Background: How the yeast proteins Nrd1 and Nab3 provoke transcription termination is poorly understood.
Results: An essential part of Nab3 contains a self-assembly domain that appears unstructured. Nrd1/Nab3 double mutants disrupt the function of this higher order complex, causing lethality.
Conclusion: A large network of molecular interactions is needed for termination.
Significance: A new essential function of Nab3 has been identified.
Keywords: Gene Regulation, Polyglutamine, RNA, RNA-binding Protein, RNA Polymerase II, Transcription Elongation Factors, Transcription Termination, Nab3, Nrd1
Abstract
Nab3 and Nrd1 are yeast heterogeneous nuclear ribonucleoprotein (hnRNP)-like proteins that heterodimerize and bind RNA. Genetic and biochemical evidence reveals that they are integral to the termination of transcription of short non-coding RNAs by RNA polymerase II. Here we define a Nab3 mutation (nab3Δ134) that removes an essential part of the protein's C terminus but nevertheless can rescue, in trans, the phenotype resulting from a mutation in the RNA recognition motif of Nab3. This low complexity region of Nab3 appears intrinsically unstructured and can form a hydrogel in vitro. These data support a model in which multiple Nrd1-Nab3 heterodimers polymerize onto substrate RNA to effect termination, allowing complementation of one mutant Nab3 molecule by another lacking a different function. The self-association property of Nab3 adds to the previously documented interactions between these hnRNP-like proteins, RNA polymerase II, and the nascent transcript, leading to a network of nucleoprotein interactions that define a higher order Nrd1-Nab3 complex. This was underscored from the synthetic phenotypes of yeast strains with pairwise combinations of Nrd1 and Nab3 mutations known to affect their distinct biochemical activities. The mutations included a Nab3 self-association defect, a Nab3-Nrd1 heterodimerization defect, a Nrd1-polymerase II binding defect, and an Nab3-RNA recognition motif mutation. Although no single mutation was lethal, cells with any two mutations were not viable for four such pairings, and a fifth displayed a synthetic growth defect. These data strengthen the idea that a multiplicity of interactions is needed to assemble a higher order Nrd1-Nab3 complex that coats specific nascent RNAs in preparation for termination.
Introduction
The Nrd1 and Nab3 proteins of Saccharomyces cerevisiae heterodimerize, bind RNA, and are important for transcription termination during the synthesis of small RNAs, such as snRNAs, small nucleolar RNAs, and cryptic unstable RNAs (1–3, 5). Both of these essential proteins possess a conserved RNA recognition motif (RRM),2 a feature that they have in common with hnRNPs of higher eukaryotes (6). Each protein has a preferred RNA recognition sequence, and, when paired as a heterodimer, the two RRMs interact with their corresponding Nrd1 and Nab3 target sites simultaneously and more tightly than either subunit alone (Fig. 1A, circled number 1) (1, 7–11). A Nab3 interaction site has been mapped on Nrd1 (Fig. 1A, circled number 2) (12, 13). Nrd1 also binds the Ser5-phosphorylated form of the repeated C-terminal domain of the large subunit of RNA polymerase II, and this interaction is important for the function of the Nrd1-Nab3 complex (Fig. 1A, circled number 3) (13).
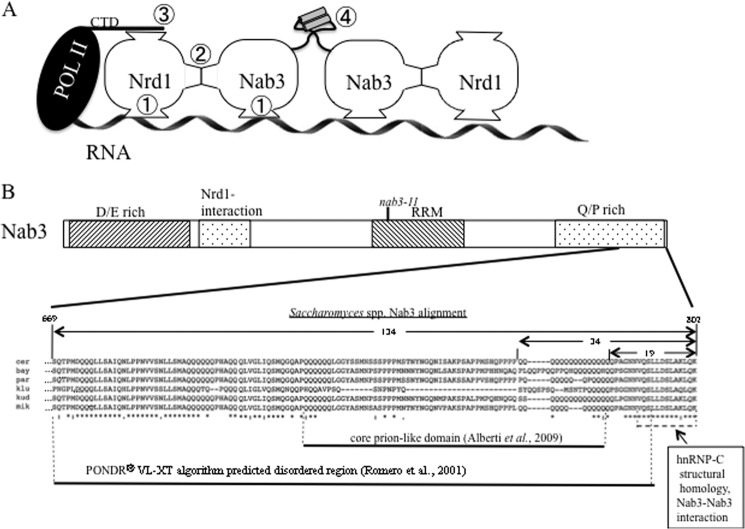
FIGURE 1.
Nab3 structure and functional domains. A, model of a termination complex extended from Carroll et al. (1). Two copies of the Nrd1-Nab3 dimer are shown. The interactions of Nrd1 with RNA, Nab3, and the RNA polymerase II CTD are indicated with circled numbers 1, 2, and 3, respectively. An interaction between two Nab3 C-terminal tails is indicated by a circled number 4, in which the potentially unstructured region of Nab3 ending in a short cylinder (hnRNP-C α-helical structural homology) is shown for two adjacent molecules. B, schematic of the Nab3 polypeptide, highlighting its C-terminal 134 amino acids (expanded). Boxes indicate the positions (not to scale) of the aspartate/glutamate (D/E)-rich region, the Nrd1-interaction domain, the RRM (with the nab3-11 mutations), and the glutamine/proline (Q/P)-rich region. The expanded sequence is output from a Clustal Omega alignment (4) of six Saccharomyces species' Nab3 orthologues (S. cerevisiae (cer), S. bayanus (bay), S. paradoxus (par), S. kluyveri (klu), S. kudriavzevii (kud), and S. mikatae (mik)). Symbols below the alignment indicate residues that are completely conserved (*) or are partially conserved with amino acids whose properties are strongly (:) or more weakly (.) similar. The previously described (16) structural alignment with hnRNP-C is indicated by a dashed line. The 19-, 34-, and 134-amino acid regions of Nab3 are indicated with horizontal arrows above the alignment. The regions of the protein predicted to be prion-like (25) or unstructured (PONDRR VL-XT algorithm (36)) are indicated.
Aside from the well studied RRM (11, 14), relatively little is known about the structure or function of the large Nab3 protein, which is proteolytically labile and has proven difficult to produce in recombinant form. It contains a non-essential, highly acidic N-terminal domain rich in glutamic and aspartic acids (Fig. 1B) (1, 6). An essential portion downstream of the RRM is a proline- and glutamine-rich segment of relatively low sequence complexity (1). These sequences may be responsible for the experimental difficulty in producing recombinant full-length protein. Using a genetic screen, we have recently discerned a function for the very C-terminal tail of Nab3 (Fig. 1, A (circled number 4) and B) (15, 16). It contains a stretch of 16 glutamines adjacent to a short peptide that shares structural homology with an α-helix of the human hnRNP-C protein (15, 16). Both elements of this “tail” region are involved in self-association, similar to the tetramerization helix of human hnRNP-C (17). Loss of either sequence component reduces the ability of Nab3 to support transcription termination (16). This newly described interaction between Nab3 tails could represent a binding interaction between adjacent Nrd1-Nab3 heterodimers that helps to assemble multiple copies of these proteins onto substrate RNAs (Fig. 1A, circled number 4).
The complex's physical contacts (Fig. 1A, circled numbers 1–4) have been shown by in vitro binding assays using recombinant proteins and cell lysates (1, 7, 12, 13, 16, 18, 19). The physiological significance of each is underscored by mutations in the relevant domains that affect cell growth and RNA metabolism. Corden and co-workers (1) proposed that short, non-coding, RNAs destined to be terminated by this system, collect multiple Nrd1-Nab3 dimers to assemble a large nucleoprotein complex.
This assemblage serves as the core of a machinery that contacts appropriately phosphorylated RNA polymerase II and recruits termination factor Sen1 and RNA processing enzymes, such as the TRAMP complex (19–21). Such a complex may resemble mammalian hnRNP-C, which packages RNA into a higher order ribonucleoprotein substrate prior to further processing (22–24).
Nab3 carries interest as well because many RNA binding proteins across phyla have unusual sequences similar to the Nab3 aspartate/glutamate- and proline/glutamine-rich regions (25–27). A variety of yeast proteins contain such low complexity sequences, some of which are intrinsically unfolded and some of which form amyloidogenic, detergent-resistant complexes (25, 28). In mammals, an expansion of polyglutamine tracts due to mutation is associated with protein aggregation and neuropathology (e.g. in amyotrophic lateral sclerosis and Huntington disease) (29, 30). The polyglutamine regions appear to be structurally dynamic, having the ability to adopt many types of secondary structure, including extended conformations, α-helices, and β-sheets (31). The protein sequence adjacent to polyglutamine stretches also plays a role in conformation because polyglutamine tracts tend to be non-randomly distributed and frequently neighbor helical coiled coil elements (32–34). Some RNA-binding proteins with low complexity sequences form cytoplasmic foci, such as stress granules that harbor enzymes and substrates of RNA metabolism. Some of these can reversibly form dynamic fibers, including amyloid-like polymers and hydrogels (26, 27).
Here we have tested the extent of the many interactions found in this higher order complex by making compound mutations affecting more than one function of the network. Crippling two functions leads to lethality in almost all cases. We also studied the unusual and undefined structure and function of the C terminus of Nab3. We found that the last 134 residues were required for cell viability. Biochemical evidence suggests that it is intrinsically unstructured up to the α-helical hnRNP-C homology region at its very end. Although the domain is essential, co-expression of Nab3 without it rescued the mutant phenotype of another Nab3 allele with an RRM mutation, consistent with the model that multiple copies of this protein function in a single complex. These results provide new evidence that there is a constellation of protein-protein and protein-RNA interactions needed to compile a termination-competent assembly and highlight the function of the Nab3 C terminus (Fig. 1A).
MATERIALS AND METHODS
Plasmid and Strain Construction
Plasmid pET32-Nab3-134 was constructed by inserting an XhoI- and BglII-digested PCR product made from pRS315Nab3FL (16) using 5′-atatagatctatctcaaactccaatggaccagc-3′ and 5′-atatctcgagctatttttgtagttttgctaaac-3′ into similarly cut pET32a. pRS315-Nab3 was made by inserting an XhoI- and BamHI-cut PCR product (made from BY4741 genomic DNA using 5′-cacgggatccagtgtaaccctgaattgtgaagag-3′ and 5′-atatctcgagcagaggaaacaaatgaagaggtgcg-3′) into similarly cut pRS315. The 34 C-terminal amino acids were deleted from Nab3 by in vitro replication of pRS315-Nab3 with Phusion DNA polymerase (New England Biolabs) and the mutagenic oligonucleotides 5′-aggtggcggaggttggtgtgac-3′ and 5′-tagactcccttttttcaatcttttccatttcttg-3′. Replicated DNA was used to transform Escherichia coli, and the deletion was confirmed by sequencing the resulting plasmid pRS315Nab3Δ34. Similarly, the 134 C-terminal amino acids of Nab3 were deleted by in vitro replication of pRS315NAB3 using mutagenic oligonucleotides 5′-aggtggttgaggaggcggacc-3′ and 5′-tagactcccttttttcaatcttttccatttcttg-3′ to yield pRS315Nab3Δ134.
The LEU2-marked plasmids pRS315 REF GFP, pRS315-Nab3, pRS315-Nab3Δ34, and pRS315-Nab3Δ134 were independently introduced into DY30229, a strain that lacks chromosomal NAB3 and contains the plasmid pRS316-nab3-11. The resulting strains (DY3031, DY3133, DY3131, and DY3130, respectively) were then tested for growth at 22 or 30 °C on SCura−leu− plates and SC-FOA plates. The lithium acetate method of transformation was used throughout. Diploid strains were generated by mating the nab3CΔ19-containing strain 1A1F (15, 16) or its otherwise isogenic wild type strain BY4742 to the nrd1Δ151–214-containing strain, YSB2078 (13). These diploids, DY2098 and DY2097, respectively, were then transformed with pRS316-NRD1 and sporulated. Tetrads were dissected, and the resulting spores were grown on SCura− or SC-FOA. Similarly, BY4742 or 1A1F were mated to YSB2064, which bears the nrd1Δ6–150 allele, and diploids were transformed with pRS316-NRD1 to yield strains DY2113 and DY2114, respectively. Tetrads were dissected, and spores were grown on selective media as indicated. The nab3-11-containing strain ACY1224 was mated to YSB2064 and YSB2078 to generate diploids that were each transformed with pRS316-NRD1 to yield the strains DY1627 and DY1628, respectively. These diploids were sporulated and dissected to yield haploid spores. The strains used in this study are shown in Table 1.
TABLE 1.
Yeast strains used in this study
| Strain | Description | Source |
|---|---|---|
| 1A1F | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 nab3CΔ19 | Loya et al. (15) |
| ACY1224 | MATα ade2 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 nab3-11 | A. Corbett (Emory University) |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Open Biosystems |
| DY1627 | MATa/α ura3-1/ura3Δ0; leu2-3,–112/leu2Δ0 his3-11,15/his3Δ1 MET15/met15Δ0 ADE2/ade2 CAN1/can1-100 TRP1/trp1-1 NAB3/nab3-11 NRD1/nrd1Δ6–150 [pRS316-NRD1-URA3] | This work |
| DY1628 | MATa/α ura3-1/ura3Δ0; leu2-3,–112/leu2Δ0 his3-11,15/his3Δ1 MET15/met15Δ0 ADE2/ade2 CAN1/can1-100 TRP1/trp1-1 NAB3/nab3-11 NRD1/nrd1Δ151–214 [pRS316-NRD1 (URA3)] | This work |
| DY2097 | MATα/a ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 his3Δ1/his3Δ1 met15Δ0/MET15 lys2Δ0/LYS2 nrd1Δ151–214/NRD1 [pRS316-NRD1 (URA3)] | This work |
| DY2098 | MATα/a ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 his3Δ1/his3Δ1 met15Δ0/MET15 lys2Δ0/LYS2 nrd1Δ151–214/NRD1 NAB3/nab3CΔ19 [pRS316-NRD1 (URA3)] | This work |
| DY2113 | MATα/a ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 his3Δ1/his3Δ1 met15Δ0/MET15; lys2Δ0/LYS2 nrdΔ16–150/NRD1 [pRS316-NRD1 (URA3)] | This work |
| DY2114 | MATα/a ura3Δ0/ura3Δ0 leu2Δ0/leu2Δ0 his3Δ1/his3Δ1 met15Δ0/MET15 lys2Δ0/LYS2 nrd1Δ6–150/NRD1 NAB3/nab3CΔ19 [pRS316-NRD1 (URA3)] | This work |
| DY3031 | MATα ura3Δ0 his3Δ1 leu2Δ0 nab3Δ0::kanMX [pRS315 REF GFP (LEU2)] [pRS316-Nab3-11 (URA3)] | This work |
| DY3130 | MATα ura3Δ0 his3Δ1 leu2Δ0 nab3Δ0::kanMX [pRS315 nab3Δ134 (LEU2)][pRS316-nab3-11 (URA3)] | This work |
| DY3131 | MATα ura3Δ0 his3Δ1 leu2Δ0 nab3Δ0::kanMX [pRS315 nab3Δ34 (LEU2)] [pRS316-nab3-11 (URA3)] | This work |
| DY3133 | MATα ura3Δ0 his3Δ1 leu2Δ0 nab3Δ0::kanMX [pRS315Nab3(LEU2)][pRS316-Nab3–11 (URA3)] | This work |
| DY30229 | MATα ura3Δ0 his3Δ1 leu2Δ0 nab3Δ0::kanMX [pRS316-Nab3-11 (URA3)] | Loya et al. (15) |
| YSB2064 | MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 nrd1Δ6–150 | Vasiljeva et al. (13) |
| YSB2078 | MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 nrd1Δ151–214 | Vasiljeva et al. (13) |
Protein Purification and Analysis
Plasmids were introduced into BL21(DE3) E. coli, and expression was induced with isopropylthiogalactoside for 3 h at 37 °C. Lysozyme-treated cells were broken by sonication, and the lysate was clarified by centrifugation for 30 min at 27,000 × g. The resulting His6-tagged thioredoxin fusion proteins were affinity-purified with immobilized nickel, dialyzed into 20 mm Tris, pH 7.5, 50 mm NaCl, 1 mm EDTA, and chromatographed onto QAE-Sephadex. Flow-through protein was collected and concentrated by centrifugal filtration as needed. The Nab3 134-amino acid polypeptide was cleaved from the C terminus of the thioredoxin fusion protein using thrombin. A second round of nickel chromatography was used to isolate the His6-tagged thioredoxin from the free Nab3 polypeptide.
Purified thioredoxin-Nab3 134-amino acid fusion protein (0.7 mg in 100 μl) was applied to a Superdex 200 10/300 column in 20 mm Tris, pH 7.5, 200 mm NaCl, using an AKTA chromatography system at a flow rate of 0.5 ml/min, and 0.5-ml fractions were collected and analyzed by SDS-PAGE. The column was calibrated with bovine thyroglobulin (670 kDa), bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), horse myoglobin (17 kDa), and vitamin B12 (1.4 kDa). To test heat stability, 40 μg of protein in 100 μl was incubated at 22 or 100 °C for 45 min. Denatured protein was pelleted for 10 min at 13,000 × g. Supernatants were removed, concentrated by precipitation with 10% (v/v) trichloroacetic acid, dissolved in SDS-sample buffer, and resolved on an 11% polyacrylamide gel. Pellets were dissolved in SDS-sample buffer and separated by PAGE. For Western blotting, yeast were lysed in boiling SDS-sample buffer, and proteins were separated on a 5% polyacrylamide gel. Gels were electrophoretically transferred to nitrocellulose, which was blocked with 5% nonfat dry milk (35), probed with 2F12 anti-Nab3 monoclonal IgG (∼1 μg/ml) and horseradish peroxidase-conjugated goat anti-mouse IgG (Invitrogen; diluted 7000-fold), developed with chemiluminescent reagents, and exposed to Kodak X-Omat film.
Protein cross-linking was carried out as described previously (16). Briefly, protein was mixed with the indicated concentration of bis(sulfosuccinimidyl) suberate (ThermoFisher Pierce) and incubated for 30 min at 22 °C. Reactions were terminated with the addition of glycine to 50 mm, and samples were precipitated with 10% trichloroacetic acid, dissolved in sample load buffer, and neutralized with sodium hydroxide for separation on SDS-PAGE.
Proteolytic digestion experiments employed 20 μg of protein and bovine α-chymotrypsin (Sigma) mixed in a 400:1 molar ratio. Following incubation at 22 °C, samples were quenched with phenylmethylsulfonyl fluoride to 1 mm, precipitated with 10% trichloroacetic acid, dissolved in sample load buffer, and neutralized with sodium hydroxide in preparation for SDS-PAGE.
For hydrogel formation, purified Nab3 134 protein was concentrated to 10 mg/ml in 20 mm Tris-HCl, pH 7.5, 0.2 m NaCl and stored at 4 °C for 6 weeks. The gel was compacted by centrifugation at 15,000 × g for 20 min.
RESULTS
A Conserved Region of Nab3 Is Biologically Essential and Biochemically Intriguing
A spontaneous mutation that removes 19 residues from the S. cerevisiae Nab3 C terminus compromises termination and self-association of the protein (15, 16). This region contains a stretch of 16 glutamines adjacent to 18 amino acids with homology to a human hnRNP α-helix responsible for hnRNP-C tetramerization (Fig. 1B). Although cells grew slowly when the Nab3 hnRNP-C homology region was removed, they remain viable (15, 16). Prior deletion analysis indicated that there was an essential region encompassed in the C-terminal 238 amino acids (1). Here we focused on the C-terminal 134 amino acids of Nab3 that are well conserved across Saccharomyces species (Fig. 1B). This region contains additional tracts of polyglutamines, which have been suggested to be prionogenic based on a bioinformatic analysis of low complexity proline/glutamine-rich domains (Fig. 1) (25). Aside from the terminal homology to the hnRNP-C α-helix, this region of Nab3 is predicted to be largely unstructured as scored by the PONDR VL-XT algorithm (Fig. 1B) (36).
To test the biological role of this region, either a plasmid encoding Nab3 lacking its terminal 34 residues (the Gln16 stretch and terminal α-helix) or one encoding Nab3 lacking its terminal 134 residues (Δ134) was introduced into a yeast strain in which the chromosomal copy of NAB3 was deleted. Nab3 function was provided via a second plasmid-borne allele called nab3-11. The nab3-11 allele is a well characterized and experimentally useful pair of point mutations in the RRM domain that result in temperature-sensitive growth (12). Cells with both a control plasmid (marked with LEU2) and the nab3-11 plasmid (marked with URA3) grew at 22 but not 30 °C, confirming the conditional phenotype (Fig. 2, left panels). Cells with a plasmid encoding wild type NAB3 covering the nab3-11 allele grew at the non-permissive temperature of 30 °C (Fig. 2, bottom left). Strains with either the nab3Δ34 or the nab3Δ134 allele in addition to the nab3-11 allele also enabled growth at the non-permissive temperature (Fig. 2, bottom left). Hence, the Nab3 protein lacking its C-terminal tail and polyglutamine stretches rescued the loss of function of nab3-11. These results suggest that the shortened Nab3 proteins accumulated and compensated for the biological function defined by the nab3-11 mutation (i.e. RNA binding). Western blotting with an anti-Nab3 monoclonal antibody confirmed that both deletion derivatives and the nab3-11 missense allele generated comparable amounts of Nab3 protein (Fig. 3A).
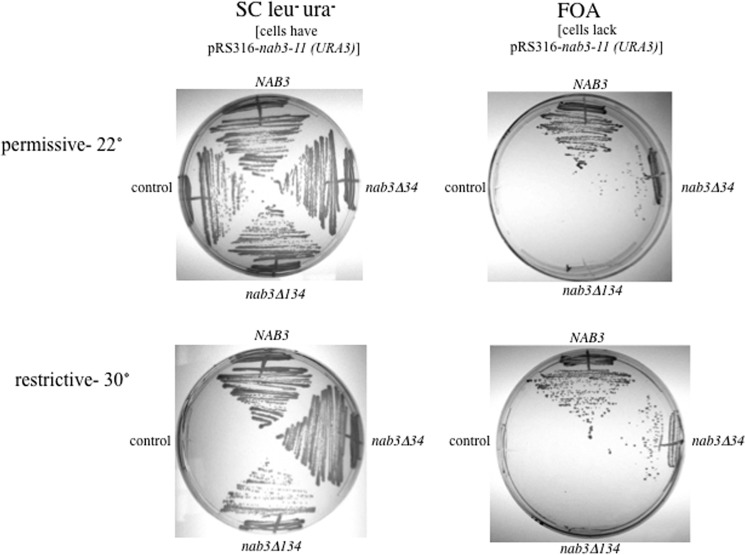
FIGURE 2.
The C-terminal 134 amino acids of Nab3 are essential. Yeast strains lacking their chromosomal copy of NAB3 and containing the nab3-11ts allele on a URA3 plasmid were transformed with a second LEU2-marked plasmid lacking NAB3 or containing NAB3 or deletion derivatives lacking its C-terminal 134 or 34 amino acids. Cells were grown on solid medium lacking uracil and leucine (left panels) or containing the drug FOA (right panels). Plates were incubated at 22 or 30 °C as indicated.
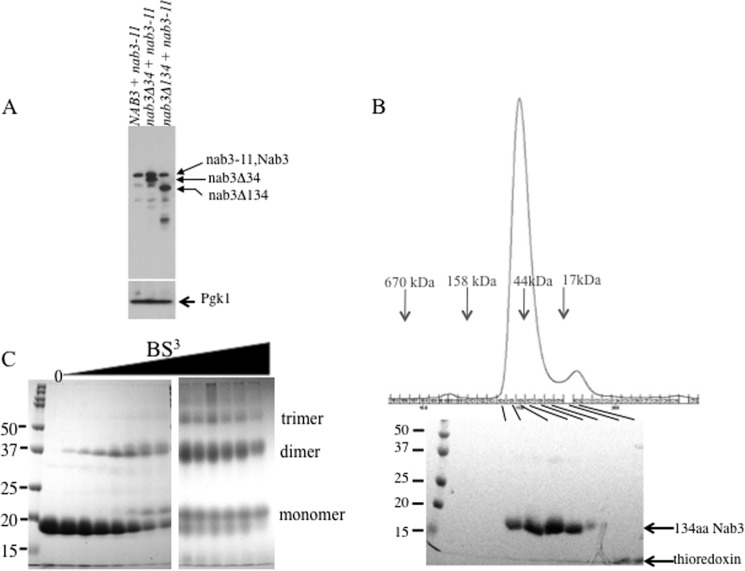
FIGURE 3.
A, expression of Nab3, Nab3-11, Nab3Δ34, and Nab3Δ134 proteins. Yeasts containing the indicated plasmid-borne versions of NAB3 were resolved by SDS-PAGE and processed for Western blotting using an anti-Nab3 monoclonal antibody. The migration positions of the respective proteins are indicated to the right. Note that wild type Nab3 and Nab3-11 proteins co-migrate. As a loading control, the lysate was probed with a monoclonal antibody against yeast 3-phosphoglycerate kinase (Pgk1). B, gel filtration and SDS-PAGE of the Nab3 134-amino acid protein. The recombinant C-terminal 134-amino acid (134aa) Nab3 peptide was purified and subjected to gel filtration on an S200 column as described under “Materials and Methods.” Protein fractions were run on an SDS gel with size markers. Note that a small amount of residual thioredoxin tag forms a small gel filtration peak and a band near the dye front of the SDS gel. C, cross-linking of the 134-amino acid peptide of Nab3. Purified recombinant peptide was incubated with increasing amounts of bis(sulfosuccinimidyl) suberate, reactions were quenched and prepared for SDS-PAGE. The following concentrations of bis(sulfosuccinimidyl) suberate (BS3) were used (from left to right after the reference marker lane): 0, 15 μm, 31 μm, 62 μm, 125 μm, 250 μm, 500 μm, 1 mm, 1.5 mm, 2 mm, 2.5 mm, 3 mm, 4 mm, and 5 mm. Samples were run on two separate gels of the same composition.
The development of this strain with both nab3 alleles allowed us to try to cure the nab3-11-containing plasmid marked with URA3, by using the counterselection agent 5-fluoroorotic acid to ask if yeast with either nab3Δ34 or nabΔ134 as the only version of NAB3 were viable. The strain with nab3Δ34 as its sole version of NAB3 grew, albeit slowly, at both the permissive and non-permissive temperatures, whereas cells with nab3Δ134 as their sole source of Nab3 protein could not grow at either the permissive or the restrictive temperatures (Fig. 2, right panels). We conclude that the terminal 134 amino acids of NAB3 are essential for yeast viability. Taken together with the ability of nab3Δ134 to rescue the nab3-11 allele, these findings support the idea that multiple copies of the Nab3 protein cooperate to provide a function important for normal yeast physiology, presumably by polymerizing on RNAs to be terminated.
To examine if the 134-amino acid peptide from the Nab3 C terminus was sufficient for self-assembly, it was produced in E. coli as a recombinant protein fused to the C terminus of thioredoxin-His6. The 134-amino acid Nab3 peptide (17 kDa) was proteolytically removed from the fusion protein, purified, and subjected to gel filtration. Its mobility was calculated to be 52 kDa relative to globular reference proteins. This apparent hydrodynamic size could be explained if it exists as an anomalously large dimer resulting from a relatively unfolded pair of partners (Fig. 3B). Alternatively, the molecules could represent a trimer.
Chemical cross-linking was previously used to show that a short coiled-coil segment of hnRNP-C and the homologous region of Nab3 could multimerize (16, 17). The 134-amino acid peptide was treated with increasing concentrations of bis(sulfosuccinimidyl) suberate, an amine-directed, bifunctional cross-linker, and separated on denaturing SDS-PAGE. Dimers were readily trapped by cross-linking, and small amounts of trimer could be observed at higher levels of cross-linking agent (Fig. 3C).
Perhaps due to its glutamine richness (53%), the C-terminal 34-amino acid peptide of Nab3 demonstrated unusual behaviors, such as migrating unusually slowly in SDS-PAGE (16) and staining poorly with Coomassie Brilliant Blue (data not shown). Full-length Nab3 in yeast lysates and a number of human hnRNP proteins also migrate anomalously slowly, which could be due in part to their low complexity regions, which may be unstructured and extended (6). Because the essential 134-amino acid piece of Nab3 contains multiple stretches of consecutive glutamines (two Gln3 stretches, two Gln6 stretches, and the Gln16 stretch) and 16 prolines and is poor in aromatic amino acids (two tyrosines, no phenylalanine or tryptophan), we pursued the hypothesis that this region is relatively unstructured. If so, the 134-amino acid Nab3 peptide might remain soluble following heating (37). The isolated peptide was heated to 100 °C for 45 min. Soluble protein (S) was separated from denatured material (P) by centrifugation and analyzed by SDS-PAGE. The 134-amino acid Nab3 peptide remained entirely soluble following this treatment (Fig. 4A). In contrast, the identical treatment of a control protein, deoxyribonuclease I, rendered it completely insoluble. This is consistent with the proposal that this part of Nab3 lacks a complex packing of secondary structural elements.
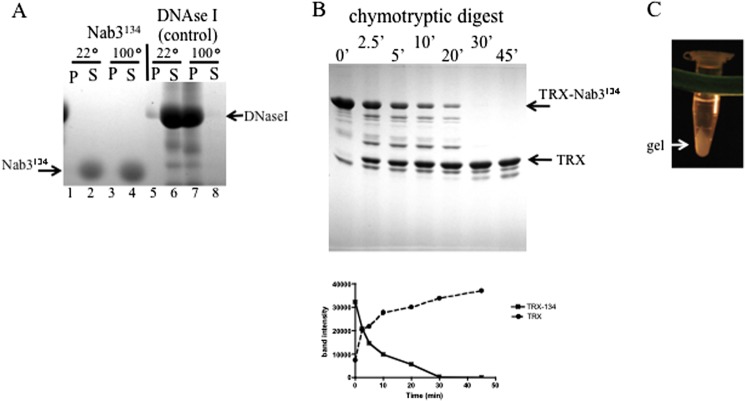
FIGURE 4.
Characterization of the 134-amino acid Nab3 peptide. A, heat stability. Forty μg of isolated Nab3 134-amino acid peptide or deoxyribonuclease I (Stratagene) were incubated at 22 or 100 °C for 45 min and separated into insoluble (P) and soluble (S) fractions by centrifugation. Samples were resolved on 11% SDS-polyacrylamide gels and stained with Coomassie Blue. B, time course of chymotryptic digestion of the Nab3 134-amino acid peptide fused to thioredoxin (TRX-Nab3134). Chymotrypsin was incubated with the target protein at a molar ratio of 1:400 for the indicated times before reactions were stopped and resolved on SDS-PAGE. Graphed below is a quantification of the stained bands using ImageJ software, showing loss of the full-length fusion protein (TRX-Nab3134) and accumulation of the thioredoxin tag (TRX) over time. C, gel formation. After incubation of purified Nab3134 at 4 °C, a hydrogel could be detected with the naked eye, and it is shown here after pelleting in a microcentrifuge.
We also used limited proteolysis to probe the unfolded nature of this region of Nab3 because unstructured domains show more susceptibility to hydrolysis than stably folded secondary structures (28, 38, 39). A recombinant fusion protein bearing the 134-amino acid Nab3 peptide was expressed and purified from E. coli, treated with chymotrypsin, and analyzed by SDS-PAGE. The Nab3 extension was digested off of the thioredoxin tag (∼14 kDa) with a half-life of ∼5 min, whereas the thioredoxin tag and a control protein (DNase I) were stable to digestion for at least 45 min (Fig. 4B) (data not shown). Similarly, the isolated Nab3134 peptide was hydrolyzed to completion more rapidly than its purified fusion partner thioredoxin when separately exposed to chymotrypsin (data not shown).
Regions of mammalian RNA binding proteins with low sequence complexity, such as one found in hnRNPA2, have been shown to undergo a phase transition at high concentration to a hydrogel-like state (27). The purified Nab3134 protein demonstrated this phenomenon when concentrated and incubated at 4 °C (Fig. 4C). This provided additional evidence for self-assembly and the hnRNP-like character of Nab3.
New Genetic Evidence for a Network of Interactions between Nrd1, Nab3, and RNA Polymerase II
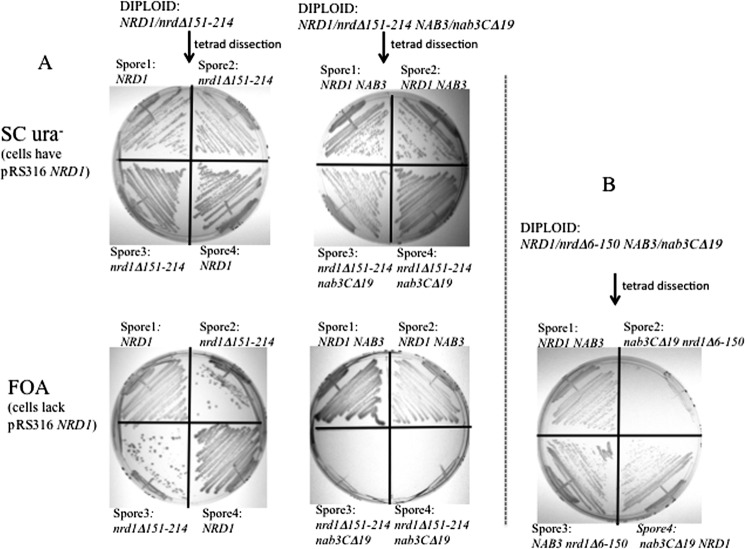
A number of interactions are important for Nrd1-Nab3-based termination, including the interactions between each of their RRM domains and the nascent transcript (Fig. 1A, circled number 1), the dimerization of these two hnRNP-like proteins through mutual contact surfaces (Fig. 1A, circled number 2), the interaction of Nrd1 with the phosphorylated C-terminal domain (CTD) of RNA polymerase II (Fig. 1A, circled number 3), and the self-assembly of Nab3 through its C-terminal tail (Fig. 1A, circled number 4). Mutations in these functions have been characterized, almost all of which lead to growth defects on their own (12, 13, 16). Apparently, the network of interactions needed for termination is redundant enough to sustain mutagenic insult. We reasoned that introducing more than one of these mutations in the same cell would be additive and potentially lethal; thus, we combined them pairwise via genetic crosses to test for synthetic lethality. First, a strain that we isolated through a screen for terminator override was examined. It has a chromosomal NAB3 mutation that encodes a termination-defective version of Nab3 lacking its 19 C-terminal amino acids (Nab3CΔ19), which is part of its self-association and hnRNP-C-like domain (Fig. 1B). It was mated to a pair of otherwise isogenic strains containing either 1) a small deletion of the Nab3-binding domain of Nrd1 (Δ151–214) or a small deletion of the RNA polymerase II CTD-binding domain of Nrd1 (Δ6–150). Both nrd1 mutations result in defects in the biogenesis of yeast snRNA as described previously by Buratowski and co-workers (13).
The diploids NAB3/NAB3 NRD1/nrd1Δ151–214 and NAB3/nab3CΔ19 NRD1/nrd1Δ151–214 were sporulated to obtain haploid cells with either the nrd1 deletion alone or both the nrd1 and nab3 mutations. Results from more than 20 tetrads showed that spores with the nrd1Δ151–214 allele were unable to germinate to form colonies even when NAB3 was wild type (data not shown). To overcome this technical difficulty, diploids were transformed with a plasmid-borne copy of NRD1 to cover the nrd1Δ151–214 defect. Such a strain could yield spores (Fig. 5A, top panels). The resulting haploids were subsequently plated on FOA-containing medium that selects for cells lacking the URA3-marked plasmid containing NRD1 used to rescue germination. In the absence of wild type NRD1, doubly mutant nrd1− nab3− spores were inviable (Fig. 5A, bottom middle panel), whereas haploid spores with only the nrd1 mutation were able to grow (Fig. 5A, bottom left panel). This showed that retention of the NRD1-containing plasmid is essential for these double mutants and that the combination of these nrd1 and nab3 mutations was lethal.
FIGURE 5.
Synthetic lethality of a nab3 tov allele (nab3CΔ19) and mutant nrd1 alleles. A, a yeast strain with the nrd1Δ151–214 (Nab3-binding domain) deletion was mated to the control strain BY4742 or an otherwise isogenic strain with the nab3CΔ19 mutation. The resulting diploids were transformed with a URA3-marked plasmid bearing wild type NRD1, sporulated, and dissected. The resulting four sister spores from a tetrad were genotyped and plated on SCura− or FOA-containing medium at 30 °C. The result from the NRD1/nrd1Δ151–214 diploid's dissection are shown in the leftmost top and bottom panels. The result from the NRD1/nrd1Δ151–214 NAB3/nab3CΔ19 diploid's dissection are shown in the right top and bottom panels of A. B, a yeast strain with the nrd1Δ6–150 (polymerase II CTD-binding domain) deletion was mated to an otherwise isogenic strain with the nab3CΔ19 mutation. Diploids were transformed with a URA3-marked plasmid bearing wild type NRD1, sporulated, and dissected. Four sister spores from the NRD1/nrd1Δ6–150 NAB3/nab3CΔ19 diploid were genotyped and plated on FOA-containing medium. A spore with both mutations failed to grow on FOA (top right quadrant). Sister spores with either mutant allele alone are viable.
Similarly, when the strain with the deletion that removes the Nrd1 CTD interaction domain (Δ6–150) was mated to the nab3CΔ19 mutant, only the spore that had both mutations failed to grow without plasmid-borne NRD1 (Fig. 5B), whereas sister spores that were wild type for NRD1 and NAB3 or those with mutations in either NRD1 or NAB3 could grow on FOA. These results suggest that the loss of either of two different functions of Nrd1, in conjunction with a version of Nab3 lacking its self-association domain, cripples the complex to the point of inviability.
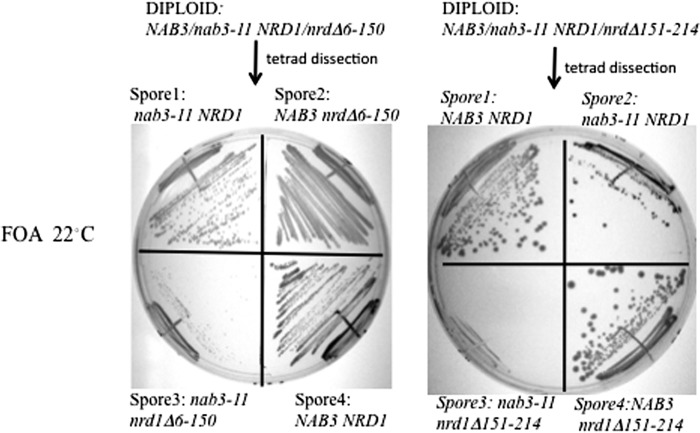
A similar pair of matings were performed between strains with the NRD1 alleles above mated to a different nab3 mutant (i.e. a previously characterized strain (nab3-11) that contains a temperature-sensitive mutation in the Nab3 RRM domain (12)). This resulted in another set of nrd1− nab3− strains in which the nab3 mutation targets a functional region of the Nab3 protein distinct from the self-assembly domain. Diploids were again transformed with a plasmid bearing NRD1 in order to facilitate germination of nrd1Δ151–214-containing spores. The resulting haploids were grown at the permissive temperature of 22 °C, which supports growth of cells with the nab3-11 allele. Spores with either the nrd1 or the nab3 mutation grew on FOA-containing medium, which selects against plasmid-borne wild type NRD1, but cells with both nab3-11 and nrd1Δ151–214 were inviable (Fig. 6, right). Cells with the nab3-11 and nrd1Δ6–150 combination were very sick, taking a number of days to yield colonies (Fig. 6, left). These synthetic genetic effects emphasize the importance of both Nab3 and Nrd1 function for an effective termination complex.
FIGURE 6.
Synthetic genetic effect of nrd1 alleles with the nab3-11 allele. A strain with a nab3-11 chromosomal allele was mated to a strain with either the nrd1Δ6–150 (left) or the nrd1Δ151–214 (right) chromosomal allele. The resulting diploids were transformed with a URA3-marked plasmid containing NRD1, sporulated, and plated onto FOA-containing medium at 22 °C. The growth phenotype for a set of tetrad sibs is shown for each with the relevant NAB3 and NRD1 genotypes indicated. Cells with the nab3-11 and nrd1Δ6–150 alleles were very slow growing (Spore3, left), and cells with both the nab3-11 and nrd1Δ151–214 mutations were not viable (Spore3, right).
The above genetic analyses tested the viability of doubly mutant cells when two different genes in the interaction network were defective. We next tested a mutant strain that lost two different functions in a single gene. It was previously shown that when two mutations in NRD1 (the aforementioned deletion alleles, Δ6–150 and Δ151–214) were present in a single copy of the gene, the double mutant could not support viability (13), suggesting that loss of both the CTD interaction and Nab3 interaction activities of Nrd1 was lethal. In this vein, we made a double mutant of NAB3 from the originally identified nab3 allele that lost its self-association domain (nab3CΔ19) and the temperature-sensitive RRM mutation (nab3-11). A LEU2-marked plasmid with these changes was transformed into a strain lacking the chromosomal copy of NAB3 but which contained a plasmid with a rescuing version of the NAB3 on a URA3-marked plasmid. At the permissive temperature, this strain's growth was comparable with that bearing only the nab3-11 mutation (data not shown). This was the only double mutant of the set that failed to show a synthetic phenotype. The results of the double mutant crosses are summarized in Table 2.
TABLE 2.
Double mutant combinations
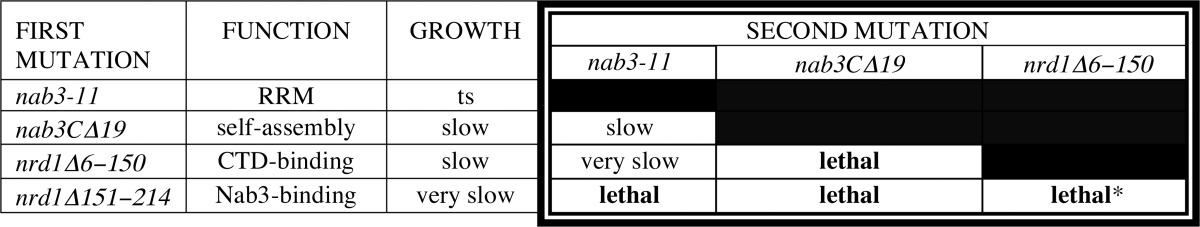
* From Vasiljeva et al. (13).
DISCUSSION
Based upon the cooperative interaction of the Nrd1-Nab3 heterodimer with target RNA as well as the known association of Nrd1 with RNA polymerase II, Corden and co-workers (1) postulated that termination involves assembly of a nucleoprotein complex on nascent RNA in order to achieve efficiency and specificity. We previously expanded this set of interactions by showing the functional importance of a small C-terminal Nab3 domain with self-assembly properties (15, 16). Here we elaborate on the Nab3 self-assembly domain's biological and physical functions and test the network of interactions that stabilize the complex by generating yeast strains with compound pairwise mutations in the complex's constellation of linkages.
Our findings emphasize that there are multiple ways in which the Nrd1-Nab3 dimer can be brought to the elongation complex in preparation for termination. These include the independent recognition of RNA by each of the proteins, recognition of polymerase by Nrd1, association of the proteins with each other, and self-association of Nab3 through its C-terminal tail (Fig. 1A). This complex network of interactions could help explain why the number, spacing, and density of Nab3 or Nrd1 recognition sites on nascent transcripts is variable between different substrate RNAs (1). Prior genetic studies have shown that crippling any one of these steps negatively impacts cell growth. Three of the nrd1-nab3 double mutant strains constructed here were not viable, and one yielded viable but very slow growing cells (Table 2). The one compound mutant that failed to show a synthetic phenotype was that harboring a doubly mutant nab3 gene containing the independently isolated RRM and self-assembly region mutations. Because the RRM mutation (nab3-11) is a tight temperature-sensitive allele, any synthetic effect could only be observed at the permissive temperature. Failure to find an exacerbation of this phenotype in the double mutant could be due to the mild impact of the nab3-11 mutation at the permissive temperature. Nevertheless, other mutations paired with nab3-11 did reveal synthetic effects, indicating that the simultaneous loss of the self-assembly and RNA binding capabilities of Nab3 has a reduced consequence in the spectrum of paired defects made to the Nrd1-Nab3 system. A more quantitative dissection of the interaction network's function must await a biochemical reconstitution of a nucleoprotein complex with full-length proteins and target RNA. It should be noted that the mutations studied here did not result in substantial losses in steady state levels of either the Nrd1 or Nab3 proteins, excluding the possibility that their cellular concentrations were impaired by these mutations.
Loss of the Nrd1 domain that interacts with Nab3 (Δ151–214; generated by Buratowski and co-workers (13)) resulted in a particularly severe phenotype. Cells with the mutation grew slowly, and pairing it with the RRM mutation or self-assembly mutation of Nab3 was lethal. In the strain used here, diploids bearing the mutation could form an ascus, but the resulting spores that received this allele failed to germinate (data not shown). This is notable because cells lacking TRF4, which encodes a component of the TRAMP complex involved in termination and degradation of the IMD2 cryptic unstable RNA as well as other cryptic unstable RNAs (15), are also defective in spore germination (40), suggesting a role for the Nab3-Nrd1 interaction in the emergence from meiosis. High copy suppression of a nrd1 allele by NAB3 provided early genetic evidence in support of the biological importance of the Nrd1-Nab3 interaction (12).
Both mutations in NRD1 studied here were lethal when combined with the nab3CΔ19 allele that removes the Nab3 self-assembly domain, adding to the evidence that this part of Nab3 is important. Interestingly, Nrd1 also contains a polyglutamine stretch (Gln8) at its C terminus (residues 567–574 of the 575-amino acid protein), and it is predicted to lie within an aggregation-prone region, similar to the Gln16 stretch of Nab3 studied previously and in this report (15, 16, 25). Loss of the terminal 60 amino acids of Nrd1, including this Gln8 tract, however, did not reveal a termination defect at the IMD2 cryptic unstable RNA, so this polyglutamine stretch does not appear to play a significant role in termination at this locus (data not shown). It is worth noting that appending a TAP tag to the C terminus of Nrd1 or Nab3 partially compromised the termination activity of each, suggesting that the addition of this relatively large extension to either protein results in steric interference that compromises their function (data not shown).
The important biological role of the curious low complexity sequences found in many RNA-binding proteins, including Nab3, has gained recent recognition (26, 27). One function of the low complexity regions is their potential to form higher order self-assemblies while escorting RNA around the cell. Indeed, low complexity sequences are associated with intrinsically disordered regions that have the propensity to form amyloid-like aggregates, including those with the potential to cause disease (25, 29). Polyglutamine stretches are often found adjacent to α-helices in domains that can multimerize, and this juxtaposition is associated with the protein's solubility, aggregation, and function (32) We found that such a region of Nab3 has properties of a class of domains/proteins categorized as intrinsically disordered, including 1) protease sensitivity, 2) solubility after heating, 3) anomalous hydrodynamic mobility, 4) anomalous migration on denaturing gels, and 5) a high score when analyzed by an algorithm trained to distinguish order from disorder in polypeptides. Intrinsically disordered proteins can undergo a transition to an ordered state upon binding to other subunits of a complex (41). Thus, we propose that this region of Nab3 in isolation is significantly unstructured, with the notable exception of the last 18 residues, which bear structural homology to a coiled coil-forming α-helix of hnRNP-C (Fig. 1, circled number 4), but it has the potential to become structured. The ability of this domain to form a hydrogel, as seen for a number of other unstructured low complexity domains, is thought to reflect such a potential to acquire structure. In this case, it would be in the context of Nrd1, RNA polymerase II, and nascent transcript in preparation for termination. This essential segment of Nab3 looks to be important for the protein's ability to achieve an assembly state with a specific three-dimensional configuration. Such a geometry appears important for termination and downstream RNA processing steps, including the recruitment of the Sen1 helicase, the TRAMP complex, and the nuclear exosome.
Acknowledgments
We thank Drs. Tanya Chernova, Xiaodong Cheng, and Charlie Moran for helpful suggestions; Anita Corbett and Steve Buratowski for the gift of yeast strains; and Maurice Swanson for anti-Nab3 antibody. We thank Dr. G. Conn and the Conn laboratory for assistance and for use of the AKTA chromatography system.
This work was supported, in whole or in part, by National Institutes of Health, NIGMS, Grant R01GM46331.
- RRM
- RNA recognition motif
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- CTD
- C-terminal domain
- SC
- synthetic complete
- FOA
- 5-fluoroorotic acid.
REFERENCES
- 1. Carroll K. L., Ghirlando R., Ames J. M., Corden J. L. (2007) Interaction of yeast RNA-binding proteins Nrd1 and Nab3 with RNA polymerase II terminator elements. RNA 13, 361–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Steinmetz E. J., Conrad N. K., Brow D. A., Corden J. L. (2001) RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413, 327–331 [DOI] [PubMed] [Google Scholar]
- 3. Arigo J. T., Eyler D. E., Carroll K. L., Corden J. L. (2006) Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol. Cell 23, 841–851 [DOI] [PubMed] [Google Scholar]
- 4. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim M., Vasiljeva L., Rando O. J., Zhelkovsky A., Moore C., Buratowski S. (2006) Distinct pathways for snoRNA and mRNA termination. Mol. Cell 24, 723–734 [DOI] [PubMed] [Google Scholar]
- 6. Wilson S. M., Datar K. V., Paddy M. R., Swedlow J. R., Swanson M. S. (1994) Characterization of nuclear polyadenylated RNA-binding proteins in Saccharomyces cerevisiae. J. Cell Biol. 127, 1173–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carroll K. L., Pradhan D. A., Granek J. A., Clarke N. D., Corden J. L. (2004) Identification of cis elements directing termination of yeast nonpolyadenylated snoRNA transcripts. Mol. Cell. Biol. 24, 6241–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Creamer T. J., Darby M. M., Jamonnak N., Schaughency P., Hao H., Wheelan S. J., Corden J. L. (2011) Transcriptome-wide binding sites for components of the Saccharomyces cerevisiae non-poly(A) termination pathway. Nrd1, Nab3, and Sen1. PLoS Genet. 7, e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Riordan D. P., Herschlag D., Brown P. O. (2011) Identification of RNA recognition elements in the Saccharomyces cerevisiae transcriptome. Nucleic Acids Res. 39, 1501–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Porrua O., Hobor F., Boulay J., Kubicek K., D'Aubenton-Carafa Y., Gudipati R. K., Stefl R., Libri D. (2012) In vivo SELEX reveals novel sequence and structural determinants of Nrd1-Nab3-Sen1-dependent transcription termination. EMBO J. 31, 3935–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hobor F., Pergoli R., Kubicek K., Hrossova D., Bacikova V., Zimmermann M., Pasulka J., Hofr C., Vanacova S., Stefl R. (2011) Recognition of transcription termination signal by the nuclear polyadenylated RNA-binding (NAB) 3 protein. J. Biol. Chem. 286, 3645–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Conrad N. K., Wilson S. M., Steinmetz E. J., Patturajan M., Brow D. A., Swanson M. S., Corden J. L. (2000) A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics 154, 557–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vasiljeva L., Kim M., Mutschler H., Buratowski S., Meinhart A. (2008) The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 15, 795–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lunde B. M., Hörner M., Meinhart A. (2011) Structural insights into cis element recognition of non-polyadenylated RNAs by the Nab3-RRM. Nucleic Acids Res. 39, 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loya T. J., O'Rourke T. W., Reines D. (2012) A genetic screen for terminator function in yeast identifies a role for a new functional domain in termination factor Nab3. Nucleic Acids Res. 40, 7476–7491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Loya T. J., O'Rourke T. W., Reines D. (2013) Yeast Nab3 protein contains a self-assembly domain found in human heterogeneous nuclear ribonucleoprotein-C (hnRNP-C) that is necessary for transcription termination. J. Biol. Chem. 288, 2111–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Whitson S. R., LeStourgeon W. M., Krezel A. M. (2005) Solution structure of the symmetric coiled coil tetramer formed by the oligomerization domain of hnRNP C. Implications for biological function. J. Mol. Biol. 350, 319–337 [DOI] [PubMed] [Google Scholar]
- 18. Steinmetz E. J., Brow D. A. (1996) Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 16, 6993–7003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vasiljeva L., Buratowski S. (2006) Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol. Cell 21, 239–248 [DOI] [PubMed] [Google Scholar]
- 20. Nedea E., Nalbant D., Xia D., Theoharis N. T., Suter B., Richardson C. J., Tatchell K., Kislinger T., Greenblatt J. F., Nagy P. L. (2008) The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol. Cell 29, 577–587 [DOI] [PubMed] [Google Scholar]
- 21. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 22. Beyer A. L., Christensen M. E., Walker B. W., LeStourgeon W. M. (1977) Identification and characterization of the packaging proteins of core 40S hnRNP particles. Cell 11, 127–138 [DOI] [PubMed] [Google Scholar]
- 23. Huang M., Rech J. E., Northington S. J., Flicker P. F., Mayeda A., Krainer A. R., LeStourgeon W. M. (1994) The C-protein tetramer binds 230 to 240 nucleotides of pre-mRNA and nucleates the assembly of 40S heterogeneous nuclear ribonucleoprotein particles. Mol. Cell. Biol. 14, 518–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barnett S. F., Friedman D. L., LeStourgeon W. M. (1989) The C proteins of HeLa 40S nuclear ribonucleoprotein particles exist as anisotropic tetramers of (C1)3 C2. Mol. Cell. Biol. 9, 492–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Alberti S., Halfmann R., King O., Kapila A., Lindquist S. (2009) A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137, 146–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han T. W., Kato M., Xie S., Wu L. C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., McKnight S. L. (2012) Cell-free formation of RNA granules. Bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 [DOI] [PubMed] [Google Scholar]
- 27. Kato M., Han T. W., Xie S., Shi K., Du X., Wu L. C., Mirzaei H., Goldsmith E. J., Longgood J., Pei J., Grishin N. V., Frantz D. E., Schneider J. W., Chen S., Li L., Sawaya M. R., Eisenberg D., Tycko R., McKnight S. L. (2012) Cell-free formation of RNA granules. Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thual C., Komar A. A., Bousset L., Fernandez-Bellot E., Cullin C., Melki R. (1999) Structural characterization of Saccharomyces cerevisiae prion-like protein Ure2. J. Biol. Chem. 274, 13666–13674 [DOI] [PubMed] [Google Scholar]
- 29. Kim H. J., Kim N. C., Wang Y. D., Scarborough E. A., Moore J., Diaz Z., MacLea K. S., Freibaum B., Li S., Molliex A., Kanagaraj A. P., Carter R., Boylan K. B., Wojtas A. M., Rademakers R., Pinkus J. L., Greenberg S. A., Trojanowski J. Q., Traynor B. J., Smith B. N., Topp S., Gkazi A. S., Miller J., Shaw C. E., Kottlors M., Kirschner J., Pestronk A., Li Y. R., Ford A. F., Gitler A. D., Benatar M., King O. D., Kimonis V. E., Ross E. D., Weihl C. C., Shorter J., Taylor J. P. (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Orr H. T. (2012) Polyglutamine neurodegeneration. Expanded glutamines enhance native functions. Curr. Opin. Genet. Dev. 22, 251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim M. W., Chelliah Y., Kim S. W., Otwinowski Z., Bezprozvanny I. (2009) Secondary structure of Huntingtin amino-terminal region. Structure 17, 1205–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fiumara F., Fioriti L., Kandel E. R., Hendrickson W. A. (2010) Essential role of coiled coils for aggregation and activity of Q/N-rich prions and PolyQ proteins. Cell 143, 1121–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schaefer M. H., Wanker E. E., Andrade-Navarro M. A. (2012) Evolution and function of CAG/polyglutamine repeats in protein-protein interaction networks. Nucleic Acids Res. 40, 4273–4287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Petrakis S., Schaefer M. H., Wanker E. E., Andrade-Navarro M. A. (2013) Aggregation of polyQ-extended proteins is promoted by interaction with their natural coiled-coil partners. BioEssays 35, 503–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reines D., Clarke M. (1985) Immunochemical analysis of the supramolecular structure of myosin in contractile cytoskeletons of Dictyostelium amoebae. J. Biol. Chem. 260, 14248–14254 [PubMed] [Google Scholar]
- 36. Romero P., Obradovic Z., Li X., Garner E. C., Brown C. J., Dunker A. K. (2001) Sequence complexity of disordered protein. Proteins 42, 38–48 [DOI] [PubMed] [Google Scholar]
- 37. Galea C. A., Pagala V. R., Obenauer J. C., Park C. G., Slaughter C. A., Kriwacki R. W. (2006) Proteomic studies of the intrinsically unstructured mammalian proteome. J. Proteome Res. 5, 2839–2848 [DOI] [PubMed] [Google Scholar]
- 38. Johnson D. E., Xue B., Sickmeier M. D., Meng J., Cortese M. S., Oldfield C. J., Le Gall T., Dunker A. K., Uversky V. N. (2012) High-throughput characterization of intrinsic disorder in proteins from the Protein Structure Initiative. J. Struct. Biol. 180, 201–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hubbard S. J. (1998) The structural aspects of limited proteolysis of native proteins. Biochim. Biophys. Acta 1382, 191–206 [DOI] [PubMed] [Google Scholar]
- 40. Kloimwieder A., Winston F. (2011) A screen for germination mutants in Saccharomyces cerevisiae. G3 1, 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wright P. E., Dyson H. J. (2009) Linking folding and binding. Curr. Opin. Struct. Biol. 19, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]