Background: Substrates of the Hippo pathway kinases Lats1/2 are largely unknown besides YAP/TAZ.
Results: Phosphorylation of angiomotin by Lats1/2 inhibits interaction with F-actin thus impairs cell migration and angiogenesis.
Conclusion: AMOTp130 is a physiological and functional substrate of Lats1/2 and the Hippo pathway.
Significance: Demonstrating how identification of novel substrates would facilitate understanding the physiology of the Hippo pathway.
Keywords: Actin, Angiogenesis, Cell Migration, Protein Kinases, Protein Phosphorylation, Lats, AMOT, Protein Kinase, the Hippo Pathway
Abstract
The Hippo tumor suppressor pathway plays important roles in organ size control through Lats1/2 mediated phosphorylation of the YAP/TAZ transcription co-activators. However, YAP/TAZ independent functions of the Hippo pathway are largely unknown. Here we report a novel role of the Hippo pathway in angiogenesis. Angiomotin p130 isoform (AMOTp130) is phosphorylated on a conserved HXRXXS motif by Lats1/2 downstream of GPCR signaling. Phosphorylation disrupts AMOT interaction with F-actin and correlates with reduced F-actin stress fibers and focal adhesions. Furthermore, phosphorylation of AMOT by Lats1/2 inhibits endothelial cell migration in vitro and angiogenesis in zebrafish embryos in vivo. Thus AMOT is a direct substrate of Lats1/2 mediating functions of the Hippo pathway in endothelial cell migration and angiogenesis.
Introduction
Organ size homeostasis is a remarkable feature of multicellular organisms. In the last decade, the Hippo pathway has been found to play a key role in control of organ size (1–14). At the center stage of this pathway is the Mst1/2-Lats1/2 kinase cascade. Upstream signals such as cell adhesion, cytoskeleton remodeling, lysophosphatidic acid (LPA)2 and its respective G-protein-coupled receptors (GPCRs) were found to regulate the Hippo pathway (15–23). YAP transcription co-activator and its paralog TAZ are the best known Hippo pathway targets mediating gene expression regulation and organ size control (22, 24–27). Nevertheless, the various upstream input signals suggest rich functions of the pathway. Indeed, Lats1/2 were reported to regulate cellular processes such as cell differentiation, cytokinesis, senescence, autophagy, centrosome duplication, and neuron dendritic tiling (28–33). It is unlikely that YAP/TAZ inactivation mediates all these functions. However, YAP/TAZ-independent functions of the Hippo pathway were poorly studied.
Here we report that the angiomotin p130 isoform (AMOTp130) is phosphorylated on a conserved HXRXXS motif by Lats1/2 downstream of GPCR signaling. Phosphorylation disrupts AMOT interaction with F-actin and correlates with reduced F-actin stress fibers and focal adhesions. Furthermore, phosphorylation of AMOT by Lats1/2 inhibits endothelial cell migration in vitro and zebrafish embryonic angiogenesis in vivo. These studies identified AMOT as a critical effector of the Hippo pathway downstream of GPCR signaling in regulation of cell migration and angiogenesis.
EXPERIMENTAL PROCEDURES
Antibodies, Plasmids, and Other Materials
We obtained anti-AMOT antibodies from Bethyl Lab; anti-Lats1 and anti-Lats2 from Cell Signaling Technologies; anti-GST from Genscript; anti-α-tubulin, anti-Flag, and anti-vinculin from Sigma; anti-thiophosphate ester from Epitomics; anti-HSP90 from BD Biosciences; anti-HA and anti-Myc from Covance; Alexa Fluor 488- or 594-conjugated secondary antibodies and Alexa Fluor 488-phalloidin from Invitrogen; Horseradish peroxidase-conjugated secondary antibodies from GE Healthcare. Anti-phosphoAMOTp130 (S175) antibody was generated by immunizing rabbits with phospho-peptide KQGHVRSLS(p)ERL. Human AMOTp130 were subcloned from other vectors into pCMV-Flag, pcDNA3-HA, pGEX-KG, and pQCXIH vectors. Human AMOTp80, mouse AMOTL1 and AMOTL2 were subcloned into pcDNA3-HA and pCMV-Flag vectors. Human AMOTp130 mutants were generated by site-directed mutagenesis. Other plasmids were described before (16, 22, 34). Phos-tag conjugated acrylamide was purchased from Wako Chemicals. All other chemicals were from Sigma.
Cell Culture, Transfection, and Viral Infection
HEK293, HEK293T, COS7, HEK293P, and HUVEC cells were cultured in DMEM (Invitrogen) containing 10% FBS (Invitrogen) and 50 μg/ml penicillin/streptomycin (P/S). Transfection with Lipofectamine (Invitrogen) was performed according to the manufacturer's instructions. For viral infection, HEK293P cells were transfected with viral constructs and packaging plasmids. 48 h later, viral supernatant was supplemented with 5 μg/ml polybrene, filtered through a 0.45 μm filter, and used to infect target cells.
Immunoprecipitation and Kinase Assay
For Lats2 kinase assays, HEK293 cells were transfected with indicated plasmids. 48 h post-transfection, cells were lysed with lysis buffer (50 mm HEPES at pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, 10 mm pyrophosphate, 10 mm glycerophosphate, 50 mm NaF, 1.5 mm Na3VO4, protease inhibitor mixture (Roche), 1 mm DTT, 1 mm PMSF) and immunoprecipitated with anti-HA antibody. The immunoprecipitates were washed three times with lysis buffer, once with wash buffer (40 mm HEPES, 200 mm NaCl), and once with kinase assay buffer (30 mm HEPES, 50 mm potassium acetate, 5 mm MgCl2). The immunoprecipitated Lats2 was subjected to kinase assay in the presence of 500 μm ATP or ATP-γS and 1 μg of recombinant GST-AMOTp130 purified from Escherichia coli as substrates. The reaction mixtures were incubated for 30 min at 30 °C. For detection by anti-thiophosphate-ester antibody, the reaction mixtures were further supplemented with 2.5 mm PNBM. Alkylating reactions were allowed to proceed for 1 h at room temperature. The reactions were terminated with SDS sample buffer and boiled before analysis by SDS-PAGE.
Actin Spin-down Assay
Actin Binding Protein Biochem Kit was obtained from Cytoskeleton. The assay was performed following the manufacturer's instructions. Briefly, F-actin was polymerized and mixed with GST-AMOTp130 proteins purified from E. coli. The mixture was then centrifuged at 150,000 × g for 1.5 h at 24 °C. Supernatants and pellets were then collected and processed for electrophoresis. Proteins were visualized by Coomassie Blue staining or Western blots.
Cell Migration Assay
Cell migration assay was performed using BD Falcon Cell culture inserts for 24-well plates with 8.0 μm pore size. Bottom sides of filters were pre-coated with 20 mg/ml fibronectin. HUVEC cells were serum-starved for 12 h and then seeded into the upper chambers of the inserts at 4 × 104 cells/well in serum-free medium, and lower chambers were filled with serum-free or 10% FBS-containing medium. After 12 h, cells were stained with 0.5% crystal violet. Cells in upper chambers were carefully removed, and bottom sides of the chambers were pictured.
Immunofluorescence Staining
Cells were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.1% Triton X-100. Cells may be treated with 100 μg/ml digitonin for 5 min as indicated. After blocking in 2% BSA for 30 min, slides were incubated with first antibody diluted in 1% BSA overnight at 4 °C. After washing with PBS, slides were incubated with Alexa Fluor 488- or 594-conjugated secondary antibodies for 1.5 h. For staining of F-actin, cells were incubated with Alexa Fluor 488-phalloidin for 1.5 h. The slides were then washed and mounted.
RNA Interference
Short interfering RNA (siRNA) oligonucleotides toward human Lats1, Lats2, AMOT, and control siRNA toward firefly luciferase were transfected into indicated cells using Lipofectamine RNAiMax Reagent (Invitrogen) following the manufacturer's instructions. Cells were analyzed 72 h post-transfection.
Zebrafish Maintenance and Angiogenesis Assay
Zebrafish embryos were produced by pairwise matings, raised at 28.5 °C and staged as described (35). A transgenic Tg(flk:GFP) line was used in this study (36). Sense-capped mRNAs were synthesized using mMESSAGE mMACHINE system (Ambion) according to the manufacturer's instructions. Plasmids encoding human AMOTp130, AMOTp130-SA, AMOTp130-S.D., mouse Lats2, or Lats2-KR were digested with XhoI and transcribed with T7 polymerase. Poly (A) tails (Takara Bio Cat. No. 2181) were added to the synthetic mRNAs. Synthesized mRNAs were purified using the MEGAclear Kit (Ambion). Antisense morpholino oligonucleotide AMOT-MO (5′-CCACTGACACAACTACCACCAAGTG-3′) (37) was synthesized by Gene Tools, LLC. Synthetic mRNAs and morpholinos were microinjected into zebrafish embryos at the one-two-cell stages as described (38). Injection doses were as following: 8.2 ng AMOT-MO; 8.2 ng AMOT-MO+400pg hAMOTp130-WT/SA/S.D.; 5.6 ng AMOT-MO+373pg hAMOTp130-WT/SA +80pg mLats2-WT/KR. Vascular phenotypes were imaged using a Zeiss Observer Z1 microscope.
RESULTS
The Hippo Pathway Phosphorylates AMOT Family Proteins
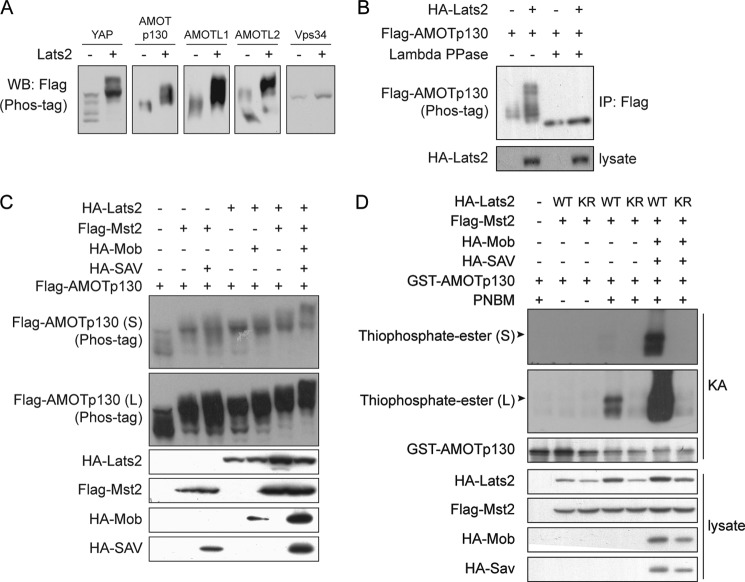
The AMOT family proteins localize to specific cellular compartments and regulate cell migration and proliferation (37, 39–41). AMOT is a component of the Hippo pathway capable of inhibiting YAP through direct binding and Lats1/2 activation (34, 42–44). Surprisingly, we found that Lats2 induced a dramatic electrophoretic mobility shift of AMOTp130 comparable to that of YAP on Phos-tag-containing gels which specifically retards phosphorylated proteins (Fig. 1A). Furthermore, Lats2 also induced dramatic up-shift of other AMOT family proteins AMOTL1 and AMOTL2 (Fig. 1A). The Lats2-induced mobility shift of AMOTp130 was eliminated by treatment of the protein with lambda protein phosphatase (Fig. 1B). In addition, co-transfection of AMOTp130 with other Hippo pathway components Mst2, Sav, Mob, and Lats2 induced AMOTp130 mobility shift in a synergistic manner (Fig. 1C). These experiments indicate that Lats1/2 in the Hippo pathway could induce phosphorylation of AMOT family proteins.
FIGURE 1.
The Hippo pathway phosphorylates angiomotin family proteins. A, Lats2 induces mobility shift of AMOT family proteins. HEK293 cells were transfected with plasmids encoding Flag-YAP or AMOT family proteins with or without Lats2 co-transfection. Cell lysates were resolved on Phos-tag-containing gels. Vps34 is a negative control. B, phosphatase eliminates AMOT mobility shift induced by Lats2. Flag-AMOTp130 was immunoprecipitated from transfected cells and treated with lambda protein phosphatase as indicated. Samples were analyzed as that in A. C, hippo pathway proteins synergistically induces AMOTp130 mobility shift. Lysates of transfected cells were resolved on Phos-tag-containing gels for mobility shift of Flag-AMOTp130. D, Lats2 phosphorylates AMOTp130 in vitro. In vitro phosphorylation of GST-AMOTp130 by immunoprecipitated Lats2 was performed. (S), short exposure; (L), long exposure.
We then examined whether Lats2 could phosphorylate AMOT in vitro. In this assay, the kinase utilizes ATP-γS as phosphate donor to generate thiophosphorylated substrate, which in turn reacts with p-nitrobenzyl mesylate (PNBM) to form a thiophosphate-ester (45). As detected by a thiophosphate-ester-specific antibody, recombinant AMOTp130 could be phosphorylated by Lats2 in vitro (Fig. 1D). The phosphorylation is more efficient when Mob and Sav were co-transfected. In contrast, the kinase inactive Lats2-KR could not phosphorylate AMOTp130. Thus, Lats2 phosphorylates AMOTp130 both in vitro and in vivo.
Lats2 Phosphorylates AMOT on Serine 175
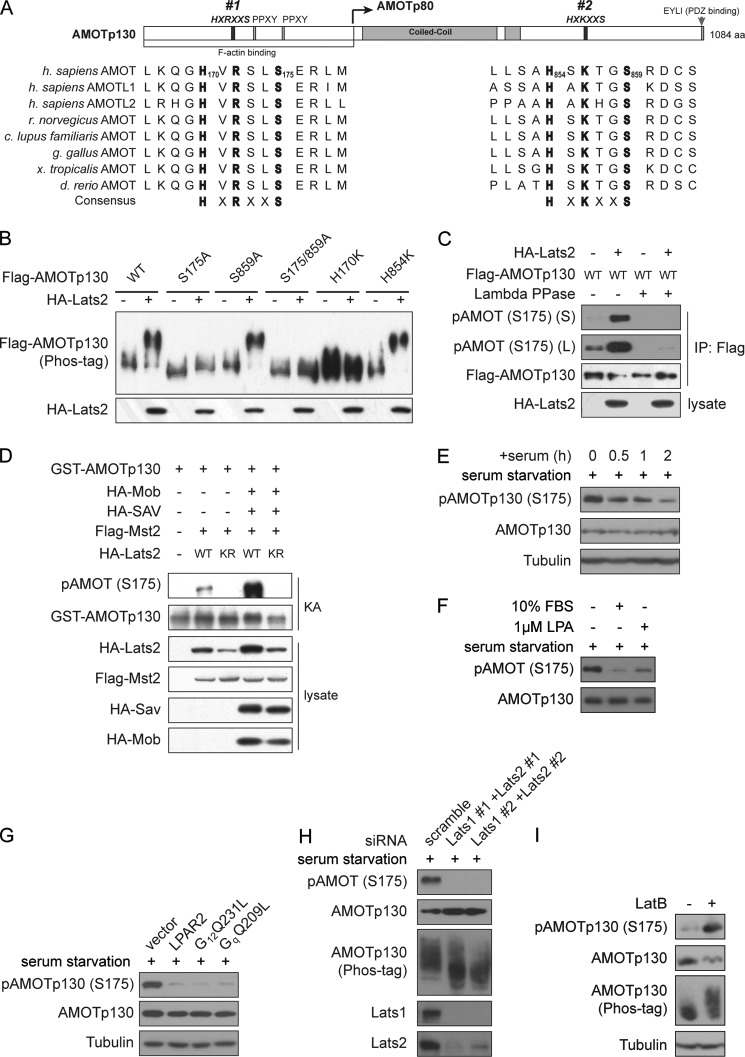
Previous studies have revealed the optimal Lats1/2 phosphorylation target consensus motif as HXRXXS (22, 25). Interestingly, we found one HXRXXS motif, and one HXKXXS motif, also permissive for phosphorylation by Lats1/2, on AMOTp130 (Fig. 2A). Both motifs are conserved in AMOT family members and across species (Fig. 2A). As shown in Fig. 2B, mutation of Ser-175 in the N-terminal motif to alanine largely repressed Lats2-induced up-shift of AMOTp130, suggesting that this reside is phosphorylated by Lats2. Further mutation of Ser-859 eliminates the mild shift of S175A, suggesting that Ser-859 is also phosphorylated by Lats2. However, individual mutation of Ser-859 did not repress the up-shift possibly due to limited sensitivity of the Phos-tag method. The histidine in the HXRXXS motif is crucial for substrate recognition. Indeed, similar to S175A, mutation of H170 largely abolished AMOTp130 up-shift in response to Lats2 (Fig. 2B). These experiments demonstrate that phosphorylation of Ser-175 is largely responsible for Lats2-induced up-shift of AMOTp130.
FIGURE 2.
AMOTp130 is phosphorylated on Ser-175 by Lats1/2 in response to GPCR signaling. A, domain organization of AMOTp130 and alignment of AMOT family proteins from different species. B, phosphorylation of Ser-175 leads to Lats2-induced AMOTp130 up-shift. Lysates of transfected HEK293 cells were analyzed by Western blots. C, Ser-175 of AMOTp130 is phosphorylated by Lats2. AMOTp130 phosphorylation was detected by pAMOT (S175)-specific antibody. D, Lats2 phosphorylates AMOTp130 on Ser-175 in vitro. Experiments were similar to that in Fig. 1D except that regular ATP was used. E, serum inhibits phosphorylation of endogenous AMOTp130 on Ser-175. Serum-starved 293T cells were stimulated with 10% FBS for the indicated time before harvest. F, LPA inhibits phosphorylation of endogenous AMOTp130 on Ser-175. Serum-starved 293T cells were stimulated with serum or LPA for 1 h before harvest. G, GPCR signaling inhibits Ser-175 phosphorylation of endogenous AMOTp130. 293T cells were transfected with indicated plasmids and serum-starved overnight before harvest. H, knockdown of Lats1 and Lats2 represses endogenous AMOTp130 phosphorylation on Ser-175. HEK293T cells were transfected with scramble or Lats1- and Lats2-specific siRNAs. Cells were serum-starved overnight before harvest. I, disruption of F-actin induces phosphorylation of AMOTp130. HEK293T cells cultured in serum-rich medium were treated with 1 μg/ml Latrunculin B (LatB) for 1 h as indicated before being harvested for Western blot analysis.
To further confirm that AMOTp130 Ser-175 is indeed phosphorylated by Lats2, we generated an anti-pAMOT (Ser-175) antibody. As expected, this antibody detected phosphorylation of ectopically expressed AMOTp130, which was markedly enhanced by Lats2 co-expression and eliminated by phosphatase treatment (Fig. 2C). Furthermore, AMOTp130 is phosphorylated on Ser-175 by Lats2 but not Lats2-KR in vitro (Fig. 2D). Thus, AMOTp130 is phosphorylated on Ser-175 by Lats2 in vitro and in vivo.
GPCR Signaling Inhibits AMOTp130 Phosphorylation Mediated by the Hippo Pathway
Following identification of the phosphorylation site we further examined the role of the Hippo pathway in physiological regulation of AMOT. Consistent with the inhibition of the Hippo pathway by serum and LPA, these stimulations inhibit endogenous AMOTp130 phosphorylation on Ser-175 (Fig. 2, E and F). LPA in serum represses the Hippo pathway via activation of GPCRs such as LPAR2, and the coupled G-proteins such as Gq and G12 (16). Consistently, expression of LPAR2 or active QL mutants of Gq or G12 potently inhibits endogenous AMOTp130 phosphorylation in serum-deprived cells (Fig. 2G). Furthermore, siRNA knockdown of Lats1 and Lats2 largely eliminates AMOTp130 phosphorylation on Ser-175 in serum-deprived cells (Fig. 2H). Integrity of the actin cytoskeleton is required for GPCR-induced repression of the Hippo pathway (16). Indeed, disruption of F-actin induced AMOTp130 phosphorylation in cells cultured in serum-rich medium (Fig. 2I). Taken together, serum and GPCR signaling regulate AMOTp130 phosphorylation on Ser-175 through Lats1/2 kinases.
Phosphorylation of AMOTp130 on Ser-175 Inhibits Its Interaction with F-actin
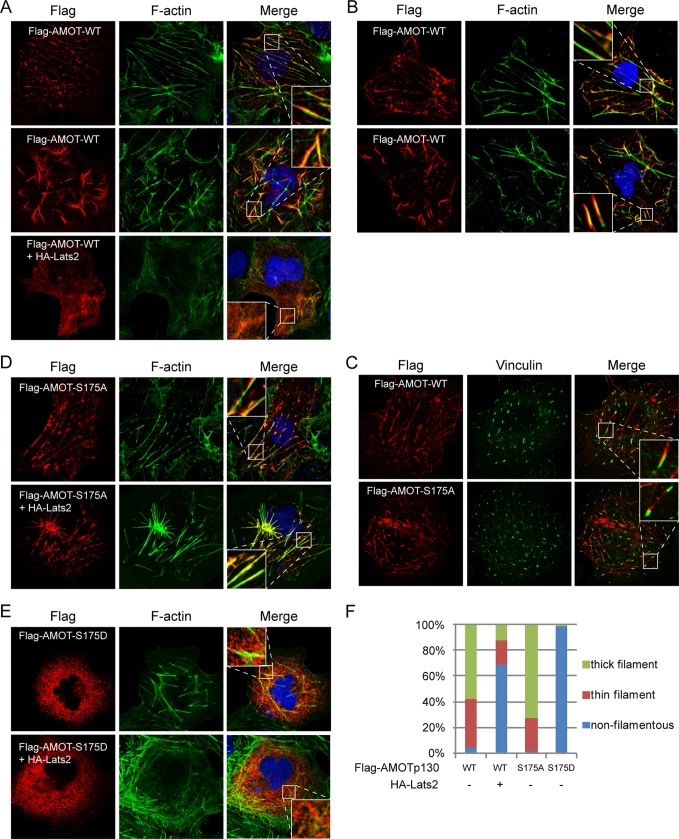
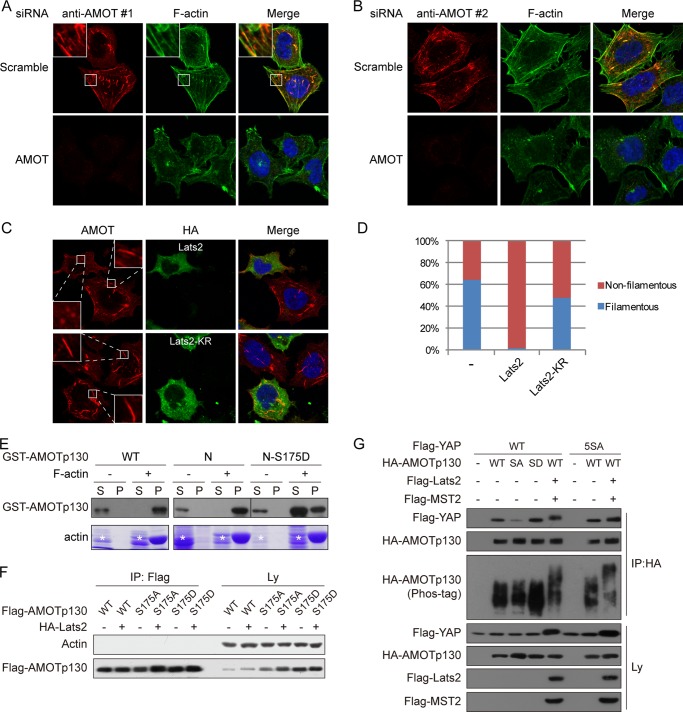
AMOTp130 is known to colocalize with F-actin in cells dependent on its N-terminal domain containing Ser-175 (46). By immunofluorescence staining, we indeed observed colocalization of ectopically expressed AMOTp130 with F-actin filaments (Fig. 3A). At lower expression level of AMOT, the AMOT-F-actin filaments closely resemble actin stress fibers (Fig. 3A). However, at higher expression level, the filaments are thicker possibly represent AMOT-F-actin filament bundles (Fig. 3A, middle panel). In support of a real stress fiber identity of AMOT-F-actin filaments, they are resistant to digitonin treatment before fixation and are anchored to focal adhesions (Fig. 3, B and C). Interestingly, expression of Lats2 disrupted the filamentous AMOTp130 and results in diffused and irregular dots-like localization of AMOT (Fig. 3, A and F). Thus phosphorylation of AMOTp130 on Ser-175 by Lats2 may disrupt AMOT-F-actin interaction. Consistently, the S175A mutant colocalizes with F-actin in a manner insensitive to Lats2 (Fig. 3, D and F). More strikingly, irrespective to Lats2 expression, the phospho-mimetic S175D mutant exhibited dots-like cytoplasmic localization (Fig. 3, E and F). Importantly, using two independent antibodies, we demonstrated that endogenous AMOT colocalizes with F-actin in 293T cells (Fig. 4, A, B, D). The filamentous localization of endogenous AMOT is also disrupted by Lats2 in a kinase-dependent manner (Fig. 4, C and D). These data demonstrate that phosphorylation of AMOTp130 by Lats2 on Ser-175 inhibits AMOTp130-F-actin interaction in vivo.
FIGURE 3.
Phosphorylation of AMOTp130 on Ser-175 inhibits interaction with F-actin. A, Lats2 inhibits colocalization of AMOTp130 with F-actin. Transfected COS7 cells were stained with anti-Flag antibody for localization of AMOTp130, AlexaFluor488-Phalloidin for F-actin, and DAPI for cell nuclei. B, colocalization of AMOTp130 with F-actin in digitonin-treated cells. Flag-AMOTp130 transfected Cos7 cells were treated with digitonin before fixation and stained as that in A. C, AMOT-F-actin filaments are anchored to focal adhesions. COS7 cells expressing wild-type or mutant AMOTs were co-stained with anti-Flag for AMOT and anti-vinculin to visualize focal adhesions. D, phospho-deficient mutant of AMOTp130 constitutively colocalizes with F-actin. Experiments were similar to these in A except the use of S175A mutant. E, phospho-mimetic mutant of AMOTp130 loses colocalization with F-actin. Experiments were similar to these in A except the use of S175D mutant. F, quantification of AMOT-F-actin colocalization. 120 cells were quantified for each transfection. Thick filament is similar to Fig. 3A, middle panel; thin filament is similar to that in Fig. 3A, top panel; non filamentous is similar to that in Fig. 3E, top panel.
FIGURE 4.
Phosphorylation inhibits endogenous AMOTp130-F-actin interaction and direct AMOTp130-F-actin interaction in vitro. A, endogenous AMOT colocalizes with F-actin. Scramble or AMOT-specific siRNA transfected HEK293T cells were stained with anti-AMOT antibody #1 for localization of endogenous AMOTp130, AlexaFluor488-Phalloidin for F-actin, and DAPI for cell nuclei. B, endogenous AMOT colocalizes with F-actin. Experiments were similar to these in A except that endogenous AMOT was stained with anti-AMOT antibody #2. C, Lats2 inhibits AMOT colocalization with F-actin in a kinase-dependent manner. HA-Lats2 wild-type or KR transfected HEK293T cells were stained with anti-AMOT antibody #1 for localization of endogenous AMOTp130 and anti-HA for Lats2 expression. D, quantification of filamentous localization of endogenous AMOT. Cells with or without filamentous AMOT in A and C were quantified. 120 cells were quantified for each sample. E, phospho-mimetic mutation inhibits AMOTp130 association with F-actin in vitro. Recombinant GST-AMOT wild-type or N-terminal fragment (N) were subjected to in vitro actin spin-down assay. (S), supernatant; (P), pellet. Actin was examined by Coomassie Blue staining and GST-AMOT was determined by Western blots. Asterisks denote nonspecific proteins. F, AMOTp130 does not co-immunoprecipitate with G-actin. HEK293 cells were transfected as indicated. Transfection amount was adjusted to make soluble AMOT levels comparable. Lysates were immunoprecipitated with anti-Flag antibody. The presence of endogenous actin and Flag-AMOTp130 in immunoprecipitates and lysates were examined by Western blots. G, phosphorylation of wild-type AMOTp130 by Lats2 does not affect interaction between AMOTp130 and YAP. HEK293 cells were transfected as indicated. AMOTp130 was immunoprecipitated with anti-HA antibody. Co-immunoprecipitation of YAP wild-type or 5SA mutant was examined by anti-Flag Western blots. AMOTp130 phosphorylation was shown by electrophoretic mobility shift on Phos-tag containing gel.
To test the possibility of a direct interaction between AMOTp130 and F-actin we performed in vitro F-actin spin-down assay. In this assay, F-actin binding proteins would co-sediment with pre-polymerized F-actin. We found that addition of F-actin brought all recombinant AMOTp130 or the N-terminal fragment into the pellet fraction indicating direct binding of AMOTp130 to F-actin (Fig. 4E). However, the phospho-mimetic S175D mutant largely remains in the supernatant (Fig. 4E). These results suggest that phosphorylation of AMOTp130 on Ser-175 would be inhibitory on the direct interaction of AMOTp130 with F-actin in vitro. In contrast, we did not observe any interaction of AMOTp130 with monomeric G-actin by co-immunoprecipitation (Fig. 4F), suggesting that AMOTp130 specifically interacts with F-actin. These results indicate that AMOTp130 directly interacts with F-actin under negative regulation by phosphorylation of Ser-175.
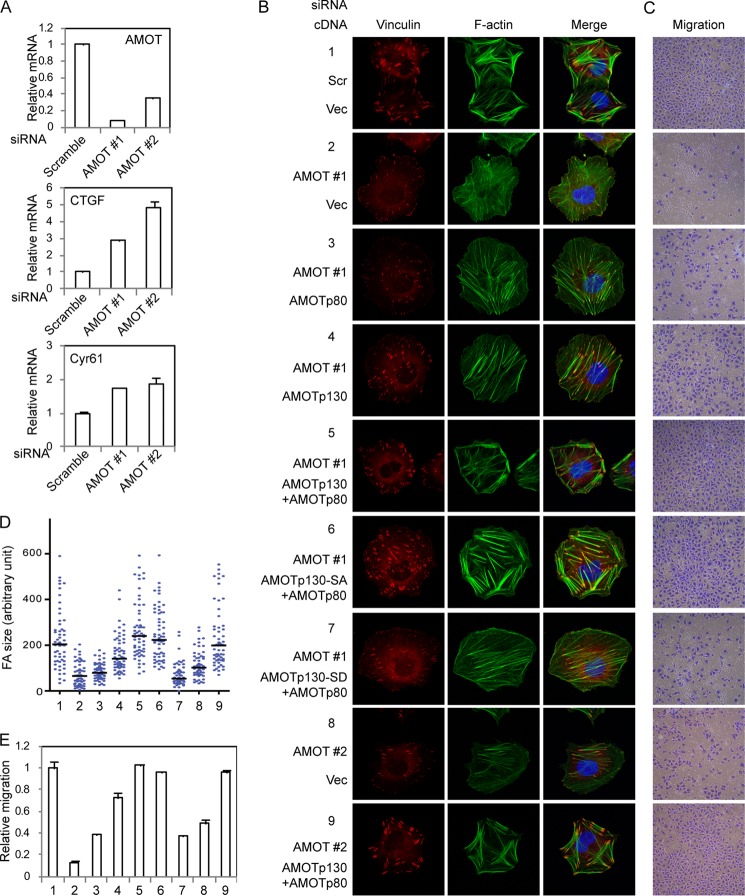
Phosphorylation of AMOTp130 on Ser-175 Interferes with Stress Fiber and Focal Adhesion Formation and Inhibits Endothelial Cell Migration
AMOTp130 is known to directly bind and inhibit YAP (34, 43, 44). However, although AMOT-SA mutant show some decreased interaction with YAP, the interaction between YAP and wild-type AMOT is not significantly affected by AMOT phosphorylation (Fig. 4G). AMOT is also known for its roles in endothelial cell morphology and migration (47). Therefore we examined the function of AMOTp130 phosphorylation by Lats1/2 in endothelial cells. In agreement with our previous observations, knockdown of AMOT induces expression of YAP target genes CTGF and Cyr61 (Fig. 5A). Interestingly, consistent with a direct association of AMOT with F-actin, knockdown of AMOT leads to reduced stress fibers in cells, which also correlates with reduced focal adhesions (Fig. 5, B and D). The defects are partially rescued by re-expression of AMOTp80 and better rescued by re-expression of AMOTp130 (Fig. 5, B and D). Interestingly, co-expression of AMOTp130 wild-type or SA mutant but not the SD mutant together with AMOTp80 rescues stress fiber and focal adhesion formation to a level similar to control cells (Fig. 5, B and D). Thus phosphorylation of AMOTp130 plays an inhibitory role on stress fiber and focal adhesion formation.
FIGURE 5.
Phosphorylation of AMOTp130 on Ser-175 inhibits endothelial cell migration. A, knockdown efficiency of AMOT siRNAs. AMOT, CTGF, and Cyr61 levels in siRNA transfected HUVEC cells were determined by realtime RT-PCR. B, phospho-mimetic mutant of AMOT is unable to compensate endogenous AMOT in actin stress fiber and focal adhesion formation. Control or AMOT-expressing stable cells transfected with siRNAs were serum-starved overnight and then seeded on fibronectin-coated coverslips for 2 h in serum-containing medium. Focal adhesions and F-actin were stained with anti-vinculin and AlexaFluor488-Phalloidin, respectively. C, phospho-mimetic AMOT mutant could not rescue cell migration defects caused by loss of AMOT. Cells the same as these in B were examined in transwell migration assay. D, size of 60 FAs from the staining of each stable cell as these in B was quantified by ImageJ and drawn into scatter plot. The median number was indicated. E, number of migrated cells in C was quantified. Experiments performed were in duplicates.
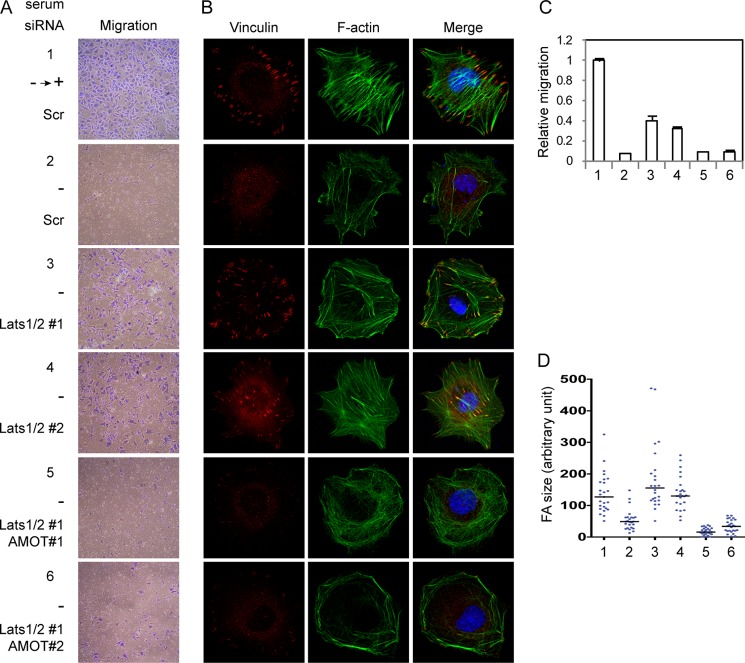
Remodeling of actin cytoskeleton and cell adhesion is crucial to cell migration. Consistent with alterations of the cytoskeleton and focal adhesions, knockdown of AMOT largely inhibits endothelial cell migration (Fig. 5, C and E). Again, we observed that co-expression of AMOTp130 wild-type or SA mutant but not the SD mutant together with AMOTp80 rescues the cell migration defects (Fig. 5, C and E). This suggests that phosphorylation of AMOT on Ser-175 inhibits cell migration. We then further investigated the role of AMOT in cell migration downstream of the Hippo signaling. Serum starvation is well known to strongly inhibit cell migration in vitro. However, we found that cell migration in starvation condition is partially but significantly rescued by knockdown of Lats1 and Lats2 (Fig. 6, A and C). This result suggests that activation of Lats1/2 by serum starvation due to blunted GPCR signaling is an important reason for repression of cell migration. More importantly, simultaneous knockdown of AMOT blocks cell migration induced by loss of Lats1/2 expression (Fig. 6, A and C). Thus inhibition of the Hippo pathway leads to cell migration in an AMOT-dependent manner. In correlation with cell migration, knockdown of Lats1/2 also induces actin stress fiber and focal adhesion formation in serum-starved cells in an AMOT-dependent manner (Fig. 6, B and D). Our data suggest that AMOTp130 is important for endothelial cell migration under negative regulation by Lats1/2-induced phosphorylation.
FIGURE 6.
Knockdown of Lats1/2 induces cell migration, actin stress fiber and focal adhesion in an AMOT-dependent manner. A, Lats1/2 plays a role in serum deprivation-induced migration retardation by repressing AMOT. siRNA-transfected HUVEC cells were serum starved and then seeded in transwell inserts in serum-free medium. The lower chambers were filled with serum-containing or serum-free medium as indicated. Cells on bottom sides of the transwells were stained after 12 h. B, Lats1/2 plays a role in serum deprivation-induced stress fiber and focal adhesion deformation by repressing AMOT. Cells were the same as these in A. Cells were serum starved overnight and then seeded on fibronectin-coated coverslips for 2 h either in serum-containing medium or serum-free medium as indicated. Focal adhesions and F-actin were stained with anti-vinculin and AlexaFluor488-Phalloidin respectively. C, number of migrated cells in A was quantified. Experiments were in duplicates. D, size of 25 FAs from the staining of each stable cell as these in B was quantified by ImageJ and drawn into scatter plot. The median number was indicated.
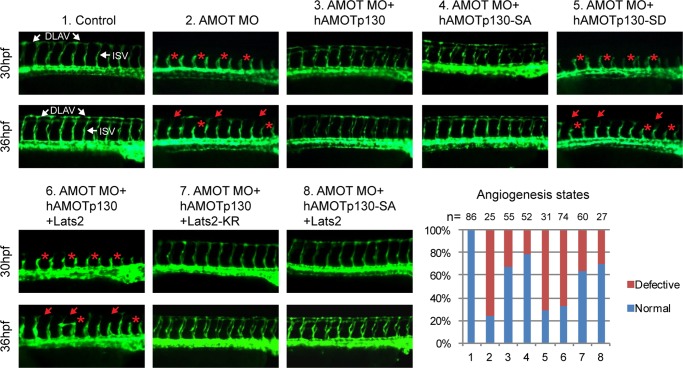
Phosphorylation of AMOTp130 on Ser-175 Inhibits Angiogenesis in Zebrafish
AMOT is known to control endothelial cell migration and angiogenesis in zebrafish (37). Thus we further investigated the effects of AMOTp130 phosphorylation on angiogenesis in zebrafish Tg(flk:GFP) transgenic embryos, in which GFP expression is driven by endothelial specific promoter flk. Consistent with the previous study (37), amot knockdown via injection of AMOT antisense morpholino-oligonucleotide (AMOT-MO) caused angiogenesis defects in AMOT morphants, in which intersegmental vessels (ISV) were disrupted and failed to reach the dorsolateral region at 30 h post-fertilization (hpf) (Fig. 7-2). At 36 hpf, some ISV had still not arrived the most dorsolateral position so that the dorsal longitudinal anastomotic vessel (DLAV) formation was disrupted (Fig. 7–2). Nevertheless, the angiogenesis defects are rescued by co-injection of mRNAs encoding human AMOTp130 (Fig. 7–3) or the phospho-deficient SA mutant (Fig. 7–4) but not the phospho-mimetic SD mutant (Fig. 7–5). Furthermore, the angiogenic activity of AMOTp130 is inhibited by co-injection of mRNAs encoding Lats2 (Fig. 7–6) but not Lats2-KR (Fig. 7–7). In addition, the angiogenic activity of AMOTp130-SA is insensitive to Lats2 (Fig. 7–8). The overall development and morphologies of injected embryos were comparable to control embryos at bud stage and 30 hpf (data not shown). These results indicate that Lats2 could inhibit the angiogenic function of AMOTp130 in vivo through phosphorylation of Ser-175.
FIGURE 7.
Phosphorylation of AMOTp130 by Lats1/2 regulates angiogenesis in zebrafish. The fluorescent microscopy analyses revealed the development of ISV and DLAV at 30 hpf and 36 hpf in the trunk region of control Tg(flk:GFP) embryos and embryos injected with AMOT-MO or a combination of AMOT-MO and mRNAs encoding human AMOTp130, Lats2 or their mutants. Red asterisk: defective ISV; red arrow: defective DLAV. ISV: intersegmental vessels. DLAV: dorsal longitudinal anastomotic vessels. Embryos were orientated with anterior to the left. Histogram depicted the quantification of normal and angiogenesis defective embryos at 30 hpf. n, embryo number.
DISCUSSION
Biological functions of kinases are largely determined by substrate selectivity. However, known substrates of the Lats1/2 kinases are very limited. Besides YAP/TAZ, several other proteins have been suggested as Lats1/2 substrates including Dyrk1a, MYPT1, and Snail1 (48–50). In this study, we identified AMOT family proteins to be new substrates of Lats1/2 with canonical target consensus motifs being phosphorylated by Lats1/2 to comparable levels as YAP/TAZ. Importantly, this phosphorylation is responsive to GPCR signaling and plays a regulatory role on AMOT interaction with F-actin thus regulates cell migration. While this report was being prepared, an independent study reported the phosphorylation of AMOT by Lats1/2 in promoting AMOT-Lats1/2 interaction, and its role in cell fate specification in preimplantation mouse embryos (51). Together, these studies suggest AMOT as an important effector of the Hippo pathway. We anticipate more substrates of Lats1/2 to be discovered to explain the ever expanding functions of the Hippo pathway.
AMOTp130 also plays a regulatory role in the Hippo pathway through both physical binding with YAP/TAZ and promotion of Lats1/2 activity (34, 42–44). In addition, AMOT also interacts with Merlin, an upstream component of the Hippo pathway (40). We demonstrated that the association between wild-type AMOTp130 and YAP is not regulated by AMOTp130 phosphorylation. However, it was shown that the phospho-mimetic AMOTp130 has elevated interaction with Lats1/2 (51). Therefore there is possibly a feedback loop consisting of the two roles of AMOTp130 as both a regulator and an effector of the Hippo pathway, although the physiological context and the exact mechanism would need further investigation. What we have found in this study also indicate that the phosphorylated and dephosphorylated AMOTp130 are both functional with differential roles. While the phosphorylated AMOT may promote Hippo signaling, the dephosphorylated AMOTp130 function as an F-actin interacting protein to promote stress fiber formation and cell migration.
YAP/TAZ are downstream effectors of the Hippo pathway and function through regulation of gene transcription (52). YAP/TAZ also promotes cell migration likely through regulation of gene expression and initiation of an EMT program (53). However, our study identified AMOTp130 as a novel mediator of the Hippo pathway in inhibiting cell migration. Phosphorylation of AMOTp130 by Lats1/2 directly inhibits F-actin binding and correlates with reduced actin stress fiber and focal adhesion formation. Considering the direct role of F-actin and focal adhesion in cell migration, the role of AMOTp130 in cell migration is likely structural. Migration promoting functions of AMOT and YAP/TAZ could be reconciled in at least two ways. First, the expression pattern of AMOT and YAP/TAZ may dictate their functions in different tissues and cell types. Second, in cells expressing both of these proteins, AMOTp130 and YAP/TAZ may mediate acute and prolonged effects of the Hippo pathway on cell migration in a cooperative manner (Fig. 8). Whether YAP/TAZ plays a role in endothelial cell migration is unknown. However, we demonstrated in this report that Lats1/2 play a role in endothelial cell migration in a manner dependent on AMOT phosphorylation. More importantly we demonstrated in vivo for the first time that the Hippo pathway may regulate developmental angiogenesis through AMOT phosphorylation. It would be interesting to investigate whether AMOT phosphorylation by the Hippo pathway also functions in pathological angiogenesis such as that in cancer.
FIGURE 8.
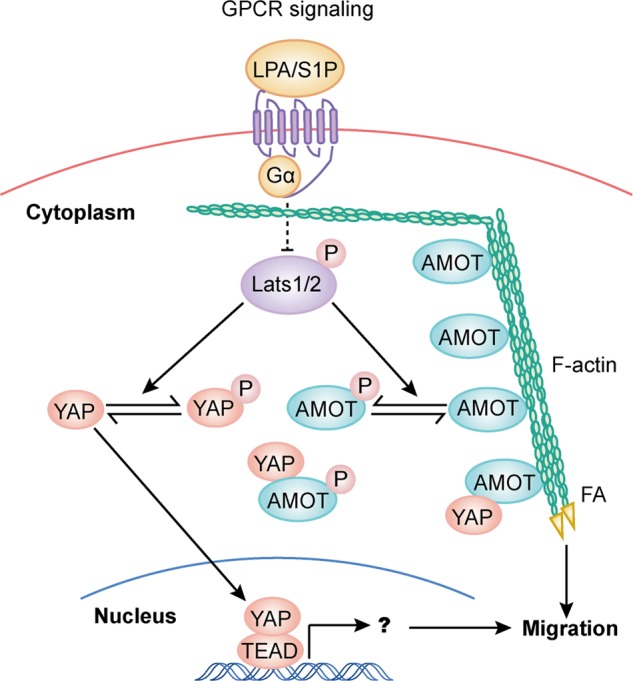
A model of Lats1/2 in regulation of cell migration through phosphorylation of AMOT and YAP. Phosphorylation of AMOT by Lats1/2 inhibits F-actin association and then actin stress fiber and focal adhesion formation. These effects of AMOT on cell structure and cell adhesion may work independently or in cooperation with YAP-dependent gene transcription alteration downstream of Lats1/2 to regulate cell migration.
Acknowledgment
We thank Dr. Bin Zhou for providing HUVEC cells.
This work was supported by grants (to B. Z.) from the National Natural Science Foundation of China (31271508), State Key Development Program for Basic Research of China (2013CB945303), Natural Science Foundation of Zhejiang (LR12C07001), and the Thousand Young Talents Plan of China.
- LPA
- lysophosphatidic acid
- AMOT
- Angiomotin
- GPCR
- G-protein-coupled receptor
- PNBM
- p-nitrobenzyl mesylate
- ATP-γS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1. Zhao B., Li L., Lei Q., Guan K. L. (2010) The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., Pan D. (2007) Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou D., Conrad C., Xia F., Park J. S., Payer B., Yin Y., Lauwers G. Y., Thasler W., Lee J. T., Avruch J., Bardeesy N. (2009) Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 16, 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Camargo F. D., Gokhale S., Johnnidis J. B., Fu D., Bell G. W., Jaenisch R., Brummelkamp T. R. (2007) YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054–2060 [DOI] [PubMed] [Google Scholar]
- 5. Wu S., Huang J., Dong J., Pan D. (2003) hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456 [DOI] [PubMed] [Google Scholar]
- 6. Jia J., Zhang W., Wang B., Trinko R., Jiang J. (2003) The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 17, 2514–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harvey K. F., Pfleger C. M., Hariharan I. K. (2003) The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell 114, 457–467 [DOI] [PubMed] [Google Scholar]
- 8. Udan R. S., Kango-Singh M., Nolo R., Tao C., Halder G. (2003) Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nature Cell Biol. 5, 914–920 [DOI] [PubMed] [Google Scholar]
- 9. Pantalacci S., Tapon N., Léopold P. (2003) The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nature Cell Biol. 5, 921–927 [DOI] [PubMed] [Google Scholar]
- 10. Tapon N., Harvey K. F., Bell D. W., Wahrer D. C., Schiripo T. A., Haber D. A., Hariharan I. K. (2002) salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467–478 [DOI] [PubMed] [Google Scholar]
- 11. Justice R. W., Zilian O., Woods D. F., Noll M., Bryant P. J. (1995) The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 9, 534–546 [DOI] [PubMed] [Google Scholar]
- 12. Xu T., Wang W., Zhang S., Stewart R. A., Yu W. (1995) Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 13. Kango-Singh M., Nolo R., Tao C., Verstreken P., Hiesinger P. R., Bellen H. J., Halder G. (2002) Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development 129, 5719–5730 [DOI] [PubMed] [Google Scholar]
- 14. Lai Z. C., Wei X., Shimizu T., Ramos E., Rohrbaugh M., Nikolaidis N., Ho L. L., Li Y. (2005) Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell 120, 675–685 [DOI] [PubMed] [Google Scholar]
- 15. Zhao B., Li L., Wang L., Wang C. Y., Yu J., Guan K. L. (2012) Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 26, 54–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu F. X., Zhao B., Panupinthu N., Jewell J. L., Lian I., Wang L. H., Zhao J., Yuan H., Tumaneng K., Li H., Fu X. D., Mills G. B., Guan K. L. (2012) Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150, 780–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mo J.-S., Yu F.-X., Gong R., Brown J. H., Guan K.-L. (2012) Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs). Genes Dev. 26, 2138–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wada K., Itoga K., Okano T., Yonemura S., Sasaki H. (2011) Hippo pathway regulation by cell morphology and stress fibers. Development 138, 3907–3914 [DOI] [PubMed] [Google Scholar]
- 19. Sansores-Garcia L., Bossuyt W., Wada K., Yonemura S., Tao C., Sasaki H., Halder G. (2011) Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 30, 2325–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fernández B. G., Gaspar P., Brás-Pereira C., Jezowska B., Rebelo S. R., Janody F. (2011) Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development 138, 2337–2346 [DOI] [PubMed] [Google Scholar]
- 21. Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., Elvassore N., Piccolo S. (2011) Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 [DOI] [PubMed] [Google Scholar]
- 22. Zhao B., Wei X., Li W., Udan R. S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., Zheng P., Ye K., Chinnaiyan A., Halder G., Lai Z. C., Guan K. L. (2007) Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21, 2747–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller E., Yang J., DeRan M., Wu C., Su A. I., Bonamy G. M., Liu J., Peters E. C., Wu X. (2012) Identification of serum-derived sphingosine-1-phosphate as a small molecule regulator of YAP. Chem. Biol. 19, 955–962 [DOI] [PubMed] [Google Scholar]
- 24. Zhao B., Li L., Tumaneng K., Wang C. Y., Guan K. L. (2010) A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(β-TRCP). Genes Dev. 24, 72–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hao Y., Chun A., Cheung K., Rashidi B., Yang X. (2008) Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 283, 5496–5509 [DOI] [PubMed] [Google Scholar]
- 26. Lei Q. Y., Zhang H., Zhao B., Zha Z. Y., Bai F., Pei X. H., Zhao S., Xiong Y., Guan K. L. (2008) TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol. 28, 2426–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chan S. W., Lim C. J., Guo K., Ng C. P., Lee I., Hunziker W., Zeng Q., Hong W. (2008) A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. Cancer Res. 68, 2592–2598 [DOI] [PubMed] [Google Scholar]
- 28. Yang X., Yu K., Hao Y., Li D. M., Stewart R., Insogna K. L., Xu T. (2004) LATS1 tumour suppressor affects cytokinesis by inhibiting LIMK1. Nature Cell Biol. 6, 609–617 [DOI] [PubMed] [Google Scholar]
- 29. Mikeladze-Dvali T., Wernet M. F., Pistillo D., Mazzoni E. O., Teleman A. A., Chen Y. W., Cohen S., Desplan C. (2005) The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell 122, 775–787 [DOI] [PubMed] [Google Scholar]
- 30. Takahashi A., Ohtani N., Yamakoshi K., Iida S., Tahara H., Nakayama K., Nakayama K. I., Ide T., Saya H., Hara E. (2006) Mitogenic signalling and the p16INK4a-Rb pathway cooperate to enforce irreversible cellular senescence. Nature Cell Biol. 8, 1291–1297 [DOI] [PubMed] [Google Scholar]
- 31. Dutta S., Baehrecke E. H. (2008) Warts is required for PI3K-regulated growth arrest, autophagy, and autophagic cell death in Drosophila. Curr. Biol. 18, 1466–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Emoto K., Parrish J. Z., Jan L. Y., Jan Y. N. (2006) The tumour suppressor Hippo acts with the NDR kinases in dendritic tiling and maintenance. Nature 443, 210–213 [DOI] [PubMed] [Google Scholar]
- 33. McPherson J. P., Tamblyn L., Elia A., Migon E., Shehabeldin A., Matysiak-Zablocki E., Lemmers B., Salmena L., Hakem A., Fish J., Kassam F., Squire J., Bruneau B. G., Hande M. P., Hakem R. (2004) Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 23, 3677–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhao B., Li L., Lu Q., Wang L. H., Liu C. Y., Lei Q., Guan K. L. (2011) Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203, 253–310 [DOI] [PubMed] [Google Scholar]
- 36. Cross L. M., Cook M. A., Lin S., Chen J. N., Rubinstein A. L. (2003) Rapid analysis of angiogenesis drugs in a live fluorescent zebrafish assay. Arteriosclerosis Thrombosis Vasc. Biol. 23, 911–912 [DOI] [PubMed] [Google Scholar]
- 37. Aase K., Ernkvist M., Ebarasi L., Jakobsson L., Majumdar A., Yi C., Birot O., Ming Y., Kvanta A., Edholm D., Aspenström P., Kissil J., Claesson-Welsh L., Shimono A., Holmgren L. (2007) Angiomotin regulates endothelial cell migration during embryonic angiogenesis. Genes Dev. 21, 2055–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhong T. P., Childs S., Leu J. P., Fishman M. C. (2001) Gridlock signalling pathway fashions the first embryonic artery. Nature 414, 216–220 [DOI] [PubMed] [Google Scholar]
- 39. Wells C. D., Fawcett J. P., Traweger A., Yamanaka Y., Goudreault M., Elder K., Kulkarni S., Gish G., Virag C., Lim C., Colwill K., Starostine A., Metalnikov P., Pawson T. (2006) A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell 125, 535–548 [DOI] [PubMed] [Google Scholar]
- 40. Yi C., Troutman S., Fera D., Stemmer-Rachamimov A., Avila J. L., Christian N., Persson N. L., Shimono A., Speicher D. W., Marmorstein R., Holmgren L., Kissil J. L. (2011) A tight junction-associated Merlin-angiomotin complex mediates Merlin's regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell 19, 527–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ranahan W. P., Han Z., Smith-Kinnaman W., Nabinger S. C., Heller B., Herbert B. S., Chan R., Wells C. D. (2011) The adaptor protein AMOT promotes the proliferation of mammary epithelial cells via the prolonged activation of the extracellular signal-regulated kinases. Cancer Res. 71, 2203–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paramasivam M., Sarkeshik A., Yates J. R., 3rd, Fernandes M. J., McCollum D. (2011) Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol. Biol. Cell 22, 3725–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oka T., Schmitt A. P., Sudol M. (2012) Opposing roles of angiomotin-like-1 and zona occludens-2 on pro-apoptotic function of YAP. Oncogene 31, 128–134 [DOI] [PubMed] [Google Scholar]
- 44. Wang W., Huang J., Chen J. (2011) Angiomotin-like proteins associate with and negatively regulate YAP1. J. Biol. Chem. 286, 4364–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shah K., Shokat K. M. (2003) A chemical genetic approach for the identification of direct substrates of protein kinases. Methods Mol. Biol. 233, 253–271 [DOI] [PubMed] [Google Scholar]
- 46. Ernkvist M., Aase K., Ukomadu C., Wohlschlegel J., Blackman R., Veitonmäki N., Bratt A., Dutta A., Holmgren L. (2006) p130-angiomotin associates to actin and controls endothelial cell shape. FEBS J 273, 2000–2011 [DOI] [PubMed] [Google Scholar]
- 47. Ernkvist M., Birot O., Sinha I., Veitonmaki N., Nyströom S., Aase K., Holmgren L. (2008) Differential roles of p80- and p130-angiomotin in the switch between migration and stabilization of endothelial cells. Biochim. Biophys. Acta 1783, 429–437 [DOI] [PubMed] [Google Scholar]
- 48. Tschöp K., Conery A. R., Litovchick L., Decaprio J. A., Settleman J., Harlow E., Dyson N. (2011) A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 25, 814–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chiyoda T., Sugiyama N., Shimizu T., Naoe H., Kobayashi Y., Ishizawa J., Arima Y., Tsuda H., Ito M., Kaibuchi K., Aoki D., Ishihama Y., Saya H., Kuninaka S. (2012) LATS1/WARTS phosphorylates MYPT1 to counteract PLK1 and regulate mammalian mitotic progression. J. Cell Biol. 197, 625–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang K., Rodriguez-Aznar E., Yabuta N., Owen R. J., Mingot J. M., Nojima H., Nieto M. A., Longmore G. D. (2012) Lats2 kinase potentiates Snail1 activity by promoting nuclear retention upon phosphorylation. EMBO J. 31, 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hirate Y., Hirahara S., Inoue K., Suzuki A., Alarcon V. B., Akimoto K., Hirai T., Hara T., Adachi M., Chida K., Ohno S., Marikawa Y., Nakao K., Shimono A., Sasaki H. (2013) Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr. Biol. 23, 1181–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao B., Ye X., Yu J., Li L., Li W., Li S., Lin J. D., Wang C. Y., Chinnaiyan A. M., Lai Z. C., Guan K. L. (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Overholtzer M., Zhang J., Smolen G. A., Muir B., Li W., Sgroi D. C., Deng C. X., Brugge J. S., Haber D. A. (2006) Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc. Natl. Acad. Sci. U.S.A. 103, 12405–12410 [DOI] [PMC free article] [PubMed] [Google Scholar]