Background: Type III polyketide synthases (PKSs) show diverse cyclization specificity.
Results: A single amino acid substitution in two Azotobacter type III PKSs reversed their cyclization specificity. Crystal structures were determined.
Conclusion: The volume of the active site cavity is a crucial determinant of the cyclization specificity.
Significance: An important insight into the cyclization specificity of type III PKSs was provided.
Keywords: Bacterial Metabolism, Crystallography, Kinetics, Polyketides, Site-directed Mutagenesis
Abstract
Type III polyketide synthases (PKSs) show diverse cyclization specificity. We previously characterized two Azotobacter type III PKSs (ArsB and ArsC) with different cyclization specificity. ArsB and ArsC, which share a high sequence identity (71%), produce alkylresorcinols and alkylpyrones through aldol condensation and lactonization of the same polyketomethylene intermediate, respectively. Here we identified a key amino acid residue for the cyclization specificity of each enzyme by site-directed mutagenesis. Trp-281 of ArsB corresponded to Gly-284 of ArsC in the amino acid sequence alignment. The ArsB W281G mutant synthesized alkylpyrone but not alkylresorcinol. In contrast, the ArsC G284W mutant synthesized alkylresorcinol with a small amount of alkylpyrone. These results indicate that this amino acid residue (Trp-281 of ArsB or Gly-284 of ArsC) should occupy a critical position for the cyclization specificity of each enzyme. We then determined crystal structures of the wild-type and G284W ArsC proteins at resolutions of 1.76 and 1.99 Å, respectively. Comparison of these two ArsC structures indicates that the G284W substitution brings a steric wall to the active site cavity, resulting in a significant reduction of the cavity volume. We postulate that the polyketomethylene intermediate can be folded to a suitable form for aldol condensation only in such a relatively narrow cavity of ArsC G284W (and presumably ArsB). This is the first report on the alteration of cyclization specificity from lactonization to aldol condensation for a type III PKS. The ArsC G284W structure is significant as it is the first reported structure of a microbial resorcinol synthase.
Introduction
Type III polyketide synthases (PKSs)2 are simple homodimeric ketosynthases that are involved in the biosynthesis of various aromatic polyketides in both plants and microorganisms (1–3). Type III PKSs typically catalyze condensation of a starter acyl thioester with several acetate units derived from malonyl-CoA. Three catalytic residues, called a Cys-His-Asn catalytic triad, are responsible for this condensation. The resulting linear polyketomethylene intermediate is subjected to cyclization in the same active site cavity. Type III PKSs synthesize a wide variety of products because they differ in their preference of starter substrate selection in the number of condensation steps and in the mechanism of intramolecular cyclization of polyketomethylene intermediates. Although the starter substrate specificity and polyketide chain extension are largely understood, the cyclization step still remains elusive. Several structural and site-directed mutagenesis studies suggested that the cyclization process is defined by the architecture of the active site cavity; several amino acid residues in the active site cavity were shown to affect the cyclization specificity (1, 4–6).
Previously we characterized two type III PKSs, ArsB and ArsC, responsible for phenolic lipids biosynthesis in a Gram-negative nitrogen-fixing soil bacterium, Azotobacter vinelandii (7, 8). Although ArsB and ArsC share 71% amino acid sequence identity, ArsB and ArsC were shown to synthesize alkylresorcinols and alkylpyrones, respectively (Fig. 1A), indicating that they are enzymatically distinct type III PKSs. Both ArsB and ArsC employ the same long-chain acyl starter substrate directly received from the type I fatty acid synthase ArsAD and subsequently catalyze three successive extensions with malonyl-CoA followed by the formation of the same tetraketide intermediate. The different catalytic properties between ArsB and ArsC depend on the cyclization specificity. ArsB catalyzes an intramolecular C2-to-C7 (numbering from the cysteine thioester) aldol condensation accompanied by thioester cleavage and decarboxylative loss of the C1 carbon as CO2 to yield alkylresorcinols (Fig. 1A). In contrast, ArsC catalyzes intramolecular C5 oxygen-to-C1 lactonization to yield tetraketide pyrones (Fig. 1A). ArsC also uses the triketide intermediate to yield triketide pyrones.
FIGURE 1.
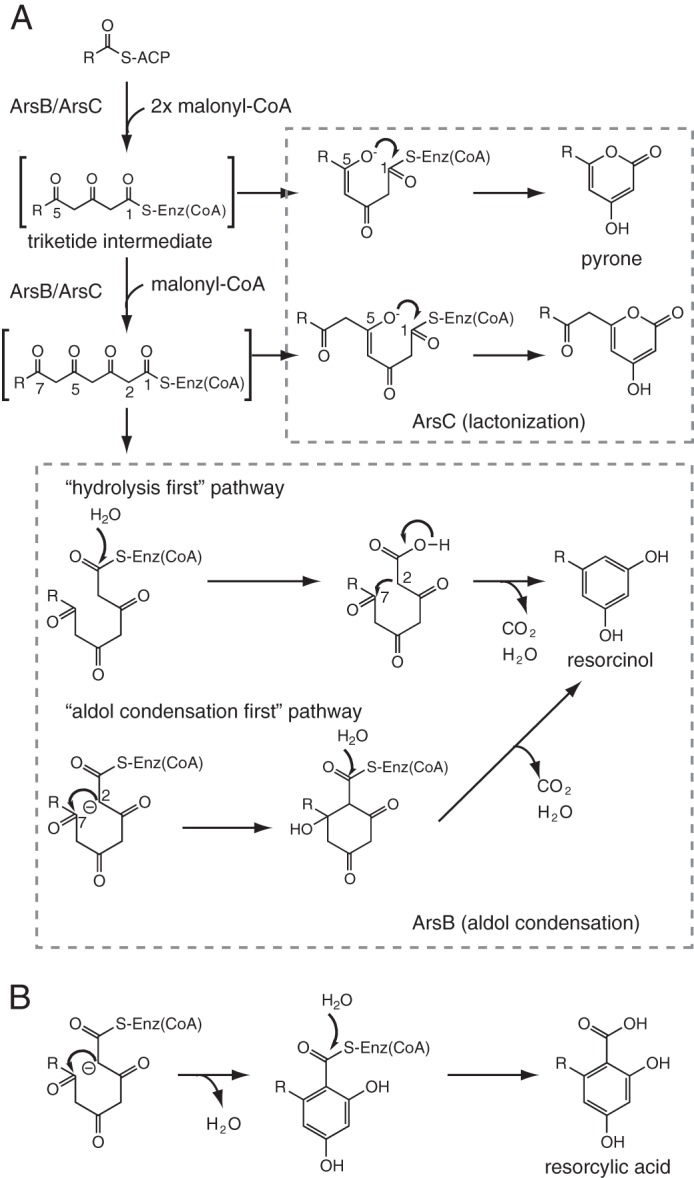
Proposed mechanism for type III PKS reactions. A, phenolic lipid synthesis by two type III PKSs in A. vinelandii. Alkylpyrone formation via an enzyme-bound linear intermediate is depicted, but alkylpyrone can also be formed from the linear intermediate that is released from the enzyme. Two pathways have been suggested for the synthesis of alkylresorcinol by ArsB. In the “hydrolysis first” pathway, aldol cyclization occurs in parallel to decarboxylation (7, 21). In contrast, during the “aldol condensation first” pathway, the cyclization is followed by sequential β-keto decarboxylation and dehydration or by coupled decarboxylation/dehydration (32). B, alkylresorcylic acid synthesis by type III PKSs, including ORAS.
Given that highly homologous arsB and arsC genes are placed adjacent to each other in the phenolic lipid biosynthetic gene cluster, these genes may have descended from the same ancestor. After duplication, they possibly gain different catalytic properties with just a few amino acid substitutions. Therefore, we assumed that these two Ars enzymes could be ideal targets to obtain an insight into the structural and mechanistic basis for functional diversity of type III PKSs. Here we report the elucidation of a key amino acid residue affecting the cyclization specificity of ArsB and ArsC. Surprisingly, only a single point mutation in both enzymes led to the swapping of their main products without a significant loss of their enzymatic activities. The crystal structures of the wild-type and G284W mutant ArsC proteins show that this single amino acid substitution drastically changes the architecture of the active site cavity. Thus our analysis reveals how type III PKSs have evolved to have different cyclization specificity.
EXPERIMENTAL PROCEDURES
Materials
Escherichia coli strains JM109, BL21 (DE3), and B834 (DE3) were purchased from Takara Biochemicals (Ohtsu, Japan). Malonyl-CoA and [2-14C]malonyl-CoA used for the enzyme assay were purchased from Sigma. Behenyl-CoA was prepared as described previously (7).
Site-directed Mutagenesis
Expression plasmids pET16b-ArsB and pET16b-ArsC (7) were used for site-directed mutagenesis. The following primers were used for site-directed mutagenesis with a Mutan-Super Express Km kit (Takara, Ohtsu, Japan): ArsB W281G mutant, 5′-ATGACCGGGGTACCGGGCTCGCAC-3′; ArsC G284W mutant, 5′-ATGACCTGGGTACCGGGCTCGCAC-3′; ArsB W281L and ArsC G284L mutants, 5′-ATGACCTTGGTACCGGGCTCGCAC-3′; ArsB W281A and ArsC G284A mutants, 5′-ATGACCGCGGTACCGGGCTCGCAC-3′; ArsB W281F and ArsC G284F mutants, 5′-ATGACCTTTGTGCCGGATTCGCAC-3′; ArsB W281I and ArsC G284I mutants, 5′-ATGACCATTGTGCCGGATTCGCAC-3′. The following primers and their complementary primers were used for site-directed mutagenesis with a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA): ArsB W281Y mutant, 5′-CGACGAGATGACCTACGTGCCGGGCTCGC-3′; ArsB W281V mutant, 5′-CGACGAGATGACCGTGGTGCCGGGCTCGC-3′; ArsC G284Y mutant, 5′-CGACACCATGACCTACGTGCCGGGCTCGC-3′; ArsC G284V mutant, 5′-CGACACCATGACCGTGGTGCCGGGCTCGC-3′. The mutations were confirmed by determining the nucleotide sequences.
Expression and Purification of Recombinant Enzymes
E. coli strain BL21 (DE3) harboring pET16b-ArsB or pET16b-ArsC was grown at 26 °C overnight in 2 liters of Luria-Bertani broth containing 50 μg/ml ampicillin. For the expression of selenomethionine-substituted ArsC, E. coli strain B834 (DE3) harboring pET16b-ArsC was grown at 26 °C overnight in M9 media supplemented with 50 mg/liter l-selenomethionine. The cells were collected by centrifugation and resuspended in buffer containing 50 mm Tris-HCl (pH 7.8), 300 mm sodium chloride, and 10% (v/v) glycerol. After sonication, cellular debris was removed by centrifugation, and the supernatant was subjected to nickel-nitrilotriacetic acid superflow resin (Qiagen, Chatsworth, CA). His-tagged enzyme was eluted with a buffer containing 200 mm imidazole. After dialysis, the N-terminal His tag was removed by the treatment with Factor Xa protease. Further purification was performed by Resource Q anion exchange column (GE Healthcare). The purified enzyme was concentrated to 10 mg/ml and stored in a solution containing 10 mm Tris-HCl (pH 7.5), 150 mm sodium chloride, and 10% (v/v) glycerol.
Enzyme Assay
The standard reaction mixture contained 100 μm extender substrate (malonyl-CoA or [2-14C]malonyl-CoA), 100 μm starter substrate (behenyl-CoA), 100 mm Tris-HCl (pH 8.0), 150 mm sodium chloride, and 6.1 μg of purified enzyme in a total volume of 100 μl. The reaction mixture was incubated at 30 °C for 30 min before being quenched with the addition of 20 μl of 6 m HCl. The products were extracted with ethyl acetate, and the organic layer was then dried out by the evaporator. The products were dissolved in 15 μl of methanol and subjected to radio-thin layer chromatography (TLC) assay, high performance liquid chromatography (HPLC), or liquid chromatography-atmospheric pressure chemical ionization mass spectrometry (LC-APCIMS) analysis. For the radio-TLC assay, Silica gel 60WF254 TLC plate (Merck, Darmstadt, Germany) was developed in benzene/acetone/acetic acid (85:15:1), and 14C-labeled products were detected by a BAS-MS imaging plate (Fuji, Tokyo, Japan). LC-APCIMS analysis was carried out using the esquire high capacity trap plus system (Bruker Daltonics, Bremen, Germany) with a Pegasil-B C4 reversed-phase column (4.6 × 250 mm) (Senshu Scientific, Tokyo, Japan) and 90% acetonitrile in water containing 0.1% acetic acid as an eluent at a flow rate of 1 ml/min. HPLC analysis was performed using the LaChrom Elite system (Hitachi, Tokyo, Japan) with the same column, eluent, and flow rate as for the LC-APCIMS analysis. UV spectra were detected using the Hitachi diode array detector L-2450.
Determination of Kinetic Parameters
The reactions, containing 100 μm malonyl-CoA, 50 mm Tris-HCl (pH 8.0), 150 mm sodium chloride, and 10 μg/ml purified enzyme were performed in a total volume of 900 μl. The concentration of behenyl-CoA was varied between 0.5 and 5 μm. After the preincubation of the reaction mixture at 30 °C for 5 min, the reactions were initiated by the addition of behenyl-CoA and continued for 180 s. The reactions were quenched with the addition of 180 μl of 6 m HCl, and the materials in the mixture were extracted with ethyl acetate. The organic layer was collected and then evaporated. The residual material was dissolved in 20 μl of methanol for HPLC analysis. 5-n-Heneicosylresorcinol, 4-hydroxy-6-(2′-oxotricosyl)-2-pyrone, and 6-heneicosyl-4-hydroxy-2-pyrone were used to generate standard curves for the quantification of products. Steady-state parameters were determined by a Hanes-wolf plot.
Crystallization, Data Collection, and Structure Determination
Crystals of wild-type ArsC, selenomethionine-labeled ArsC, and the ArsC G284W mutant were grown from a 1:1 mixture of a protein solution (7 mg/ml in 10 mm Tris-HCl (pH 7.5), 150 mm sodium chloride, and 10% (v/v) glycerol) and a reservoir solution (0.1 m imidazole-HCl (pH 7.5), 0.15 m calcium acetate, and 12.5% polyethylene glycol 3350) using the hanging-drop vapor-diffusion method at 20 °C. Before x-ray data collection, the crystal was transferred into the reservoir solution supplemented with 20% (v/v) glycerol as a cryoprotectant and flash-frozen in the liquid nitrogen stream. X-ray data were collected using beamlines BL-5A and AR-NW12A (Photon Factory, Tsukuba, Japan). All diffraction data were indexed, integrated, and scaled using the program HKL2000 (9). Selenomethionine-labeled protein crystals were used for the single-wavelength anomalous dispersion phasing method. Selenium positions and initial phases were calculated by the program autoSHARP (10). After density modification, clearly interpretable electron density maps were obtained. The program ARP/wARP (11) was used for automatic initial protein model building and refinement. The initial model of ArsC G284W was determined by the molecular replacement method using the program MOLREP (12) with the coordinates of the refined wild-type ArsC structure as a template. Coot (13) was used for visual inspection and manual rebuilding of the model. Refmac (14) was used for refinement. The figures were prepared using PyMOL (15). Each active site volume was calculated using the program CASTp (16). The geometries of the final ArsC structures were evaluated using the program Rampage (17).
RESULTS
In Silico Analysis of Key Residues Affecting Cyclization Diversity between ArsB and ArsC
For the identification of amino acid residues contributing to cyclization diversity between ArsB and ArsC, we first compared the amino acid sequences between ArsB and ArsC on the basis of residues important for functional diversity, as proposed by Austin and Noel (1). The sequence alignment analysis revealed replacement of four amino acid residues in the active site cavity. Ser-157, Leu-219, Thr-235, and Trp-281 of ArsB were replaced with Ala-160, Met-222, Ser-238, and Gly-284 in ArsC (Fig. 2A). To narrow down key residue candidates, we also aligned the sequences with other type III PKS sequences. Besides ArsB and ArsC, several microbial type III PKSs have been identified as alkylresorcylic acid synthases, alkylresorcinol synthases, and alkylpyrone synthases. For example, Mycobacterium tuberculosis PKS18 and Bacillus subtilis BpsA were reported to produce alkylpyrone via lactonization (18, 19). In contrast, Actinoplanes missouriensis AgqA was reported to produce both alkylresorcinol and alkylresorcylic acid via decarboxylative aldol condensation and aldol condensation, respectively (20). Neurospora crassa ORAS (2′-oxoalkylresorcylic acid synthase), Streptomyces griseus SrsA, and Myxococcus xanthus FtpA were also reported to produce alkylresorcylic acid via aldol condensation (Fig. 1B) (6, 21–24). Trp-281 of ArsB is a conserved aromatic residue among these alkylresorcinol- and/or alkylresorcylic acid-producing type III PKSs although other residues, Ser-157, Leu-219, and Thr-235, show no significant conservation. This Trp residue is replaced by a smaller residue in alkylpyrone-producing enzymes such as PKS18 and ArsC. However, BpsA, which was identified as a pyrone synthase according to in vitro and heterologous expression studies (19), has a Trp residue in this position just like alkylresorcinol- and/or alkylresorcylic acid-producing enzymes. We assume that BpsA is a potential alkylresorcinol- and/or alkylresorcylic acid-producing enzyme. In fact, BpsA was also reported to produce alkylresorcinol, although its main product is alkylpyrone (19). Recently, Rubin-Pitel et al. (6) focused on this aromatic residue of ORAS and showed that the substitution of the aromatic residue Phe by Gly in ORAS altered product profiles from alkylresorcylic acids to alkylpyrones. Therefore, we hypothesized that this residue diversifies cyclization specificity between aldol condensation and lactonization in microbial type III PKSs.
FIGURE 2.
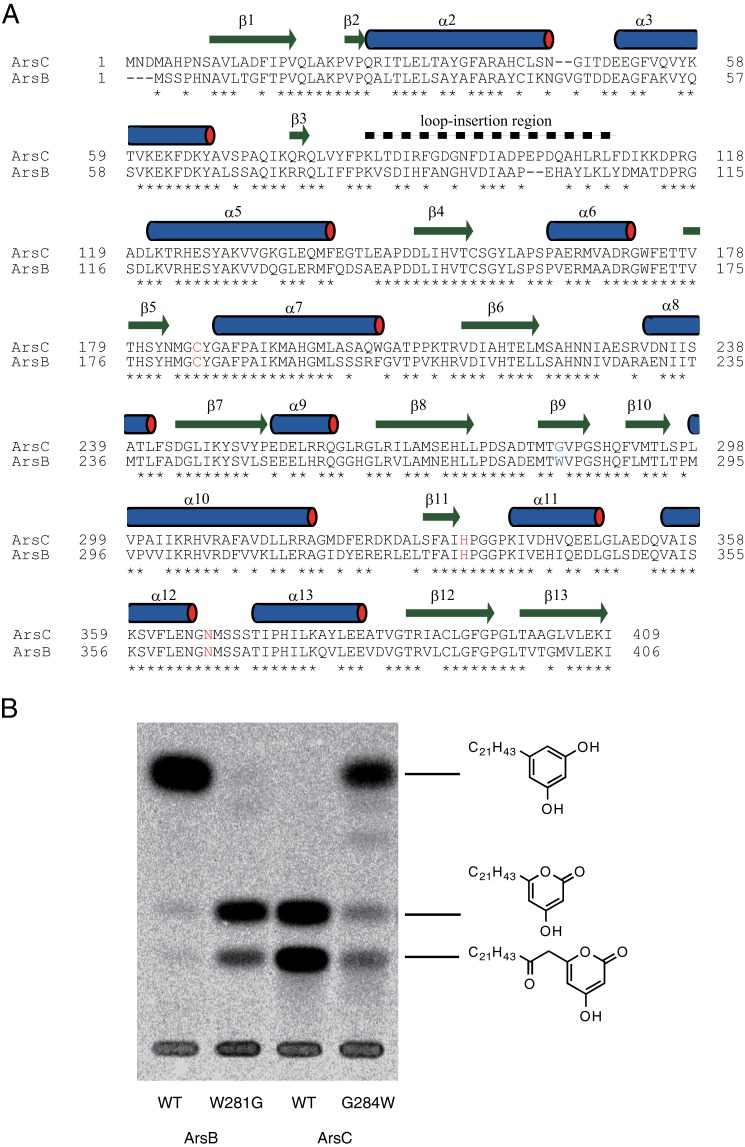
Identification of a key amino acid residue for the cyclization specificity of type III PKSs. A, amino acid sequence comparison of two type III PKSs from A. vinelandii. The amino acid residues consisting of the catalytic triad are shown in red. The residues responsible for cyclization specificity are shown in blue. The secondary structural elements of ArsC, which are identical both in the wild-type and the G284W mutant enzymes, are indicated by bars above the sequence. For the secondary structural element numbering of ArsC, we adopted the numbering system used for known type III PKS family proteins, such as ORAS. Therefore, helices α1 and α4 are missing in the ArsC structure. B, radio-TLC analysis of in vitro reaction products synthesized by the ArsB and ArsC proteins (wild-type or mutant) from behenyl-CoA and [2-14C]malonyl-CoA.
Identification of Trp-281 of ArsB and Gly-284 of ArsC as Key Residues for Their Cyclization Specificity
To prove this hypothesis, we constructed ArsB W281G and ArsC G284W mutants and analyzed their reaction products. Radio-TLC analysis showed that G284W substitution of ArsC resulted in the considerable reduction of alkylpyrone production and the concomitant increase of alkylresorcinol production (Fig. 2B). Although alkylpyrones appeared to be produced in a somewhat larger amount in the ArsC G284W reaction than in the ArsB reaction, the ArsC G284W mutant showed a comparable kcat/Km value for alkylresorcinol synthesis to that for alkylpyrone synthesis catalyzed by wild-type ArsC (Table 1). This result showed that G284W mutation altered the cyclization specificity of ArsC from lactonization to aldol condensation without a significant loss of the polyketide chain elongation activity. In contrast, W281G substitution of ArsB resulted in the complete loss of alkylresorcinol production and the simultaneous increase of alkylpyrone production (Fig. 2B). We note that triketide pyrone was produced in a larger amount than tetraketide pyrone in the ArsB W281G reaction, whereas triketide and tetraketide pyrones were produced in a similar amount in the ArsC reaction. This result is in agreement with the steady-state kinetic parameters of ArsB W281G; the kcat/Km value of triketide pyrone production was 4.2 times higher than that of tetraketide pyrone production (Table 1). Because the wild-type ArsB produced only tetraketide resorcinol, W281G mutation affected not only cyclization specificity but also chain elongation; ArsB W281G had a stronger tendency to release the triketide before additional chain elongation to generate the tetraketide compared with wild-type ArsB and ArsC. Thus, the enzymatic properties of ArsB W281G are different from those of ArsC. However, the result of our “reciprocal” mutational analysis of ArsB and ArsC (which produced ArsB-W281G and ArsC-G284W) clearly demonstrates that Trp-281 of ArsB or Gly-284 of ArsC occupies a critical position for cyclization of polyketomethylene intermediates.
TABLE 1.
Kinetic analysis of ArsB, ArsC, and their mutants
| Enzymes | kcat | Km | kcat/Km | Products |
|---|---|---|---|---|
| ×10−3 s−1 | μm | s−1m−1 | ||
| ArsB | 66 ± 21 | 6.2 ± 1.8 | 11000 | Resorcinol |
| ArsBW281G | 6.9 ± 2.1 | 0.70 ± 0.26 | 11000 | Triketide pyrone |
| ArsBW281G | 2.1 ± 0.7 | 0.95 ± 0.43 | 2600 | Tetraketide pyrone |
| ArsC | 12 ± 7 | 1.5 ± 0.5 | 7600 | Triketide pyrone |
| ArsC | 14 ± 7 | 1.1 ± 0.3 | 12000 | Tetraketide pyrone |
| ArsCG284W | 5.0 ± 1.5 | 0.29 ± 0.12 | 27000 | Resorcinol |
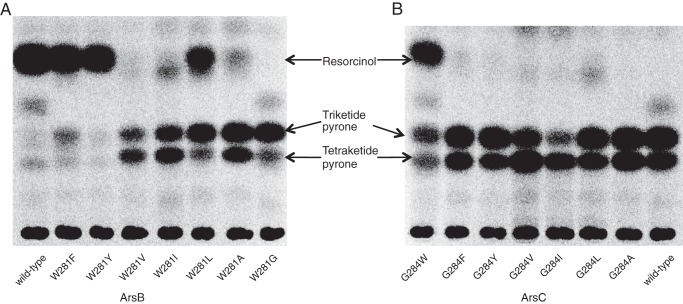
Next, we substituted Trp-281 of ArsB and Gly-284 of ArsC to other aromatic and aliphatic residues (Phe, Tyr, Val, Ile, Leu, and Ala) to explore the effect of steric and aromatic properties of the amino acid residue at this position (Fig. 3). The ArsB W281F and W281Y mutants retained alkylresorcinol-producing activity and did not produce alkylpyrone. The ArsB W281L mutant produced alkylpyrone as well as alkylresorcinol. Other ArsB mutants such as W281V, W281I, and W281A severely decreased alkylresorcinol-producing activity, and their major products were alkylpyrone, as observed for the ArsB W281G mutant. These results indicate that alkylresorcinol/alkylpyrone production roughly depends on the size of the amino acid residue at position 281 in ArsB. The presence of a bulky amino acid residue seems to allow the generation of alkylresorcinol and exclude the production of alkylpyrone in ArsB. In contrast, in the case of ArsC, mutants other than G284W, in which Gly-284 was substituted to Phe, Tyr, Val, Ile, Leu, and Ala, did not acquire the alkylresorcinol-producing activity. Thus, some substitution pairs such as ArsB W281Y and ArsC G284Y mutants did not share product profiles. This inconsistency may be explained by the other three amino acid variations in the active site cavities of ArsB and ArsC, as described above (see “Discussion”).
FIGURE 3.
Radio-TLC analysis of in vitro reaction products synthesized by ArsB (A) and ArsC (B) from behenyl-CoA and [2-14C]malonyl-CoA.
Overall Structures of ArsC and ArsC G284W
We have determined the crystal structure of the wild-type and G284W mutant ArsC proteins at 1.76 and 1.99 Å resolution, respectively (Table 2). The overall structure of the ArsC G284W mutant is almost identical to that of the wild-type enzyme (root mean square deviation (r.m.s.d.) of Cα atoms between the A chain of the wild-type protein and the A chain of the G284W mutant is 0.21 Å). Each of the ArsC structures has four dimers per crystallographic asymmetric unit. The ArsC structure adopts the conserved homodimeric αβαβα thiolase fold observed in other crystal structures of type III PKSs (Fig. 4A). A search for structurally related proteins using the program Dali (25) revealed that the closest protein is Dictyostelium discoideum Steely, a hybrid enzyme composed of a type I fatty acid synthase and a type III PKS (26) (Z score = 43.0, r.m.s.d. of 2.2 Å, sequence identity of 21%). We note that ArsC also exhibits structural similarity with PKS18 (18) (Z score = 41.8, r.m.s.d. of 2.3 Å, sequence identity of 20%) and ORAS (6, 21) (Z score = 40.7, r.m.s.d. of 2.4 Å, sequence identity of 17%). Thus ArsC is structurally similar to other type III PKSs; however, the N-terminal 120-residue region of ArsC shows structural deviations. The unique structural element is the loop-insertion region (84–110) between helix α3 and helix α5 (Fig. 4A). This loop is a disordered region in five of the eight molecules in the crystallographic asymmetric unit, indicating that this region is flexible.
TABLE 2.
Data collection, phasing, and refinement statistics
| Datasets | ArsC |
||
|---|---|---|---|
| Wild type | G284W mutant | SeMeta | |
| Data collection statistics | |||
| Beamline | PF BL-5A | PF AR-NW12A | PF BL-5A |
| Wavelength (Å) | 1.00000 | 1.00000 | 0.97915 |
| Space group | P21 | P21 | P21 |
| Unit-cell parameters | |||
| a (Å) | 102.32 | 102.89 | 102.53 |
| b (Å) | 143.39 | 142.76 | 143.55 |
| c (Å) | 129.62 | 129.77 | 129.92 |
| β (degree) | 110.33 | 110.45 | 110.20 |
| Resolution (Å) | 50.00-1.76 | 50.00-1.99 | 50.00-2.53 |
| (outer shell) | (1.82-1.76) | (2.02-1.99) | (2.62-2.53) |
| Unique reflections | 345,492 (37,894) | 244,208 (15,807) | 115,095 (9,308) |
| Redundancy | 3.8 (2.7) | 5.4 (4.8) | 7.4 (7.2) |
| Completeness (%) | 97.2 (80.3) | 96.9 (61.6) | 99.3 (96.8) |
| Rmerge | 0.120 (0.546) | 0.140 (0.821) | 0.086 (0.114) |
| Mean 〈I/σ(I)〉 | 21.3 (2.2) | 19.9 (1.9) | 39.4 (22.5) |
| Refinement statistics | |||
| Rfac (%) | 18.3 | 20.5 | |
| Rfree (%) | 23.0 | 27.3 | |
| No. of protein atoms | 24,691 | 24,661 | |
| No. of waters | 2,676 | 2,369 | |
| No. of sodium ions | 8 | 8 | |
| r.m.s.d. | |||
| Bond lengths (Å) | 0.027 | 0.023 | |
| Bond angles (ο) | 2.110 | 1.872 | |
| Ramachandran plot | |||
| Favored region (%) | 96.6 | 96.1 | |
| Allowed region (%) | 3.3 | 3.5 | |
| Outer region (%) | 0.1 | 0.4 | |
a ArsC selenomethionine (SeMet) derivative data were used only for initial phase determination.
FIGURE 4.
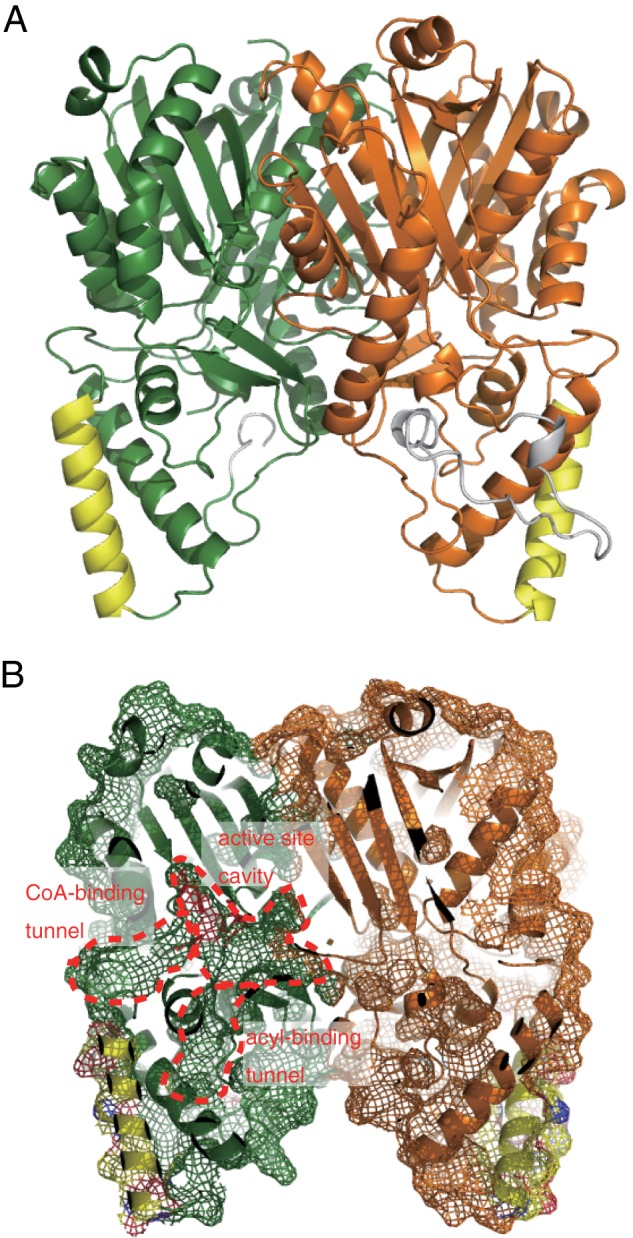
Overall structure of ArsC. A, ribbon drawing diagram of the ArsC dimer. Each monomer is shown in green and orange. Helix α3 and the flexible insertional loop region are shown in yellow and gray, respectively. B, molecular surface of the ArsC dimer focusing on the inside of the enzyme. The CoA binding tunnel, active site cavity, and acyl binding tunnel are indicated.
Structure of the Substrate-binding Site of ArsC
Previous structural studies on plant type III PKSs, alfalfa chalcone synthase (27) and daisy 2-pyrone synthase (4), revealed that several residues of the helix α3 are involved in interactions with two phosphates of CoA at the entrance of the CoA binding tunnel, suggesting that helix α3 plays an important role in CoA substrate binding. In microbial type III PKSs using acyl-CoA substrates, helix α3 occupies a similar position (Fig. 5). Even Steely, which accepts acyl-ACP substrates, has helix α3 at the equivalent position (Fig. 5). However, in the ArsC structure, helix α3 shifts away from the CoA-binding site by >8 Å (Fig. 5), resulting in the rearrangement of the CoA binding region. Although ArsB and ArsC were shown to react with the acyl-CoA substrate in vitro, they are supposed to interact with the ACP domain of ArsA during substrate transfer in vivo (8). Given this unique catalytic property, this conformational difference of helix α3 may enable the formation of a unique ACP-binding site of ArsC.
FIGURE 5.
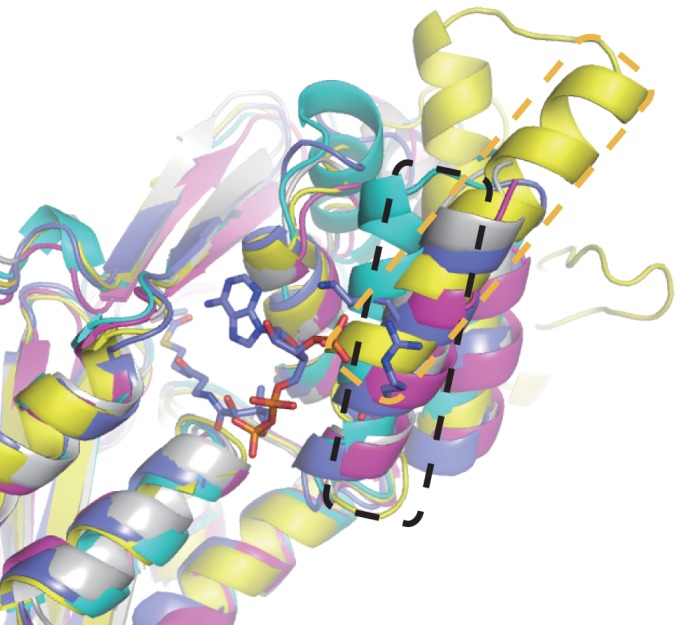
Overlay of the structure of ArsC with structures of other type III PKSs. ArsC, ORAS, PKS18, 2-pyrone synthase, and Steely are shown in yellow, cyan, magenta, purple, and gray, respectively. The sticks model represents the acetoacetyl-CoA in the complex structure of 2-pyrone synthase with acetoacetyl-CoA. Helix α3 of ArsC and other type III PKSs are highlighted with orange and black boxes, respectively.
Structure of the Acyl Binding Tunnel of ArsC
ArsC possesses a hydrophobic acyl binding tunnel flanked with an active site cavity, as observed in the structures of microbial type III PKSs (6, 18, 22, 28) (Figs. 4B and 6). In the structure of the ArsC G284W mutant, a weak electron density that probably corresponds to polyethylene glycol derived from the crystallization buffer is observed in the acyl binding tunnel as reported in the structures of Streptomyces coelicolor tetrahydroxynaphthalene synthase (28) and the ORAS F252G mutant (6). The amino acid residues that consist of the acyl binding tunnel are not conserved in type III PKSs, and therefore, the architectures of the acyl binding tunnel are diverse. For example, the acyl binding tunnel of PKS18 extends out to the surface of protein (18). In the ORAS structure, the tunnel does not extend to solvent and is inwardly directed into the protein (6). ArsC has a straight acyl binding tunnel extending toward the surface of the protein, as observed for PKS18. However, the unique flexible loop between helix α3 and helix α5 forms the base of this acyl binding tunnel (Fig. 6). The side chains of Phe-95 and Ile-97 in this flexible loop block solvent access to the acyl binding tunnel. This flexible loop may function as a lid during substrate binding.
FIGURE 6.
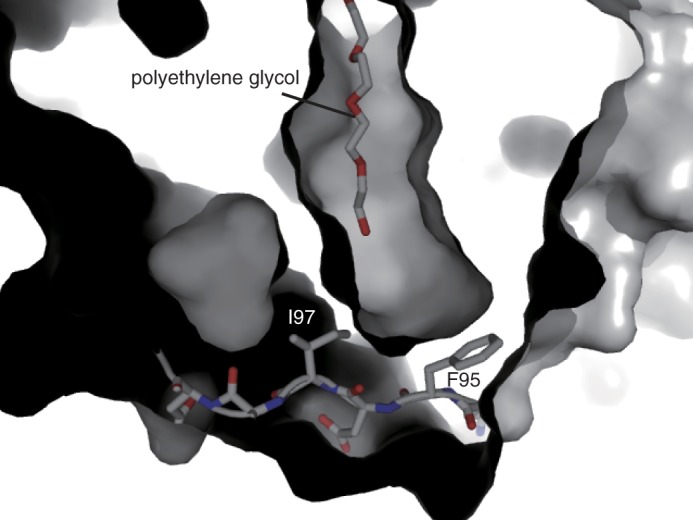
The molecular surface of the ArsC G284W mutant focusing on the acyl binding tunnel. The flexible loop between helix α3 and helix α5 and polyethylene glycol are shown in stick models.
Structural Basis for the Cyclization Specificity
The active site cavity containing the catalytic triad Cys-186–His-334–Asn-367 is placed inside the ArsC molecule, similar to those of other type III PKSs. Gly-284, the key residue for cyclization specificity, is located at the center of the active site cavity in the ArsC structure (Fig. 7A). The active site cavity extends beyond the Gly-284 position and reaches to the dimeric interface. Ala-160 of ArsC participates in the formation of the active site cavity of the neighboring monomer. Plant type III PKSs have a similar cross-subunit interaction. Met-137 of chalcone synthase corresponding to Ala-160 of ArsC is highly conserved in plant type III PKSs and makes cross-subunit interactions by protruding into the second monomer and forming the base of the active site cavity (27). Chalcone synthase also has a small residue, Gly-256, at the equivalent position as Gly-284 of ArsC, enabling the opening of the active site cavity to the dimeric interface.
FIGURE 7.
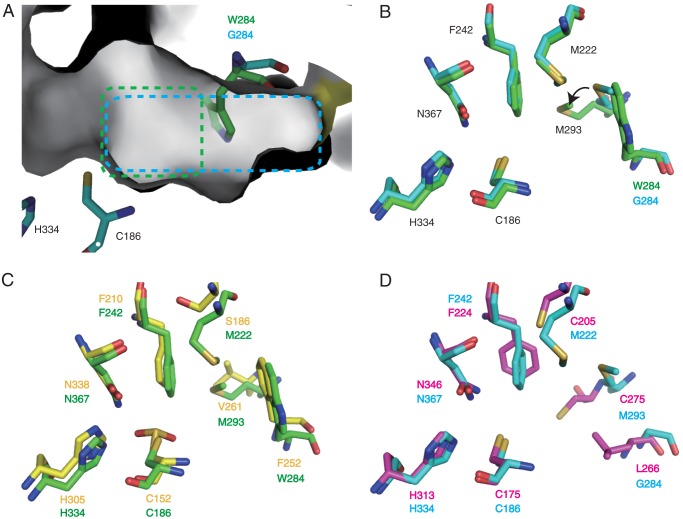
The active site cavity of ArsC. A, molecular surface of the active site cavity of ArsC. Residues of wild-type ArsC and the G284W mutant are shown in cyan and green sticks, respectively. The active site cavities of wild-type ArsC and the G284W mutant are highlighted in cyan and green boxes. B, the superimposed structures of wild-type ArsC and the ArsC G284W mutant. The whole A chain atoms of the wild-type ArsC structure were superimposed onto those of the ArsC G284W mutant structure. Residues of wild-type ArsC and the G284W mutant are shown in cyan and green sticks, respectively. C, the superimposed structures of ArsC G284W and ORAS. Residues of the ArsC G284W mutant and ORAS (PDB code 3EUO) are shown in green and yellow sticks, respectively. D, the superimposed structures of wild-type ArsC with PKS18. Residues of wild-type ArsC and PKS18 (PDB code 1TED) are shown in cyan and magenta sticks, respectively.
In the structure of the ArsC G284W mutant, the bulky side chain of the mutated residue protrudes toward the space of the active site cavity and blocks the connection to the dimeric interface (Fig. 7A). Thus, this substitution causes a significant reduction of cavity volume from 592 Å3 in wild-type ArsC to 293 Å3 in the ArsC G284W mutant (Fig. 8, A and B). This change of cavity volume does not affect the elongation reaction with malonyl-CoA but strikingly affects the cyclization reaction. This substitution also gives rise to the following three minor structural arrangements. First, the polypeptide backbone between Trp-284 and Pro-286 moves 0.8 Å toward the catalytic triad. Second, the presence of the bulky side chain of Trp-284 causes a conformational movement of Met-293 (Fig. 7B). The side chain of Met-293 swings by 40° toward the acyl binding tunnel, thereby narrowing the entrance of the acyl binding tunnel. Third, the presence of the aromatic side chain affects the positioning of water molecules. In the structure of wild-type ArsC, a water molecule network is present in the active site cavity, and Ser-369 holds a water molecule with a hydrogen bond (Fig. 9A). Judging from the short distance (3.7 Å) between this water molecule and the catalytic cysteine, this water molecule possibly plays an important role in catalysis. However, in the structure of the ArsC G284W mutant, the water molecule network is disrupted by the bulky side chain of Trp-284, and Ser-369 loses the ability to retain the water molecule (Fig. 9B).
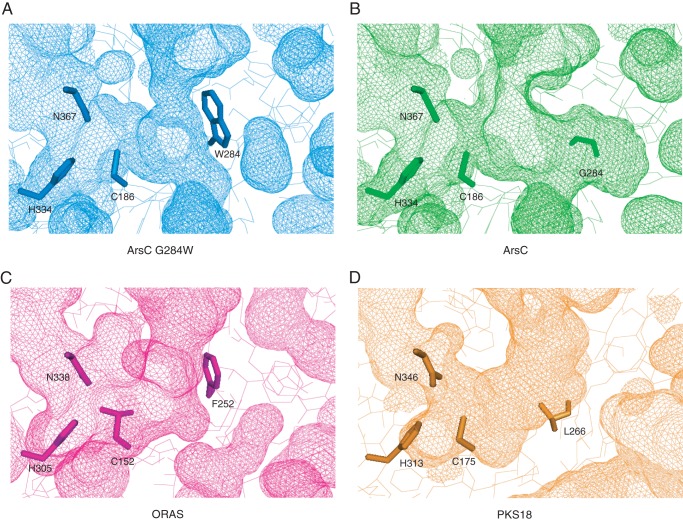
FIGURE 8.
Molecular surfaces of the active site cavities in the ArsC G284W mutant (A), wild-type ArsC (B), ORAS (C), and PKS18 (D). Residues of the catalytic triad and the proposed important residue for the cyclization specificity are shown in stick models. Cys152 of ORAS was reported to be oxidized into sulfinic acid (6).
FIGURE 9.
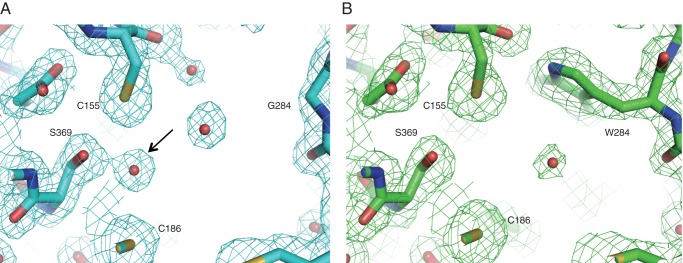
The comparison of active site of wild-type ArsC (A) with the ArsC G284W mutant (B). Water molecules are shown in red spheres. 2Fo − Fc electron density maps contoured at 1.0 σ are shown around the active sites. An arrow indicates the water molecule present adjacent to Ser-369 in the wild-type ArsC structure.
Although the overall sequence identity is relatively low between the ArsC G284W mutant and ORAS (sequence identity, 17%) or between wild-type ArsC and PKS18 (sequence identity, 20%), their cyclization manners are similar to each other (7, 21). To find out whether alkylresorcinol synthases and alkylpyrone synthases have a common structural property, we compared the active site cavities of ArsC with other type III PKSs. The structural comparison with ORAS revealed that ORAS has a slightly larger cavity volume (392 Å3) than the ArsC G284W mutant (293 Å3). This may enable ORAS to catalyze an additional fourth polyketide extension step for the synthesis of pentaketide compounds. ORAS has an aromatic residue, Phe-252, corresponding to Trp-284 in the ArsC G284W mutant (Figs. 7C and 8, A and C). The position and orientation of Trp-284 in the structure of the ArsC G284W mutant are well superimposed with those of Phe-252 in the structure of ORAS. In ORAS, the F252G substitution does not rearrange adjacent residues including Val-261 corresponding to Met-293 in ArsC. ORAS lacks a cross-subunit effect for the formation of the active site cavity because of the occlusion by the large aromatic residue Phe-252, as observed in the ArsC G284W mutant. This aromatic residue in microbial resorcinol-producing and resorcylic acid-producing enzymes is likely to be important in providing a steric wall for polyketomethylene intermediates to be folded into a suitable form for cyclization via aldol condensation.
In contrast, active site cavities of pyrone-forming ArsC and PKS18 do not share a common feature. PKS18 has a decreased cavity volume (256 Å3) compared with wild-type ArsC (592 Å3). PKS18 contains a larger residue, Leu-266, corresponding to Gly-284 in ArsC (Figs. 7D and 8, B and D). In addition, the polypeptide backbone between Val-273 and Glu-276 in PKS18 corresponding to that between Phe-291 and Thr-294 in ArsC moves 2.5 Å toward the catalytic triad. These arrangements contribute to the reduction of the cavity volume in PKS18. Most type III PKSs were reported to produce pyrones as byproducts, as a result of non-enzymatic, spontaneous lactonization of polyketide intermediates (2). Substitution of Trp-281 by Gly, Val, Ile, or Ala in ArsB probably leads to a derailment of aldol condensation, allowing spontaneous lactonization for the synthesis of alkylpyrone (see “Discussion”).
DISCUSSION
Crystal structures of many type III PKSs have been determined. As for microbial type III PKSs, structures of PKS18 and ORAS were determined as a pyrone synthase and a resorcylic acid synthase, respectively (6, 18, 22). However, no structure of a microbial resorcinol synthase has been available. In this study we succeeded in the determination of structures of wild-type ArsC (pyrone synthase) and an ArsC G284W mutant (“artificial” resorcinol synthase). Because the amino acid sequence of ArsB (resorcinol synthase) is highly homologous to that of the ArsC G284W mutant, the structure of the ArsC G284W mutant could provide important insights into the catalytic mechanism of microbial resorcinol-producing type III PKSs, as described below.
Both lactonization and aldol condensation reactions should start from the elimination of an active methylene proton between two carbonyl groups to form an enolate anion. In the lactonization reaction, the resulting anion at the oxygen directly attacks a carbonyl group. In the case of aldol condensation, the active anion reacts with a carbonyl group via the olefinic carbon to form a C-C bond. Our mutagenic and structural studies clearly showed that Trp-281 of ArsB and Gly-284 of ArsC should occupy a critical position for the cyclization specificity of each enzyme. Trp-281 appears to be important in providing a steric wall for the proper folding of the polyketomethylene intermediate to aldol condensation in ArsB. The existence of an aromatic group may restrict the conformation of the growing polyketide chain and probably excludes spontaneous lactonization by preventing the oxygen of enolate from getting close enough to react with a carbonyl group. Thus, Trp-281 in ArsB properly controls the number of condensation reactions during the polyketide extension and the conformation of the intermediates during the cyclization so that ArsB produces only tetraketide alkylresorcinol. In contrast, ArsC, which has Gly-284 at the equivalent position, produces triketide and tetraketide alkylpyrones via lactonization. The large cavity of ArsC may give flexibility to the polyketomethylene intermediate chain, giving a derailment product. Therefore, ArsC cannot exclude the formation of a triketide product by spontaneous cyclization caused from the flexibility of the triketide intermediate, allowing the formation of both triketide and tetraketide pyrones. Thus the condensation and cyclization steps seem not to be strictly controlled in the ArsC reaction. We assume that arsB and arsC genes may have descended from the same ancestor and that ArsB and ArsC probably acquired different catalytic properties by amino acid substitutions. Replacement of Gly by Trp (at position 281) in ArsB or replacement of Trp by Gly (at position 284) in ArsC would be critical evolutionary events to generate a different specificity in cyclization (aldol condensation or lactonization). It is difficult to predict, however, the original cyclization specificity of the ancestral enzyme.
We showed that substitution of Trp-281 by Gly in ArsB altered its cyclization specificity from aldol condensation to lactonization to produce triketide and tetraketide pyrones, which are common derailment products of PKSs. Several similar results in which pyrone-producing activity was conferred on type III PKSs by amino acid substitution have been previously reported (4, 6, 29, 30). To date, however, there has been no report on the alteration of cyclization specificity of a type III PKS from lactonization to aldol condensation. In this context, there is only one report to confer a resorcinol-producing activity on a plant type III PKS; mutation of eight amino acid residues was required for altering the cyclization specificity of alfalfa chalcone synthase from a chalcone-producing Claisen condensation to a stilbene (resorcinol)-producing aldol condensation (5). In the present study we showed for the first time that substitution of Gly-284 by Trp in ArsC altered its cyclization specificity from lactonization to aldol condensation. It should be emphasized that only one amino acid residue is a critical determinant of different cyclization specificity between ArsB and ArsC.
As described above, our results clearly showed that Trp-281 of ArsB plays a critical role in aldol condensation. There are other microbial type III PKSs that catalyze aldol condensations that also have aromatic residues such as Trp and Phe at the corresponding position. Thus our results contribute to prediction of cyclization specificity based on amino acid sequence of microbial type III PKSs. However, our results also suggest that differences in three other amino acid residues in the active site cavity (Ser-157, Leu-219, and Thr-235 in ArsB and Ala-160, Met-222, and Ser-238 in ArsC) caused several differences in catalytic properties between recombinant (wild-type or mutant) ArsB and ArsC enzymes that have the same amino acid residue at the critical position (281 of ArsB and 284 of ArsC). First, the enzymatic properties of ArsC G284W differed from those of ArsB; ArsC G284W produced resorcinol as the major product with a small, but significant amount of pyrone, whereas ArsB produced resorcinol with trace amounts of pyrone. This result suggests that ArsC G284W is somewhat inferior to ArsB as a resorcinol synthase and indicates that the shape of the active site cavity of ArsC G284W is less suited for aldol condensation than that of ArsB. Second, the enzymatic properties of ArsB W281G also differed from those of ArsC; ArsB W281G produced ∼4-fold greater amounts of triketide pyrone than tetraketide pyrone, whereas ArsC produced both triketide and tetraketide pyrones in comparable amounts. This indicates that the shape of the ArsB W281G active site cavity is not as ideally suited to the three rounds of chain-elongation compared to the active site of ArsC. Third, some of the substitution pairs between ArsB and ArsC, such as ArsB W281Y and ArsC G284Y mutants, did not share product profiles; ArsB W281Y and W281F produced alkylresorcinol, but ArsC G284Y and G284F produced alkylpyrone but not alkylresorcinol. In other words, when the amino acid residue at position 281 is an aromatic residue, the resorcinol synthase activity is retained in the alkylresorcinol synthase ArsB. In contrast, in the case of substitution of Gly-284 in the alkylpyrone synthase ArsC, only Trp, but not Tyr or Phe, can confer the resorcinol synthase activity to ArsC. This result also implies the entire shape of the active site cavity of ArsC G284W is less suited for the aldol condensation than the active site cavity of ArsB. All three results suggest that not only is one amino acid residue (for example, Trp-281 of ArsB and Gly-284 of ArsC) critical but also other amino acid residues in the active site cavity are important and can affect the enzymatic properties including cyclization specificity of microbial type III PKSs. This notion should be considered in the sequence-based prediction of cyclization specificity of microbial type III PKSs. In fact, BpsA mainly produces alkylpyrone, not alkylresorcinol, even though it has a Trp residue at the critical position (19). This indicates the shape of the active site cavity of BpsA is also not likely ideal for aldol condensation.
As for the plant type III PKS, Austin et al. (5) determined the structure of resorcinol-producing stilbene synthase (STS) and proposed the reaction mechanism. STS is proposed to employ an “aldol switch” mechanism with the thioesterase-like hydrogen bond network composed of Thr-132, Glu-192, and Ser-338, allowing the activation of a water molecule for the hydrolysis of the C1-cysteine thioester bond. The resulting free carboxylic acid intermediate is cyclized via aldol condensation and then decarboxylated to form a resorcinol product. However, the structure of the ArsC G284W mutant suggests that ArsB does not use such a reaction mechanism. ArsB contains Cys-155 at the position corresponding to Thr-132 in STS. The presence of this Cys residue hinders the formation of the hydrogen bond network in the ArsC G284W mutant. ORAS also has Cys-120 at the corresponding position (6). Moreover, in the ArsC G284W mutant, no water molecule for the hydrolysis of the thioester bond is present adjacent to Ser-369, which is equivalent to Ser-338 in STS. Thus, the aldol switch network is not conserved in microbial resorcinol- and resorcylic acid-producing type III PKSs. Another difference is that STS does not have a steric wall, as observed in ArsB. Trp-281 of ArsB is replaced by Gly in STS. In other plant alkylresorcylic acid synthases, Oryza sativa ARAS1 and ARAS2 (31), this aromatic residue is replaced by Met. Consequently, several amino acid variations and structural differences are observed between microbial and plant aldol condensation-catalyzing type III PKSs, indicating that different reaction mechanisms are employed. In the reaction catalyzed by plant type III PKSs, the aldol switch triggers the formation of an acidic intermediate by cleavage of the thioester bond of the Cys-bound linear intermediate. The acidic intermediate is then cyclized via aldol condensation (5). This study and that of the ORAS structure (6) indicate, however, that microbial type III PKSs do not make use of the aldol switch mechanism but instead provide a steric wall so that polyketomethylene intermediates may be folded into suitable forms for cyclization via aldol condensation. Interestingly, Posehn et al. (32) suggested that aldol cyclization catalyzed by ArsB occurs before hydrolysis of the thioester bond. The identification of amino acid residues directly responsible for aldol condensation in ArsB and the elucidation of the molecular mechanism for the reaction are our future challenges.
In conclusion, we showed that substitution of a single amino acid resulted in the alteration of the cyclization specificity of two Azotobacter type III PKSs, ArsB and ArsC; Trp-281 of ArsB and Gly-284 of ArsC are crucial residues for the specificity of cyclization. Structural analysis revealed that the volume of the active site cavity was a crucial determinant of the cyclization specificity of each enzyme. Thus this study provides a vital element for predicting cyclization specificity of microbial type III PKSs on the basis of their amino acid sequence. It is noteworthy that we succeeded for the first time in altering the cyclization specificity of a type III PKS from lactonization to aldol condensation by replacement of only a single amino acid residue.
This work was supported in part by the Targeted Proteins Research Program of the Ministry of Education, Culture, Sports, Science, and Technology, Japan and a Funding Program for Next Generation World-leading Researchers from the Bureau of Science, Technology, and Innovation Policy, Cabinet Office, Government of Japan. This work was performed under the approval of the Photon Factory Program Advisory Committee (proposal 2008S2-001).
The atomic coordinates and structure factors (codes 3VS8 and 3VS9) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PKS
- polyketide synthase
- LC-APCIMS
- liquid chromatography-atmospheric pressure chemical ionization mass spectrometry
- r.m.s.d.
- root mean square deviation
- ORAS
- 2′-oxoalkylresorcylic acid synthase
- STS
- stilbene synthase.
REFERENCES
- 1. Austin M. B., Noel J. P. (2003) The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20, 79–110 [DOI] [PubMed] [Google Scholar]
- 2. Katsuyama Y., Horinouchi S. (2010) Comprehensive Natural Products II: Chemistry and Biology, Vol. 1, pp. 147–170, Elsevier Science Publishing Co., Inc., New York [Google Scholar]
- 3. Morita H., Abe I., Noguchi H. (2010) Comprehensive Natural Products II: Chemistry and Biology, Vol. 1, pp. 171–225, Elsevier Science Publishing Co., Inc., New York [Google Scholar]
- 4. Jez J. M., Austin M. B., Ferrer J., Bowman M. E., Schröder J., Noel J. P. (2000) Structural control of polyketide formation in plant-specific polyketide synthases. Chem. Biol. 7, 919–930 [DOI] [PubMed] [Google Scholar]
- 5. Austin M. B., Bowman M. E., Ferrer J. L., Schröder J., Noel J. P. (2004) An aldol switch discovered in stilbene synthases mediates cyclization specificity of type III polyketide synthases. Chem. Biol. 11, 1179–1194 [DOI] [PubMed] [Google Scholar]
- 6. Rubin-Pitel S. B., Zhang H., Vu T., Brunzelle J. S., Zhao H., Nair S. K. (2008) Distinct structural elements dictate the specificity of the type III pentaketide synthase from Neurospora crassa. Chem. Biol. 15, 1079–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Funa N., Ozawa H., Hirata A., Horinouchi S. (2006) Phenolic lipid synthesis by type III polyketide synthases is essential for cyst formation in Azotobacter vinelandii. Proc. Natl. Acad. Sci. U.S.A. 103, 6356–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miyanaga A., Funa N., Awakawa T., Horinouchi S. (2008) Direct transfer of starter substrates from type I fatty acid synthase to type III polyketide synthases in phenolic lipid synthesis. Proc. Natl. Acad. Sci. U.S.A. 105, 871–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 10. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 11. Morris R. J., Perrakis A., Lamzin V. S. (2002) ARP/wARP's model-building algorithms. I. The main chain. Acta Crystallogr. D. Biol. Crystallogr. 58, 968–975 [DOI] [PubMed] [Google Scholar]
- 12. Vagin A., Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 13. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 14. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 15. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 16. Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. (2006) CASTp. Computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 34, W116–W118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry. φ, ψ, and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 18. Sankaranarayanan R., Saxena P., Marathe U. B., Gokhale R. S., Shanmugam V. M., Rukmini R. (2004) A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat. Struct. Mol. Biol. 11, 894–900 [DOI] [PubMed] [Google Scholar]
- 19. Nakano C., Ozawa H., Akanuma G., Funa N., Horinouchi S. (2009) Biosynthesis of aliphatic polyketides by type III polyketide synthase and methyltransferase in Bacillus subtilis. J. Bacteriol. 191, 4916–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Awakawa T., Fujita N., Hayakawa M., Ohnishi Y., Horinouchi S. (2011) Characterization of the biosynthesis gene cluster for alkyl-O-dihydrogeranyl-methoxyhydroquinones in Actinoplanes missouriensis. Chembiochem 12, 439–448 [DOI] [PubMed] [Google Scholar]
- 21. Funa N., Awakawa T., Horinouchi S. (2007) Pentaketide resorcylic acid synthesis by type III polyketide synthase from Neurospora crassa. J. Biol. Chem. 282, 14476–14481 [DOI] [PubMed] [Google Scholar]
- 22. Goyal A., Saxena P., Rahman A., Singh P. K., Kasbekar D. P., Gokhale R. S., Sankaranarayanan R. (2008) Structural insights into biosynthesis of resorcinolic lipids by a type III polyketide synthase in Neurospora crassa. J. Struct. Biol. 162, 411–421 [DOI] [PubMed] [Google Scholar]
- 23. Funabashi M., Funa N., Horinouchi S. (2008) Phenolic lipids synthesized by type III polyketide synthase confer penicillin resistance on Streptomyces griseus. J. Biol. Chem. 283, 13983–13991 [DOI] [PubMed] [Google Scholar]
- 24. Hayashi T., Kitamura Y., Funa N., Ohnishi Y., Horinouchi S. (2011) Fatty acyl-AMP ligase involvement in the production of alkylresorcylic acid by a Myxococcus xanthus type III polyketide synthase. Chembiochem 12, 2166–2176 [DOI] [PubMed] [Google Scholar]
- 25. Holm L., Sander C. (1995) Dali. A network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 26. Austin M. B., Saito T., Bowman M. E., Haydock S., Kato A., Moore B. S., Kay R. R., Noel J. P. (2006) Biosynthesis of Dictyostelium discoideum differentiation-inducing factor by a hybrid type I fatty acid-type III polyketide synthase. Nat. Chem. Biol. 2, 494–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ferrer J. L., Jez J. M., Bowman M. E., Dixon R. A., Noel J. P. (1999) Structure of chalcone synthase and the molecular basis of plant polyketide biosynthesis. Nat. Struct. Biol. 6, 775–784 [DOI] [PubMed] [Google Scholar]
- 28. Austin M. B., Izumikawa M., Bowman M. E., Udwary D. W., Ferrer J. L., Moore B. S., Noel J. P. (2004) Crystal structure of a bacterial type III polyketide synthase and enzymatic control of reactive polyketide intermediates. J. Biol. Chem. 279, 45162–45174 [DOI] [PubMed] [Google Scholar]
- 29. Funa N., Ohnishi Y., Ebizuka Y., Horinouchi S. (2002) Alteration of reaction and substrate specificity of a bacterial type III polyketide synthase by site-directed mutagenesis. Biochem. J. 367, 781–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shimokawa Y., Morita H., Abe I. (2010) Structure-based engineering of benzalacetone synthase. Bioorg. Med. Chem. Lett. 20, 5099–5103 [DOI] [PubMed] [Google Scholar]
- 31. Matsuzawa M., Katsuyama Y., Funa N., Horinouchi S. (2010) Alkylresorcylic acid synthesis by type III polyketide synthases from rice Oryza sativa. Phytochemistry 71, 1059–1067 [DOI] [PubMed] [Google Scholar]
- 32. Posehn S. E., Kim S. Y., Wee A. G. H., Suh D.-Y. (2012) Mapping the mechanism of the resorcinol ring formation catalyzed by ArsB, a type III polyketide synthase from Azotobacter vinelandii. Chembiochem 13, 2212–2217 [DOI] [PubMed] [Google Scholar]