Background: Irisin, which corresponds to the FNDC5 receptor ectodomain, is a purported exercise-induced myokine produced by cleavage.
Results: Irisin forms an extensive intersubunit β-sheet dimer with an FNIII-like fold.
Conclusion: Irisin forms a continuous β-sheet dimer not observed previously for any FNIII protein and may be representative of a new FNIII class.
Significance: The preformed irisin dimer suggests mechanisms for ligand and receptor activation.
Keywords: Exercise, Hormones, Muscle, Receptors, X-ray Crystallography
Abstract
Irisin was recently identified as a putative myokine that is induced by exercise. Studies suggest that it is produced by cleavage of the FNDC5 (fibronectin domain-containing protein 5) receptor; irisin corresponds to the extracellular receptor ectodomain. Data suggesting that irisin stimulates white-to-brown fat conversion have led to the hypothesis that it does so by binding an unknown receptor, thus functioning as a myokine. As brown fat promotes energy dissipation, myokines that elicit the transformation of white to brown fat have potentially profound benefits in the treatment of obesity and metabolic disorders. Understanding the molecular basis for such exercise-induced phenomena is thus of considerable interest. Moreover, FNDC5-like receptors are highly conserved and have been shown to be critical for neuronal development. However, the structural and molecular mechanisms utilized by these proteins are currently unknown. Here, we describe the crystal structure and biochemical characterization of the FNDC5 ectodomain, corresponding to the irisin myokine. The 2.28 Å structure shows that irisin consists of an N-terminal fibronectin III (FNIII)-like domain attached to a flexible C-terminal tail. Strikingly, the FNIII-like domain forms a continuous intersubunit β-sheet dimer, previously unobserved for any FNIII protein. Biochemical data confirm that irisin is a dimer and that dimerization is unaffected by glycosylation. This finding suggests a possible mechanism for receptor activation by the irisin domain as a preformed myokine dimer ligand or as a paracrine or autocrine dimerization module on FNDC5-like receptors.
Introduction
More than 1000 genes are activated in skeletal muscle in response to exercise and contribute to enhanced health (1–5). Although the molecular mechanisms involved in this response have been largely unknown, studies carried out in the last decade showing that skeletal muscle functions as a secretory organ have started to shed light on these processes. Indeed, skeletal muscle represents ∼40% of the body weight of lean men and women and hence constitutes a large reservoir for the production of signaling molecules. Several hundred such molecules that are secreted by skeletal muscle have recently been identified, prompting Pedersen et al. (6) to coin the term “myokine.” Myokines are specifically defined as cytokines or other peptides that are produced, expressed, and released by muscle fibers and that exert endocrine effects (6).
One beneficial outcome of exercise is the “browning” of white adipose tissue (fat) to brown fat. Brown fat cells possess large numbers of mitochondria that contain a protein called UCP1 (uncoupling protein 1), which functions to dissipate the proton-motive force normally used to drive ATP synthesis (7). As a consequence of UCP1 action, the energy in the mitochondrial electrochemical gradient is released in the form of heat rather than being stored as fat. Studies in rodents have unequivocally demonstrated that brown fat profoundly influences body weight (8). In addition to a role in obesity reduction, high brown fat levels have been associated with many positive health effects, such as resistance to metabolic diseases (2). Hence, factors that may stimulate brown fat production have been highly sought after. Recently, irisin was identified as a putative myokine secreted by muscle in response to exercise (9).
Irisin, named for the Greek messenger goddess, was discovered in a screen looking for factors secreted by muscle in response to PGC-1α (peroxisome proliferator-activated receptor-gamma coactivator-1α) activation (9). PGC-1α is known to stimulate many of the well characterized beneficial effects of exercise in muscle, including white-to-brown fat conversion (10). Moreover, increasing PGC-1α expression improves metabolic parameters, such as insulin sensitivity and signaling (11). Boström et al. (9) demonstrated that irisin is produced by proteolytic processing of a transmembrane receptor, FNDC5 (fibronectin domain-containing protein 5). FNDC5 is a 209-residue protein with an N-terminal 29-residue signal sequence, followed by the irisin or putative fibronectin III (FNIII)2 domain, a linking peptide, a transmembrane domain, and a 39-residue cytoplasmic segment. Their data indicated that cleavage in the linking peptide releases soluble irisin into the extracellular milieu (9). FNDC5, which is expressed in skeletal muscle, pericardium, heart, and brain, was originally discovered as a receptor and shown to be critical for the differentiation of myoblasts and neurons (12–14).
The function of FNDC5 as a receptor has not been explored. Indeed, since its discovery, multiple studies have focused on the physiological role(s) of irisin or the FNDC5 ectodomain in metabolism (15–20). Boström et al. (9) proposed that soluble irisin signals by binding to an as yet unidentified receptor. Subsequently, the potential for irisin as a chemotherapeutic in the treatment of obesity and metabolic diseases has caused significant excitement (3, 21). However, subsequent studies on irisin have produced conflicting results (1, 22–25), not consistent with those of Boström et al. Hence, more studies are clearly needed to define the role(s) of irisin in metabolism. Moreover, FNDC5 is completely conserved among vertebrates, yet FNDC5-like receptors have not been well characterized cellularly, and nothing is known about these receptors at the molecular and biochemical levels. Thus, to gain insight into irisin and FNDC5 structure and function, we performed biochemical studies and determined the structure of irisin by x-ray crystallography to 2.28 Å resolution. The structure reveals that irisin contains a fold similar to FNIII proteins. However, quite unexpectedly and distinct from any previously solved FNIII structure, irisin forms a continuous intersubunit β-sheet dimer, which has important implications for receptor activation and signaling.
EXPERIMENTAL PROCEDURES
Purification and Crystallization of Irisin
An artificial gene encoding irisin (residues 30–140) was codon-optimized for Escherichia coli expression and was purchased from GenScript Corp. (Piscataway, NJ). The gene encodes human irisin (irisin is 100% conserved from mouse to human) and results in the production of the mature processed protein, which lacks the N-terminal signal sequence (9). The gene was subcloned into the pET15b vector such that the N-terminal hexahistidine tag was included in the protein for purification. The vector was transformed into E. coli BL21(DE3) cells for expression, and the protein was purified in a single step by nickel-nitrilotriacetic acid chromatography. The N-terminal His tag was removed by thrombin using a thrombin capture cleavage kit (Sigma). The protein was buffer-exchanged into 50 mm Tris (pH 7.5), 300 mm NaCl, 5% glycerol, and 1 mm DTT for crystallization. Crystals were grown via hanging drop vapor diffusion by mixing the protein (at 50 mg/ml) 1:1 with a reservoir of 0.68 m citrate and 0.1 m cacodylate (pH 6.5). Crystals took 2 weeks to grow to maximum size and were cryopreserved directly from the drop.
Structure Determination and Refinement of the Irisin Structure
The irisin crystals are in space group P41212, with a = b = 93.4 and c = 285.6 Å. Native data were originally collected on in-house x-ray sources at 3.1 Å, and a thimerosal heavy atom derivative was obtained at 3.3 Å for phasing using MIRAS (multiple isomorphous replacement with anomalous scattering). There are eight irisin subunits in the crystallographic asymmetric unit, and all subunits interact to form four identical dimers. The experimental electron density map was readily traced, and a final native data set was collected at 2.28 Å on beamline 8.3.1 of the Advanced Light Source for refinement. All x-ray intensity data were processed with MOSFLM (see Table 1). The final model includes residues 30–123 of six subunits, residues 30–127 and 30–128 of two subunits, and 320 solvent molecules. The final refinement statistics are provided in Table 1.
TABLE 1.
Data collection and refinement statistics for irisin
| Irisin | |
|---|---|
| Data collection | |
| Space group | P41212 |
| Cell dimensions | |
| a, b, c (Å) | 93.40, 93.40, 285.60 |
| α, β, γ | 90.0°, 90.0°, 90.0° |
| Resolution (Å) | 142.5-2.28 |
| Rsym or Rmerge | 6.7 (44.3)a |
| I/σI | 11.2 (2.1) |
| Completeness (%) | 92.4 (88.8) |
| Redundancy | 4.2 (4.0) |
| Refinement | |
| Resolution (Å) | 142.5-2.28 |
| No. of reflections | 57,048 |
| Rwork/Rfree (%) | 22.7/24.5 |
| No. of atoms | |
| Protein | 5968 |
| Water | 350 |
| B-Factors | |
| Protein | 52.3 |
| Water | 52.5 |
| r.m.s.d.b | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.16 |
a Values in parentheses are for the highest resolution shell.
b r.m.s.d., root mean square deviation.
Production and Purification of Glycosylated Irisin from HEK293 Cells
To produce soluble glycosylated irisin, we amplified irisin cDNA fragments by PCR using mouse Fndc5 cDNA (Addgene) as a template. The PCR-amplified fragments were inserted into the pHLSec2 vector. When expressed in mammalian cells, this construct generates protein with the amino acid sequence egsADSPSAPVNVTVRHLKANSAVVSWDVLEDEVVIGFAISQQKKDVRMLRFIQEVNTTTRSCALWDLEEDTEYIVHVQAISIQGQSPASEPVLFKTPREAEKMASKNKDEVTMKEefhhhhhhhh (where the lowercase letters indicate sequence derived from the vector).
For expression, we followed a transient expression method using HEK293 cells and serum-free culture medium adapted from recombinant fibronectin expression (26). The conditioned medium was collected after 6–7 days of transfection. Secreted protein was purified with a cobalt column using standard procedures. That irisin is modified via N-linked glycosylation was verified by a molecular weight shift upon SDS-PAGE after peptide N-glycosidase F treatment (which specifically removes N-linked glycans).
Irisin Mutagenesis
Irisin R75E and I77W mutations were created using the Stratagene site-directed mutagenesis protocol. Primers containing the desired mutations were used to PCR amplify the DNA. The amplified DNA was incubated with DpnI, which digests the methylated parental DNA strands. The DNA was then transformed into DH5α cells and plated onto Luria broth-agar plates. Plasmids were transformed into BL21(DE3) cells, and the mutations were confirmed by sequencing. The resulting transformed cells were used for protein expression and purification as described for the wild-type protein.
Gel Filtration Analyses of Irisin and Glycosylated Irisin
Gel filtration was used to determine the molecular weights of the irisin proteins (non-glycosylated, mutant non-glycosylated, and glycosylated). All gel filtration experiments were performed using a HiLoad 16/600 Superdex 200 prep grade column. Experiments were performed in a buffer containing 300 mm NaCl, 5% glycerol, and 20 mm Tris HCl (pH 7.5).
RESULTS AND DISCUSSION
Overall Structure of Irisin
For structural studies, irisin was expressed in E. coli as the mature myokine (residues 30–140), which lacks the N-terminal 29-residue signal sequence, and purified to homogeneity (see “Experimental Procedures”). The protein was crystallized, and the structure was solved and refined to Rwork/Rfree = 22.7/24.5% and to 2.28 Å resolution (see “Experimental Procedures” and Table 1). The structure contains an N-terminal domain (residues 30–123) with homology to FNIII domains and a mostly disordered C-terminal tail composed of residues 124–140 (Fig. 1, A and B). The FNIII domain is one of the most commonly occurring protein domains and is typically used as a building block for modular proteins. For example, several FNIII modules are linked in tandem to form extracellular matrix proteins, such as fibronectin and tenascin, and FNIII domains are found in the ectodomains of many receptors (27). FNIII domains typically share only 15–20% sequence identity; however, despite this limited homology, their structures have surprisingly similar folds, composed of a β-sandwich with three β-strands on one side and four on the other. Database searches show that irisin has the strongest structural similarity to the third FNIII domain of tenascin (termed TNfnIII3) and the 10th FNIII module of fibronectin (termed FNfnIII10). Superimposition of 86 and 85 related Cα atoms of irisin onto those of fnIII10 and TNfnIII3 results in root mean square deviations of 1.49 and 1.46 Å, respectively (28–29). These structural similarities are limited to the β-strand regions of the proteins, with the loop regions showing little structural homology. Irisin contains the typical FNIII arrangement with a four-stranded β-sheet that packs tightly against a three-stranded β-sheet, with the following structural topology (according to FNIII nomenclature): βA(35–43)-βB(46–52)-βC(60–69)-βC′(73-81)-βE(85–91)-βF(95–106)-βG(108–116) (Fig. 1A).
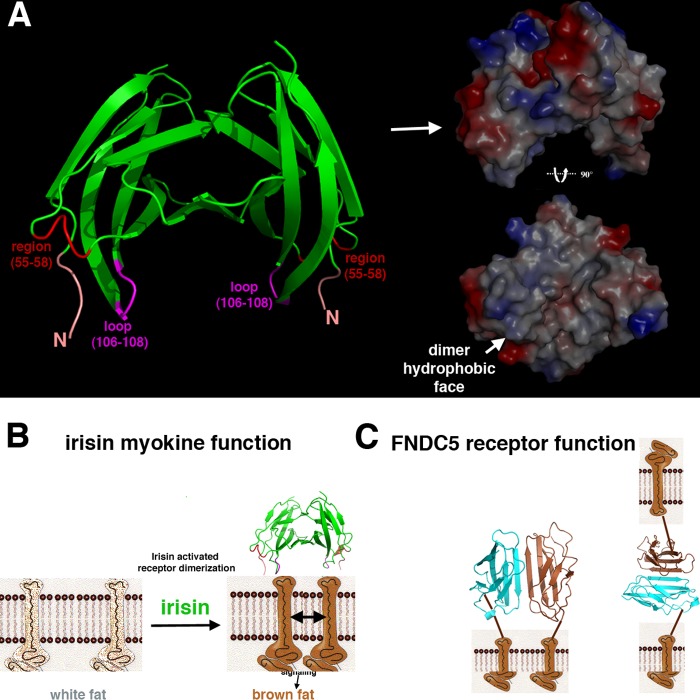
FIGURE 1.
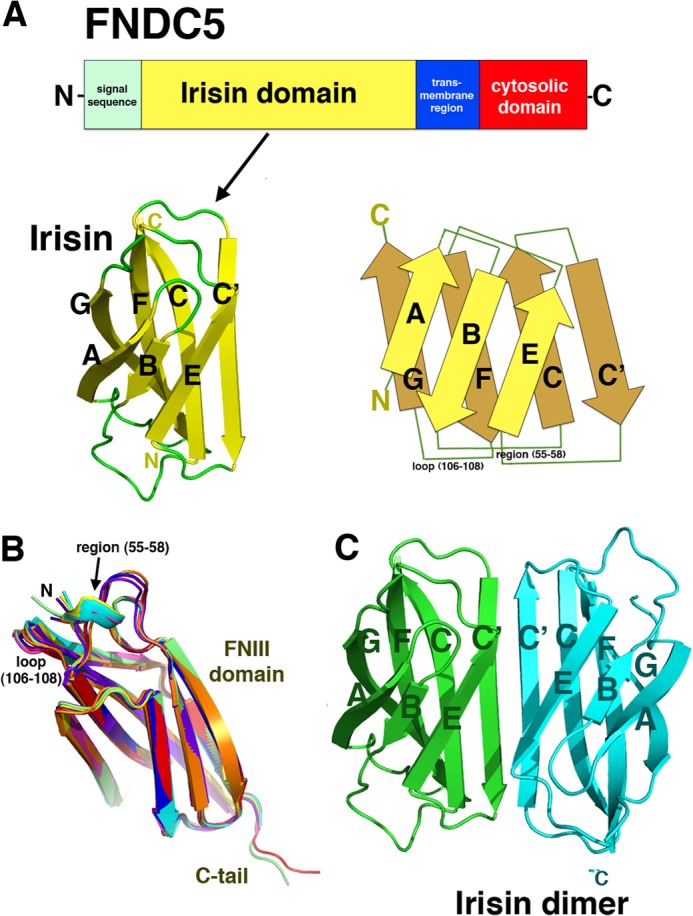
Crystal structure of irisin. A, irisin is a proteolytic product of FNDC5. The domain organization of the FNDC5 receptor is shown (upper). It contains an N-terminal signal sequence, which provides proper membrane insertion of the receptor and is subsequently cleaved. This is followed by the irisin domain, which contains an N-terminal FNIII-like region and a flexible C-terminal tail. The irisin domain is connected to a short transmembrane region, which is followed by the cytosolic region. The irisin domain is putatively produced following proteolytic cleavage of mature FNDC5 (with the signal sequence removed) (9). The FNDC5 schematic is the irisin subunit structure, showing only the FNIII domain and a topology diagram (lower). B, superimpositions of eight subunits in the crystallographic asymmetric unit showing the regions of flexibility that are found on the same face and that may be candidates for protein-protein interaction sites. Also indicated are the FNIII domain and the C-terminal tail, which is observed in two subunits. C, structure of the irisin dimer. Figs. 1 (B and C), 2A, 3 (A and B), and 4A were made using PyMOL (35).
Irisin Forms a Continuous Eight-stranded β-Sheet Dimer
An unexpected and remarkable finding from the irisin structure is that, unlike any previously characterized FNIII domain, it forms a tight dimer in which the C′ strands of the four-stranded β-sheets combine to create a continuous antiparallel eight-stranded β-sheet (Fig. 1C). Most FNIII domains are linked in tandem with other FNIII repeats or other modular domains to create beads on a string-like structures. Although somewhat flexible, the linkages between FNIII repeats (from the G strand to the A strand), such as those found in fibronectin, could hinder the formation of the intimate intersubunit β-sheet dimer observed in irisin. In fact, despite the fact that hundreds of FNIII domains have been structurally characterized, only the alternatively spliced variant of oncofetal fibronectin, in which an extra FNIII domain is inserted between fnIII7 and fnIII8, has been shown to dimerize via its FNIII domains (30). However, the resultant oncofetal fibronectin dimers form in an extended head-to-tail fashion, in which interchain contacts are formed by side chains in loops or strands between subunits and not via intimate backbone H-bonding interactions. This dimerization mode buries only 400–500 Å2 of two FNIII subunit modules and is completely different from that observed for irisin. Indeed, to our knowledge, the structure of irisin reveals the first case of a continuous β-sheet dimer formed between two FNIII domains. This resulting dimerization interface is extensive, burying 1400 Å2 of the two subunits. The continuous β-sheet interactions form the core of the irisin dimer and contribute 10 backbone H-bonds between the two interacting four-stranded β-sheets. This type of intersubunit H-bonding between backbone atoms of separate subunits has been implicated previously in the protein stability for other protein oligomers. An example is the thermostability imparted by two intersubunit β-sheets in the Pyrobaculum aerophilum Nudix hydrolase dimer (31).
Although the intersubunit H-bonds between the four-stranded β-sheets form the foundation of the irisin dimer, interactions between side chains on neighboring subunits provide further stability. Specifically, two salt bridges between Arg-75 and Glu-79′ (where the prime indicates the other subunit in the dimer) secure the ends of the dimer (Fig. 2A). Further fastening the dimer together are contacts between the three-stranded β-sheets found in each subunit, which are locked together by a Trp-90/Trp-90′ “tryptophan zipper”-like interaction. Tryptophan zipper interactions have been shown to greatly stabilize β-strand interactions in short peptides (32). The presence of the small side chains of Ala-88/Ala-88′ are essential in permitting the tight stacking of the Trp-90/Trp-90′ side chains in irisin. Thus, the irisin FNIII structure displays multiple structural attributes that favor dimerization and hence may define a new dimeric class of FNIII proteins. Indeed, our analyses show that the C′ β-strands of FNIII proteins characterized so far, such as TNfnIII3 and fnIII10, have twists, bulges, and prolines, which are design features that prevent extension of the β-sheet (Fig. 3, A and B) (33). Hence, the FNIII domains characterized to date appear to have selected for structural characteristics that prevent dimerization, whereas irisin has done the opposite, acquiring features that are optimal for dimerization.
FIGURE 2.
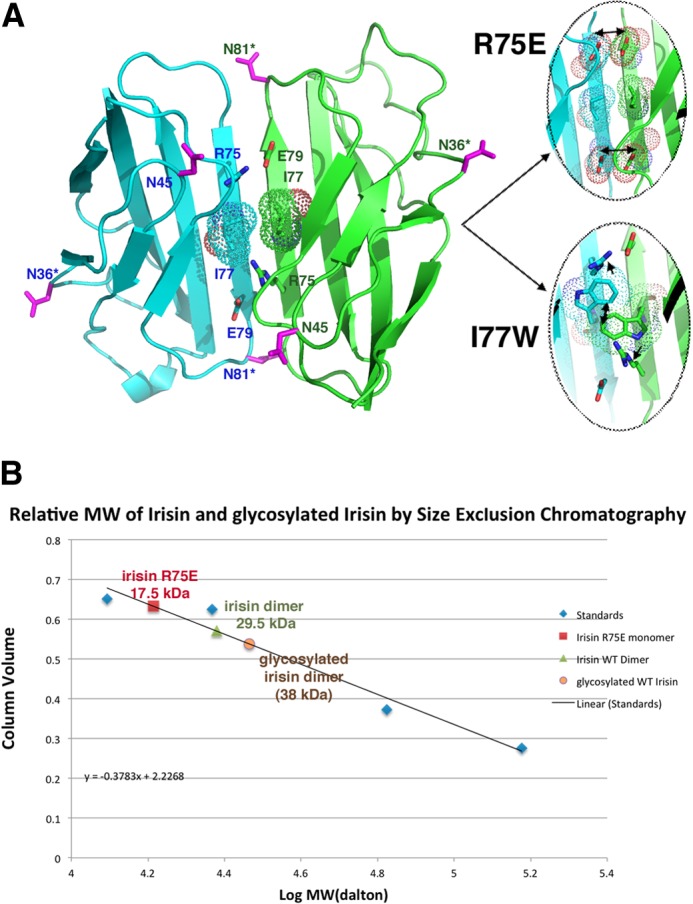
Irisin dimer contacts and mutagenesis experiments. A, ribbon diagram showing key cross-strand-specific salt bridges that fasten the ends of the irisin intersubunit β-sheet dimer and the location of Ile-77, which is positioned in the center of the dimer interface and was selected as a site for mutagenesis to disrupt the dimer. Also shown are the locations of the asparagine residues (magenta). Asterisks denote the two asparagines contained within NXT motifs and modified by glycosylation. The locations of these residues are notably surface-exposed and in positions in which modification would not be predicted to hinder dimerization. The magnified images are of the locations (modeling) where mutations were made to disrupt the dimer (R75E and I77W). The R75E mutation resulted in a clash with the cross-strand Glu-79, whereas the I77W mutation was predicted to prevent the formation of the hydrophobic interface in the dimer as well as disrupt the Arg-75–Glu-79 salt bridge due to its large size. B, size exclusion chromatography experiments showed that bacterially expressed (non-glycosylated) and glycosylated irisin proteins are dimers, whereas the R75E mutation is monomeric. The I77W mutant was unstable and could not be produced in soluble form. The y axis is the elution volume normalized for column volume, and the x axis is the log of the molecular weight (MW).
FIGURE 3.
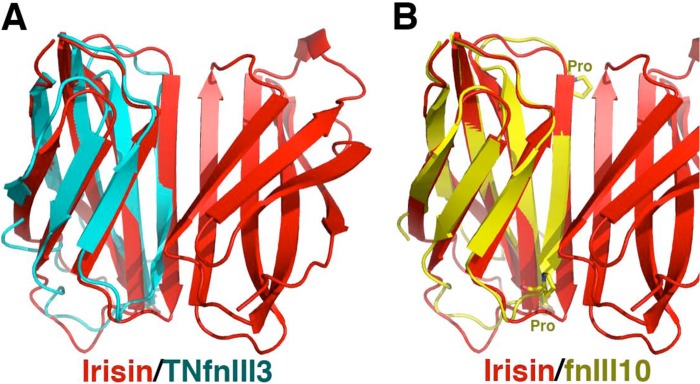
Superimposition of irisin (red) with the third FNIII domain of tenascin (TNfnIII3, cyan) and the 10th FNIII domain of fibronectin (fnIII10, yellow). A, overlay of TNfnIII3 onto irisin showing the highly twisted nature of the C′ strand of TNfnIII3 at the dimer interface, making dimer formation by this domain impossible. B, overlay of fnIII10 onto irisin highlighting that not only does fnIII10 have a highly twisted and bulged structure but that it also contains two prolines that prevent optimal dimeric β-sheet H-bonding potential.
Biochemical Data Support Dimerization of Non-glycosylated and Glycosylated Irisin
Our structural data provide strong support that irisin is a dimer. In fact, the crystal structure contains eight independent subunits in the crystallographic asymmetric unit, all of which combine to form the same dimer. Irisin has two asparagines contained within NXT motifs that are glycosylated in mammalian cells (see “Experimental Procedures”). The structure shows that these glycosylation sites are surface-exposed and are on regions not likely to affect dimerization (Fig. 2A). However, to assess the oligomeric states of glycosylated and non-glycosylated irisin, we performed size exclusion chromatography experiments. We first produced glycosylated irisin in HEK293 cells (see “Experimental Procedures”). N-Linked glycosylation was verified by a molecular weight shift upon SDS-PAGE following peptide N-glycosidase F treatment, which specifically removes N-linked glycans. The purified non-glycosylated and glycosylated irisin proteins were applied to a gel filtration column to assess their molecular weights. The experiments clearly demonstrated that both forms of the protein are dimers (Fig. 2B).
Mutagenesis Data Support the Irisin Dimer
The structural and biochemical data support that irisin is dimeric. However, to directly assess if the dimer observed in the structure is that found in solution, we performed mutagenesis studies, followed by gel filtration analyses. Two mutants were constructed, R75E and I77W. The structure predicts that an R75E mutation should impair dimer formation because Arg-75 forms the only salt bridge in the dimer: between Arg-75 and Glu-79 (Fig. 2A). An R75E mutation would not only eliminate the favorable salt bridge but also introduce a clash between the now two proximally located negatively charged residues. Ile-77 is in a key position of the dimer (Fig. 2A), as it sits in the center of the dimer, forming the nexus of the hydrophobic dimer core. Modeling showed that substitution of Ile-77 with tryptophan would be particularly problematic for dimer formation, as the large size of the tryptophan side chain cannot be accommodated within the dimer core, even with side chain rotations, and would also impinge on the Arg-75–Glu-79 salt bridge, forcing the residues to move away from the unfavorable steric clash and preventing their optimal placement for electrostatic contacts with each other (Fig. 2A). As predicted by the structure, the R75E mutation led to the production of monomeric protein. Indeed, although the mutant did not express as well as the wild-type protein, the protein was in the monomeric form (Fig. 2B). As the structure predicts, the I77W mutation was particularly harmful, and we could not obtain enough soluble pure protein for biochemical studies. Thus, the structural, mutagenesis, and biochemical data all support that irisin is a dimer.
Although the irisin protomers that comprise the crystallographic asymmetric unit all form dimers with essentially identical structures, superimposition of the eight subunits results in root mean square deviations of 0.24–1.2 Å. The β-strand regions are essentially identical in all of the subunits, and the slightly elevated root mean square deviations result from flexibility in two loop regions composed of residues 55–58 and 106–108, which adopt altered structures depending on the crystal-packing environment (Fig. 1B). Interestingly, the flexible loops (residues 55–58 and 106–108) and the protein N terminus lie on a hydrophobic face of the dimer, suggesting these regions as possible candidates for interactions with other proteins, such as a putative receptor (Fig. 4A). In fact, loop 106–108 in irisin corresponds to the RGD loop in FNfnIII10 that contacts its integrin receptor.
FIGURE 4.
Speculative models for irisin signaling. A, location of the flexible loop regions in the irisin dimer. Left, the irisin dimer, with the N terminus colored salmon, the flexible region of residues 55–58 colored red, and the flexible loop at residues 106–108 colored magenta. Right, electrostatic surface representation of the irisin dimer (blue and red indicate electropositive and electronegative regions, respectively) (upper) and the dimer rotated by 90° showing the hydrophobic face (white) of the loop-containing regions (lower). B, model for irisin functioning as a myokine cleaved from FNDC5. This model shows how a preformed irisin dimer acting as a myokine could facilitate dimerization and activation of an as yet unidentified receptor, leading to signaling events that stimulate white-to-brown fat conversion. C, model for FNDC5 receptor function. Shown are possible modes of signaling and cell adhesion processes affected by dimerization of the extracellular irisin domain of the FNDC5 receptor. Left, scenario in which FNDC5 molecules exist in the same cell. Here, irisin domains may aid in dimerization of the receptors, leading to signaling events, or a dimerized receptor may subsequently bind a ligand that would induce structural changes within the dimer, leading to downstream signaling. Right, FNDC5 molecules on proximally located cells could dimerize either to initiate signaling programs or to facilitate cell adhesion processes.
Irisin Is a Preformed Dimer and Models for Myokine and Receptor Activation
The finding that irisin exists as a preformed dimer has important implications for its function as a highly conserved ectodomain of FNDC5-like receptors as well as a putative myokine ligand. Most well known cases of ligand-mediated receptor signaling involve either ligand- or receptor-mediated dimerization (34). Two general classes of receptor-activated dimerization involve binding of a single ligand that stimulates dimerization of two receptors or binding of two monomeric ligands that facilitate dimer interactions between monomeric receptors. The structure of irisin suggests a mechanism for myokine ligand signaling via binding of a preformed dimer (Fig. 4B). Critically, this tight dimerization would be predicted to take place in the context of the full-length FNDC5 receptor, as the irisin domain is flexibly attached to the membrane-spanning domain. Hence, dimerization of the FNDC5 ectodomain may form intracellular or even intercellular dimers at the cell surface, leading to autocrine or paracrine signaling (Fig. 4C). In addition to signaling, FNDC5 dimerization between two cells could function in a cell-cell adhesion mode (Fig. 4C). In conclusion, our findings reveal a heretofore unseen FNIII intersubunit β-sheet dimer formed by the ectodomain of a novel receptor and putative myokine and should stimulate future studies aimed at understanding the signaling networks involving these receptor proteins.
Acknowledgments
We thank the Advanced Light Source (ALS) and the support staff. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, and Material Science Division of the United States Department of Energy at the Lawrence Berkeley National Laboratory.
This work was supported, in whole or in part, by National Institutes of Health Grants GM074815 (to M. A. S.) and CA047056 (to H. P. E.). This work was also supported by an MD Anderson Trust fellowship and a Burroughs Wellcome career development award.
The atomic coordinates and structure factors (code 4LSD) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- FNIII
- fibronectin III.
REFERENCES
- 1. Keller P., Vollaard N. B., Gustafsson T., Gallagher I. J., Sundberg C. J., Rankinen T., Britton S. L., Bouchard C., Koch L. G., Timmons J. A. (2011) A transcriptional map of the impact of endurance exercise training on skeletal muscle phenotype. J. Appl. Physiol. 110, 46–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Melov S., Tarnopolsky M. A., Beckman K., Felkey K., Hubbard A. (2007) Resistance exercise reverses aging in human skeletal muscle. PLoS ONE 2, e465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pedersen B. K., Febbraio M. A. (2012) Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat. Rev. Endocrinol. 8, 457–465 [DOI] [PubMed] [Google Scholar]
- 4. Pinto A., Di Raimondo D., Tuttolomondo A., Buttà C., Milio G., Licata G. (2012) Effects of physical exercise on inflammatory markers of atherosclerosis. Curr. Pharm. Des. 18, 4326–4349 [DOI] [PubMed] [Google Scholar]
- 5. Thompson D., Karpe F., Lafontan M., Frayn K. (2012) Physical activity and exercise in the regulation of human adipose tissue physiology. Physiol. Rev. 92, 157–191 [DOI] [PubMed] [Google Scholar]
- 6. Pedersen B. K., Steensberg A., Fischer C., Keller C., Keller P., Plomgaard P., Febbraio M., Saltin B. (2003) Searching for the exercise factor: is IL-6 a candidate? J. Muscle Res. Cell Motil. 24, 113–119 [DOI] [PubMed] [Google Scholar]
- 7. Cannon B., Nedergaard J. (2004) Brown adipose tissue: function and physiological significance. Physiol. Rev. 84, 277–359 [DOI] [PubMed] [Google Scholar]
- 8. Seale P., Kajimura S., Spiegelman B. M. (2009) Transcriptional control of brown adipocyte development and physiological function–of mice and men. Genes Dev. 23, 788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boström P., Wu J., Jedrychowski M. P., Korde A., Ye L., Lo J. C., Rasbach K. A., Boström E. A., Choi J. H., Long J. Z., Kajimura S., Zingaretti M. C., Vind B. F., Tu H., Cinti S., Højlund K., Gygi S. P., Spiegelman B. M. (2012) A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481, 463–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Handschin C., Spiegelman B. M. (2008) The role of exercise and PGC1α in inflammation and chronic disease. Nature 454, 463–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wenz T., Rossi S. G., Rotundo R. L., Spiegelman B. M., Moraes C. T. (2009) Increased muscle PGC-1α expression protects from sarcopenia and metabolic disease during aging. Proc. Natl. Acad. Sci. U.S.A. 106, 20405–20410 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Ferrer-Martínez A., Ruiz-Lozano P., Chien K.R. (2002) Mouse PeP: a novel peroxisomal protein linked to myoblast differentiation and development. Dev. Dyn. 224, 154–167 [DOI] [PubMed] [Google Scholar]
- 13. Hashemi M. S., Ghaedi K., Salamian A., Karbalaie K., Emadi-Baygi M., Tanhaei S., Nasr-Esfahani M. H., Baharvand H. (2013) Fndc5 knockdown significantly decreased neural differentiation rate of mouse embryonic stem cells. Neuroscience 231, 296–304 [DOI] [PubMed] [Google Scholar]
- 14. Teufel A., Malik N., Mukhopadhyay M., Westphal H. (2002) Frcp1 and Frcp2, two novel fibronectin type III repeat containing genes. Gene 297, 79–83 [DOI] [PubMed] [Google Scholar]
- 15. Dun S. L., Lyu R. M., Chen Y. H., Chang J. K., Luo J. J., Dun N. J. (2013) Irisin immunoreactivity in neural and non-neural cells of the rodent. Neuroscience 240, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huh J. Y., Panagiotou G., Mougios V., Brinkoetter M., Vamvini M. T., Schneider B. E., Mantzoros C. S. (2012) FNDC5 and irisin in humans: 1. predictors of circulating concentrations in serum and plasma II. mRNA expression and circulating concentration in response to weight loss and exercise. Metabolism 61, 1725–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu J. J., Wong M. D., Toy W. C., Tan C. S., Liu S., Ng X. W., Tavintharan S., Sum C. F., Lim S. C. (2013) Lower circulating irisin is associated with type 2 diabetes mellitus. J. Diabetes Complications 27, 365–369 [DOI] [PubMed] [Google Scholar]
- 18. Moon H. S., Dincer F., Mantzoros C. S. (2013) Pharmacological concentrations of irisin increase cell proliferation without influencing markers of neurite outgrowth and synaptogenesis in mouse H19–7 hippocampal cell lines. Metabolism 62, 1131–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Roca-Rivada A., Castelao C., Senin L. L., Landrove M. O., Baltar J., Belén Crujeiras A., Seoane L. M., Casanueva F. F., Pardo M. (2013) FNDC5/irisin is not only a myokine but also an adipokine. PLoS ONE 8, e60563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stengel A., Hofmann T., Goebel-Stengel M., Elbelt U., Kobelt P., Klapp B. F. (2013) Circulating levels of irisin in patients with anorexia nervosa and different stages of obesity–correlation with body mass index. Peptides 39, 125–130 [DOI] [PubMed] [Google Scholar]
- 21. Elbelt U., Hofmann T., Stengel A. (2013) Irisin: what promise does it hold? Curr. Opin. Clin. Nutr. Metab. Care 16, 541–547 [DOI] [PubMed] [Google Scholar]
- 22. Pekkala S., Wiklund P., Hulmi J. J., Ahtiainen J. P., Horttanainen M., Pöllänen E., Mäkelä K. A., Kainulainen H., Häkkinen K., Nyman K., Alén M., Herzig K. H., Cheng S. (2013) Are skeletal muscle FNDC5 gene expression and irisin release regulated by exercise and related to health? J. Physiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raschke S., Elsen M., Gassenhuber H., Sommerfeld M., Schwahn U., Brockmann B., Jung R., Wisløff U., Tjønna A. E., Raastad T., Hallén J., Norheim F., Drevon C. A., Romacho T., Eckardt K., Eckel J. (2013) Evidence against a beneficial effect of irisin in humans. PLoS ONE 8, e73680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Timmons J. A., Baar K., Davidsen P. K., Atherton P. J. (2012) Is irisin a human exercise gene? Nature 488, E9–E10 [DOI] [PubMed] [Google Scholar]
- 25. Erickson H. P. (2013) Irisin and FNDC5 in retrospect: an exercise hormone or a transmembrane receptor? Adipocyte 2, 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ohashi T., Erickson H. P. (2011) Fibronectin aggregation and assembly: the unfolding of the second fibronectin type III domain. J. Biol. Chem. 286, 39188–39199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwarzbauer J. E., DeSimone D. W. (2011) Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb. Perspect. Biol. 3, A005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Leahy D. J., Hendrickson W. A., Aukhil I., Erickson H. P. (1992) Structure of a fibronectin type III domain from tenascin phased by MAD analysis of the selenomethionyl protein. Science 258, 987–991 [DOI] [PubMed] [Google Scholar]
- 29. Leahy D. J., Aukhil I., Erickson H. P. (1996) 2.0 Ä crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 84, 155–164 [DOI] [PubMed] [Google Scholar]
- 30. Schiefner A., Gebauer M., Skerra A. (2012) Extra-domain B in oncofetal fibronectin structurally promotes fibrillar head-to-tail dimerization of extracellular matrix protein. J. Biol. Chem. 287, 17578–17588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang S., Mura C., Sawaya M. R., Cascio D., Eisenberg D. (2002) Structure of a Nudix protein from Pyrobaculum aerophilim reveals a dimer with two intersubunit β-sheets. Acta Crystallogr. D. Biol. Crystallogr. 58, 571–578 [DOI] [PubMed] [Google Scholar]
- 32. Santiveri C. M., Jiménez M. A. (2010) Tryptophan residues: scarce in proteins but strong stabilizers of β-hairpin peptides. Biopolymers 94, 779–790 [DOI] [PubMed] [Google Scholar]
- 33. Richardson J. S., Richardson D. C. (2002) Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl. Acad. Sci. U.S.A. 99, 2754–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hollenberg M. D. (1991) Structure-activity relationships for transmembrane signaling: the receptor's turn. FASEB J. 5, 178–186 [DOI] [PubMed] [Google Scholar]
- 35. DeLano W. L. (2008) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]