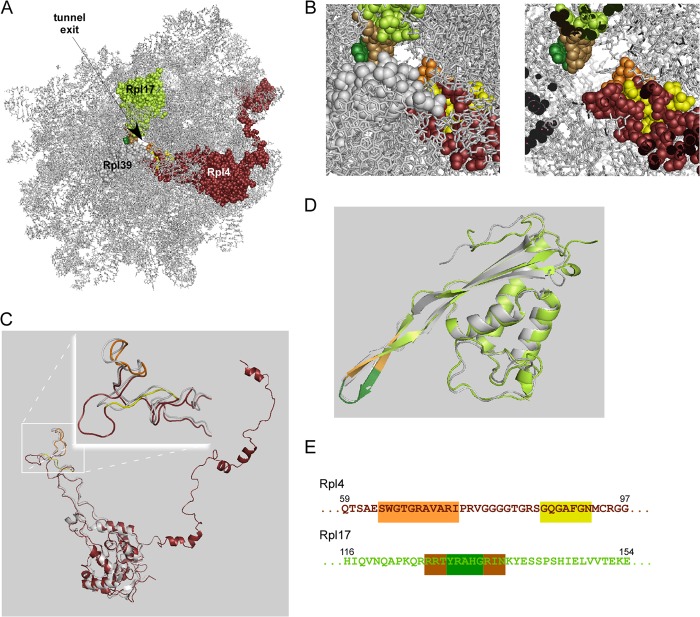
FIGURE 1.
The ribosomal tunnel proteins of yeast. A, view of the Saccharomyces cerevisiae large ribosomal subunit based on PDB code 3U5H/3U5I (7, 10). Shown is the ribosomal RNA (gray) of the large subunit and the three tunnel proteins Rpl4 (purple), Rpl17 (lime), and Rpl39 (gray). B, enlarged image section of the tunnel exit region. Left, exterior view; right, same image section but with a clipping plane at the constriction site of the tunnel. Rpl4 (purple, flexible residues in orange, insertion segment in yellow); Rpl17 (lime, flexible tip of the β-hairpin in green, hinges in brown). C, superposition of S. cerevisiae Rpl4 (colors are as described in A, PDB code 3U5I) and Haloarcula marismortui L4 (gray, PDB code 2QA4). The inset shows a close-up of the tunnel exposed segments of Rpl4/L4. The flexible segment of S. cerevisiae Rpl4 (orange) is structurally similar to the corresponding segment of H. marismortui L4. The insertion segment (yellow) leads to the formation of an extended loop in Rpl4. D, superposition of S. cerevisiae Rpl17 (color code as described in A, PDB code 3U5I) and H. marismortui L22 (gray, PDB code 2QA4) reveals structural similarity. E, partial amino acid sequences of S. cerevisiae Rpl4 (amino acids 59–97) and S. cerevisiae Rpl17 (amino acids 116–154), containing the tunnel exposed flexible segments. Rpl4, orange, flexible residues; yellow, insertion segment; Rpl17, green, flexible tip, brown, hinges.
The ribosomal tunnel has important functions in the regulation of translation and in early protein biogenesis (9, 14, 15). On the one hand, the conformation of a nascent polypeptide is strongly affected by the tunnel environment. On the other hand, the nascent polypeptide conformation impacts the tunnel components, which in turn may transmit the information to distant sites of the ribosome. The exact mechanisms of the interplay between nascent chains, tunnel components, and other functionally important sites of the ribosome are not understood.
Some nascent polypeptides can adopt an α-helical structure within the ribosomal tunnel. This was determined with two different approaches that make use of the fact that polypeptide segments are more compact when they adopt an α-helical rather than an extended conformation. In the tunnel of wheat germ ribosomes, α-helix formation was monitored by incorporating two fluorescent dyes into the same nascent chain and monitoring separation of the dyes via FRET (16, 17). In the tunnel of rabbit ribosomes, α-helix formation was tested via the availability of cysteine residues within the nascent chain for modifying reagents, which cannot enter the ribosomal tunnel (18–20). In both systems, it was found that transmembrane (TM) segments can adopt an α-helical conformation within the eukaryotic ribosomal tunnel. A later cryo-EM study revealed that a hydrophilic nascent chain segment with a strong propensity to from an α-helix even free in solution indeed adopted α-helical conformation in the vestibule of the wheat germ tunnel (21).
Until now, studies of contacts between eukaryotic tunnel proteins and nascent chains are confined to the wheat germ system. In a previous study, it was shown that ribosomal proteins corresponding in size to Rpl4 (∼40 kDa), Rpl17 (∼20 kDa), and Rpl39 (∼6 kDa) contact a TM segment inside the tunnel (16). In a later study, the consequences of these interactions for translocation of multimembrane-spanning proteins into the endoplasmic reticulum membrane were characterized (17, 22). In this context, Lin and co-workers (22) established the contact between TM segments and Rpl17 using an Rpl17-specific antibody. Due to the lack of antibodies, similar experiments were not performed with respect to Rpl4 or Rpl39 (17). To date, contacts between the tunnel proteins and non-TM segments have not been analyzed in similar detail.
Here, we have employed a homologous yeast translation system, a newly developed FLAG-exposure assay, and site-specific UV cross-linking to identify and characterize interactions of nascent TM and non-TM segments with the ribosomal tunnel proteins Rpl4, Rpl17, and Rpl39 from yeast. As model nascent chains, we employed Dap2 and Dap2α, which differ in a 16-residue segment only. Dap2 contains a hydrophobic SA, which forms an α-helical TM anchor, and Dap2α contains a hydrophilic segment with the propensity to form an α-helix. Using this set-up, we asked: does the SA segment fold in the ribosomal tunnel of yeast? Does folding occur in the vestibule or already in the narrower regions of the tunnel? Does a hydrophilic nascent chain segment with the propensity to form an α-helix also fold? How do the contacts of the TM and non-TM segments to the three tunnel proteins compare?
EXPERIMENTAL PROCEDURES
Strains and Plasmids
MH272–3fα (ura3, leu2, his3, trp1, ade2) (23) was used as a wild type strain. To generate a strain expressing exclusively His6-Rpl4a, we generated a Δrpl4aΔrpl4b strain, in which both copies of RPL4 (RPL4A and RPL4B) were deleted. To that end, RPL4A and RPL4B plus 300 base pairs up- and downstream of the open reading frames were cloned into pSP65 (Promega). In the case of RPL4a, a HindIII/StuI fragment within the coding region was replaced with the LEU2 marker gene. In the case of RPL4b, a BstEII/MfeI fragment within the coding region was replaced with the HIS3 gene. The resulting disruption constructs were used to generate the diploid MH272–3f rpl4a::LEU2 rpl4b::HIS3 strain. Tetrad dissection revealed that haploid strains containing both mutations rpl4a::LEU2 and rpl4b::HIS3 were unviable (data not shown). The N-terminally His6-tagged version of RPL4a contains a 42-base pair insertion following the third codon of the RPL4a open reading frame encoding for the amino acids GSSHHHHHHSSGLV. In addition, codons four and five (numbering corresponding to wild type Rpl4a) were changed to GCTAGC to introduce a Nhe1 site. This results in the exchange of Q4L and V5A at the amino acid level. Expression of His6-Rpl4 from plasmid pYCPlac22 in a Δrpl4aΔrpl4b background rescued the lethal phenotype of the double deletion strain (data not shown).
Plasmids encoding Dap2 (pSPUTK-Dap2), Dap2α (pSPUTK-Dap2α-E2K), and the FLAG-tagged versions, pSPUTK-FLAG-Dap2, pSPUTK-FLAG-Dap2α and pSPUTK-FLAG-Pgk1, which contain the DYKDDDDK peptide inserted behind the initiator methionine were described previously (24, 25). Plasmids pSPUTK-FLAG-Dap2-amb39, pSPUTK-Dap2-amb39, pSPUTK-Dap2α-amb39, pSPUTK-Dap2-amb38, pSPUTK-Dap2α-amb38, pSPUTK-Dap2-amb37, pSPUTK-Dap2α-amb37, pSPUTK-Dap2-amb36, pSPUTK-Dap2α-amb36, pSPUTK-Dap2-amb35 contain a TAG codon at codon position 39, 38, 37, 36, or 35 of the respective gene. pSPUTK-FLAG-Dap2ΔN is a version of Dap2, in which the N-terminal 23 amino acids were replaced with the FLAG tag and pSPUTK-FLAG-Dap2αΔN is a version of Dap2α, in which the N-terminal 23 amino acids were replaced with the FLAG tag (Fig. 2A).
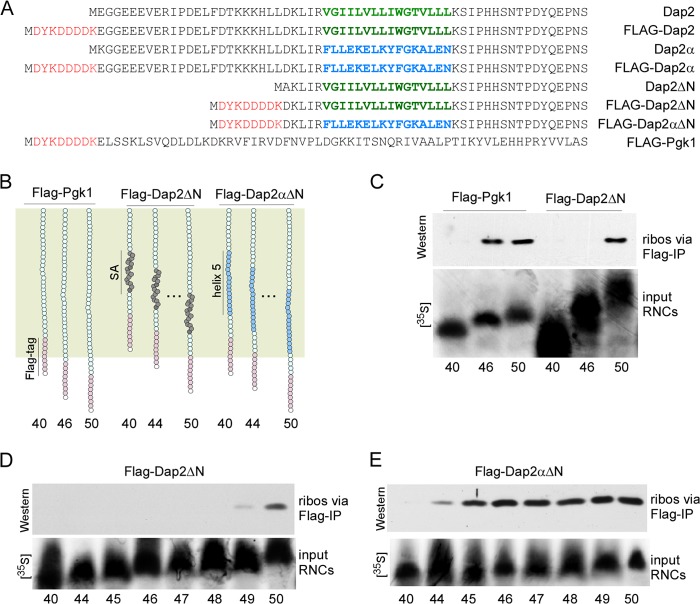
FIGURE 2.
Determination of the effective length of the yeast ribosomal tunnel. A, amino acid sequences of the nascent chains employed in this study. The FLAG tag is shown in red, the SA segment of Dap2 is shown in green, and helix 5 of Pgk1, which replaces the SA in Dap2α, is shown in blue. B, schematic representation of nascent FLAG-Pgk1, FLAG-Dap2ΔN, and FLAG-Dap2αΔN. The ribosomal tunnel is indicated in palegreen. The SA of Dap2 is shown in dark gray; helix 5 of Pgk1, which replaces the SA of Dap2 in FLAG-Dap2αΔN, is shown in light blue; the FLAG tag is shown in pink. The length of nascent chains, including the FLAG tag, is indicated below the schematic. The amino acid sequence of the nascent chains is shown in A. C, FLAG exposure assay with RNCs carrying 40-, 46-, or 50-residue FLAG-Pgk1 or FLAG-Dap2ΔN. Only RNCs exposing the FLAG tag can bind to anti-FLAG beads under native conditions. The amount of RNCs bound to anti-FLAG beads was determined via Western blotting using an antibody recognizing Rps9 as a ribosomal marker (ribos via FLAG-IP). To compare the amount of RNCs applied to the FLAG exposure assays, parallel translation reactions were performed in the presence of [35S]methionine ([35S]) (lower panel, input RNCs). Numbers indicate the length of the nascent chains, including the FLAG tag. D, FLAG exposure assay with RNCs carrying 40-, and 44–50-residue FLAG-Dap2ΔN. The analysis was as described in C. E, FLAG exposure assay with RNCs carrying 40-, 44–50-residue FLAG-Dap2αΔN. The analysis was as described in C.
In Vitro Transcription and Translation Reactions
Yeast translation extracts were prepared as described previously (26) from MH272–3fα (wild type strain) or Δrpl4aΔrpl4b + pYCPlac22-His6-Rpl4 (His6-Rpl4 strain). DNA templates for transcription reactions were generated by PCR using TaqDNA polymerase. The lengths of the nascent chains was determined by the reverse primers that generated truncated, stop codon-less templates as described previously (25). mRNAs were generated by in vitro transcription using SP6 polymerase (26). Translations for UV cross-linking reactions were performed in the presence of [35S]methionine (PerkinElmer Life Science) (24). Translations for the FLAG exposure experiments were performed in the presence of cold methionine. FLAG-tagged nascent chains were used to isolate ribosome nascent chain complexes (RNCs) under native conditions in the FLAG exposure assay, or to enrich UV cross-linking products prior to analysis under denaturing conditions.
FLAG Exposure Assay
FLAG-tagged RNCs were generated and isolated as described (25, 26). In brief, 80-μl translation reactions were incubated at 20 °C for 80 min and were subsequently terminated by the addition of cycloheximide to a final concentration of 200 μg/ml. In UV cross-linking experiments, samples were photolyzed prior to purification as described (24). Translation reactions were centrifuged at 95,000 rpm, at 4 °C in a TLA100 rotor for 20 min. The pellet after centrifugation, containing ribosomes and RNCs, was carefully resuspended in 150 μl of IP buffer (20 mm HEPES-KOH, pH 7.4, 120 mm potassium acetate, 2 mm magnesium acetate, 50 μg/ml trypsin inhibitor, 1 mm PMSF, protease inhibitor mix (1.25 μg/ml leupeptin, 0.75 μg/ml antipain, 0.25 μg/ml chymostatin, 0.25 μg/ml elastinal, 5 μg/ml pepstatin A). Forty μl of anti-FLAG® M2 affinity gel (anti-FLAG beads, Sigma) were added, and the mixture was incubated for 1 h at 4 °C on a shaker. Anti-FLAG beads were separated from the supernatant by centrifugation and were washed twice with 500 μl of ice-cold IP buffer. Bound material was released by incubation in SDS-PAGE sample buffer for 10 min at 95 °C, and subsequently, aliquots were run on 10% Tris-Tricine gels (27), transferred to nitrocellulose filters, and were analyzed by Western blotting using the antibodies indicated in the figure legends. Antibodies employed for decoration of Western blots and affinity purification of UV cross-links were raised in rabbits and described previously (25).
UV Cross-linking
For site-specific UV cross-linking experiments, the TAG amber mutation was introduced at specific positions of Dap2, FLAG-Dap2, and Dap2α and the photoactivatible cross-linker ϵANB-Lys-tRNAamb (tRNA Probes; termed “probe” hereafter) was incorporated at the TAG codon during in vitro translation as previously described (24). The position of the amber mutation indicates the distance of the probe from the N terminus of the nascent chain (see above). By changing the length of the nascent chain, probes were positioned at different distances from the PTC. As an example, 60-residue nascent Dap2 with the amber mutation at position 39 is termed Dap2-amb39-60; the distance of the probe from the PTC in this nascent chain is 21 residues (PTC-Δ21). In some experiments, UV cross-linking was performed with FLAG-tagged nascent chains to enrich nascent chain cross-links via FLAG affinity purification (FLAG-IP). Cross-linking reactions were not affected by the presence of the FLAG tag (data not shown). In UV cross-linking experiments, the residues of the FLAG tag were omitted in the calculation of the length of the nascent chain.
A standard 80 μl of UV cross-linking reaction contained 48 pmol of ϵANB-Lys-tRNAamb. Translations were performed in the dark for 80 min at 20 °C. Samples were then photolyzed in an ice-water bath for 10 min using a 500-watt mercury arc lamp (24). To identify cross-linking partners, samples were denatured, and subsequently, immunoprecipitations were performed under denaturing conditions using protein A-Sepharose beads coated with anti-Rpl17, anti-Rpl39, or anti-Srp54 as indicated (24). His6-Rpl4a extract was employed for the identification of the cross-link between the nascent chain and Rpl4. To this end, translations were performed in a translation extract derived from Δrpl4aΔrpl4b expressing His6-Rpl4a (His6-Rpl4 extract). After UV cross-linking, translation reactions were denatured in dissociation buffer (200 mm Tris-HCl, 4% SDS, 100 μg/ml bovine serum albumin, protease inhibitor mix (as described above), 1 mm PMSF, pH 7.5). Twenty-five μl of the denatured samples were added to 30 μl of Ni-NTA (Qiagen) in a volume of 600 μl buffer N1 (20 mm Tris-HCl, 1% Triton X-100, 300 mm NaCl, pH 8.0). Samples were incubated for 2 h at 4 °C, washed once with 600 μl of buffer N1, once with 600 μl of buffer N2 (20 mm Tris-HCl, 1% Triton X-100, 300 mm NaCl, 50 mm imidazole, pH 8.0), and finally were washed twice with 50 μl of buffer N3 (20 mm Tris-HCl, 300 mm NaCl, pH 8.0). Nascent chains and UV-adducts bound to Ni-NTA were released by incubation in SDS sample buffer for 10 min at 95 °C. Samples were analyzed on 10% Tris-Tricine gels followed by autoradiography. In some experiments RNCs carrying radiolabeled FLAG-tagged nascent chains were isolated via anti-FLAG beads. In this case, the IP buffer was supplemented with 0.1% v/v Triton X-100. Anti-FLAG beads were washed once with 500 μl of ice-cold IP buffer supplemented with 0.1% Triton X-100 and twice with 50 μl ice-cold IP buffer lacking Triton X-100.
RESULTS
To test for α-helix formation within nascent chain segments inside the ribosomal tunnel, we developed the FLAG exposure assay. To that end, a sequence encoding for the eight-residue FLAG tag was inserted 3′ of the start codon of the gene of interest (Fig. 2A). Then, mRNAs of different lengths, each lacking a stop codon, were transcribed in vitro and were subsequently employed to produce RNCs in a yeast translation extract (25, 28). Because affinity purification of RNCs under native conditions works only if the FLAG antibody can access the nascent FLAG tag, the approach allows determination of the effective length of the yeast ribosomal tunnel.
Nascent chains of the soluble, cytosolic enzyme Pgk1 from yeast (29) were employed to determine the length of an extended nascent chain protected by the yeast ribosomal tunnel. To that end, [35S]methionine-labeled RNCs carrying different lengths of FLAG-Pgk1 (Fig. 2B) were generated and similar quantities of RNCs were applied to FLAG-IP reactions (Fig. 2C, input RNCs). Western blotting with a ribosomal marker protein was then employed to determine whether RNCs were able to bind to the anti-FLAG beads (Fig. 2C, ribos via FLAG-IP). Using this FLAG exposure assay, RNCs carrying 46- as well as 50-residue FLAG-Pgk1 were efficiently isolated, however, RNCs carrying 40-residue FLAG-Pgk1 were not (Fig. 2C). Thus, the FLAG epitope was not sufficiently exposed on 40-residue nascent FLAG-Pgk1 but was exposed at a length of 46 amino acids or more.
Dap2 is a type II membrane protein, which contains a short, but strongly hydrophobic, SA localized between amino acids 30–45 (Fig. 2A). During the biogenesis of Dap2, the SA serves as the signal recognition particle (SRP)-dependent targeting signal and later serves as the single TM segment of Dap2 (24, 30–33). To test for compaction of the SA with the FLAG exposure assay, the N-terminal 23 residues of Dap2 were replaced with a FLAG tag (FLAG-Dap2ΔN) (Fig. 2, A and B). As a result, 50 residues, but not 46 residues of FLAG-Dap2ΔN, were sufficient for the binding to the anti-FLAG beads (Fig. 2C). Mapping of residues one by one revealed that the minimal length of nascent FLAG-Dap2ΔN required for efficient exposure of the FLAG tag was 50 residues (Fig. 2D). Thus, FLAG-Dap2ΔN was more compact inside the tunnel than FLAG-Pgk1, most likely because the SA of Dap2 adopted an α-helical structure. The results confirm previous findings obtained with different experimental systems (16–19, 21).
We next wanted to test whether secondary structure formation inside the tunnel also occurred in a hydrophilic segment, which can form a stable α-helix in a folded protein structure. To that end, we used the chimera Dap2α, in which the 16-residue SA sequence was replaced with the complete 16-residue helix 5 of Pgk1 (Fig. 2, A and B) (24, 29). The N-terminal 23 residues of Dap2α were replaced with a FLAG tag, such that FLAG-Dap2αΔN differed from FLAG-Dap2ΔN in only 16 residues (Fig. 2, A and B). Ribosome-bound FLAG-Dap2αΔN was efficiently isolated at a length of 45 residues (Fig. 2E). Thus, replacing the SA of Dap2 with helix 5 of Pgk1 changed the length of the nascent chain required for the exposure of the FLAG tag by five amino acids. Remarkably, the nascent chain length required for FLAG exposure showed a sharp cut-off for both FLAG-Dap2ΔN or FLAG-Dap2αΔN (Fig. 2, D and E). This indicates that the bulk of nascent chains of one kind had adopted the same, or very similar, conformation within the tunnel. Although the SA of FLAG-Dap2ΔN was compact, helix 5 of FLAG-Dap2αΔN was extended (Fig. 2, D and E).
To characterize the interactions of nascent chains with tunnel proteins in more detail, we wanted to unequivocally establish interactions of nascent chains with Rpl4, Rpl17, and Rpl39 in the yeast system. To that end, a UV-activatable probe was introduced at defined positions of nascent chains. This allowed us to test for the proximity of specific nascent chain residues with tunnel components (24).
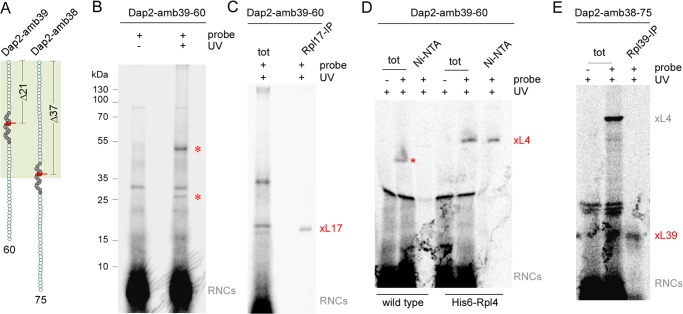
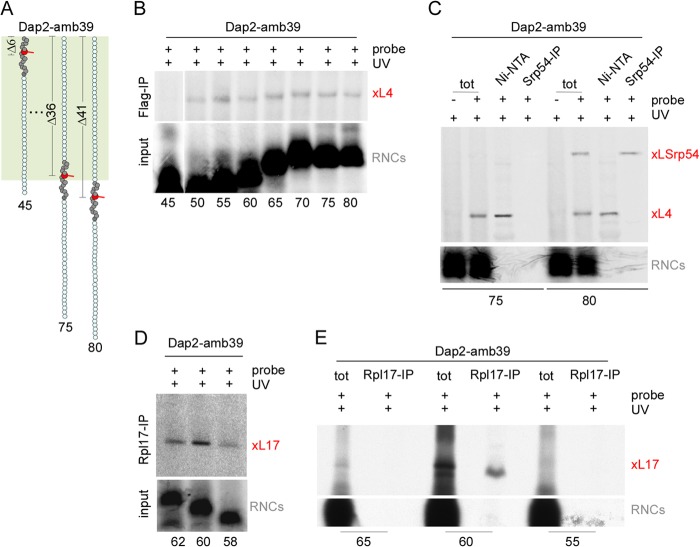
First, the probe was introduced at position 39 of Dap2, and 60-residue radiolabeled nascent chains were generated (Dap2-amb39-60, Fig. 3A). The probe in Dap2-amb39-60 is localized 21 residues distal of the PTC (PTC-Δ21). After UV illumination, two specific cross-link bands were detected via autoradiography (Fig. 3B, red asterisks). The molecular mass of the upper band was consistent with a cross-link of Dap2-amb39-60 to Rpl4 that of the lower band was consistent with a cross-link to Rpl17. Indeed, the faster migrating cross-link was immunoprecipitated with anti-Rpl17 under denaturing conditions (Fig. 3C). Because the available Rpl4 sera did not efficiently immunoprecipitate Rpl4 under denaturing conditions (data not shown) a yeast strain was generated, in which both copies of Rpl4 (Rpl4a and Rpl4b) were replaced with hexahistidine-tagged Rpl4a (Δrpl4aΔrpl4b + His6-Rpl4). RNCs carrying Dap2-amb39-60 were then generated in a wild type or in a translation extract derived from the strain expressing His6-Rpl4 (His6-Rpl4 extract). The molecular mass of the cross-link obtained with His6-Rpl4 extract was increased compared with wild type (Fig. 3D). Moreover, the cross-link to His6-Rpl4, but not to Rpl4, was isolated via Ni-NTA affinity purification (Fig. 3D). Thus, a probe in position PTC-Δ21 within the SA was in contact either with Rpl17 or Rpl4. No cross-link to Rpl39 was detected. To test for the interaction of the SA with Rpl39 in the vestibule of the tunnel, we employed Dap2-amb38-75, in which the probe is positioned at PTC-Δ37 (Fig. 3A). In this nascent chain, the probe was in close contact to a protein of ∼6 kDa, which was identified as Rpl39 via immunoprecipitation using anti-Rpl39 (Fig. 3E). Thus, all three tunnel proteins of yeast make contact to a TM segment, such as the SA of Dap2.
FIGURE 3.
Identification of contacts between a nascent SA and the tunnel proteins Rpl4, Rpl17, and Rpl39. A, schematic representation of 60-residue Dap2-amb39 containing a probe in position PTC-Δ21, and 75-residue Dap2-amb38 containing a probe in position PTC-Δ37. The ribosomal tunnel is indicated in palegreen, the SA of Dap2 is shown in dark gray, and the probe is shown in red. The length of the nascent chains and the distance to the PTC is indicated. B, UV cross-linking pattern of nascent Dap2-amb39-60. Wild type RNCs carrying radiolabeled FLAG-tagged nascent chains, were photolyzed (+ UV), or, as a control, were kept in the dark (−UV). Subsequently, cross-linked products were purified via anti-FLAG beads and were analyzed on Tris-Tricine gels followed by autoradiography. The two major cross-linking products are labeled with a red asterisks. Positions of prestained molecular mass marker proteins are indicated. Note that the N-terminal FLAG tag on the nascent chain, which was employed to enrich the cross-link products, was not included in the calculation of the nascent chain length. C, identification of a cross-link between Rpl17 and Dap2-amb39-60. Wild type RNCs carrying radiolabeled Dap2-amb39-60 were photolyzed as described in B. Subsequently, cross-link products to Rpl17 were immunoprecipitated under denaturing conditions using anti-Rpl17 coupled to protein A-Sepharose beads (Rpl17-IP). xL17 indicates the cross-link product between Dap2-amb39-60 and Rpl17. D, identification of a cross-link between Rpl4 and Dap2-amb39-60. RNCs carrying radiolabeled Dap2-amb39-60 were generated in a wild type (wild type) or His6-Rpl4 (His6-Rpl4) translation extract. As a control, unmodified Lys instead of ϵANB-Lys was incorporated into the nascent chains (−probe). After photolysis, samples were subjected to Ni-NTA affinity purification. xL4 indicates the cross-link product between Dap2-amb39-60 and His6-Rpl4. The red asterisk indicates the cross-link to wild type Rpl4, which runs at a lower molecular mass. E, identification of cross-link between Rpl39 and Dap2-amb38-75. RNCs carrying radiolabeled Dap2-amb38-75 were generated in a wild type translation extract. After photolysis, cross-links to Rpl39 were immunoprecipitated using anti-Rpl39 coupled to protein A-Sepharose beads (Rpl39-IP). xL39 indicates the cross-link product between Dap2-amb39-60 and Rpl39. Analysis of the samples in C–E was as described in B. The total (tot) represents 10% of material employed for immunoprecipitation reactions (C and E) or Ni-NTA purification reactions (D).
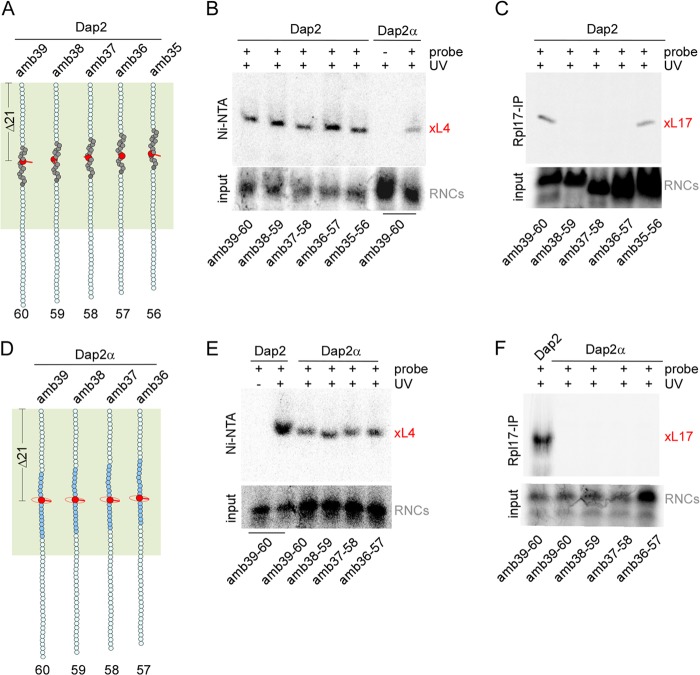
If a nascent chain segment inside of the tunnel adopts α-helical conformation, different scenarios with respect to its contact with tunnel proteins can be envisaged. At one extreme, a stable helical segment might move without changing its orientation with respect to the tunnel wall. Alternatively, the adjacent extended nascent chain segments might allow a helix to rotate around its axis, such that its orientation with respect to the tunnel wall changes. To distinguish between these possibilities, we employed a series of constructs in which the probe was placed at five successive positions between residue 39 (amb39) and residue 35 (amb35) of Dap2. The length of the nascent chains was then shortened successively such that the probe was at PTC-Δ21 in all nascent chains (Fig. 4A). If the SA adopted a stable α-helical conformation (Fig. 2), the probes would stick out to different sides, covering approximately one turn of an α-helix (Fig. 4A). This set of probes was used to test for proximity to Rpl4 and Rpl17, respectively. The results show that all of the probes at PTC-Δ21 were in close proximity to Rpl4 (Fig. 4B). However, only two probes, oriented to the same side of the helix, were in close proximity to Rpl17 (Fig. 4, A and C, Dap2-amb39-60 and Dap2-amb35-56). Thus, Rpl4 was in contact with all sides of the α-helical SA, whereas Rpl17 was in contact with only one side of the same helix.
FIGURE 4.
Sequence context and orientation of a probe at position PTC-Δ21 affects cross-linking to Rpl17 but not to Rpl4. A, schematic representation of Dap2-amb39-60, Dap2-amb38–59, Dap2-amb37–58, Dap2-amb36–57, and Dap2-amb35-56. The color code is as described in the legend to Fig. 3. B, contacts between Rpl4 and a probe in the SA at position PTC-Δ21. RNCs carrying radiolabeled nascent chains as shown in A were generated in a His6-Rpl4 translation extract. For direct comparison, RNCs carrying Dap2α-amb39-60 were analyzed in parallel. As a control, unmodified Lys instead of ϵANB-Lys was incorporated into the nascent chains (−probe). After UV cross-linking samples were denatured, cross-link products were purified via Ni-NTA and were analyzed on a Tris-Tricine gel followed by autoradiography. The lower panel shows the relative amount of RNCs applied to the cross-linking reactions (input). The cross-link between nascent chains and Rpl4 is indicated in red (xL4). C, contacts between Rpl17 and a probe in the SA at position PTC-Δ21. RNCs carrying radiolabeled nascent chains shown as in A were generated in a wild type translation extract. After UV cross-linking samples were denatured, cross-link products were immunoprecipitated via anti-Rpl17-coupled beads and were analyzed on a Tris-Tricine gel followed by autoradiography. The cross-link between nascent chains and Rpl17 is indicated in red (xL17). D, schematic representation of Dap2α-amb39-60, Dap2α-amb38–59, Dap2α-amb37–58, and Dap2α-amb36–57. The color code is as described in the legend to Fig. 3, and helix 5 is indicated in blue. E, contacts between Rpl4 and a probe in helix 5 at position PTC-Δ21. Nascent chains shown in D were analyzed as described in B. For direct comparison, RNCs carrying Dap2-amb39-60 were analyzed in parallel. As a control, one sample was kept in the dark (−UV). F, contacts between Rpl17 and a probe in helix 5 at position PTC-Δ21. RNCs as shown in D were generated in a wild type translation extract and were analyzed as described in C. The UV cross-link between Rpl17 and Dap2-amb39-60 (lane 1) served as a positive control.
Dap2α was employed to test for interactions of Rpl4 and Rpl17 with extended, hydrophilic nascent chain segments. To that end, the probe was successively incorporated at position PTC-Δ21 between residues 39–36 of Dap2α (Fig. 4D). All of these probe positions were in contact with Rpl4 (Fig. 4E). In contrast, none of the probe positions were in contact with Rpl17 (Fig. 4F). Next, we wanted to establish the region of the tunnel in which the nascent chain was in contact with the three tunnel proteins. In the case of Rpl4 and Rpl17, we employed Dap2-amb39 because Dap2-amb39-60 was in contact with both of the two tunnel proteins (Fig. 3, C and D, and Fig. 4, B and C). The length of Dap2-amb39 was increased from 45 to 80 in steps of five residues (Fig. 5A). Rpl4 was not close to the probe at PTC-Δ6 (Dap2-amb39-45) proximal of the tunnel constriction (Fig. 5, A and B). However, Rpl4 was close to the probe starting at position PTC-Δ11 up to position PTC-Δ41 at the very exit of the tunnel (Fig. 5, A and B). To confirm that Rpl4 was able to contact a probe emerging from the tunnel, we made use of the well characterized interaction between the SA of Dap2 and the Srp54 subunit of the SRP (24, 33). As expected, Srp54 was in contact with the SA in Dap2-amb39-80, but not in Dap2-39-75 (Fig. 5, A and C). In the same experiment, Rpl4 was in close proximity to both nascent chains (Fig. 5C). Thus, Rpl4 can indeed contact a probe accessible also for SRP. In contrast, Rpl17 was in contact with the probe only in a narrow and confined region of the tunnel. Contact of Rpl17 to a probe in the SA was detected between PTC-Δ19 and PTC-Δ23 (Fig. 5D), but was not detected for a probe at PTC-Δ16 or PTC-Δ26 (Fig. 5E).
FIGURE 5.
Rpl17 contacts the SA in a confined region, whereas Rpl4 contacts the SA throughout the tunnel. A, schematic representation of nascent Dap2-amb39 between 45 to 80 residues. The color code is as described in the legend to Fig. 3. B, Rpl4 is close to a probe between positions PTC-Δ11–PTC-Δ41. RNCs carrying FLAG-tagged radiolabeled nascent chains of the indicated length were generated in wild type translation extract. UV cross-linking and enrichment of the cross-link products was performed as described in the legend to Fig. 3. Shown is an autoradiography of the material isolated via FLAG-IP (upper panel) and of the RNCs employed for the cross-linking reactions (lower panel, input). The cross-link between the nascent chains and Rpl4 is indicated in red (xL4). C, a probe at position PTC-Δ41 at the exit of the ribosomal tunnel is in close contact to either Rpl4 or Srp54. RNCs carrying radiolabeled nascent Dap2-amb39 of 75 or 80 residues were generated in a His6-Rpl4 translation extract and UV cross-linking was performed as described in the legend to Fig. 3. As a control, unmodified Lys instead of ϵANB-Lys was incorporated into the nascent chain (−probe). Cross-link products were purified via Ni-NTA as described in the legend to Fig. 3. Cross-links between nascent chains and Srp54 were immunoprecipitated using αRpl54 coupled to protein A-Sepharose beads (Srp54-IP). Samples were analyzed on a Tris-Tricine gel followed by autoradiography. Total (tot) represents 10% of the material employed in the immunoprecipitation reactions. The cross-link between the nascent chain and Rpl4 (xL4), the cross-link between the nascent chain and Srp54 (xLSrp54) are indicated in red. The lower panel shows the RNCs on a shorter exposure of the same gel. D, contacts between Rpl17 and probes between positions PTC-Δ19–PTC-Δ23. RNCs carrying radiolabeled nascent Dap2-amb39 were generated in a wild type translation extract and were analyzed as described in the legend to Fig. 4. The cross-link between the nascent chain and Rpl17 (xL17) is indicated in red. The lower panel shows the relative amount of RNCs employed for the cross-linking reactions (input). E, contacts between Rpl17 and probes between positions PTC-Δ16–PTC-Δ26. The experiment was performed and analyzed as described in the legend to Fig. 4, the cross-link between the nascent chain and Rpl17 (xL17) is indicated in red. The lower panel shows the RNCs on a shorter exposure of the same gel.
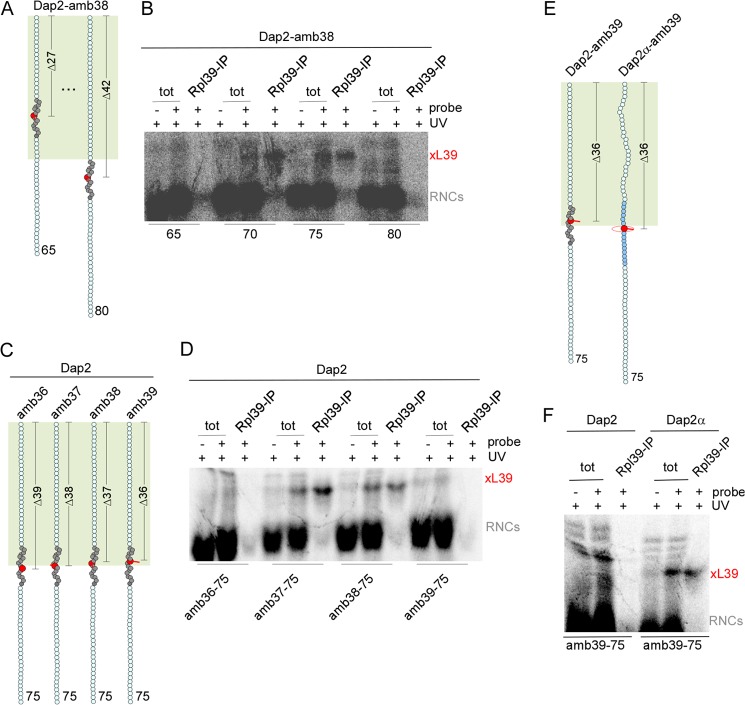
To define the region of the tunnel in which nascent chains were in contact with Rpl39, we employed Dap2-amb38 (Fig. 6A, compare with Fig. 3E). The probe in 70- and 75-residue Dap2-amb38, but not in 65- or 80-residue Dap2-amb38, was in close proximity of Rpl39 (Fig. 6B). Thus, Rpl39 was in contact with SA residues between PTC-Δ32 to PTC-Δ37, in the vestibule region of the tunnel. Proximity of Rpl39 to the probe in the SA was orientation dependent (Fig. 6C). Only a probe at position 37 or 38 but not at position 36 or 39 within the SA was in contact with Rpl39 (Fig. 6D). However, in contrast to Rpl17, the interaction of Rpl39 was not confined to residues within the SA, but Rpl39 was also in close contact with helix 5 of Dap2α (Fig. 6, E and F).
FIGURE 6.
Rpl39 contacts nascent chains in the vestibule region of the ribosomal tunnel. A, schematic representation of Dap2-amb38-65 and Dap2-amb38-80. The color code is described in the legend to Fig. 3. B, contacts between Rpl39 and probes in the SA between PTC-Δ27–PTC-Δ42 as shown in A. RNCs carrying radiolabeled nascent Dap2-amb38 were generated in a wild type translation extract, and UV cross-linking and analysis was performed as described in the legend to Fig. 3. 10% of the material employed in the reaction (tot) and the immunoprecipitated (Rpl39-IP). The cross-link between the nascent chain and Rpl39 (xL39) is indicated. The cross-link between nascent chains and Rpl4 was cut off to improve the facility of inspection. C, schematic representation of Dap2-amb36-75, Dap2-amb37-75, Dap2-amb38-75, and Dap2-amb39-75. D, Rpl39 contacts only specific residues within the SA. Cross-linking, affinity purification, and analysis was performed with the RNCs shown in C as described in B. E, schematic representation of Dap2-amb39-75 and Dap2α-amb39-75. F, Rpl39 contacts probes in the SA and also in helix 5. Cross-linking, affinity purification, and analysis was performed with RNCs as shown in E as described in the legend to Fig. 3.
DISCUSSION
The minimal binding epitope of the FLAG M2 antibody consists of DYKD, whereas the four extraneous residues of the FLAG tag (Fig. 2A) are thought to provide a spacer, improving accessibility and flexibility of the binding epitope (34). The calculations below are based on the assumption that the FLAG M2 antibody can bind efficiently to the DYKD epitope, when the full-length eight-amino acid FLAG tag (M-DYKDDDK) has emerged from the tunnel. Under this assumption, the yeast ribosomal tunnel covers 36 residues of a predominantly extended nascent chain. This is in good agreement with structural data of the eukaryotic ribosomal tunnel (9). A study using a bulky mass-tagging reagent revealed that 33 residues of an extended nascent chain were protected by the tunnel (19). Most likely, the difference of three residues is owed to the different experimental systems: the FLAG exposure assay may require a spacer longer than four residues (see above), the mass-tagging approach might detect nascent chains prior to their exit from the tunnel. Looking at it from a different angle, one may view the tunnel exit as an extended opening region.
We observed a sharp transition from non-binding to binding with the FLAG exposure assay. This observation is best explained by the assumption that the bulk of nascent chains of one kind had adopted the same or similar conformation and that this conformation was stable. Otherwise, one would expect that nascent chains too short to bind when folded but long enough to bind when unfolded, had been trapped by binding to the FLAG antibody, driving the equilibrium to the unfolded state.
There is debate about the tunnel region(s), in which secondary structure formation can occur. Using the mass-tagging approach, poly-alanine segments were found to compact very close to the PTC. This compaction was lost at the constriction but was regained in the vestibule (11). A later study revealed that α-helix formation close to the PTC was confined to artificial segments such as the poly-Ala, however, was not observed for example for a natural TM segment, which adopted α-helical structure only in the vestibule (20). A cryo-EM study visualized a hydrophilic nascent chain segment with strong propensity to form an α-helix in the vestibule but no other region of the tunnel. Of note, 90-residue nascent Dap2 (DPAP-B) was not visible in the tunnel, suggesting that it had not adopted a defined conformation. However, in these experiments, the SA of Dap2 was far outside of the tunnel (21, 35). In contrast, a FRET approach revealed that a TM segment already adopted compact structure close to the PTC prior to passage of the tunnel constriction (17). We find that compaction of the nascent chain occurred when the most C-terminal residue of the SA was at PTC-Δ14. At this distance, the SA just passed the constriction (36) and faced the β-hairpin of Rpl17. Thus, folding of the SA in the yeast ribosomal tunnel occurred significantly before the SA had reached the vestibule (Fig. 7A).
FIGURE 7.
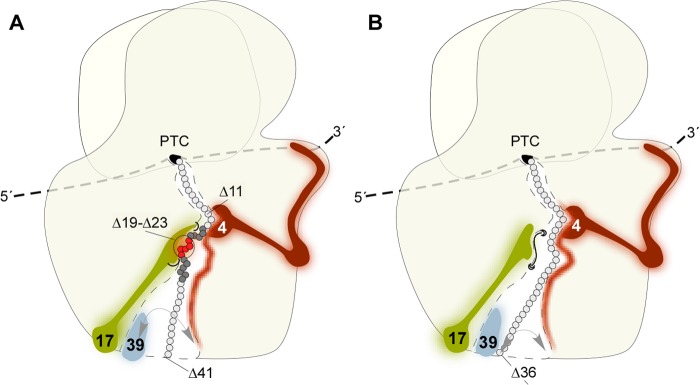
Model of nascent chain conformations and nascent chain interactions with the ribosomal tunnel proteins. A view of the ribosome (beige) translating an mRNA molecule into a polypeptide chain is shown. The entrance of the polypeptide tunnel at the PTC is shown in black. The localization of Rpl17 (17, lime green), Rpl4 (4, purple), Rpl39 (39, gray), roughly reflects structural information (compare with Fig. 1). Nascent chain residues within hydrophilic segments are shown in light gray, and residues within the hydrophobic signal anchor segment in dark gray or red. In the vestibule region of the tunnel, the nascent chain possesses flexibility. This is indicated by the gray arrows. A, contacts of a nascent chain containing a hydrophobic SA segment. Rpl4 contacts residues from PTC-Δ11 to PTC-Δ41 (Δ11-Δ41). This extended zone of Rpl4 contacts (purple shadow in the tunnel lumen) suggests that the tunnel exposed domain of Rpl4 is flexible. In addition, the nascent chain and the probe within the nascent chain may adopt different conformations. Rpl17 contacts amino acid residues within the folded SA segment in only a narrow region of the tunnel (Δ19-Δ23). Rpl39 contacts residues within the SA in the vestibule region (not shown in the model). A nascent chain containing an α-helical domain becomes accessible from the outside 41 residues apart from the PTC (Δ41). B, contacts of a hydrophilic nascent chain. Contacts of Rpl4 and Rpl39 to a hydrophilic nascent chain are similar to those of a nascent chain containing a hydrophobic SA (compare with A). In contrast, Rpl17 is excluded from contacts to a hydrophilic nascent chain (black barrier). This suggests nascent chain-induced conformational changes inside the tunnel, which change the accessibility of Rpl17. A mostly extended hydrophilic nascent chain becomes accessible from the outside 36 residues apart from the PTC (Δ36). The nascent chain is flexible in the vestibule (gray arrow) and e.g. a residue at PTC-Δ36 can contact either Rpl4 or Rpl39. For more details, see “Discussion.”
Considering that Rpl4 and Rpl17 expose domains to the very same region of the tunnel (Fig. 1B), they interact with nascent chains surprisingly differently. What allows the β-hairpin of Rpl17 to be strongly discriminative with respect to its contacts, whereas the patch-like domain of Rpl4 is not? The UV probe used to analyze contacts between nascent chains and tunnel proteins is highly reactive and forms cross-links with any adjacent amino acid. The probe was attached to a lysine, the side chain of which is flexible and can be anywhere in a half-sphere with the radius equal to a fully stretched side chain centered at its Cα. This radius is 7.8 A in the case of lysine, covalent linkage with the probe extends the radius to ∼12 Å (37, 38). Thus, probes at the exit may loop back into the vestibule. However, even then, the probe is far from the patch-like Rpl4 domain seen in the structures of the eukaryotic tunnel (Fig. 1, A and B, and Fig. 7) (7, 8). Also, the cross-linking pattern of Rpl17, which is thought to transduce nascent peptide-specific information to other parts of the ribosome, is peculiar (5, 39). Most of the β-hairpin is rigid and attached to the tunnel wall. Only a short tip between two hinges is thought to possess high flexibility (Fig. 1, D and E) (5, 39). These properties may explain why Rpl17 does not contact a probe in the lower part of the tunnel. However, due to the flexibility and reactivity of the probe, it is surprising that Rpl17 did not contact any of the probes in nascent Dap2α (Fig. 7B). How Rpl17 is excluded from nearby residues, whereas Rpl4 promiscuously contacts residues throughout the tunnel is not understood. However, the surprising difference between Rpl17 and Rpl4 is not confined to yeast. Johnson and co-workers (17) observed similar cross-linking patterns of TM segments to Rpl17 and a protein corresponding in size to Rpl4 in the wheat germ system. The authors suggested that each sample contained folded and unfolded TM segments and that the probe was therefore positioned at different tunnel locations (17). However, the FLAG exposure assay suggests that in the yeast system, nascent chains of one kind adopted a uniform and stable conformation (see above). Moreover, the lack of contacts between Rpl17 and a variety of positions within nascent Dap2α cannot be accounted for with multiple nascent chain conformations.
We speculate that the tunnel exposed domain of Rpl4 is flexible. Indeed, analysis of the static properties of four different bacterial tunnels revealed that the tunnel-exposed loop of bacterial L4 is flexible and may move freely in the lumen (39). The glycine-rich loop of yeast Rpl4 contains an insertion of 8 residues compared with its bacterial counterpart (Fig. 1, C and D). Possibly, the whole segment is more flexible in translating ribosomes than suggested by the crystal structures (Fig. 1, A–C, and Fig. 7). The data suggest that structural changes in a translating ribosome may also affect the β-hairpin of Rpl17. Indeed, specific regions of the tunnel seem to be more flexible than originally thought (39). Specifically the region at PTC-Δ18–22, in which we observe contacts between the SA and Rpl17, was recently found to undergo nascent chain induced conformational changes (36) and nascent chain interactions with ribosomes also alter the conformation of tunnel-exposed ribosomal RNA nucleotides (40).
Johnson and co-workers (17) found that a protein, corresponding in size to Rpl39, formed UV cross-links with the same probe positions that also formed cross-links to Rpl17. In our analysis, none of the probes was in contact with both Rpl17 and Rpl39. Consistent with its localization in the vestibule, Rpl39 was in contact only with probes between PTC-Δ32 to -Δ37 (Fig. 7). In the vestibule, the folded SA possessed a defined orientation with respect to Rpl39, whereas its orientation was random with respect to Rpl4. This observation is consistent with the idea that domains of Rpl4 possessed high flexibility, whereas Rpl17 and Rpl39 occupied more defined positions inside the ribosomal tunnel.
This work was supported by SFB 746, Forschergruppe 967, and by the Excellence Initiative of the German federal and state governments (EXC 294) (to S. R).
- PTC
- peptidyl transferase center
- SA
- signal anchor
- RNC
- ribosome-nascent chain complex
- TM
- transmembrane
- Ni-NTA
- nickel-nitrilotriacetic acid
- SRP
- signal recognition particle
- PDB
- Protein Data Bank.
REFERENCES
- 1. Nissen P., Hansen J., Ban N., Moore P. B., Steitz T. A. (2000) The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 [DOI] [PubMed] [Google Scholar]
- 2. Ban N., Nissen P., Hansen J., Moore P. B., Steitz T. A. (2000) The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289, 905–920 [DOI] [PubMed] [Google Scholar]
- 3. Yusupov M. M., Yusupova G. Z., Baucom A., Lieberman K., Earnest T. N., Cate J. H., Noller H. F. (2001) Crystal structure of the ribosome at 5.5 A resolution. Science 292, 883–896 [DOI] [PubMed] [Google Scholar]
- 4. Harms J., Schluenzen F., Zarivach R., Bashan A., Gat S., Agmon I., Bartels H., Franceschi F., Yonath A. (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107, 679–688 [DOI] [PubMed] [Google Scholar]
- 5. Berisio R., Schluenzen F., Harms J., Bashan A., Auerbach T., Baram D., Yonath A. (2003) Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat. Struct. Biol. 10, 366–370 [DOI] [PubMed] [Google Scholar]
- 6. Armache J. P., Jarasch A., Anger A. M., Villa E., Becker T., Bhushan S., Jossinet F., Habeck M., Dindar G., Franckenberg S., Marquez V., Mielke T., Thomm M., Berninghausen O., Beatrix B., Söding J., Westhof E., Wilson D. N., Beckmann R. (2010) Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution. Proc. Natl. Acad. Sci. U.S.A. 107, 19748–19753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., Yusupov M. (2011) The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334, 1524–1529 [DOI] [PubMed] [Google Scholar]
- 8. Klinge S., Voigts-Hoffmann F., Leibundgut M., Arpagaus S., Ban N. (2011) Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334, 941–948 [DOI] [PubMed] [Google Scholar]
- 9. Wilson D. N., Beckmann R. (2011) The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr. Opin. Struct. Biol. 21, 274–282 [DOI] [PubMed] [Google Scholar]
- 10. Jenner L., Melnikov S., Garreau de Loubresse N., Ben-Shem A., Iskakova M., Urzhumtsev A., Meskauskas A., Dinman J., Yusupova G., Yusupov M. (2012) Crystal structure of the 80S yeast ribosome. Curr. Opin. Struct. Biol. 22, 759–767 [DOI] [PubMed] [Google Scholar]
- 11. Lu J., Deutsch C. (2005) Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 12, 1123–1129 [DOI] [PubMed] [Google Scholar]
- 12. Dresios J., Derkatch I. L., Liebman S. W., Synetos D. (2000) Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry 39, 7236–7244 [DOI] [PubMed] [Google Scholar]
- 13. Peisker K., Braun D., Wölfle T., Hentschel J., Fünfschilling U., Fischer G., Sickmann A., Rospert S. (2008) Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol. Biol. Cell 19, 5279–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rospert S. (2004) Ribosome function: How to govern the fate of a nascent polypeptide. Curr. Biol. 14, R386-R388 [DOI] [PubMed] [Google Scholar]
- 15. Mankin A. S. (2006) Nascent peptide in the “birth canal” of the ribosome. Trends Biochem. Sci. 31, 11–13 [DOI] [PubMed] [Google Scholar]
- 16. Woolhead C. A., McCormick P. J., Johnson A. E. (2004) Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116, 725–736 [DOI] [PubMed] [Google Scholar]
- 17. Lin P. J., Jongsma C. G., Pool M. R., Johnson A. E. (2011) Polytopic membrane protein folding at L17 in the ribosome tunnel initiates cyclical changes at the translocon. J. Cell Biol. 195, 55–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kosolapov A., Tu L., Wang J., Deutsch C. (2004) Structure acquisition of the T1 domain of Kv1.3 during biogenesis. Neuron 44, 295–307 [DOI] [PubMed] [Google Scholar]
- 19. Lu J., Deutsch C. (2005) Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry 44, 8230–8243 [DOI] [PubMed] [Google Scholar]
- 20. Tu L. W., Deutsch C. (2010) A folding zone in the ribosomal exit tunnel for Kv1.3 helix formation. J. Mol. Biol. 396, 1346–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bhushan S., Gartmann M., Halic M., Armache J. P., Jarasch A., Mielke T., Berninghausen O., Wilson D. N., Beckmann R. (2010) α-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 17, 313–317 [DOI] [PubMed] [Google Scholar]
- 22. Lin P. J., Jongsma C. G., Liao S., Johnson A. E. (2011) Transmembrane segments of nascent polytopic membrane proteins control cytosol/ER targeting during membrane integration. J. Cell Biol. 195, 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Heitman J., Movva N. R., Hiestand P. C., Hall M. N. (1991) FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 88, 1948–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berndt U., Oellerer S., Zhang Y., Johnson A. E., Rospert S. (2009) A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc. Natl. Acad. Sci. U.S.A. 106, 1398–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Raue U., Oellerer S., Rospert S. (2007) Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem. 282, 7809–7816 [DOI] [PubMed] [Google Scholar]
- 26. Garcia P. D., Hansen W., Walter P. (1991) In vitro protein translocation across microsomal membranes of Saccharomyces cerevisiae. Methods Enzymol. 194, 675–682 [DOI] [PubMed] [Google Scholar]
- 27. Schägger H., von Jagow G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 [DOI] [PubMed] [Google Scholar]
- 28. Fünfschilling U., Rospert S. (1999) Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell 10, 3289–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watson H. C., Walker N. P., Shaw P. J., Bryant T. N., Wendell P. L., Fothergill L. A., Perkins R. E., Conroy S. C., Dobson M. J., Tuite M. F. (1982) Sequence and structure of yeast phosphoglycerate kinase. EMBO J. 1, 1635–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ng D. T., Brown J. D., Walter P. (1996) Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 134, 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cheng Z., Gilmore R. (2006) Slow translocon gating causes cytosolic exposure of transmembrane and lumenal domains during membrane protein integration. Nat. Struct. Mol. Biol. 13, 930–936 [DOI] [PubMed] [Google Scholar]
- 32. Roberts C. J., Pohlig G., Rothman J. H., Stevens T. H. (1989) Structure, biosynthesis, and localization of dipeptidyl aminopeptidase B, an integral membrane glycoprotein of the yeast vacuole. J. Cell Biol. 108, 1363–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y., Berndt U., Gölz H., Tais A., Oellerer S., Wölfle T., Fitzke E., Rospert S. (2012) NAC functions as a modulator of SRP during the early steps of protein targeting to the ER. Mol. Biol. Cell 23, 3027–3040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roosild T. P., Castronovo S., Choe S. (2006) Structure of anti-FLAG M2 Fab domain and its use in the stabilization of engineered membrane proteins. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Halic M., Becker T., Pool M. R., Spahn C. M., Grassucci R. A., Frank J., Beckmann R. (2004) Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427, 808–814 [DOI] [PubMed] [Google Scholar]
- 36. Lu J., Hua Z., Kobertz W. R., Deutsch C. (2011) Nascent peptide side chains induce rearrangements in distinct locations of the ribosomal tunnel. J. Mol. Biol. 411, 499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Strandberg E., Killian J. A. (2003) Snorkeling of lysine side chains in transmembrane helices: how easy can it get? FEBS Lett. 544, 69–73 [DOI] [PubMed] [Google Scholar]
- 38. Sadlish H., Pitonzo D., Johnson A. E., Skach W. R. (2005) Sequential triage of transmembrane segments by Sec61α during biogenesis of a native multispanning membrane protein. Nat. Struct. Mol. Biol. 12, 870–878 [DOI] [PubMed] [Google Scholar]
- 39. Fulle S., Gohlke H. (2009) Statics of the ribosomal exit tunnel: implications for cotranslational peptide folding, elongation regulation, and antibiotics binding. J. Mol. Biol. 387, 502–517 [DOI] [PubMed] [Google Scholar]
- 40. Vázquez-Laslop N., Ramu H., Klepacki D., Kannan K., Mankin A. S. (2010) The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 29, 3108–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]