Background: Understanding glucocorticoid receptor (GR)-regulated gene transcription requires detailed description of cofactor actions.
Results: NELF subunits have new activities altering GR induction properties of exogenous and endogenous genes.
Conclusion: NELF-A and NELF-B are decelerators functioning at two positions after GR and before/at the reporter gene site of action.
Significance: Functional ordering of NELF-A and NELF-B relative to other factors in GR transactivation is determined.
Keywords: Gene Transcription, Glucocorticoid Receptor, Mathematical Modeling, Steroid Hormone, Transcription Factors, EC50, Gene Induction, NELF-A and NELF-B, Cofactor Action, Competitive Decelerator
Abstract
NELF-B is a BRCA1-interacting protein and subunit (with NELF-A, -C/D, and -E) of the human negative elongation factor (NELF) complex, which participates in RNA polymerase II pausing shortly after transcription initiation, especially for synchronized gene expression. We now report new activities of NELF-B and other NELF complex subunits, which are to attenuate glucocorticoid receptor (GR)-mediated gene induction, reduce the partial agonist activity of an antagonist, and increase the EC50 of an agonist during nonsynchronized expression of exogenous and endogenous reporters. Stable knockdown of endogenous NELF-B has the opposite effects on an exogenous gene. The GR ligand-binding domain suffices for these biological responses. ChIP assays reveal that NELF-B diminishes GR recruitment to promoter regions of two endogenous genes. Using a new competition assay, NELF-A and NELF-B are each shown to act independently as competitive decelerators at two steps after the site of GR action and before or at the site of reporter gene activity. A common motif in each NELF was identified that is required for full activity of both NELF-A and NELF-B. These studies allow us to position the actions of two new modulators of GR-regulated transactivation, NELF-A and NELF-B, relative to other factors in the overall gene induction sequence.
Introduction
The process of gene induction and repression by steroid receptors has three properties that are relevant for differential control of transcription during development, differentiation, and homeostasis. The total amount of gene expression, or Amax, is the maximal response with ligand concentrations sufficient to saturate the receptor. The EC50 is the concentration of ligand needed for half-maximal response and determines the potency of gene expression by a given steroid. Interestingly, the EC50 is not determined by the steroid binding affinity for receptor and can be highly variable even for the same gene under various settings (1–3). The partial agonist activity (PAA)5 of an antisteroid is the reduced activity that most antisteroids display under assorted conditions. The original thought that antisteroids, or antagonists, have no intrinsic activity and block all activity of agonists has given way to the realization that an “antagonist” can display a partial amount of full agonist activity. Furthermore, differing cellular conditions, acting like a rheostat, can cause an antisteroid to display varying amounts of full agonist activity that may approach 100%. Thus, like the EC50, the PAA of an antisteroid with an individual gene is rarely constant (1–3). Clearly, a better grasp of why all genes do not display the same Amax, EC50, and PAA with the same receptor-steroid complex is critical not only to understand the molecular actions of steroid hormones but also for selective treatment of human endocrine pathologies.
One clue about the variations of Amax, EC50, and PAA is that they are modulated by many transcription cofactors (e.g. coactivators, corepressors, and comodulators). This behavior appears to be general in that all classical steroid receptors (androgen, estrogen, glucocorticoid, mineralocorticoid, and progesterone) and some nuclear receptors are sensitive to these factors (1–3). Furthermore, the ability of factors to adjust Amax, EC50, and PAA is seen for both gene induction and gene repression (4–7) and is not restricted to synthetic reporter genes but also occurs in a gene-selective manner with endogenous genes of primary human cells (i.e. peripheral blood mononuclear cells) (8). It thus is likely that alterations of the parameters of receptor-regulated gene expression are widespread and physiologically relevant.
Our understanding of how these parameters are modified is complicated by the fact that few landmarks in the sequence of steroid-regulated reactions have been determined other than steroid binding to its cognate receptor, direct or indirect binding of receptor-steroid complex to biologically active DNA sequences such as hormone response elements, and recruitment of various cofactors to increase or decrease gene transcription rates. Although the details of steroid hormone action are still poorly defined, most modulatory factors are thought to act shortly after receptor-steroid binding/tethering to DNA. However, any step can, under the appropriate conditions, influence the Amax, EC50, and/or PAA (9, 10). Not all factors will alter all three parameters (8, 11). In fact, the ability to modulate Amax and/or EC50, along with the direction of the modulation, conveys otherwise inaccessible mechanistic information (12–14). Consequently, we were attracted by reports that cofactor of BRCA1 (COBRA1 = NELF-B) represses gene transcription by estrogen receptors (ERs) and androgen receptors (15–17) and binds glucocorticoid receptors (GRs) (16). NELF-B has been suggested to be a tumor suppressor (18) and an oncogene (19). NELF-B is also an integral subunit of the human negative elongation factor (NELF) complex (20, 21) that is instrumental in RNA polymerase II pausing (22–26). Recent studies suggest that a major function of polymerase pausing is to enforce synchronized gene expression during differentiation and development (27). Chromatin immunoprecipitation experiments revealed that NELF knockdown can influence which transcriptional start site of a gene is used (28). These observations suggested that NELF-B could modify ER and androgen receptor actions at steps well downstream of receptor-steroid complex binding to hormone response elements.
The objectives of this study were to determine whether NELF-B interacts with glucocorticoid receptors (GRs) to repress transactivation and alter the Amax, EC50, and PAA. To this end, we initially examined GR induction of a transiently transfected reporter gene, which would not be encumbered by mechanisms to ensure synchronized gene expression. Positive results with both synthetic and endogenous GR-regulated genes prompted us to investigate NELF-B effects on GR binding to enhancers. Our recently developed competition assay (13, 14, 29) was used to determine the kinetically determined mode and site of action of wild type and mutant NELF-B and NELF-A. We conclude that NELF-B and NELF-A are new GR cofactors that act independently, but additively, as a competitive decelerator (14), each at two different steps.
EXPERIMENTAL PROCEDURES
Unless otherwise indicated, all cell growth was at 37 °C, and all other operations were performed at 0 °C.
Chemicals
Dexamethasone (Dex) was purchased from Sigma. Dex-21-mesylate (Dex-Mes) was synthesized as described previously (30). RU486 was a gift from Etienne Baulieu (Paris, France). Restriction enzymes and T4 DNA ligase were from New England Biolabs (Beverly, MA), and the Dual-Luciferase reporter assay was from Promega (Madison, WI).
Plasmids
Renilla-TS reporter, rat GR (pSG5-GR), GREtkLUC, pSG5-TIF2, and GAL/GR525C have been previously described (31). FR-LUC reporter is from Stratagene (La Jolla, CA). Human NELF-A (missing the first 11 residues), FLAG/NELF-B (32), FLAG/NELF-C/D, and FLAG/NELF-E were from the University of Texas Health Science Center at San Antonio. FLAG/NELF-B (in pcDNA3) has the C-terminal 30 amino acids deleted and replaced by the C-terminal 52 amino acid of the neomycin gene. Full-length wild type (wt) NELF-B, also with an N-terminal FLAG tag, was constructed by removing the FLAG/full-length wtNELF-B from IRES-COBRA with EcoR1/BamH1 digestion and then inserting it into the pcDNA3.1(−) vector (Invitrogen).
The 4mtNELF-B was first generated using QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) in the context of the chimeric NELF-B with the following primers: L283A/L287A forward: 5′-GAGGGCGCGGGAGGCCCAGGGGTTTGCCGATGGCGTCAAG-3′ and reverse: 5′-CTTGACGCCATCGGCAAACCCCTGGGCCTCCCGCGCCCTC-3′; K291A/K292A forward: 5′-GGGGTTTCTCGATGGCGTCGCCGCCGGCCAGGAGCAGGTGCTGG-3′ and reverse: 5′-CCAGCACCTGCTCCTGGCCGGCGGCGACGCCATCGAGAAACCCC-3′. The above full-length wtNELF-B plasmid was digested with EcoR1/SbfI, and the larger of two fragments was used as the vector for ligation. The shorter fragment (1023 bp) was redigested with SacII to generate two fragments, of which the second shorter fragment 226 bp from EcoR1 to SacII was isolated. The chimeric 4mtNELF-B was digested by SacII/SbfI to produce two DNA fragments. The shorter 797-bp species containing the four mutated amino acids was incubated with the 226-bp EcoR1/ScaII fragment and the above EcoRI/SbfI vector fragment in a ratio of 1:1:3 overnight at 16 °C to afford the desired plasmid.
Double mutant I470A/M474A NELF-A was generated by using QuikChange II XL site-directed mutagenesis kit. The forward primer is 5′-GAGAAGGCCCTCGCCCTGGGCTTCGCGGCCGGCTCCCG-3′, and the reverse primer is 5′-CGGGAGCCGGCCGCGAAGCCCAGGGCGAGGGCCTTCTC-3′.Introducing R478A/E479A into the double mutant I470A/M474A NELF-A plasmid was achieved by overlapping PCR. Sites for XmalI and EcoRV were found located 353 bp upstream and 231 bp downstream of amino acid Arg-478, respectively. The first round PCR amplified from XmalI to R478A/E479A, and the second round was from R478A/E479A to EcoRV. These two amplified DNA fragments were purified, mixed with the ratio of 1:1, and used as a template to generate a third DNA fragment from XmalI to EcoRV, which contains all four mutants. The third PCR product and the wild type NELF-A plasmid were then digested by XmalI/EcoRV, purified, and processed for DNA ligation. The primers used for overlapping PCR are XmalI forward 5′-CCATCTTCCCGGGAAGCCAGC-3′, EcoRV reverse 5′-GGCAGCCTGCACCTGAGGAGTG-3′, R478A/E479A forward 5′-CTTCGCGGCCGGCTCCGCTGCTAACCCGTGCCAGGAGC-3′, and R478A/E479A reverse 5′-GCTCCTGGCACGGGTTAGCAGCGGAGCCGGCCGCGAAG-3′.
IP6K3 intron 1 GRE, IGFBP1 promoter, and intron 1 GRE region were amplified from U2OS genomic DNA using primers (with SalI and MluI restriction site, gene sequences are underlined) as follows: IP6K3 intron 1 GRE, forward: 5′-ACGGTCGACTGCCTGGAGCCCTCTCACTT-3′ and reverse: 5′-ACGACGCGTACTAGGGTACCTAGAAGAGT-3′; IGFBP1 promoter, forward: 5′-ACGGTCGACATGGGCATCAGAAATGTGAA-3′ and reverse: 5′-ACGACGCGTACACAGCGCGCACCTTATA-3′; IGFBP1 intron 1 GRE, forward: 5′-ACGGTCGACGAGACTCAAGGAGGAAGCT-3′ and reverse: 5′-ACGACGCGTTGGAAGTCCTAATTCCTCCA-3′. IP6K3-GRE-tkLUC, IGFBP1-promoter-tkLUC, and IGFBP1-Intron1 GRE-tkLUC were constructed by insertion of the SalI/MluI fragment of the above PCR products into the SalI/MluI fragment of tkLUC vector. The site-directed mutagenesis kit (Stratagene) was used with the following primers (mutated nucleotides are underlined) to make mutant IP6K3-GRE-tkLUC and IGFBP1-Intron1 GRE-tkLUC: IP6K3-GRE-tkLUC Mut, 5′-ATCCCCTGGGCACAGGAATAGCAACTCCTAGCCCAGCAGC-3′; IGFBP1-Intron1 GRE-tkLUC Mut, 5′-TTCCCAGATGTTTACAGAATATAAACTGAGAGTTGAGGCTAAAAG-3′. All the mutants were verified by sequencing.
Antibodies
Anti-GR mouse and rabbit monoclonal antibodies (MA1-510 and PA1-516A; Affinity BioReagents), anti-NELF-B rabbit polyclonal antibody (ab48336; Abcam), anti-NELF-A rabbit polyclonal antibody (sc-23599; Santa Cruz Biotechnology), anti-β-actin mouse monoclonal antibody (A2228; Sigma), and anti-FLAG mouse monoclonal antibody (F3165; Sigma) are commercially available. Anti-NELF-C/D rabbit polyclonal antibody was raised by us using bacterially expressed NELF-C/D.
Cell Culture, Transient Transfection, and Reporter Analysis
Monolayer cultures of U2OS, U2OS.rGR, COS-7, and CV-1 cells were grown as described previously (31, 33). T47D cells were cultured in Dulbecco's modified minimum essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS). NELF-B stable knockdown T47D cell line was described previously (15). Triplicate samples of cells were seeded into 24-well plates at 20,000 cells/well and transiently transfected the following day with luciferase reporter and DNA plasmids by using 0.7 μl of Lipofectamine (Invitrogen) or FuGENE 6 (Roche Applied Science) per well according to the manufacturer's instructions. The total transfected DNA was adjusted to 300 ng/well of a 24-well plate with pBlueScriptII SK+ (Stratagene). The molar amount of plasmids expressing different protein constructs was kept constant with added empty plasmid or plasmid expressing human serum albumin (31). Renilla-TS (10 ng/well of a 24-well plate) was included as an internal control. After transfection (32 h), cells were treated with medium containing appropriate hormone dilutions. The cells were lysed 20 h later and assayed for reporter gene activity using Dual-Luciferase assay reagents according to the manufacturer's instructions (Promega, Madison, WI). Luciferase activity was measured by an EG&G Berthold luminometer (Microlumat LB 96P). The data were normalized to Renilla null luciferase activity and expressed as a percentage of the maximal response with Dex before being plotted ± S.E., unless otherwise noted.
Total RNA Extraction and Reverse Transcription-PCR (RT-PCR)
For quantitative real-time PCR (qRT-PCR), total RNA was extracted by TRIzol reagent (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was synthesized by SuperScript II reverse transcriptase (Invitrogen). The relative levels of target mRNAs were quantitated using SYBR Green and the ABI 7900HT real-time PCR system. The primers for insulin-like growth factor-binding protein 1 (IGFBP1), glucocorticoid-induced leucine zipper (GILZ), ladinin 1 (LAD1), inositol hexaphosphate kinase 3 (IHPK3 or IP6K3), and β-actin are as shown in Table 1. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was quantitated by TaqMan using primer from ABI (4310884E).
TABLE 1.
RT-PCR primers
| Human gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| NELF-B | GACTTCTGCAGCAGCCTGTT | GATCCAGCTGTTCCAGCTTC |
| GILZ | AATGCGGCCACGGATG | GGACTTCACGTTTCAGTGGACA |
| IGFBP1 | CCAAGGGACAGGAGACATCAG | AGGGTAGACGCACCAGCAGAGT |
| IP6K3 | TTCTCGCTGGTGGAAGACAC | CAGCAACAAGAACCGATGC |
| LAD1 | AGCATGAAGCTCCCAGACAAC | ACCCACAGGAGCCACGAATAA |
| β-Actin | GCGAGAAGATGACCCAGATCATG | GTCACCGGAGTCCATCACGAT |
Western Blotting
Cytosols for Western blots were prepared as described below for immunoprecipitation assays, probed with rabbit anti-GR, FLAG, NELF-B, NELF-A, and NELF-C/D antibodies (described above), and visualized by ECL detection reagents as described by the manufacturer (Amersham Biosciences).
Immunoprecipitation Assays
Immunoprecipitations were conducted as described previously (31) with the following modifications. The day before transfection, U2OS.rGR or COS-7 cells were seeded into 150-mm dishes at 2,000,000 cells/dish containing 20 ml of medium. On the next day, DNA was transfected. After 2 days of growth, cells were treated with ethanol, Dex, or RU486 and washed once with 20 ml of PBS 1 h later. The cells were lysed for 5 min at room temperature with 1.6 ml of CytoBuster protein extraction buffer (EMD Biosciences, La Jolla, CA) containing protease inhibitor cocktail (PIC, Roche Applied Science) and clarified by centrifugation at 13,000 × g for 10 min at 4 °C. Samples for sodium molybdate treatment were prepared with the modification that the cell lysates were adjusted to contain either 20 mm Na2MoO4 or 20 mm Na2SO4 immediately after rupturing the cells followed by treatment with ethanol, Dex, or RU486 and incubation at 20 °C for 20 min. Aliquots (700 μl) of supernatant were incubated on a roller drum (3 rpm) with 50 μl of FLAG-M2 agarose (Sigma, 4 °C for overnight) to immobilize the complexes with FLAG-tagged proteins. On the next day, the antibody complexes were centrifuged (1 min at 16,000 × g at 4 °C), washed four times with 1 ml of lysis buffer (50 mm Tris, 150 mm NaCl, 0.1% Nonidet P-40, pH 7.5), extracted with 20 μl of 2× SDS loading buffer (95 °C for 5 min), and separated by 10–12% SDS-PAGE.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP was performed as described (34) with minor modifications. Two days before harvest, U2OS.rGR or U2OS cells were seeded into 150-mm dishes at 3,000,000 cells/dish in DMEM with 10% FBS. On the next day, DNA (including GR plasmid with U2OS cells) was transfected. On the third day, after ligand Dex (100 nm) or Dex-Mes (1 μm) treatment for 1 h, cells were cross-linked with 1 mm dimethyl 3,3′-dithiobispropionimidate in 1× PBS at 37 °C for 30 min. Then formaldehyde (37%) was added directly into the solution to a final concentration of 1% (room temperature for 10 min with shaking), which was followed by glycine to a final concentration of 0.125 m (room temperature for 5 min with shaking). Cells were washed with ice-cold PBS twice and harvested by scraping into 2 ml of ice-cold PBS with freshly added PIC. After centrifugation (at 2500 × g for 10 min), the cell pellet was resuspended in 1 ml of cell lysis buffer (50 mm HEPES, 140 mm NaCl, 1 mm EDTA, 10% glycerol, 0.5% Nonidet P-40, 0.25% Triton X-100, freshly added PIC, pH 7.5) and incubated on ice for 10 min.
The nuclei were collected by centrifugation (at 2500 × g for 10 min) and resuspended in 1 ml of protein extraction buffer (200 mm NaCl, 1 mm EDTA, 0.5 mm EGTA, 10 mm Tris-HCl, freshly added PIC, pH 8.1) and rotated 10 min at 4 °C. After centrifugation (2500 × g, 10 min, 4 °C), the pellet was resuspended in 300 μl of chromatin extraction buffer (1 mm EDTA, 0.5 mm EGTA, 10 mm Tris-HCl, freshly added PIC, pH 8.1). The 300-μl sample was sheared to 200–400-bp fragments with a Bioruptor UCD-200TM (Diagenode; Sparta, NJ; high power, 5 min, six cycles, total 30 min). After removing the insoluble debris by centrifugation (13,000 × g, 10 min, 4 °C), 2.7 ml of chromatin extraction buffer, 390 μl of 10% Triton X-100, 39 μl of 10% sodium deoxycholate, and fresh PIC were added to the supernatant.
All of the sample was treated with 200 μl of preblocked protein A beads (Amersham Biosciences) with gentle mixing (at 4 °C for 2 h). After centrifugation (3,000 × g, 1 min), 100 μl of supernatant was taken as input. Aliquots (1 ml) of supernatant were treated with 1–2 μg of antibody at 4 °C overnight. The next morning, 30 μl of preblocked protein A beads was added for 1 h. The pellet was centrifuged (2000 × g, 1 min) and washed sequentially by 1 ml of low salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, 150 mm NaCl, pH 8.1), 1 ml of high salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, 500 mm NaCl, pH 8.1), 1 ml of LiCl wash buffer (250 mm LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, 10 mm Tris-HCl pH 8.1), and three times with 1 ml of Tris-EDTA buffer.
Chelex-100 (100 μl of a 10% slurry) was added directly to the washed beads pellet and boiled for 10 min at 99 °C to reverse cross-link. After centrifugation (13,000 × g, 5 min, 4 °C), the supernatant was transferred to a new tube. The 100-μl input sample was precipitated by 10 μl of 3 m sodium acetate (pH 5.2) and 250 μl of 96% ethanol at −80 °C for 30 min followed by centrifugation (20,000 × g, 15 min, 4 °C) and washing with 500 μl of 70% ethanol. The input DNA pellet was reverse-cross-linked by adding 100 μl of 10% Chelex-100 as described above. The 100-μl input sample was purified with MinElute® PCR purification kit (Qiagen) in 300 μl of EB buffer (10 mm Tris-Cl, pH 8.5). The immunoprecipitated DNA was quantified by real-time PCR using the primers listed in Table 2.
TABLE 2.
ChIP-qPCR primers used in this study
| Site (bp) | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| IP6K3 TSS/pause (−27 to 160) | AGCAGAACACCCAGCTGAC | CACCATGTACAGGCATCAGG |
| IP6K3 3′ of pause (141–303) | CCTGATGCCTGTACATGGTG | GACCCTCTCTCCCCTCAGTC |
| IP6K3 GRE (534–686) | CTGGAGCCCTCTCACTTCAG | CTGGGCTAGGACATGCTGTT |
| IGFBP1 IG2 (−1101 to −935) | ATGGGCATCAGAAATGTGAAT | TCCTTTAGGAGTGGTGTTG |
| IGFBP1 IG1/TSS (−165 to 46) | GACCTGGGCTGTCTTTTTGA | CGCTAGGAGCTGAGTGTTCA |
| IGFBP1 pause (−2 to 229) | TGGGTGCACTAGCAAAACAA | AAACTCTGGGCAAGTGATGG |
| IGFBP1 intron 1 GRE (1105 to 1269) | CCAGGAGGTGTTTGGAATGT | TCATGTTCTTAGGGGGCAAC |
Two-factor Competition Assays
A full description is found in Refs. 13 and 29. Briefly, the effect of four concentrations of each of two factors (total = 16 combinations) on the maximum induced activity (Amax) and EC50 is determined from directly fitting a Michaelis-Menten curve to the average (n = 3, S.D. is usually <10% for more than 95% of the triplicates) value of induced luciferase activity from transiently transfected reporter (GREtkLUC) with EtOH and three subsaturating concentrations of Dex (total samples = 192). Graphs of 1/EC50 and Amax/EC50 (and EC50/Amax for a decelerator) versus the concentration of one cofactor were constructed at each of the concentrations of the second factor. A linear plot for EC50/Amax versus factor 1 indicates that the plots in Amax/EC50 approach a zero asymptote at infinite factor 1, in which case factor 1 would be a competitive decelerator (C) before or at the concentration-limiting step (CLS). A competitive decelerator acts similarly to a competitive inhibitor in enzyme kinetics. A downward curving, nonlinear plot that becomes linear after first subtracting an estimated nonzero asymptote from the Amax/EC50 values, and then plotting EC50/Amax, indicates that factor 1 is a C after the CLS. The case of a nonlinear, upward curving plot is described in the text. In all cases, however, it is critical to make corrections if Western blots show that expression of the transiently transfected protein (at constant levels of either total cellular protein or an internal standard, such as β-actin) is nonlinear. This is because the interpretation of the graphs is predicated on the x axis being a linear scale. To determine the linear equivalent of expressed plasmid, the nonlinear plot of optical density versus ng of transfected plasmid is first fit to a Michaelis-Menten plot of
The functional equivalent of the transfected plasmid that gives a linear optical density versus plasmid plot is then obtained from the formula of
 |
The x axis value of the amount of plasmid in the various graphs is then this “corrected plasmid” value. These same Western blots are used to determine the relative amount of endogenous factor (in units of ng of factor plasmid) when dealing with an accelerator. It is not necessary to quantitate the relative amount of an endogenous factor that displays decelerator activity. If an experiment used n concentrations for each cofactor, then there would be a total of four to six graphs, each with n separate curves. The shape of the curves and how they change with the other cofactor are then compared with Table 1B of Ref. 13 to determine the kinetically defined mechanism of action and site of action, relative to each other and to the CLS. Many of the entries in Table 1B of Ref. 13 require an estimate of the intersection point of a set of linear regression fits to the graphs. For a family of lines of the form y = ai + bix, an unbiased estimate of the intersection can be obtained from “a versus b plots,” which are a linear regression on the graph of a versus b to give a new plot of the form y′ = c + dx, where c is the y axis value of the intersection point of the family of lines in the original graph and the negative value of d corresponds to the x axis value of the intersection point.
Statistical Analysis
Unless otherwise noted, all experiments were performed in triplicate multiple times. KaleidaGraph 3.5 (Synergy Software, Reading, PA) was used to determine a least-squares best fit (R2 was almost always >0.95) of the experimental data to the theoretical dose-response curve, which is given by the equation derived from Michaelis-Menten kinetics of y = (free steroid)/(free steroid + dissociation constant (Kd)) (where the concentration of total steroid is approximately equal to the concentration of free steroid because only a small portion is bound), to yield a single EC50 value. The values of n independent experiments were then analyzed for statistical significance by the two-tailed Student's t test using InStat 2.03 for Macintosh (GraphPad Software, San Diego, CA). The Mann-Whitney test or the Alternate Welch t test is used when the difference between the S.D. values of two populations is statistically significant.
RESULTS
NELF-B Modulates GR Induction Parameters with Exogenous Reporter Gene
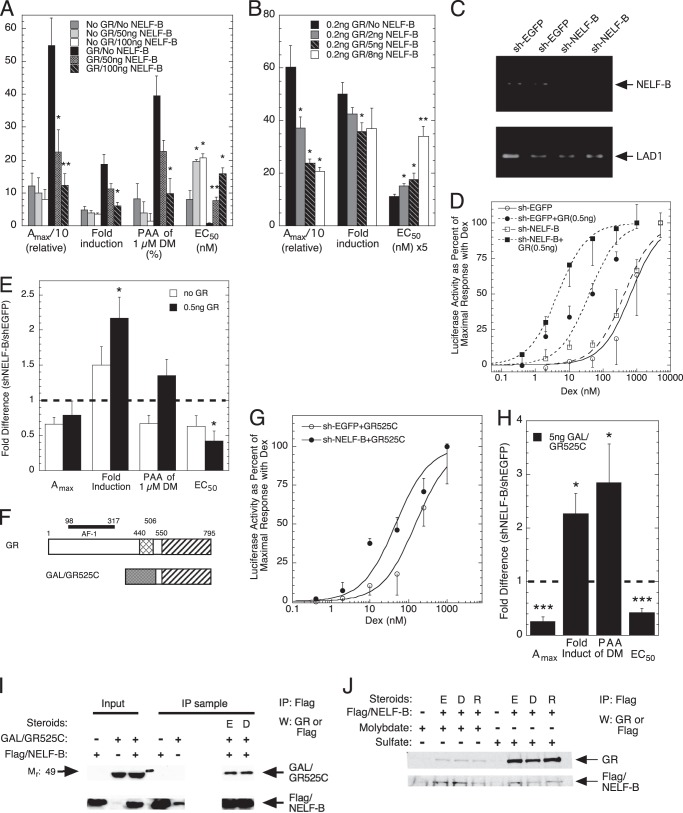
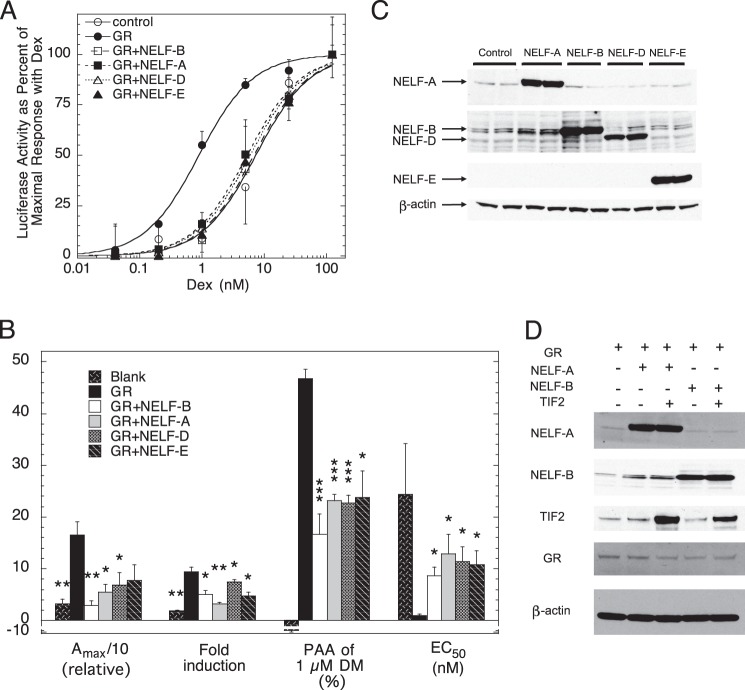
Human osteosarcoma (U2OS) cells were transiently transfected with the synthetic reporter gene (GREtkLUC) and a GR-expressing plasmid plus varying concentrations of two different NELF-B plasmids: a chimera lacking the C-terminal 30 amino acids (Fig. 1A) and the full-length wild type NELF-B (Fig. 1B) (see “Experimental Procedures”). In each case, cells were induced with a range of steroid concentrations for 20 h as described under “Experimental Procedures.” The low levels of endogenous GR in U2OS cells give a weak response to exogenous NELF-B that is greatly enhanced with added GR (Fig. 1A). In the presence of increased GR, the Amax with saturating concentrations of the agonist Dex is dramatically decreased by NELF-B. At the same time, the -fold induction by Dex is lowered, the PAA of the antiglucocorticoid Dex-Mes is reduced, and the EC50 of Dex is significantly increased. Similar results are obtained with lower amounts of all exogenous factors: the full-length NELF-B, GR, and GREtkLUC (Fig. 1B). This indicates that the C-terminal 30 amino acids of NELF-B are unnecessary for activity and that wt and chimeric NELF-B can be used interchangeably (see also Fig. 6 below). In all cases, the responses are directly dependent upon the concentration of added NELF-B. Thus, NELF-B is a very active modulator of GR induction parameters over a range of factor concentrations and has the hallmark of a corepressor, which is to reduce the Amax (35).
FIGURE 1.
Characteristics of NELF-B modulation of GR induction properties of exogenous reporter. A and B, effect of exogenous NELF-B on Amax, -fold induction, PAA, and EC50 of transfected reporter in U2OS cells. Triplicate wells of cells were transiently transfected with GREtkLUC reporter (100 ng in A, 12 ng in B) and the indicated amounts of chimeric NELF-B plasmid (A) or full-length NELF-B plasmid (B), without or with GR plasmid (0.5 ng in A, 0.2 ng in B), induced by steroid, and analyzed as described under “Experimental Procedures.” *, p < 0.05, **, p < 0.01 versus no NELF-B. y axis numbers are the quantitative values for the categories of the x axis (average ± S.E., n = 3 and 4 independent experiments in A and B, respectively). DM, Dex-Mes. C–E, modulation of GR activity with exogenous reporter upon reducing endogenous NELF-B. C, levels of NELF-B and control (LAD1) mRNAs in T47D cells stably transfected with shEGFP or shNELF-B RNA. Duplicate mRNA samples were prepared as described under “Experimental Procedures” and separated on agarose gels. The quantitative abundance was determined by qRT-PCR of the same original samples. D, representative dose-response curves for Dex induction of GREtkLUC in T47D cells stably transfected with shEGFP or shNELF-B RNA after transient transfection without or with GR plasmid. E, -fold changes in GR induction parameters from 5 independent experiments (± S.E.) in T47D cells stably transfected with shEGFP or shNELF-B RNA after transient transfection without or with GR plasmid. Thick horizontal dashed line represents no difference between shEGFP and shNELF-B cells. *, p < 0.02 versus no GR. F, graphic representation of domains of full-length rat GR and GR C terminus fused to GAL4 DBD. Cross hatching = GR DBD; shading = GAL4 DBD; striped = GR LBD. G, intracellular levels of NELF-B affect the EC50 of gene induction by GAL/GR525C. Representative dose-response curves for Dex induction of GREtkLUC in T47D cells stably transfected with shEGFP or shNELF-B RNA after transient transfection without or with GAL/GR525C plasmid are shown. H, -fold changes in GR induction parameters from 7 independent experiments (± S.E.) in T47D cells stably transfected with shEGFP or shNELF-B RNA after transient transfection without or with GAL/GR525C plasmid. Thick horizontal dashed line represents no difference between shEGFP and shNELF-B cells. *, p < 0.05, ***, p < 0.0005 versus -fold difference = 1. I, binding of NELF-B to overexpressed GAL/GR525C. Cytosolic extracts of Cos-7 cells that had been transiently transfected with GAL/GR525C plus FLAG/NELF-B were treated with sodium sulfate, with or without EtOH (E) or Dex (D). Receptors were then analyzed for co-immunoprecipitation (IP) with FLAG/NELF-B using anti-FLAG antibody as described under “Experimental Procedures.” W, Western blot. J, NELF-B binds only to activated endogenously expressed GRs. A cytosolic solution of U2OS.rGR cells with endogenous GR and transiently transfected FLAG-tagged NELF-B was split in half, treated with sodium molybdate (to block activation) or sodium sulfate, and then incubated with EtOH (E), Dex (D), or RU486 (R) before being immunoprecipitated (IP) with anti-FLAG antibody as described under “Experimental Procedures.” W, Western blot.
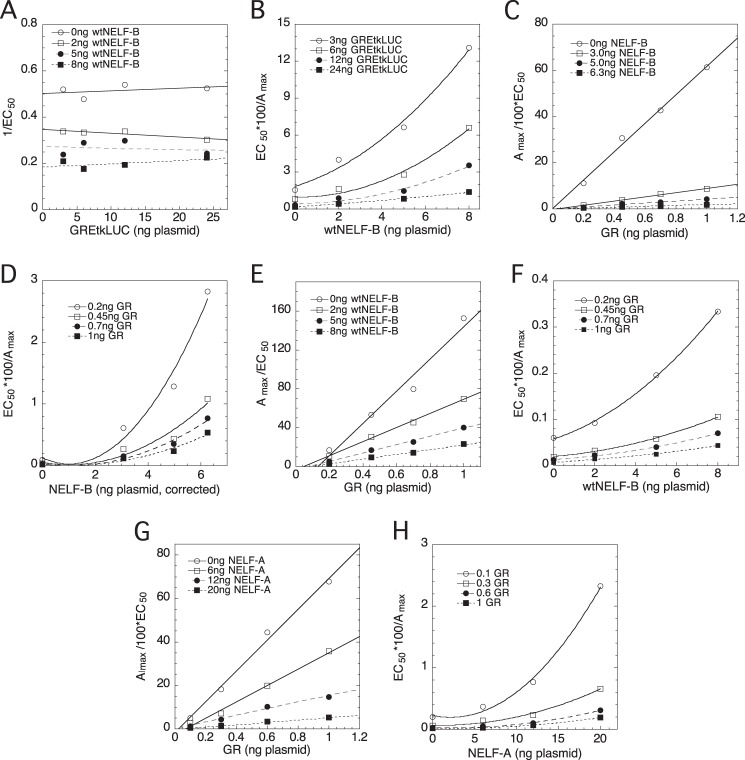
FIGURE 6.
Full-length NELF-B and NELF-A act at two steps in competition assays with GREtkLUC and with GR. U2OS cells were transiently transfected with the indicated varying amounts of GREtkLUC reporter, NELF-B, or NELF-A plasmid or GR plasmid as described under “Experimental Procedures.” The Amax and EC50 were determined as described for Fig. 5D. Plots, after correcting for nonlinear expression of chimeric NELF-B as described under “Experimental Procedures,” of 1/EC50 versus GREtkLUC for NELF-B (A), EC50/100 × Amax versus NELF-B (B, D, and F) or versus NELF-A (H), and Amax/100 × EC50 versus GR with NELF-B (C and E) and with NELF-A (G) were made and interpreted as described under “Results.” Similar results were seen in 3 or 4 additional independent experiments.
To examine the effects of reduced levels of NELF-B, we used T47D cells in which NELF-B was decreased by stable transfection with short helical (sh) RNAs (18). T47D cells expressing a shEGFP oligonucleotide were used as a control. qRT-PCR revealed that the levels of NELF-B mRNA in the NELF-B knockdown (KD) cells are reduced by 80% relative to the control cells (Fig. 1C). Interestingly, transient transfection of NELF-B plasmid increases NELF-B mRNA in U2OS cells by a factor of 2 to a level that is only 40% of that in T47D cells expressing the shEGFP oligonucleotide (data not shown). Due to the low -fold induction by endogenous GR of the T47D cells, GR plasmid was cotransfected with GREtkLUC reporter. Under these conditions, decreased NELF-B causes a significant left-shift in the dose-response curve (Fig. 1D), consistent with added NELF-B moving the EC50 to higher steroid concentrations in Fig. 1, A and B. At the same time, the Amax goes down. However, the -fold induction increases in NELF-B KD versus control cells (Fig. 1E) because the basal level is much lower in the NELF-B KD cells (22.1 ± 6.7 versus 59.6 ± 7.8 (S.E., n = 5)). The PAA for the antiglucocorticoid Dex-Mes is proportionately higher in the NELF-B KD cells with exogenous GR, but the increase is not statistically significant. We conclude that NELF-B has a consistent effect in two different cell lines regardless of whether the intracellular level of NELF-B is increased by transfected NELF-B plasmid or decreased by siRNA-mediated degradation of endogenous NELF-B mRNA.
GR Ligand-binding Domain Binds NELF-B and Mediates NELF-B Modulatory Actions
We previously found that the GR ligand-binding domain (LBD) is sufficient to respond to the modulatory actions of the coactivator TIF2 and the comodulator Ubc9 (36–38). Using transiently transfected GAL/GR chimera (GAL/GR525C, Fig. 1F) driving the synthetic GAL-regulated reporter (FRLuc) in the same knockdown T47D cells as in Fig. 1, D and E, reduced levels of NELF-B are seen to shift the dose-response curve to the left as compared with the EGFP KD cells, thereby reducing the EC50 (Fig. 1, G and H). As with full-length GR, diminished NELF-B decreases the Amax but increases the -fold induction while augmenting the PAA of the antiglucocorticoid Dex-Mes (Fig. 1H). The quantitative changes in all induction parameters with reduced NELF-B are very similar for GAL/GR525C (Fig. 1H) and full-length GR (Fig. 1E). We conclude that the GR LBD is sufficient to mediate most of the actions of NELF-B under these conditions.
Transiently transfected human GR and human NELF-B are reported to bind to each other (16). This was confirmed by showing that NELF-B binds to both ligand-free and -bound GAL/GR525C (Fig. 1I). Thus, the GR LBD is sufficient for binding NELF-B. We next asked whether NELF-B binding to endogenous full-length GR required activation by examining the interactions of transiently transfected human FLAG-tagged NELF-B with endogenous GR in U2OS cells stably transfected with rat GR (U2OS.rGR cells) (39). To distinguish between unactivated and activated receptors, cell-free lysates were first treated with either sodium molybdate, to block activation of the receptor to the DNA-binding form (40), or the iso-electronic but inactive control, sodium sulfate. Vehicle, agonist steroid, or antiglucocorticoid was then added followed by brief warming to 20 °C to activate the receptors. Finally, anti-FLAG antibody was added to immunoprecipitate the FLAG/NELF-B and associated proteins. When receptor activation to the DNA-binding form of the receptor is blocked by the addition of sodium molybdate, GR co-immunoprecipitation with FLAG/NELF-B is almost totally prevented as compared with samples containing sodium sulfate (Fig. 1J). The ability of ligand-free GR to be activated is consistent with earlier studies (41). Thus, we deduce that NELF-B binds to endogenously expressed GR but only after activation.
NELF-B Modifies the Induction Properties of Endogenous Genes
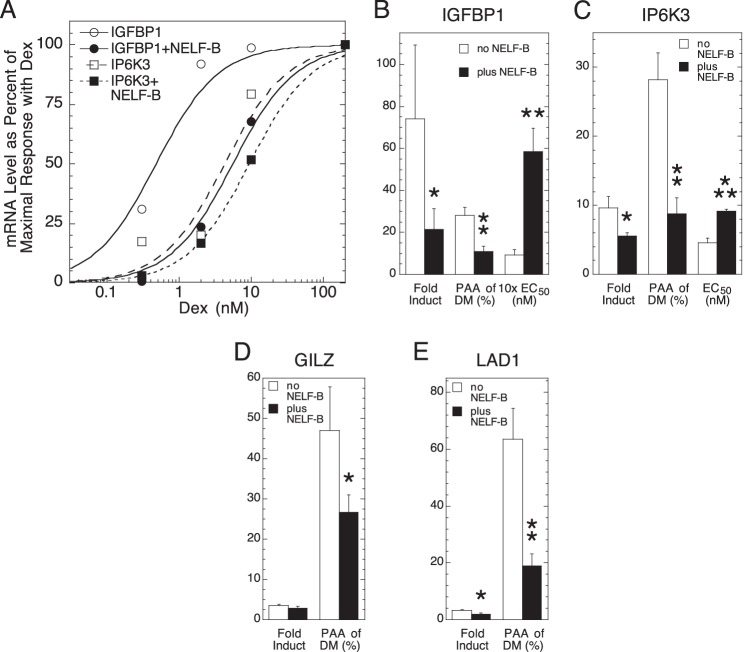
To determine the generality of the above observations, we asked whether NELF-B altered the GR induction parameters of four endogenous genes with low (basal) levels of expression in the absence of added steroid: GILZ (42), IP6K3 (IHPK3) (43), IGFBP1 (44), and LAD1 (45). The effect of NELF-B on GR induction was examined in U2OS cells because the extensive squelching that was usually seen with these four genes in U2OS.rGR cells prevents an accurate assessment of GR transactivation parameters (data not shown). In U2OS cells, NELF-B reduces the -fold induction of Dex and the PAA of Dex-Mes while increasing in EC50 for IGFBP1 and IP6K3 (Fig. 2, A–C). The low -fold induction of GILZ and LAD1 with added NELF-B precluded an assessment of changes in EC50, but the -fold induction and PAA of Dex-Mes are each decreased (Fig. 2, D and E). Thus, the effects of NELF-B with synthetic reporters are recapitulated for endogenous genes in a manner that depends quantitatively upon the gene examined.
FIGURE 2.
NELF-B modulates GR induction parameters for endogenous genes. A, NELF-B increases the EC50 for Dex induction of IGFBP1 and IP6K3. Representative dose-response curves for Dex induction of IGFBP1 or IP6K3 mRNA in U2OS cells after transfection with GR and NELF-B or control plasmid (hSA/pSG5) were determined by qRT-PCR as described under “Experimental Procedures.” B–E, summary of effect of NELF-B versus control plasmid transfection on -fold induction by saturating concentrations of Dex, PAA with 1 μm Dex-Mes (DM), and EC50 (when determinable) for IGFBP1 (B; n = 7), IP6K3 (C; n = 5), GILZ (D; n = 6), and LAD1 (E; n = 6). Error bars represent S.E. *, p < 0.05, **, p < 0.005, ***, p < 0.0005 versus no NELF-B.
Binding of GR and NELF-B to IP6K3 Gene Elements
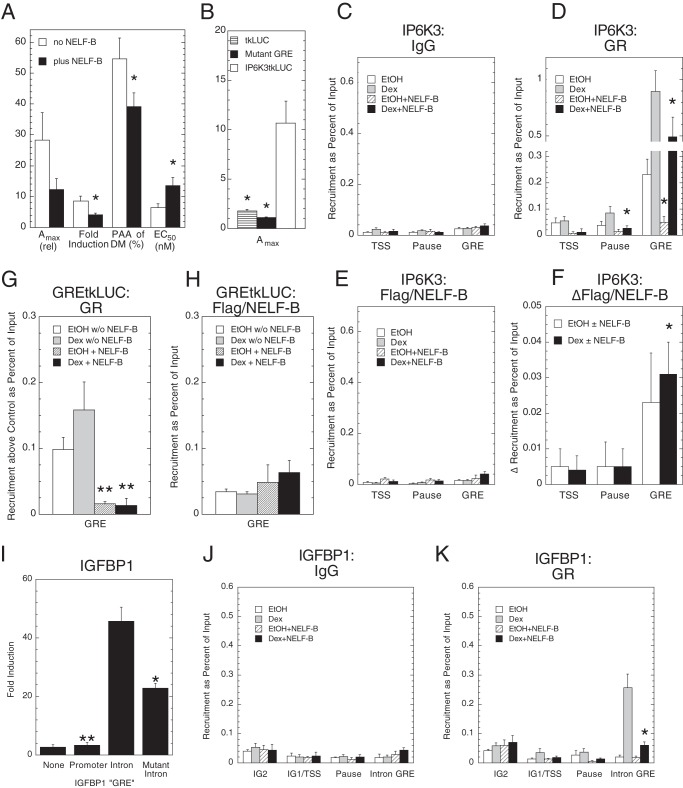
To obtain more information about how NELF-B modulates GR induction parameters, we asked whether NELF-B affected GR binding to the IP6K3 GRE. Guided by ChIP-seq data on GR regulation of gene expression in 3134 murine mammary adenocarcinoma cells,6 in silico analysis revealed a putative GRE at +665 in intron 1 of the human IP6K3 gene (see also 57). Fusion of this region upstream of tkLUC afforded a GR-inducible reporter (“IP6K3tkLUC”) that is appropriately modulated by NELF-B in CV-1 cells (Fig. 3A), whereas mutation of the candidate GRE (ggaaCagcaTGtcct to ggaaTagcaACtcct; capital letters indicate affected nucleotides) eliminates GR inducibility (Fig. 3B). This argues that the IP6K3 GRE is in the middle of intron 1 and is active in different gene settings and cells.
FIGURE 3.
Binding of GR and NELF-B to elements of IP6K3 and IGFBP1 genes and GREtkLUC reporter. A, the intron 1 region of IP6K3 gene contains a GR-inducible enhancer. A summary of effects of exogenous NELF-B on Amax, -fold induction, PAA, and EC50 of transfected reporter (IP6K3 intron 1 fused upstream of tkLUC plasmid) in CV-1 cells from 5 independent experiments (± S.E., *, p < 0.05) is shown. B, GR inducibility of IP6K3 intron 1 fragment is due to a GRE. The average Amax (n = 4, ± S.E., *, p < 0.05 versus no NELF-B) with 1 μm Dex for the empty tkLUC reporter, the IP6K3 intron 1/tkLUC reporter with a mutation in the putative GRE, and the wild type IP6K3 intron 1/tkLUC reporter was determined in transiently transfected CV-1 cells. C–F, recruitment control (C) of GR (D) and of FLAG/NELF-B (E) to regions of endogenous IP6K3 gene in U2OS cells. ChIP assays were performed as described under “Experimental Procedures.” The recruitment of FLAG/NELF-B above background (F) was calculated as the signal with anti-FLAG antibody ± added FLAG/NELF-B. The average values from 5 (GR) and 4 (NELF-B) experiments are plotted (± S.E., *, p < 0.05 versus no NELF-B in D, versus 0 in F). G and H, recruitment of GR (G) and FLAG/NELF-B (H) to regions of exogenous GREtkLUC reporter in CV-1 cells. The average recruitment of GR (above IgG controls) and FLAG/NELF-B from 5 and 3 independent ChIP assays, respectively, is plotted (± S.E., **, p < 0.008 versus no NELF-B). I, biological activity of different IGFBP1 GRE sequences. -Fold induction of luciferase activity by 1 μm Dex in U2OS cells transiently transfected by GR plasmid and no reporter (None), tkLUC (Promoter), tkLUC with upstream fusions of either IGFBP1 intron 1 sequence (data not shown) (Intron), or the same IGFBP1 intron 1 sequence with the mutant palindromic GRE (wt = AGAACATAATGTGAG; mutant = AGAAtATAAacTGAG). *, p < 0.05, **, p < 0.005 versus intron (n = 4; n = 2 for None) is shown. J and K, ChIP assays of endogenous IGFBP1 gene in U2OS cells. Experiments were conducted as for C and D with primers directed toward the two previously reported GREs (IG2 and IG1, which are just 5′ of the TSS) (47), the putative pause site at +50 bp, and the intron 1 GRE. *, p < 0.05 versus no NELF-B.
The possible binding of GR and FLAG-tagged NELF-B to the TSS and to the usual location of paused polymerase at about +50 bp (26, 46), to the region 3′ of the TSS/pause, and to the intronic GRE at +665 was examined by ChIP assays in U2OS cells that had been transiently transfected with FLAG/NELF-B and GR and treated with Dex. FLAG/NELF-B was used because anti-NELF-B antibody was unable to detect any recruitment of NELF-B (data not shown). As indicated in Fig. 3, C–E, both strong GR binding to the GRE ± steroid and weak binding of GR to the regions of the TSS and possible pause sites for RNA polymerase II are reduced by NELF-B. This response appears relatively specific because the signal at the GRE seen with IgG is much weaker and unaffected by NELF-B (Fig. 3C). Conversely, exogenous NELF-B produces a slightly greater increase in the binding of NELF-B to the GRE as compared with the TSS or pause regions (Fig. 3E). This is clearer when the data are plotted as increase in FLAG/NELF-B binding above the background of no added NELF-B (Fig. 3F). These results suggest that the modulatory effects of NELF-B on the GR induction parameters for IP6K3 are due to reduced association of GR with the GRE that may be accompanied by increased NELF-B binding.
A similar behavior was seen for GR and NELF-B recruitment to the GRE of the exogenous GREtkLUC reporter (Fig. 3, G and H). Thus, the parallel capacity of NELF-B to modulate the Amax, EC50, and PAA for GR induction of exogenous GREtkLUC (Fig. 1A) and endogenous IP6K3 gene (Fig. 2C) is mirrored in the comparable lower binding of GR, and marginally higher NELF-B binding, to the GRE sequences of each target gene.
Binding of GR and NELF-B to IGFBP1 Gene Elements
Two GREs upstream of the TSS of the IGFBP1 gene have been reported (47). In silico scanning revealed the presence of a cluster of a possible tandem GRE and three half-sites in the first intron of the human IGFBP1 gene. When these GREs are fused upstream of the tkLUC plasmid, transiently transfected into CV-1 cells, and assayed for steroid inducibility in the presence of cotransfected GR plasmid, the intronic GRE is found to be much more responsive to the agonist Dex than the GREs in the promoter region (Fig. 3I). Mutation of the single palindromic GRE in intron 1 greatly decreases the activity with Dex. We conclude that the most potent GRE of the IGFBP1 gene is in intron 1.
ChIP assays could not detect any binding of NELF-B (in the form of FLAG/NELF-B) to various regions of the IGFBP1 gene in U2OS cells (data not shown). However, GR binding was significantly decreased by added NELF-B (Fig. 3, J and K), just as for the IP6K3 gene. The recruitment of GR to the intronic GREs, as well as the decrease in specific GR binding with added NELF-B, is much more robust than to either of the promoter GREs (IG1 and IG2). Similar results were seen in U2OS.rGR cells, with higher levels of GR recruitment resulting from the higher levels of stably transfected GR (data not shown). Thus, as compared with the promoter GREs, the behavior of GR binding to the intronic GREs more closely mirrors the modulatory properties of NELF-B in the gene activation assays of Fig. 2B.
All NELF Subunits Have the Same Modulatory Activity as NELF-B
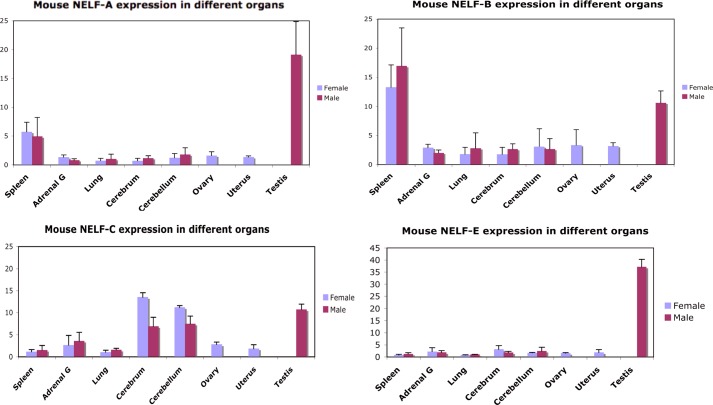
The facts that NELF-B binds to endogenously expressed GRs after activation (Fig. 1J) and that the recruitment of NELF-B to biologically active GREs is slightly greater than to the potential pause regions (Fig. 3F) suggest that the modulatory activity of NELF-B on GR induction parameters in our system may be separate from the role of NELF-B in polymerase pausing as part of the NELF complex. This possibility is supported both by the varying ratios of NELF proteins in several tissue culture cell lines (48) and by different tissue distributions of each of the NELF subunit mRNAs in assorted mouse tissues (Fig. 4 and Table 3). All NELF subunits have the same effect on Amax, EC50, and PAA in the transient transfection assay of Fig. 1, although some appear to be more active (Fig. 5, A and B). It has been reported that decreasing the levels of one NELF component often affects the levels of other NELF components (48). We do not see this when overexpressing NELF components in U2OS cells (Fig. 5C), nor does overexpressed NELF-B alter GR levels (Fig. 5D). These results suggest that each NELF is independently active because they cannot all be limiting for NELF complex action. In other words, if the individual NELF components act solely through the NELF complex, then the only way for them to affect Amax, EC50, and PAA is through the formation of more complexes. This implies that under any given circumstance, only one of the components is limiting in that adding either of the other three would not increase the amount of NELF complex. Thus, the addition of only one of the NELF components will affect the Amax, EC50, and PAA at any given time. Although the identity of this component can change as the relative concentrations of the components change, the addition of each of the four components to one set of concentrations, as in Fig. 5, A and B, cannot result in more complex formation.
FIGURE 4.
Levels of NELF subunits in different mouse tissues. cDNA was prepared from the indicated mouse tissues. The amount of each NELF subunit in each tissue was then determined by qRT-PCR using the primers in Table 3. Similar results are seen on the BioGPS website. Adrenal G, adrenal gland; error bars represent S.D. of triplicates.
TABLE 3.
RT-PCR primers for mouse NELF subunit genes
| Mouse gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| NELF-A | AACAAGAATGCCCTGACCAC | CTTCGGAATGCCTTTGAGAG |
| NELF-B (COBRA) | TGGATCCCTGCCATAAGTTC | CAGGGACAGTGTGTTGATGG |
| NELF-C | AGGAGGAACAGGGTGGTTCT | TGATGTCTGCAGGATTCAGG |
| NELF-E | GGACCCTGGAAGGGAAGTTA | CCGAAGCCGATCACTAGAAG |
FIGURE 5.
Modulatory activity of NELF subunits. A, all NELF subunits reduce the EC50 of gene induction by GR. Representative dose-response curves for Dex induction of GREtkLUC by each NELF subunit in transiently transfected U2OS cells are shown. B, the average -fold changes in GR induction parameters from 4 independent experiments (± S.E.) in U2OS cells treated as in panel B with vector alone (blank), GR and vector (GR), or GR and one NELF subunit. *, p < 0.05, **, p < 0.005, ***, p < 0.0005 versus GR alone. DM, Dex-Mes. C, overexpression of one NELF subunit does not affect the level of the other subunits. Western blots of NELF subunits in U2OS cytosols after transient transfection of each NELF subunit as described under “Experimental Procedures” are shown. FLAG/NELF-E was detected with anti-FLAG antibody. D, levels of GR, TIF2, NELF-B, and NELF-A protein are not affected during overexpression of various factors. U2OS cells were transiently transfected with different combinations of factors under the conditions of the competition assays of Fig. 6. Cytosolic extracts were then analyzed by Western blotting to show that only the level of the transfected protein changes, with β-actin being used as an internal control.
Application of Competition Assay to Determine Actions of NELF-A and -B versus GR and GREtkLUC
To further define the actions of NELF subunits, we employed a discriminating competition assay that compares the actions of two factors in a competition assay. This assay defines factor actions in kinetic terms of either an enzyme-kinetics activator (49, 50), which we call an accelerator, or one of five types of enzyme-kinetics inhibitors, or decelerators, relative to the CLS (10, 12–14, 29).
Given the sensitivity, and novel information provided by the competition assay, we wanted to reinvestigate the importance of the C-terminal residues that are missing in the chimeric NELF-B. Therefore, we first compared the properties of wtNELF-B with chimeric NELF-B in a competition assay with GREtkLUC. Briefly, several subsaturating concentrations of Dex were used to induce the GREtkLUC reporter in the presence of 16 combinations of factors (four of GREtkLUC with four of NELF-B). Exact fits of GR induction of luciferase activity to a first order Hill plot yield the Amax and EC50 for each combination of GREtkLUC and NELF-B. Graphs (1/EC50, Amax/EC50, and EC50/Amax) versus factor 1 with the four concentrations of factor 2 are plotted, and their characteristics as compared with those in Table 1B of Dougherty et al. (13). As previously reported for the chimeric NELF-B (14), graphs of 1/EC50 versus GREtkLUC give horizontal plots that decrease in position with added NELF-B (Fig. 6A). Critically, as with the chimeric NELF-B, the graphs of EC50/Amax versus wtNELF-B are again best fit by a power function with an exponent of n = 2 (Fig. 6B). The expression of full-length NELF-B was found to be linear up to 8 ng of plasmid (data not shown). Therefore, no correction for nonlinear expression is needed to plot Fig. 6B. This means, as described previously (29), that NELF-B is a competitive decelerator acting at n = 2 locations at or before the CLS. Furthermore, because two competitive decelerators cannot act at the same site according to the theory on which the assay is based (10, 12, 13), NELF-B must either act at the CLS and at another site before the CLS or act at two different sites before the CLS. Thus, the wtNELF-B and chimeric NELF-B have identical properties with GREtkLUC; GREtkLUC is an accelerator at the CLS, whereas both forms of NELF-B are competitive decelerators acting at two sites before or at the CLS.
We next examined the properties of wt and chimeric NELF-B in the competition assay with GR. The graph of Amax/EC50 versus GR is uniquely categorized (13) as linear plots that intersect at x = y = 0, with the slopes decreasing with added NELF-B (Fig. 6C). This plot identifies GR as an accelerator at or before the CLS and NELF-B as either a competitive decelerator (C) or a mixed decelerator before or at the CLS. This last possibility for NELF-B is eliminated by the graph of EC50/Amax versus NELF-B (Fig. 6D), which displays the characteristic upward curving quadratic plot of a factor that is a competitive decelerator acting at n = 2 locations at or before the CLS. Finally, from the linear 1/EC50 versus GR plot, where the curves intersect at the origin with slopes that decrease with increasing chimeric NELF-B (data not shown) (13), we can further identify the actions of GR as an accelerator before both the CLS and the chimeric NELF-B. Assays with the full-length wild type NELF-B gave virtually identical results (Fig. 6, E and F). This is evident in plots of Amax/EC50 versus GR, which are linear and intersecting close to the origin (Fig. 6E versus Fig. 5D) and particularly in the EC50/Amax versus NELF-B plots, which give the same upward curvature (Fig. 6F versus Fig. 5C). Thus, wtNELF-B and chimeric NELF-B remain indistinguishable in being a competitive decelerator acting at two sites before or at the CLS, whereas GR is an accelerator acting before the CLS. This further supports the conclusions from Fig. 1, A and B, that wt and chimeric NELF-B are interchangeable and that the C-terminal 30 residues of NELF-B are extraneous.
We next determined the properties of NELF-A in a competition assay with GR. The results (Fig. 6, G and H) are nearly identical to those with NELF-B. Thus, NELF-A, like NELF-B, acts as a competitive decelerator at two positions before or at the CLS. At the same time, both sites of action of NELF-A are after GR, which is again an accelerator. However, these graphs give no information as to whether the two sites of action of NELF-A are the same or different from those of NELF-B.
Comparison of Actions of NELF-B and NELF-A in Competition with TIF2
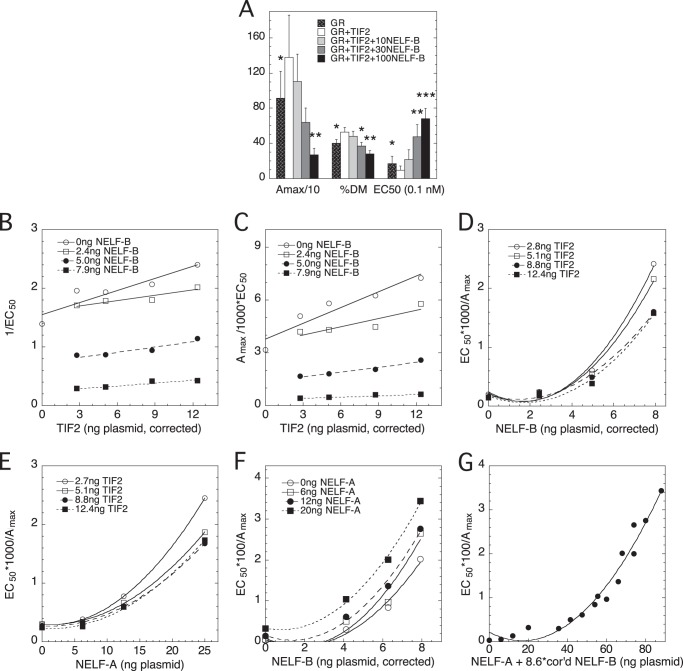
To gain further mechanistic information about NELF-A and NELF-B actions, and to determine whether there are any differences between NELF-A and NELF-B, we conducted competition assays with other factors. First, we asked whether NELF-B can reverse the effects of the accelerator TIF2 (14, 29). As shown in Fig. 7A, increasing amounts of NELF-B effectively counteract the actions of TIF2. We therefore asked whether NELF-B and NELF-A are equally capable of reversing the accelerator effects of TIF2. With NELF-B, the positive linear slopes of 1/EC50 versus TIF2 (Fig. 7B) and Amax/EC50 versus TIF2 (Fig. 7C) intersecting at a point of y ≈ 0 and x less than the amount of endogenous TIF2 (as determined by Western blots (data not shown) and expressed in terms of ng of TIF2 plasmid that give the same amount of TIF2 protein), combined with the decreasing slopes with increasing NELF-B, are diagnostic of TIF2 being an accelerator after the CLS and NELF-B being a decelerator before or at the CLS (13). Finally, as seen above for GR and NELF-B in Fig. 6, B, D, and F, the plot of EC50/Amax versus NELF-B (Fig. 7D) is decidedly nonlinear. This argues that NELF-B is again acting as a competitive decelerator at two sites, before or at the CLS.
FIGURE 7.
NELF-B and NELF-A function additively as competitive decelerators at two steps with different cofactors. Competition assays were conducted as in Fig. 6 with GR, GREtkLUC, NELF-A, full-length wild type NELF-B, and/or TIF2. A, NELF-B reverses effects of TIF2 on GR induction parameters. The average -fold changes in GR induction parameters from 5 independent experiments (± S.E.) in U2OS cells transiently transfected with GR ± TIF2 plasmid and the indicated amounts (ng) of NELF-B plasmid are plotted in the same manner as Fig. 1A. *, p < 0.05, **, p < 0.005, ***, p < 0.0005 versus GR plus TIF2. B–E, graphical analysis of competition of NELF-B or NELF-A, with TIF2 for modulation of GR induction parameters. Experiments were performed with the indicated amounts of NELF-B (after correction for nonlinear expression) or NELF-A and TIF2 (after correction for nonlinear expression) and then analyzed as above in Fig. 6. The “a versus b plots” for Amax/EC50 versus TIF2 graphs such as in panel C (see “Experimental Procedures”), which yield an unbiased determination of a common intersection point, gave average values for the x and y axis coordinates of the intersection point as −12.8 ± 6.4 and 0.73 ± 1.20 (S.D., n = 4), respectively. Western blots determined that endogenous TIF2 protein is equivalent to 4.1 ng of TIF2 plasmid, so −4.1 ng TIF2 plasmid is where there is no TIF2 in the cells, and the intersection point of the Amax/EC50 versus TIF2 graphs is less than this. F and G, competition assay with NELF-A and NELF-B. A graph of EC50/Amax versus corrected (cor'd) amount of NELF-B with indicated amounts of NELF-A (F) was constructed as for panel E. The graph of EC50/Amax versus the combined amounts of NELF-A and NELF-B (G) was compiled as described under “Experimental Procedures.” Similar results were seen in 2 additional independent experiments.
We next examined the competition of NELF-A with TIF2. As with NELF-B, the plots of 1/EC50 and Amax/EC50 (data not shown) versus TIF2 give positive slopes that decrease with added NELF-A and intersect at values of TIF2 that are more negative than the calculated point of zero endogenous TIF2. Importantly, the plot of EC50/Amax is again best fit by a quadratic function (Fig. 7E). Therefore, NELF-A, like NELF-B, exerts its effects as a competitive decelerator of TIF2 actions by acting at two distinct steps before or at the CLS. Furthermore, with both NELF-A and NELF-B, TIF2 is an accelerator that functions after the CLS.
Actions of NELF-A and NELF-B Are Additive
The above similar actions of NELF-A and NELF-B with TIF2 cannot determine whether the two NELFs act individually or via a common complex. To answer this question, we asked whether the effects of two NELF subunits are additive by competing NELF-A with NELF-B. If the effects are additive, then the two NELFs must be completely interchangeable and act independently. Additivity also precludes them from exclusively acting in a common complex because both cannot be limiting simultaneously, i.e. only one of either NELF-A or NELF-B can affect Amax, EC50, and PAA at any given time if they only act through the NELF complex. Plots of EC50/Amax versus each factor display qualitatively identical properties, with the best fits being with quadratic curves (Fig. 7F and data not shown). As described above, such plots are diagnostic of both factors acting as a competitor at two sites before or at the CLS. Importantly, in each case, the addition of the second factor increases the responses of the first factor to give higher positioned curves in all graphs. This behavior is consistent with the two factors acting additively. As further support for this conclusion, a plot of EC50/Amax versus the combined total of functionally active NELF-A and NELF-B was constructed. To do so, a comparison of the amount (in ng) of each transfected NELF plasmid by itself that is required for half-maximal reduction of Amax/EC50 and 1/EC50 (data not shown) revealed that 8.6 ± 1.4-fold (S.E., n = 6) more activity is expressed per ng of NELF-B plasmid (corrected for nonlinear expression) than from the NELF-A plasmid. Therefore, the total amount of active NELF proteins in each of the 16 combinations is calculated as ng of NELF-A plasmid plus 8.6 times the ng of corrected NELF-B plasmid. The graph of EC50/Amax versus the combined amount of active NELFs is nicely fit by a single upward curving quadratic plot (Fig. 7G). Together, the graphs of Fig. 7, F and G, strongly argue that the combined effects of NELF-A and NELF-B under these conditions are additive, with both factors acting independently as competitive decelerators at two sites before or at the CLS.
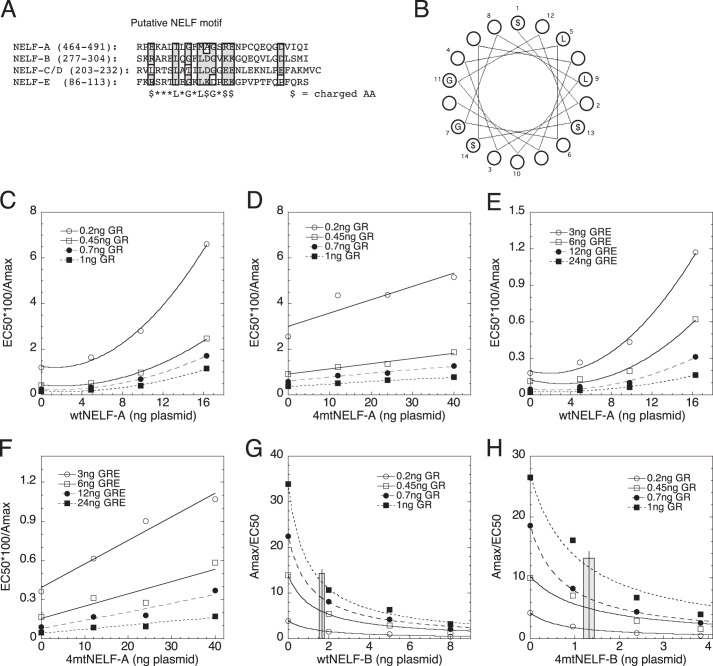
A Common Motif Is Involved in NELF-A and NELF-B Actions
The binding of NELF-B to GR (Fig. 1, I and J), the weak recruitment of NELF-B to the GRE of the IP6K3 gene (Fig. 3, E and F), and the additive activity of NELF-A and NELF-B (Fig. 7, F and G) suggest that both NELFs have activities independent of the NELF complex. Interestingly, an in silico screen of all four NELF subunits revealed a common motif of $XXXLXGXL$GX$$, where $ = charged amino acid (Fig. 8A). Hydropathy plots of the surrounding region in each NELF suggested that this “NELF motif” is part of an α-helical structure, with local hydrophobic and nonbulky groups at opposite sides and neighboring charged residues (Fig. 8B). This motif does not contain any phosphorylatable residues and thus is not a target of Cdk9 or other kinases. No mutants of this region in any NELF were found in the literature. Interestingly, however, this region of NELF-A was found to be dispensable for NELF complex formation, binding to RNA polymerase II, and activity (21). Furthermore, the region containing this sequence is highly conserved in each NELF from humans to zebrafish (and to Drosophila for NELF-A and NELF-B) (data not shown), which suggests a biological function. To test this hypothesis, we made a quadruple alanine mutant in NELF-A and NELF-B by converting $XXXLXGXL$GX$$ to $XXXAXGXA$GXAA (bold letters indicate changed amino acid residues) and testing the activity of the mutant versus wt protein in the above competition assays. These experiments were corrected for differences in protein expression as detected by Western blots. Specifically, wtNELF-A is expressed 2.45 times more efficiently than 4mtNELF-A, whereas 4mtNELF-B is produced 2.09-fold better than wtNELF-B (data not shown).
FIGURE 8.
Conserved NELF motif is required for full activity of NELF-A and NELF-B. A, putative NELF motif. A motif identified by manual scanning of regions of NELF-B against the other NELF-proteins is aligned for each NELF. Numbers in parentheses indicate the amino acid residues shown. Shaded residues indicate ≥75% similarity ($ = charged amino acid). B, spatial distribution of NELF motif residues in an α-helical structure. C–F, comparison of plots of EC50/Amax for wtNELF-A (C and E) or 4mtNELF-A (D and F) competition assays with GR (C and D) and with GREtkLUC (E and F) performed as in Fig. 6D. G and H, potency of NELF-B is reduced by mutations in NELF motif. Average potency of wtNELF-B and 4mtNELF-B (± S.E., n = 16) in reducing Amax/EC50 in competition assays with GR was determined as described under “Experimental Procedures” and displayed as a vertical line, with shaded area indicating ± S.E.
Mutation of NELF-A (4mtNELF-A) does not alter the ability of either GR to act as an accelerator before the CLS or GREtkLUC to be an accelerator at the CLS (data not shown). What has changed is that the curvilinear quadratic fit for EC50/Amax versus wtNELF-A with increasing GR (Fig. 8C) is now a linear fit with 4mtNELF-A (Fig. 8D). Similarly, in competition assays with GREtkLUC, the four-amino acid mutation of the NELF motif converts the normal curvilinear plot of EC50/Amax versus wtNELF-A (Fig. 8E) to a linear plot (Fig. 8F). These results mean that the mutations have inactivated the ability of wtNELF-A to act at one of the two sites before or at the CLS and that the 4mtNELF-A now acts as a competitive decelerator at just one site before or at the CLS. Furthermore, these mutations have not simply disabled the wtNELF-A because the 4mtNELF-A still exhibits partial wild type activity.
With NELF-B, the effect of mutating the NELF-motif is less dramatic but nonetheless significant. Graphs of EC50/Amax versus both wt and 4mtNELF-B, with increasing GR or GREtkLUC, still display upward curvature that is fit by quadratic plots, but the curvature is less with 4mtNELF-B (data not shown). This was quantitated by determining the amount of NELF-B that is required for a 50% reduction in the Amax/EC50 versus NELF plots (Fig. 8, G and H), each of which eventually goes to zero. After correcting for more efficient expression of 4mtNELF-B, the amount of plasmid needed for 50% decrease is raised from 1.68 ± 0.13 ng of plasmid with wtNELF-B to 2.76 ± 0.25 ng of plasmid with 4mtNELF-B (S.E., n = 16, p = 0.0007). Similarly, in competition assays with GREtkLUC, 1.68 ± 0.12 ng of wtNELF-B is sufficient to inhibit the Amax/EC50 by 50%, whereas 2.37 ± 0.24 ng of 4mtNELF-B (after adjustment for higher expression; S.E., n = 12, paired p = 0.0007) is needed. These results argue that the mutated NELF domain in NELF-B causes a decrease in the activity of at least one site of NELF-B action. Collectively, an intact NELF domain is required for full activity of both NELF-A and NELF-B.
DISCUSSION
NELF-A, -B, -C/D, and -E are best known for their role as subunits of the NELF complex, which is involved in the pausing of RNA polymerase II. We now find that each subunit has the additional ability to increase the EC50 and decrease the Amax of GR-mediated gene transactivation. The properties of NELF-A, and especially NELF-B, were extensively investigated. NELF-B is equally active with exogenous and endogenous genes. NELF-B both reduces GR recruitment to the GREs of target genes and may weakly bind to GREs in a manner that depends upon the endogenous gene. A target of NELF-B is the GR LBD, as witnessed by the binding of NELF-B to the GR LBD and by the ability of a GAL/GR-LBD chimera to respond similarly to the full-length GR either with added NELF-B or in cells in which endogenous NELF-B had been knocked down by NELF-B shRNA. Competition assays of NELF-B or NELF-A with GR, GREtkLUC reporter, or TIF2 reveal that both NELFs each act as competitive decelerators at two sites before or at the CLS. However, the action of both NELF-A and NELF-B at one site is attenuated by the mutation of an evolutionarily conserved motif that is shared by all four NELF proteins.
Several lines of evidence suggest that the above new properties of NELF-A and NELF-B are independent of the NELF complex and result instead from their actions either as individual proteins or in complexes with other proteins that are unique for each NELF. First, the ability of exogenous NELF-A and NELF-B to produce additive effects (Fig. 7, F and G) is incompatible with both factors acting through a common complex because both NELF-A and NELF-B cannot be limiting at the same time, i.e. both factors affecting Amax, EC50, and PAA at the same time. Likewise, the ability of each NELF subunit to evoke similar changes in GR transactivation properties (Fig. 5, A and B) is incompatible with action via a common complex because all four factors cannot be simultaneously limiting. Second, a NELF domain has been identified in all four NELF proteins. Surprisingly, this domain is irrelevant for NELF-A activity in the NELF complex (21). However, mutation of this domain alters the activity of NELF-A in our assays, which strongly indicates that an activity independent of the NELF complex has been affected. Third, several cell lines are reported to contain unequal amounts of the four proteins (48), and all of the NELF subunit mRNAs are present at different levels in many tissues (Fig. 4). In Drosophila, ChIP-chip assays revealed that 80% of the binding sites of NELF-B and NELF-E overlap but 20% do not (26). In both cases, nonequivalent abundances or binding of NELF proteins mean that some NELF components are in excess and could have NELF complex-independent activities. Fourth, both NELF-A and NELF-B are found to act at two steps. Thus, to the extent that the NELF complex is thought to cause polymerase pausing at a single step (51, 52), at least one step of NELF-A and NELF-B action would be different from that of the NELF complex. Fifth, recent results suggest that polymerase pausing is restricted to those genes requiring synchronized expression during development and differentiation (27), which is not the case for our transiently transfected reporter gene system. The NELF complex has also been proposed to alter chromatin organization (53, 54). However, this activity is again unlikely with the exogenous GREtkLUC reporter in our studies. Transfected genes are thought to possess little if any organized chromatin structure (55). Furthermore, recent ChIP-Seq experiments indicate that 80–95% of endogenous GR-regulated genes already have an open chromatin conformation (56–58), thereby minimizing the need for chromatin reorganizations. Finally, each constituent of the NELF complex is listed on the protein interaction database BioGRID to interact with 8–32 proteins, many of which are unique to each specific NELF subunit. It is likely that several of these complexes evoke biological responses. To summarize, our results indicate that NELF-A and NELF-B have inhibitory actions on steroid-mediated gene expression that are independent of the NELF complex. This does not preclude that NELF-A and NELF-B are still involved in the NELF complex; instead, it says that the NELF complex does not play a quantitative role in controlling the amount of GR-induced gene product in the present system.
We have used the recently developed competition assay (13, 14) to identify both NELF-A and NELF-B as each being competitive decelerators at two kinetically defined steps at or before the CLS, which is the equivalent in a steady-state system of the rate-limiting step in enzyme kinetics. This assay does not require any prior knowledge of the mechanism of action of the competing factors (29). In fact, any mechanism by which the factor influences GR activity will be detected. In this manner, this assay is a high-throughput assay to detect GR activity-modulating mechanisms. These assays tell us that both NELFs act after the site of GR action and before the site of TIF2 action, which is after the CLS, although the precise molecular mechanisms remain unknown. These results also confirm our earlier findings that GREtkLUC is an accelerator at the CLS (14) and that TIF2 is an accelerator after the CLS with both endogenous and exogenous genes (12, 13). Thus, we can construct the following sequence of cofactor action that applies for GR-regulated transactivation in this system and, we propose, in several other systems.
With NELF-B, at least one step results in a decrease of GR binding to the GRE.
The competition assay positions the kinetically defined mode of factor activity relative to a benchmark step (the CLS) but is silent regarding where or when factor binds. Therefore, we cannot yet incorporate the NELF-B-induced reduction of GR binding to the GRE, as revealed by ChIP assays, into the above kinetically defined sequence of NELF actions. It should also be realized that the step at which a factor binds is not always equivalent to when that factor exerts its biological effect. For example, the number of genomic binding sites for GRs is ∼10-fold greater than the number of induced genes (59, 60), there is no temporal correlation between GR binding to enhancer regions and the transcriptional response (61), p300 is recruited to the androgen-responsive elements of the TMPRSS2 and FKBP5 genes but is required for androgen induction of only TMPRSS2 (62), and paused RNA polymerase II is bound well before it is active in transcriptional elongation.
Co-immunoprecipitation and ChIP assays suggest that NELF-B associates with the LBD of activated GRs in a manner that decreases the binding GR-steroid complexes to those GREs controlling gene expression (Figs. 1 and 3). This contrasts with the behavior of ER (15). NELF-B binds to the ER LBD and decreases ER-mediated gene transactivation, just as seen for GR. However, the recruitment of ER to the promoters of ER-regulated genes is not decreased by excess NELF-B. Thus, the major actions of NELF-B with GR and ER appear different, with NELF-B having a greater effect on promoter-bound ERs while reducing either the amount of GR that binds to the promoter region or the strength of GRE-bound receptors.
Interestingly, the decrease in GR recruitment to the GRE in the presence of added NELF-B is similar with exogenous and endogenous reporter genes (Fig. 3, D and G). It is generally believed that transiently transfected genes are not good models for receptor-regulated gene expression because they do not possess the same level of chromatin organization (55, 63). However, the similar effects of factor on gene induction and GR recruitment to endogenous and exogenous genes both in the present study and in earlier studies (29) suggest that situations exist where the two systems may be studied interchangeably.
In conclusion, NELF-A and NELF-B are new cofactors for GRs that possess additive activities for altering the Amax and EC50 of GR-regulated transactivation of exogenous and endogenous genes in different cells in addition to their involvement in the NELF complex. These activities rely, in part, on the integrity of an evolutionarily conserved motif shared not only by NELF-A and NELF-B but also by NELF-C/D and NELF-E. Evidence is presented that at least some these activities are independent of the NELF complex composed of all four proteins. Because NELF-C/D and NELF-E display the same modulatory activity as NELF-A and NELF-B with GR-induced transcription, we propose that this NELF motif is intimately involved in the individual actions of each protein. Competition assays have identified the kinetically defined mode and site of action of NELF-A and NELF-B as competitive decelerators at two steps relative to several factors during GR transactivation. Such dual site action has also been found for another GR cofactor, PA1 (29), and thus may be more prevalent than currently appreciated. Further studies are required to identify the precise steps that NELF-A and NELF-B are inhibiting (which may be different), to determine whether NELF-C/D and NELF-E also act at two steps before or at the CLS and if so whether they are the same or different from those affected by NELF-A and NELF-B, and to further define the role of the NELF motif in these new actions of each NELF protein. Such studies of this novel family of cofactors should significantly increase our molecular understanding of GR-regulated gene transcription and open new windows for modulating GR induction properties.
Acknowledgments
We thank Geun-Shik Lee (Steroid Hormones Section, NIDDK, National Institutes of Health) for quantitating the NELF mRNAs in different tissues, Brian Lewis (NCI, National Institutes of Health) for critical review of this paper, and Gordon Hager (NCI, National Institutes of Health) for sharing unpublished ChIP-seq data.
This research was supported by the Intramural Research Program of the National Institutes of Health, the NIDDK, and a Voelcker Scholar Award (to R. L.).
G. Hager, personal communication.
- PAA
- partial agonist activity
- GR
- glucocorticoid receptor
- GRE
- glucocorticoid response elements
- NELF
- negative elongation factor
- ER
- estrogen receptor
- GILZ
- glucocorticoid-induced leucine zipper
- Dex
- dexamethasone
- Dex-Mes
- Dex-21-mesylate
- qRT-PCR
- quantitative real-time PCR
- PIC
- protease inhibitor cocktail
- LBD
- ligand-binding domain
- DBD
- DNA-binding domain
- KD
- knockdown
- CLS
- concentration-limiting step
- TSS
- transcription start site
- sh
- short helical
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1. Simons S. S., Jr. (2003) The importance of being varied in steroid receptor transactivation. Trends Pharmacol. Sci. 24, 253–259 [DOI] [PubMed] [Google Scholar]
- 2. Simons S. S., Jr. (2008) What goes on behind closed doors: physiological versus pharmacological steroid hormone actions. Bioessays 30, 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simons S. S., Jr. (2010) Glucocorticoid receptor cofactors as therapeutic targets. Curr. Opin. Pharmacol. 10, 613–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kunz S., Sandoval R., Carlsson P., Carlstedt-Duke J., Bloom J. W., Miesfeld R. L. (2003) Identification of a novel glucocorticoid receptor mutation in budesonide-resistant human bronchial epithelial cells. Mol. Endocrinol. 17, 2566–2582 [DOI] [PubMed] [Google Scholar]
- 5. Zhao Q., Pang J., Favata M. F., Trzaskos J. M. (2003) Receptor density dictates the behavior of a subset of steroid ligands in glucocorticoid receptor-mediated transrepression. Int. Immunopharmacol. 3, 1803–1817 [DOI] [PubMed] [Google Scholar]
- 6. Sun Y., Tao Y. G., Kagan B. L., He Y., Simons S. S., Jr. (2008) Modulation of transcription parameters in glucocorticoid receptor-mediated repression. Mol. Cell. Endocrinol. 295, 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van der Laan S., Lachize S. B., Vreugdenhil E., de Kloet E. R., Meijer O. C. (2008) Nuclear receptor coregulators differentially modulate induction and glucocorticoid receptor-mediated repression of the corticotropin-releasing hormone gene. Endocrinology 149, 725–732 [DOI] [PubMed] [Google Scholar]
- 8. Luo M., Simons S. S., Jr. (2009) Modulation of glucocorticoid receptor induction properties by cofactors in peripheral blood mononuclear cells. Hum. Immunol. 70, 785–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim Y., Sun Y., Chow C., Pommier Y. G., Simons S. S., Jr. (2006) Effects of acetylation, polymerase phosphorylation, and DNA unwinding in glucocorticoid receptor transactivation. J. Steroid Biochem. Mol. Biol. 100, 3–17 [DOI] [PubMed] [Google Scholar]
- 10. Ong K. M., Blackford J. A., Jr., Kagan B. L., Simons S. S., Jr., Chow C. C. (2010) A theoretical framework for gene induction and experimental comparisons. Proc. Natl. Acad. Sci. U.S.A. 107, 7107–7112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Awasthi S., Simons S. S., Jr. (2012) Separate regions of glucocorticoid receptor, coactivator TIF2, and comodulator STAMP modify different parameters of glucocorticoid-mediated gene induction. Mol. Cell. Endocrinol. 355, 121–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chow C. C., Ong K. M., Dougherty E. J., Simons S. S., Jr. (2011) Inferring mechanisms from dose-response curves. Methods Enzymol. 487, 465–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dougherty E. J., Guo C., Simons S. S., Jr., Chow C. C. (2012) Deducing the temporal order of cofactor function in ligand-regulated gene transcription: theory and experimental verification. PLoS One 7, e30225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blackford J. A., Jr., Guo C., Zhu R., Dougherty E. J., Chow C. C., Simons S. S., Jr. (2012) Identification of location and kinetically defined mechanism of cofactors and reporter genes in the cascade of steroid-regulated transactivation. J. Biol. Chem. 287, 40982–40995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aiyar S. E., Sun J. L., Blair A. L., Moskaluk C. A., Lu Y. Z., Ye Q. N., Yamaguchi Y., Mukherjee A., Ren D. M., Handa H., Li R. (2004) Attenuation of estrogen receptor α-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 18, 2134–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun J., Blair A. L., Aiyar S. E., Li R. (2007) Cofactor of BRCA1 modulates androgen-dependent transcription and alternative splicing. J. Steroid Biochem. Mol. Biol. 107, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kininis M., Isaacs G. D., Core L. J., Hah N., Kraus W. L. (2009) Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol. Cell. Biol. 29, 1123–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aiyar S. E., Cho H., Lee J., Li R. (2007) Concerted transcriptional regulation by BRCA1 and COBRA1 in breast cancer cells. Int. J. Biol. Sci. 3, 486–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McChesney P. A., Aiyar S. E., Lee O. J., Zaika A., Moskaluk C., Li R., El-Rifai W. (2006) Cofactor of BRCA1: a novel transcription factor regulator in upper gastrointestinal adenocarcinomas. Cancer Res. 66, 1346–1353 [DOI] [PubMed] [Google Scholar]
- 20. Yamaguchi Y., Takagi T., Wada T., Yano K., Furuya A., Sugimoto S., Hasegawa J., Handa H. (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97, 41–51 [DOI] [PubMed] [Google Scholar]
- 21. Narita T., Yamaguchi Y., Yano K., Sugimoto S., Chanarat S., Wada T., Kim D. K., Hasegawa J., Omori M., Inukai N., Endoh M., Yamada T., Handa H. (2003) Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 23, 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu C. H., Yamaguchi Y., Benjamin L. R., Horvat-Gordon M., Washinsky J., Enerly E., Larsson J., Lambertsson A., Handa H., Gilmour D. (2003) NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 17, 1402–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muse G. W., Gilchrist D. A., Nechaev S., Shah R., Parker J. S., Grissom S. F., Zeitlinger J., Adelman K. (2007) RNA polymerase is poised for activation across the genome. Nat. Genet. 39, 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Core L. J., Lis J. T. (2008) Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319, 1791–1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gilchrist D. A., Nechaev S., Lee C., Ghosh S. K., Collins J. B., Li L., Gilmour D. S., Adelman K. (2008) NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 22, 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee C., Li X., Hechmer A., Eisen M., Biggin M. D., Venters B. J., Jiang C., Li J., Pugh B. F., Gilmour D. S. (2008) NELF and GAGA factor are linked to promoter-proximal pausing at many genes in Drosophila. Mol. Cell. Biol. 28, 3290–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lagha M., Bothma J. P., Esposito E., Ng S., Stefanik L., Tsui C., Johnston J., Chen K., Gilmour D. S., Zeitlinger J., Levine M. S. (2013) Paused Pol II coordinates tissue morphogenesis in the Drosophila embryo. Cell 153, 976–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun J., Li R. (2010) Human negative elongation factor activates transcription and regulates alternative transcription initiation. J. Biol. Chem. 285, 6443–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Z., Sun Y., Cho Y.-W., Chow C. C., Simons S. S., Jr. (2013) PA1 protein, a new competitive decelerator acting at more than one step to impede glucocorticoid receptor-mediated transactivation. J. Biol. Chem. 288, 42–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Simons S. S., Jr., Pons M., Johnson D. F. (1980) α-Keto mesylate: A reactive, thiol-specific functional group. J. Org. Chem. 45, 3084–3088 [Google Scholar]
- 31. Wang Q., Blackford J. A., Jr., Song L.-N., Huang Y., Cho S., Simons S. S., Jr., and (2004) Equilibrium interactions of corepressors and coactivators with agonist and antagonist complexes of glucocorticoid receptors. Mol. Endocrinol. 18, 1376–1395 [DOI] [PubMed] [Google Scholar]
- 32. Ye Q., Hu Y. F., Zhong H., Nye A. C., Belmont A. S., Li R. (2001) BRCA1-induced large-scale chromatin unfolding and allele-specific effects of cancer-predisposing mutations. J. Cell Biol. 155, 911–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He Y., Simons S. S., Jr. (2007) STAMP, a novel predicted factor assisting TIF2 actions in glucocorticoid receptor-mediated induction and repression. Mol. Cell. Biol. 27, 1467–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nelson J. D., Denisenko O., Bomsztyk K. (2006) Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1, 179–185 [DOI] [PubMed] [Google Scholar]
- 35. Horwitz K. B., Jackson T. A., Bain D. L., Richer J. K., Takimoto G. S., Tung L. (1996) Nuclear receptor coactivators and corepressors. Mol. Endocrinol. 10, 1167–1177 [DOI] [PubMed] [Google Scholar]
- 36. Cho S., Kagan B. L., Blackford J. A., Jr., Szapary D., Simons S. S., Jr. (2005) Glucocorticoid receptor ligand binding domain is sufficient for the modulation of glucocorticoid induction properties by homologous receptors, coactivator transcription intermediary factor 2, and Ubc9. Mol. Endocrinol. 19, 290–311 [DOI] [PubMed] [Google Scholar]
- 37. Tao Y.-G., Xu Y., Xu H. E., Simons S. S., Jr. (2008) Mutations of glucocorticoid receptor differentially affect AF2 domain activity in a steroid-selective manner to alter the potency and efficacy of gene induction and repression. Biochemistry 47, 7648–7662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee G.-S., Simons S. S., Jr. (2011) Ligand binding domain mutations of the glucocorticoid receptor selectively modify the effects with, but not binding of, cofactors. Biochemistry 50, 356–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rogatsky I., Trowbridge J. M., Garabedian M. J. (1997) Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol. Cell. Biol. 17, 3181–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leach K. L., Dahmer M. K., Hammond N. D., Sando J. J., Pratt W. B. (1979) Molybdate inhibition of glucocorticoid receptor inactivation and transformation. J. Biol. Chem. 254, 11884–11890 [PubMed] [Google Scholar]
- 41. Willmann T., Beato M. (1986) Steroid-free glucocorticoid receptor binds specifically to mouse mammary tumour virus DNA. Nature 324, 688–691 [DOI] [PubMed] [Google Scholar]
- 42. Shi X., Shi W., Li Q., Song B., Wan M., Bai S., Cao X. (2003) A glucocorticoid-induced leucine-zipper protein, GILZ, inhibits adipogenesis of mesenchymal cells. EMBO Rep. 4, 374–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saiardi A., Nagata E., Luo H. R., Snowman A. M., Snyder S. H. (2001) Identification and characterization of a novel inositol hexakisphosphate kinase. J. Biol. Chem. 276, 39179–39185 [DOI] [PubMed] [Google Scholar]
- 44. Undén A. L., Elofsson S., Brismar K. (2005) Gender differences in the relation of insulin-like growth factor binding protein-1 to cardiovascular risk factors: a population-based study. Clin. Endocrinol. (Oxf.) 63, 94–102 [DOI] [PubMed] [Google Scholar]
- 45. Santin A. D., Zhan F., Bellone S., Palmieri M., Cane S., Bignotti E., Anfossi S., Gokden M., Dunn D., Roman J. J., O'Brien T. J., Tian E., Cannon M. J., Shaughnessy J., Jr., Pecorelli S. (2004) Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int. J. Cancer 112, 14–25 [DOI] [PubMed] [Google Scholar]
- 46. Sun J., Pan H., Lei C., Yuan B., Nair S. J., April C., Parameswaran B., Klotzle B., Fan J. B., Ruan J., Li R. (2011) Genetic and genomic analyses of RNA polymerase II-pausing factor in regulation of mammalian transcription and cell growth. J. Biol. Chem. 286, 36248–36257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen W., Rogatsky I., Garabedian M. J. (2006) MED14 and MED1 differentially regulate target-specific gene activation by the glucocorticoid receptor. Mol. Endocrinol 20, 560–572 [DOI] [PubMed] [Google Scholar]
- 48. Sun J., Watkins G., Blair A. L., Moskaluk C., Ghosh S., Jiang W. G., Li R. (2008) Deregulation of cofactor of BRCA1 expression in breast cancer cells. J. Cell. Biochem. 103, 1798–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fromm H. J. (1975) Initial Rate Enzyme Kinetics, Springer-Verlag New York Inc., New York [Google Scholar]
- 50. Segel I. H. (1993) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems, John Wiley & Sons, Inc., New York [Google Scholar]
- 51. Adelman K., Lis J. T. (2012) Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat. Rev. Genet. 13, 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamaguchi Y., Shibata H., Handa H. (2013) Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim. Biophys. Acta 1829, 98–104 [DOI] [PubMed] [Google Scholar]
- 53. Aida M., Chen Y., Nakajima K., Yamaguchi Y., Wada T., Handa H. (2006) Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol. Cell. Biol. 26, 6094–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gilchrist D. A., Dos Santos G., Fargo D. C., Xie B., Gao Y., Li L., Adelman K. (2010) Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 143, 540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Archer T. K., Lefebvre P., Wolford R. G., Hager G. L. (1992) Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science 255, 1573–1576 [DOI] [PubMed] [Google Scholar]
- 56. Biddie S. C., John S., Sabo P. J., Thurman R. E., Johnson T. A., Schiltz R. L., Miranda T. B., Sung M. H., Trump S., Lightman S. L., Vinson C., Stamatoyannopoulos J. A., Hager G. L. (2011) Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol. Cell 43, 145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. John S., Sabo P. J., Thurman R. E., Sung M. H., Biddie S. C., Johnson T. A., Hager G. L., Stamatoyannopoulos J. A. (2011) Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43, 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wiench M., John S., Baek S., Johnson T. A., Sung M. H., Escobar T., Simmons C. A., Pearce K. H., Biddie S. C., Sabo P. J., Thurman R. E., Stamatoyannopoulos J. A., Hager G. L. (2011) DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 30, 3028–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Biddie S. C., John S., Hager G. L. (2010) Genome-wide mechanisms of nuclear receptor action. Trends Endocrinol. Metab. 21, 3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rao N. A., McCalman M. T., Moulos P., Francoijs K. J., Chatziioannou A., Kolisis F. N., Alexis M. N., Mitsiou D. J., Stunnenberg H. G. (2011) Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome Res. 21, 1404–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Luca F., Maranville J. C., Richards A. L., Witonsky D. B., Stephens M., Di Rienzo A. (2013) Genetic, functional and molecular features of glucocorticoid receptor binding. PLoS One 8, e61654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ianculescu I., Wu D. Y., Siegmund K. D., Stallcup M. R. (2012) Selective roles for cAMP response element-binding protein binding protein and p300 protein as coregulators for androgen-regulated gene expression in advanced prostate cancer cells. J. Biol. Chem. 287, 4000–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chinenov Y., Sacta M. A., Cruz A. R., Rogatsky I. (2008) GRIP1-associated SET-domain methyltransferase in glucocorticoid receptor target gene expression. Proc. Natl. Acad. Sci. U.S.A. 105, 20185–20190 [DOI] [PMC free article] [PubMed] [Google Scholar]