Background: Homologous recombination is regulated both positively and negatively in eukaryotic cells to suppress genomic instability.
Results: FBH1 can disrupt RAD51 filaments in vitro and suppresses formation of spontaneous RAD51 foci in mammalian cells. In cells defective for FBH1, hyper-recombination is observed.
Conclusion: FBH1 is a negative regulator of homologous recombination.
Significance: RAD51 activity must be carefully controlled to preserve genomic integrity.
Keywords: DNA Damage, DNA Helicase, DNA Recombination, DNA Repair, DNA Replication
Abstract
Efficient repair of DNA double strand breaks and interstrand cross-links requires the homologous recombination (HR) pathway, a potentially error-free process that utilizes a homologous sequence as a repair template. A key player in HR is RAD51, the eukaryotic ortholog of bacterial RecA protein. RAD51 can polymerize on DNA to form a nucleoprotein filament that facilitates both the search for the homologous DNA sequences and the subsequent DNA strand invasion required to initiate HR. Because of its pivotal role in HR, RAD51 is subject to numerous positive and negative regulatory influences. Using a combination of molecular genetic, biochemical, and single-molecule biophysical techniques, we provide mechanistic insight into the mode of action of the FBH1 helicase as a regulator of RAD51-dependent HR in mammalian cells. We show that FBH1 binds directly to RAD51 and is able to disrupt RAD51 filaments on DNA through its ssDNA translocase function. Consistent with this, a mutant mouse embryonic stem cell line with a deletion in the FBH1 helicase domain fails to limit RAD51 chromatin association and shows hyper-recombination. Our data are consistent with FBH1 restraining RAD51 DNA binding under unperturbed growth conditions to prevent unwanted or unscheduled DNA recombination.
Introduction
Homologous recombination (HR)5 is an evolutionarily conserved pathway for the repair of DNA double strand breaks, DNA interstrand cross-links, and collapsed replication forks. DNA repair mediated by HR requires a homologous template, which in proliferating cells is generally the sister chromatid generated in S-phase (1). In diploid organisms, HR can also utilize the homologous chromosome as a template, but this is disfavored wherever possible because it can lead to loss of heterozygosity (LOH), a potential driver of tumorigenesis (2). Indeed, many of the homozygous mutations found in tumor suppressor genes in neoplastic cells probably arose via LOH following an initial, monoallelic mutational event. A pivotal player in HR is the RAD51 protein (RecA in bacteria), a DNA-dependent ATPase that polymerizes on ssDNA to form a nucleoprotein filament. This structure is a central intermediate in HR and is necessary for DNA strand invasion to occur as part of the search for a homologous DNA sequence (1, 3).
Because of the potential for HR to generate LOH or other forms of genome rearrangements, it is critical that cells carefully regulate the process. Several factors have been identified that actively assist RAD51 at the DNA strand invasion step of HR, including BRCA2, RAD54, and the so-called RAD51 paralogs in eukaryotes (4–6). Similarly, RAD51 is subject to negative regulation in order to prevent inadvertent or unwanted HR reactions. One form of regulation is via the disruption of RAD51 filaments. The best characterized factor to perform this task is the yeast Srs2 protein, a superfamily 1 DNA helicase belonging to the highly conserved UvrD family (7). Srs2 utilizes the energy derived from ATP hydrolysis to translocate on ssDNA, a process that serves to progressively disrupt the Rad51 filament (8). Srs2 also binds directly to yeast Rad51. Moreover, genetic and biochemical evidence in yeast is fully consistent with Srs2 playing a role in Rad51 filament disruption and in restraining HR in the cell (9, 10).
In addition to Srs2, there are other UvrD family members expressed in eukaryotic cells. A recently identified family member in human cells is PARI (11). This protein also binds to RAD51, but it contains amino acid substitutions in critical residues required for ATPase function, and hence PARI is not an active helicase/translocase. How PARI regulates RAD51 function at a mechanistic level is not known. Another eukaryotic UvrD family member is the F-box helicase 1 (FBH1), which is found in fission yeast and higher eukaryotes but not in budding yeast (12). FBH1 is a bifunctional protein comprising an N-terminal F-box domain and a centrally located helicase domain. Previous biochemical studies have indicated that human FBH1 is an active DNA helicase/translocase (12). The F-box domain is involved in the post-translational modification of proteins as part of the so-called SCF complex (Skp1-Cul1-Fbh1), which serves as an E3 ubiquitin ligase (13). The targets for the FBH1 F-box in human cells remain to be identified.
Genetic studies in the fission yeast, S. pombe, have provided strong evidence that Fbh1 regulates Rad51 function in that organism. Deletion of S. pombe fbh1 leads to mild sensitivity to certain DNA-damaging agents as well as to the formation of spontaneous foci containing Rad51 (called Rhp51 in S. pombe) (14, 15). Fbh1 is essential for viability in the absence of anti-recombinases, such as Rqh1 and Srs2, and this synthetic lethality can be suppressed by inactivation of HR factors that promote presynaptic filament assembly (14). Moreover, Fbh1 limits Rad51-dependent recombination at blocked replication forks in a manner dependent on its helicase/translocase activity (16). Taken together, these findings suggest that Fbh1 regulates HR through directly regulating Rad51 function, at least in S. pombe.
Disruption of the Fbh1 gene in the chicken DT40 cell line leads to elevated levels of sister chromatid exchange, a marker of crossover during HR-mediated DNA repair (17). In contrast, mouse Fbh1-deficient cells display no clear defect in HR-mediated repair of DSBs but do show a moderate increase in RAD51 focus formation at sites of DNA damage (17, 18). In human cells, FBH1 accumulates at sites of DNA damage and replication stress in a manner dependent on its helicase activity (19).
In order to gain insight into the role of FBH1 in mammalian cells, we have taken a combined biochemical, biophysical, and molecular genetic approach. We show that purified human FBH1 is able to bind directly to RAD51 and to disrupt RAD51 filaments. Consistent with this, analysis of a mouse cell line with a defective FBH1 helicase indicates that the association of RAD51 with chromatin is normally restrained by FBH1. Our data provide mechanistic insight into the role of FBH1 as a negative regulator of RAD51 function in order to prevent unwanted HR.
EXPERIMENTAL PROCEDURES
Propagation of Mouse ES Cells
A totipotent, adherent mouse ES cell line, designated R1, was obtained from Yusa et al. (20). A derivative of R1, containing both copies of the Fbh1 gene inactivated, was generated by the use two targeting vectors differing only in their drug resistance cassette. These vectors were designed to delete helicase domains II and III, along with the insertion of either a puromycin (pur)- or neomycin (neo)-resistant gene plus an HSV domain. Confluent Buffalo rat liver (BRL) cells were grown for 7 days at 37 °C in a humidified atmosphere containing 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), 2.4 mm glutamine, and non-essential amino acids. The medium was collected and used as conditioned medium for ES cell culture. ES cells were grown in 50% DMEM and 50% medium conditioned by BRL cells, supplemented with 10% FCS, 2.4 mm glutamine, 1,000 units/ml murine leukemia inhibitory factor required by ES cells to maintain their pluripotent state, non-essential amino acids, and 0.1% β-mercaptoethanol.
Cell Cycle Analysis
Cells were treated with 10 μg/ml BrdU for 20 min. Then cells were harvested and fixed in 70% ethanol. Fixed cells were stained with BrdU antibody (1:10; BD Biosciences), 40 μg/ml propidium iodide, and 100 μg/ml RNase A in PBS. Cells were then labeled with secondary anti-mouse AF488 antibody (1:200; Invitrogen). Cell cycle distributions and BrdU population were measured using a FACSCalibur flow cytometer (BD Biosciences).
Clonogenic Survival Assays
The sensitivity of mouse ES cells to increasing doses of DNA-damaging agents was determined using a clonogenic assay. Cells were trypsinized, counted, and plated into 60-mm dishes coated with 0.1% porcine skin type A gelatin at a density of 1,000 cells/dish. Incubation was for a further 24 h, following which the cells were either exposed to ionizing radiation or treated with drug-containing medium for 2 h (cisplatin) or 24 h (camptothecin). After the treatment period, cells were washed in PBS and then grown in fresh medium. Incubation was for 6–8 days, following which cells were washed in PBS, fixed, and stained with crystal violet. Colonies were then counted using a Stuart Scientific Colony Counter. Colonies containing more than 50 cells were counted as survivors, and the number of colonies at each time point or drug dose was expressed as a percentage of the colony number in the untreated control. All time points were performed in triplicate, and the experiment was repeated three times.
Sister Chromatid Exchange (SCE) Analysis
Mouse ES cells were grown in medium containing 10 μg/ml BrdU for 24 h. 100 ng/ml KaryoMAX Colcemid was added for the final 30 min of the treatment period. The cells were then washed in PBS and harvested by trypsinization. Cells were swelled in hypotonic buffer containing 50 mm KCl before being incubated at 37 °C for 30 min. Fixing was in freshly made ice-cold fixative solution (methanol/acetic acid, 3:1), and fixed cells were stored at −20 °C. To prepare the slides, the stored cells were resuspended in 1 ml of fresh, ice-cold fixative solution, and single droplets from a Gilson P200 pipette were dropped from a height of ∼30 cm onto glass coverslips, followed by incubation overnight in the dark at 42 °C. The slides were then stained with 20 μg/ml bis-benzimide for 30 min, followed by exposure to 254-nm UV light in phosphate buffer, pH 6.8, for 3 h. The slides were then incubated in prewarmed 2× SSC at 65 °C for 2 h before staining with 0.8% MSG Giemsa for 3.5 min. Slides were scored blindly using a Nikon Eclipse 80i microscope equipped with Lucia G software. 800–1,000 metaphase chromosomes/cell line were examined, and an unpaired t test was used to determine the significance in any difference in the distribution of SCEs.
Immunofluorescence Analysis
Cells were seeded onto glass coverslips coated with 0.5% gelatin at ∼25% confluence. 24 h later, cells were left untreated or were treated with 100 nm camptothecin and incubated at 37 °C. Cells were then fixed in 4% paraformaldehyde in PBS for 10 min, washed in PBS, and stored at 4 °C. After permeabilization with 0.1% Triton X-100 in PBS, cells were washed and exposed to the primary antibody (anti-Rad51 (1:5,000) (kindly provided by Dr. R. Kanaar) or anti-γH2AX (1:500) (Upstate)) for 16 h. Following this, the samples were washed with 0.1% Triton X-100 in PBS and then exposed for 30 min to secondary antibody. After final washes in 0.1% Triton X-100 in PBS, the nuclei were counterstained with DAPI Vectashields® before the coverslips were sealed onto glass slides. Nuclear staining patterns were visualized with a confocal laser-scanning microscope (Zeiss LSM 510 Meta), and images were stored and analyzed using Zeiss LSM Image Browser software. Scoring was done either blindly by counting the number of nuclear foci or alternatively by using ImageJ software to measure the relative total nuclear fluorescence from RAD51 staining. The differences between the cell lines were evaluated statistically using unpaired t tests.
DR-GFP Assay
The puromycin-resistant gene in the DR-GFP plasmid was excised using EcoRV and BspEI and blunt end-ligated with a hygromycin-resistant gene under the control of a PGK promotor to generate the DR-GFP-Hyg plasmid. Mouse ES cells were electroporated with the DR-GFP-Hyg by using a Bio-Rad electroporator (GenePulser XcellTM) at a single pulse of 800 V and 3.0 microfarads. Stable transfected clones were selected with 50 μg/ml hygromycin. Cells containing the DR-GFP-Hyg plasmid were subjected into the DR-GFP assay as described previously (21).
Plasmid Constructs and Proteins
The plasmid hFBH1/pAS2-1 encoding for human FBH1 isoform 4 (2.91 kb, 969 amino acids) served as a source of FBH1 cDNA in this study (12). FBH1 was subcloned as a fusion with glutathione S-transferase (GST) (N terminus) and hexahistidine tags (C terminus) in pFastBac1 via SalI and KpnI sites to yield a transfer vector for bacmid production. The D698N mutant of FBH1 was generated using the QuikChange site-directed mutagenesis kit (Stratagene). To produce antigen for anti-FBH1 antibody preparation, the N-terminal part of FBH1 (isoform 4) including amino acids 1–484 was subcloned into the pTYB12 vector via NheI/XhoI sites, creating a fusion with an N-terminal self-cleavable affinity tag containing a chitin-binding domain (CBD-FBH1(1–484)).
Antibodies
A polyclonal antibody against an FBH1 fragment spanning amino acids 1–484 was raised in a chicken. The antibody was extracted from egg yolk and affinity-purified using a column conjugated with the antigen. A polyclonal antibody against the full-length human RAD51 was raised in rabbits and affinity-purified using a RAD51-conjugated column.
Protein Purification
GST-FBH1-His6 (or its D698N mutant) was produced in Sf9 insect cells by means of a baculovirus system. Typically, 1.5 × 108 cells were infected by appropriate virus and incubated for 2 days at 28 °C. Harvested cells were resuspended in GST-lysis buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 1 mm EDTA, 10% (v/v) glycerol, 2 mm DTT, 0.1% (v/v) Triton X-100) and disrupted by Dounce homogenization. The soluble protein extract obtained by centrifugation (47,000 × g for 45 min at 4 °C) was incubated with 2 ml of 50% GSH-agarose for 2–4 h at 4 °C on a rotary shaker. Beads were washed three times with 30 ml of GST-lysis buffer (centrifugation at 300 × g for 5 min; 4 °C). FBH1 protein was eluted from the beads by GST elution buffer (40 mm Tris-HCl, pH 8.1, 50 mm NaCl, 10% (v/v) glycerol) containing 10 mm glutathione. Elution fractions containing FBH1 protein were pooled and incubated with 0.5 ml of 50% heparin beads for 1 h at 4 °C. After binding, beads were washed three times with 20 ml of Hep-buffer A (40 mm Tris-HCl, pH 7.5, 0.1 m NaCl, 1 mm EDTA, 10% (v/v) glycerol, 1 mm β-mercaptoethanol), and protein was eluted with Hep-buffer B (40 mm Tris-HCl, pH 7.5, 1 m NaCl, 1 mm EDTA, 10% glycerol (v/v), 1 mm β-mercaptoethanol). The heparin-elution fractions (1 ml) containing FBH1 were pooled and dialyzed overnight at 4 °C against 1 liter of dialysis buffer (40 mm Tris-HCl, pH 7.5, 50 mm NaCl, 1 mm EDTA, 40% (v/v) glycerol, 1 mm β-mercaptoethanol). The final protein preparation was aliquoted, snap-frozen in liquid nitrogen, and stored at −80 °C. Protein concentration was measured by a Bradford assay. RAD51, RAD51K133R, RPA, and RECQ5 were produced and purified as described previously (22–24).
Separation of Soluble and Chromatin-associated Proteins
Semiconfluent cells were washed in PBS, trypsinized, and counted. A pellet of ∼2 × 106 cells was washed three times in PBS and resuspended in 250 μl of freshly made protein buffer A (10 mm HEPES, pH 7.0, 10 mm KCl, 1.5 mm MgCl, 0.34 m sucrose, 10% (v/v) glycerol, 1 mm DTT, 0.1% (v/v) Triton X-100, and protease inhibitors). The sample was centrifuged at 210 × g for 4 min at 4 °C, and the supernatant was removed and stored (cytoplasmic soluble fraction). The pellet was washed in 200 μl of protein buffer A and centrifuged at 210 × g for 4 min at 4 °C before the nuclei were lysed in 175 μl of freshly made buffer B (3 mm EDTA, 0.2 mm EGTA, 1 mm DTT, protease inhibitors). Incubation was for 10 min on ice, followed by centrifugation at 270 × g for 4 min at 4 °C. The supernatant was removed and stored (nuclear soluble fraction). The pellet was resuspended in 175 μl of buffer B and centrifuged at 16,000 × g for 1 min at 4 °C before the supernatant was discarded, and the pellet was resuspended (chromatin-associated fraction) in 175 μl of Laemmli buffer (4% SDS, 20% glycerol, 120 mm Tris-HCl, pH 6.8), boiled, and sheared with a 25-gauge needle. Storage of all samples was at −80 °C. For analysis, 10× BFB-2-ME (0.1% (w/v) bromphenol blue, 5% (w/v) β-mercaptoethanol) was added, the samples were boiled, and the soluble fractions were combined. Proteins were separated on a 10% BisTris gel with MOPS buffer. Proteins were transferred to a membrane and probed with Rad51 antibody (1:200; Abcam), ORC4L antibody (1:1,000; Abcam), or GRB2 antibody (1:1,000; BD Transduction Laboratories).
GST Pull-down Assays
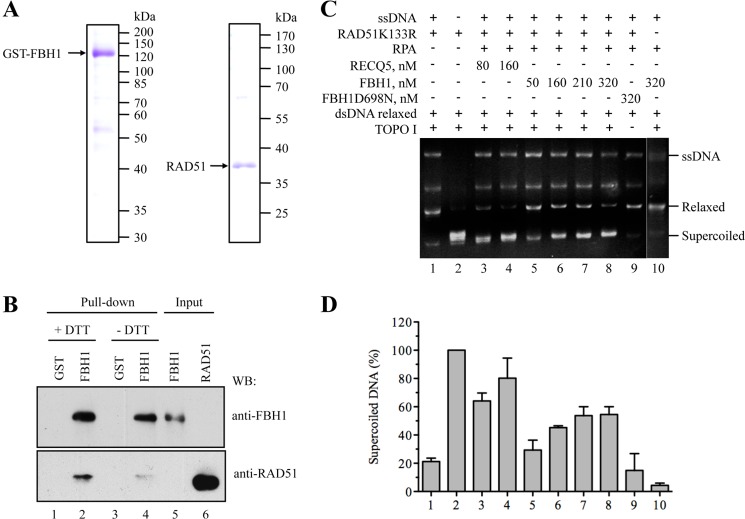
100 ng of purified RAD51 and 200 ng of GST-FBH1-His (or 200 ng of GST) were added to 0.4 ml of NET-N150 buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 0.2% (v/v) Triton X-100) and incubated for 4 h at 4 °C on a rotary shaker. Where required, DTT was present at a concentration of 1 mm. 15 μl of GSH-agarose slurry was added to the protein mixtures, followed by incubation for another 1 h at 4 °C. Beads were collected by centrifugation (300 × g for 3 min at 4 °C), washed four times with 1 ml of NET-N150 buffer, and incubated with 12 μl of 2× Laemmli buffer at 95 °C for 5 min. Samples were subjected to SDS-PAGE on a 9% gel, followed by Western blot analysis.
DNA Topology Modification Assays
Reactions were carried out at 37 °C in a final volume of 25 μl of buffer R (25 mm Tris-HCl, pH 7.5, 50 mm KCl, 1 mm DTT, 1 mm MgCl2, 100 μg/ml BSA) supplemented with an ATP-regenerating system consisting of 10 units/ml creatine phosphokinase, 12 mm creatine phosphate, and 2 mm ATP. 2 μm RAD51K133R was preincubated with circular M13mp8.32 ssDNA (9 μm nucleotides) for 6 min, followed by the addition of 150 nm RPA and a DNA helicase (RECQ5 (80 and 160 nm), FBH1 (50, 160, 210, and 320 nm), or FBH1D698N (320 nm)). After a 6-min incubation, topologically relaxed pGEM-7Zf(+) DNA (7 μm base pairs) and 3 units of wheat germ topoisomerase I (Promega) were added to complete the reaction, which was then incubated for a further 8 min. Reactions were terminated by the addition of 5 μl of STOP buffer (6% (w/v) SDS, 0.1 m EDTA, 5 mg/ml proteinase K), followed by a 25-min incubation at 37 °C. Deproteinized DNA species were resolved by electrophoresis in a 1% agarose gel run in 0.5× TBE buffer at 100 V for 2 h. Gels were stained with ethidium bromide (0.5 μg/ml) and photographed on a UV transilluminator. DNA bands were quantified using ImageQuant software, and the quantity of supercoiled DNA products was expressed as a percentage of the amount of product generated in the reaction containing only RAD51K133R and dsDNA (100%). To generate topologically relaxed dsDNA, 4 μg of supercoiled pGEM-7Zf(+) DNA were incubated with 1.6 μg of Escherichia coli topoisomerase I at 37 °C for 30 min in 40 μl of buffer R supplemented by 100 μg/ml BSA. Reaction was stopped by heat inactivation at 65 °C for 10 min.
Single-molecule Fluorescence Resonance Energy Transfer (smFRET)
All experiments were performed at room temperature. Reactions were carried out in buffer composed of 50 mm Tris-HCl, pH 8.0, 50 mm KCl, and 10 mm MgCl2 and supplemented by an oxygen-scavenging system (1 mg/ml glucose oxidase, 0.4% (w/v) d-glucose, 0.02 mg/ml catalase, and 2 mm Trolox) (25). smFRET assays were preformed according to previously described protocols (25, 26). Briefly, DNA FRET constructs (30 pm) were specifically tethered to the surface of a biofunctionalized microfluidic chamber through a biotin-neutravidin link, followed by the addition of proteins.
smFRET Data Analysis
Data were recorded and analyzed by scripts written in IDL to generate fluorescence intensity time trajectories of individual molecules. A time resolution of 0.03 s was used for all experiments. Single-molecule fluorescence time trajectories were viewed and analyzed using programs written in Matlab. FRET efficiency (EFRET) was approximated as the ratio between the acceptor intensity and the sum of the acceptor and donor intensities. Histograms were generated using a sample size of over 100 individual molecular trajectories.
RESULTS
To investigate the function of the FBH1 helicase in the maintenance of genome stability, we undertook a multidisciplinary study combining cellular, biochemical, and biophysical approaches.
Characterization of a Mouse ES Cell Line with a Helicase Domain Deletion
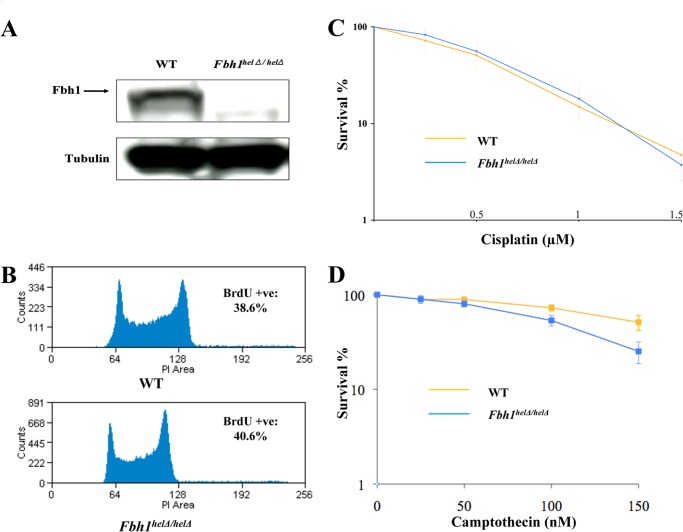
To investigate the functional consequences of inactivating the helicase activity of FBH1, we characterized a previously described mouse ES cell line with a deletion in the FBH1 helicase domain (27). In this cell line, a replacement of each allele of the mouse Fbh1 gene region encoding helicase domains II and III was made using a drug resistance cassette, leading to termination of both Fbh1 open reading frames. Helicase domain II contains the Walker B box that is required for binding the ATP-Mg2+ complex and is essential, therefore, for ATPase activity and hence DNA translocation-unwinding. The correct targeting of the two Fbh1 alleles was verified previously by PCR using multiple primer pairs along the Fbh1 gene (27). To confirm that the targeted cells, hereafter called Fbh1helΔ/helΔ, had a gene disruption that impacted on FBH1 protein levels, we performed Western blot analysis. This analysis confirmed that no detectable full-length FBH1 protein was expressed in the Fbh1helΔ/helΔ cells (Fig. 1A). The doubling time, plating efficiency, and cell cycle distribution of the Fbh1helΔ/helΔ cells were all similar to those of the wild-type cells, indicating that there is no gross disturbance in cell cycle or cell growth properties due to loss of FBH1 helicase function in mouse ES cells (Fig. 1B) (data not shown).
FIGURE 1.
Characterization of Fbh1helΔ/helΔ cells. A, Fbh1helΔ/helΔ cells do not express any full-length FBH1 protein. Western blot of an extract from WT and Fbh1helΔ/helΔ cells was immunoblotted for FBH1 and tubulin as a loading control. B, cell cycle distribution is only marginally altered in Fbh1helΔ/helΔ cells compared with WT cells by FACS analysis. C, clonogenic survival of WT and Fbh1helΔ/helΔ cells following exposure to cisplatin. D, clonogenic survival of WT and Fbh1helΔ/helΔ cells following exposure to camptothecin. The means and the S.E. (error bars) are shown for three independent experiments.
Next, we asked if the Fbh1helΔ/helΔ cells showed any abnormal survival response following exposure to DNA-damaging agents. For this, clonogenic survival analyses were performed. We tested a number of agents that damage DNA via different mechanisms and produce lesions that are repaired by different pathways. Of particular interest were agents that induce DNA cross-linking or generate DNA DSBs, either directly or through replication fork collapse, because HR is normally required for repair of these types of DNA damage. There was no difference in survival between the wild-type and mutant cells for most agents studied, including cisplatin and ionizing radiation (Fig. 1C) (data not shown). Nevertheless, there was a mildly increased sensitivity to camptothecin in the mutant cells, which was statistically significant (IC50 of 150 nm for wild-type versus 105 nm for the Fbh1helΔ/helΔ cells; p = 0.0035; Fig. 1D).
Fbh1helΔ/helΔ Cells Have Increased RAD51 Focus Formation and Chromatin Association
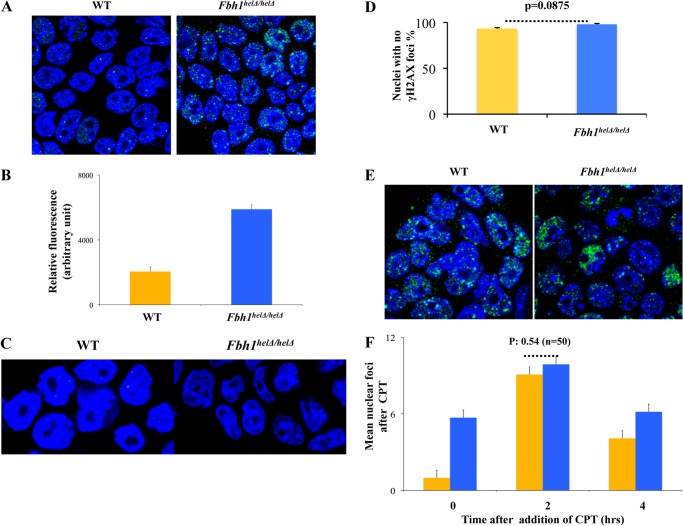
Previous studies in fission yeast cells have indicated that FBH1 is able to regulate the function of RAD51. To investigate whether this was conserved in mammalian cells, we first quantified the level of nuclear RAD51 foci in the wild-type and mutant cells that had not been exposed to any exogenous DNA damage stress. This revealed that the Fbh1helΔ/helΔ cells exhibited a substantial excess of spontaneous RAD51 foci compared with the wild-type cells (Fig. 2A). Only 2% of the wild-type cells exhibited five or more foci (mean = 0.98; n = 50), whereas 50% of the mutant cells contained at least five RAD51 foci (mean = 5.7; n = 50; p < 0.0001 compared with wild-type cells). As an alternative method of quantification, which took into account the observation that many nuclei in the Fbh1helΔ/helΔ cell population contained extensive RAD51 staining that was not limited to discrete focal sites, we quantified the total nuclear fluorescence intensity for the stained RAD51. This showed that loss of Fbh1 helicase function led to a highly significant increase in the level of overall nuclear RAD51 staining (n = 75; p < 0.0001; Fig. 2B). One possible explanation for these data is that the staining pattern in Fbh1helΔ/helΔ cells reflects a general increase in the level of DNA DSBs. However, we consider this to be very unlikely, because the number of spontaneous γ-H2AX foci was not altered in the mutant cells (Fig. 2, C and D).
FIGURE 2.
Fbh1helΔ/helΔ cells cannot regulate RAD51 focus formation. A, examples of RAD51 foci (green) in WT and Fbh1helΔ/helΔ cells. Nuclei were stained with DAPI (blue). B, relative total nuclear fluorescence for RAD51 staining in WT and Fbh1helΔ/helΔ cells. Data were analyzed using ImageJ. The means and S.E. (error bars) are shown for three independent experiments. C, examples of nuclei stained for γ-H2AX (green). Nuclei were stained with DAPI (blue). D, quantification of the data from C. E, images of WT and Fbh1helΔ/helΔ cells 2 h after exposure to 100 nm camptothecin. RAD51 foci are shown in green. Nuclei were stained with DAPI (blue). F, quantification of the data from E, along with equivalent data from time 0 and 4 h. The mean number of nuclear RAD51 foci is shown. S.E. values from at least three independent experiments are indicated.
Next, we asked if this excessive RAD51 focus formation in undamaged Fbh1helΔ/helΔ cells was also seen following exogenously induced DNA damage. Following a dose of camptothecin that reduced cell survival by 25–50%, the number of nuclear RAD51 foci was quantified at two time points after the addition of the drug (2 and 4 h). We found that loss of Fbh1 helicase function was not associated with an increase in the maximal number of RAD51 foci. After 2 h of camptothecin exposure, RAD51 focus formation was increased relative to the spontaneous level in both wild-type and mutant cells, but the level was very similar in the two cell lines (Fig. 2, E and F). After 4 h, the level of RAD51 nuclear fluorescence was declining in both cell lines, and the excess number of foci characteristic of unperturbed Fbh1helΔ/helΔ cells compared with wild-type cells was reestablished. Hence, FBH1 deficiency is associated with a failure to suppress RAD51 focus formation but apparently only in unperturbed or undamaged ES cells.
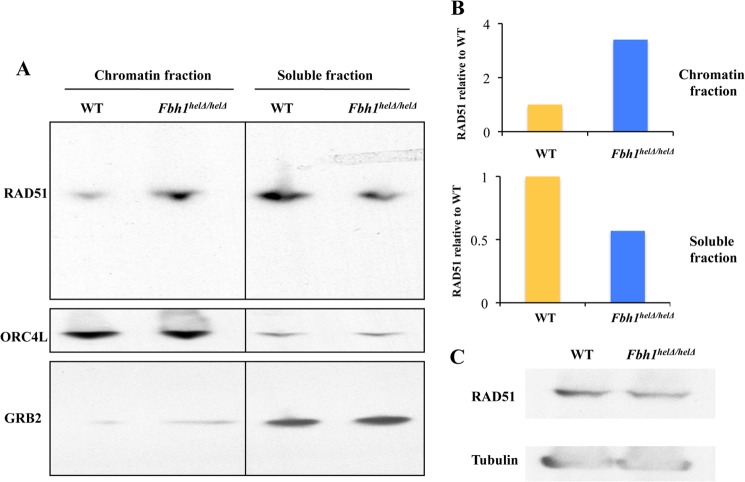
If the role of FBH1 is to prevent spontaneous RAD51 focus formation in mammalian cells, as our data imply, then it might be expected that the subnuclear distribution of RAD51 would be abnormal in the Fbh1helΔ/helΔ cells. Consistent with this proposal, we found that loss of FBH1 helicase function was associated with an increase in the proportion of RAD51 that is chromatin-associated (Fig. 3, a and b). This difference was not accounted for by any change in the overall cellular level of RAD51 (Fig. 3c).
FIGURE 3.
Abnormal subcellular distribution of RAD51 in Fbh1helΔ/helΔ cells. A, Western blot of the chromatin fraction and soluble protein fraction of RAD51 (top), ORC4L (a chromatin marker; middle), and GRB2 (a soluble fraction marker; bottom). B, quantification of the Western blots from A showing the relative distribution of RAD51 in the chromatin and soluble factions. In each case, the WT cells were given a value of 1. C, Western blot comparing total cellular RAD51 levels to those of a loading control (tubulin).
Fbh1helΔ/helΔ Cells Show Hyper-recombination and Resistance to Poly(ADP-ribose) Polymerase (PARP) Inhibitors
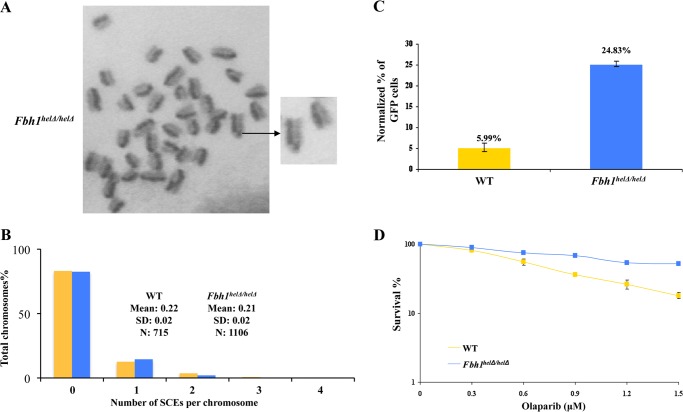
Next, we investigated whether the excess chromatin-bound RAD51 was able to alter the spontaneous level of HR in the Fbh1helΔ/helΔ. To study this, we initially quantified the frequency of SCEs, which reflect crossover during HR events. However, we found no significant difference in SCE levels between the wild-type and Fbh1helΔ/helΔ mutant cells (p > 0.05; Fig. 4, A and B). Next, we quantified the frequency of HR more generally using the widely used DR-GFP assay (see “Experimental Procedures”). In this, HR can be stimulated by the cleavage of the GFP substrate with the I-SceI endonuclease to generate a DNA double strand break. We found that I-SceI-induced HR was significantly elevated in the Fbh1helΔ/helΔ cells compared with the wild-type ES cell control (around 4-fold; p < 0.01; Fig. 4C). Hence, we conclude that the excessive RAD51 focus formation and chromatin binding associated with loss of FBH1 helicase activity has functional consequences and is capable of activating at least some forms of HR.
FIGURE 4.
Hyper-recombination in Fbh1helΔ/helΔ cells. A, representative image of metaphase chromosomes from Fbh1helΔ/helΔ cells stained for SCEs. The enlargement on the right shows an SCE. B, quantification of SCE data for WT (yellow bars) and Fbh1helΔ/helΔ (blue bars) cells. The number of SCEs per chromosome is shown. The key indicates the mean and S.D. from over 700 chromosomes in each case (p > 0.05). C, hyper-recombination in Fbh1helΔ/helΔ cells as determined using the DR-GFP assay. The percentage of cells expressing GFP was normalized to a transfection control (a plasmid expressing RFP). The values above the bars indicate the average percentages. The means and S.E. (error bars) for three independent experiments are shown. D, clonogenic survival of WT and Fbh1helΔ/helΔ cells following exposure to olaparib. The means and the S.E. are shown for three independent experiments.
Cells deficient in homologous recombination factors, such as BRCA2, have been shown to exhibit hypersensitivity to inhibitors of PARP (28–30). We analyzed, therefore, whether the Fbh1helΔ/helΔ cells might show an altered response to PARP inhibition. Consistent with the Fbh1helΔ/helΔ cells displaying hyper-recombination rather than recombination deficiency, we found that they were significantly more resistant to the PARP inhibitor olaparib than the wild-type cells (p < 0.05; Fig. 4D).
Direct Physical Interaction between Human FBH1 and RAD51
Given that our cellular data are consistent with FBH1 acting as a negative regulator of the association of RAD51 with chromatin, we investigated whether there might be a functional interaction between RAD51 and FBH1 proteins in vitro. To test whether FBH1 can physically interact with RAD51, we purified recombinant GST-tagged FBH1 and untagged RAD51 (Fig. 5A) and then performed affinity pull-down assays. The RAD51 was incubated with GST-FBH1 or with GST alone under either reducing or non-reducing conditions. The GST-FBH1 and GST proteins were subsequently captured on glutathione-agarose beads, and any additional bound proteins were eluted and analyzed by Western blotting. We found that RAD51 could bind to the GST-FBH1 beads but not to control GST beads (Fig. 5B, lanes 1–4). Notably, reducing conditions increased the extent of binding of GST-FBH1 and RAD51 (Fig. 5B, compare lanes 2 and 4). Collectively, these data indicate that FBH1 forms a complex with RAD51 through a direct interaction, as might be expected of a regulator of RAD51 function.
FIGURE 5.
FBH1 forms a complex with RAD51 and disrupts RAD51 filaments. A, SDS-PAGE analysis of purified FBH1 (left) and RAD51 (right). Gels were stained with Coomassie Blue. The molecular masses of protein standards are indicated on the right. B, GST pull-down assay. Purified RAD51 protein (100 ng) was incubated with GST-FBH1-His (FBH1) protein or with GST alone (each 200 ng) in the presence or absence of 1 mm DTT (+DTT/−DTT). Proteins were affinity-captured using GSH-agarose beads and analyzed by Western blotting (WB) using chicken anti-FBH1 and rabbit anti-RAD51 antibodies. Lane 5, FBH1 (40% of input); lane 6, RAD51 (5% of input). C, RAD51K133R was assembled on M13 ssDNA (9 μm nucleotides) in the presence of 2 mm ATP and an ATP-regenerating system and was then incubated with increasing concentrations of FBH1 (50, 160, 210, and 320 nm) or RECQ5 (80 and 160 nm) for 6 min before the addition of relaxed DNA (7 μm bp) and wheat germ DNA topoisomerase I. After deproteinization, reactions were analyzed by electrophoresis in a 1% agarose gel, followed by ethidium bromide staining. Gels were quantified using ImageQuant software, and the concentration of supercoiled DNA products was calculated as a percentage of the amount of product generated in the reaction carried out in the absence of ssDNA, FBH1/RECQ5, and RPA (lane 2). D, quantification of the data. The plot shows the average values from three independent experiments performed as described in C. The numbers under the bars correspond to the numbers of lanes in C. Error bars, S.D.
FBH1 Disrupts RAD51 Nucleoprotein Filaments in Vitro
Next, we investigated whether purified FBH1 protein could disrupt RAD51 filaments in vitro. In these experiments, we used an ATPase-defective mutant of RAD51, RAD51K133R, which has been shown to form stable filaments in the presence of ATP, which wild-type RAD51 does not (31). RAD51K133R-ssDNA filaments were preformed on M13 ssDNA and were then incubated with increasing concentrations of GST-FBH1 in the presence of RPA and ATP. Following this, a previously validated DNA topology assay was performed that permits detection of free RAD51 molecules in solution (10). In this assay, binding of RAD51 to topologically relaxed, covalently closed circular dsDNA induces lengthening of the DNA and the formation of positive supercoils. This effect can be monitored as a reduction in the DNA linking number upon treatment with eukaryotic topoisomerase I, giving rise to negatively supercoiled DNA (10, 32). We observed that FBH1 promoted the formation of the negatively supercoiled DNA product in a concentration-dependent manner, indicative of the presence of a filament-disrupting activity (Fig. 5, C and D). Formation of supercoiled DNA was not detected in reactions containing an ATPase-defective mutant of FBH1, FBH1-D698N, indicating that filament disruption by wild-type FBH1 is dependent on its ssDNA translocase activity, which is driven by ATP hydrolysis (Fig. 5, C and D). Moreover, FBH1 alone did not induce DNA supercoiling in a DNA topology modification assay (Fig. 5, C and D, lane 10). We also carried out control reactions in which the FBH1 protein was substituted by the RECQ5 helicase, which is also known to bind RAD51 (33). We found that RECQ5 disrupted RAD51K133R-ssDNA filaments in a manner similar to that of FBH1 (Fig. 5, C and D).
Real Time Observation of RAD51 Filament Disruption by FBH1
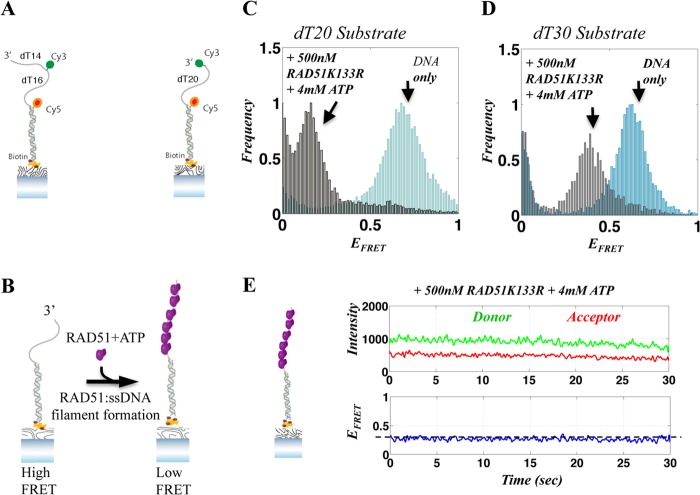
To substantiate and extend the findings described above, we used smFRET-based assays to monitor FBH1-mediated disruption of RAD51-ssDNA filament in real time. To probe RAD51 filament assembly and disassembly on ssDNA, we used two partial duplex FRET constructs. Both contained an 18-bp duplex region but differed in whether they had a 20- or a 30-nt poly(dT) 3′ tail, as illustrated in Fig. 6A. The 3′-terminus of the dT20 tail construct was labeled with a Cy3 (donor) dye molecule, whereas the dT30 tail was labeled internally 14 nt away from the 3′-end. In both cases, the 5′-end of the ssDNA/dsDNA duplex junction was labeled with a Cy5 (acceptor) dye molecule. These FRET constructs exhibited a steady state FRET efficiency (EFRET) of ∼0.6 (Fig. 6, C and D, blue histograms; illustrated in Fig. 6B), making them suitable for monitoring RAD51 filament-mediated extension of the contour length of the ssDNA, a known feature of such filaments. We established that these partial duplex substrates possess, as expected, an ssDNA 3′-tail that is longer than the experimentally determined minimal length (16 nt) required for stable RAD51-ssDNA filament formation (34). The DNA-FRET substrates were specifically tethered to the surface of the flow chamber via a biotin moiety on the blunt end of the duplex (Fig. 6A). We utilized two different probes to rule out the possibility that our observations were specific for a particular DNA substrate. However, because both of these FRET substrates yielded very similar data, we focused on the results obtained with the 30-nt tailed substrate. For consistency, the concentration of surface-tethered DNA was fixed at 30 pm in all experiments.
FIGURE 6.
Single-molecule FRET assay for RAD51-ssDNA filament formation. A, illustration of the DNA FRET substrates used in our assays, consisting of partial duplex DNA with a dT30 or dT20 tail. B, illustration of single-molecule RAD51-ssDNA filament assembly assays. The DNA substrate shown on the left yields high FRET at steady state. Upon the addition of RAD51 and ATP, a RAD51-ssDNA filament forms, extending the ssDNA, resulting in a decrease in FRET (right). C and D, FRET histograms of a population of individual DNA molecules for the 20-nt (C) and 30-nt (D) substrates, showing the resulting FRET values when ssDNA is relaxed (DNA only; blue histogram) and after the assembly of a RAD51K133R-ssDNA filament, resulting in a FRET decrease (gray). E, representative smFRET trajectory of a 30-nt DNA substrate with a RAD51K133R-ssDNA filament. The top panel shows the donor and acceptor intensities, whereas the corresponding FRET efficiency is shown in the bottom panel. The FRET efficiency associated with stable RAD51K133R-ssDNA is marked with a horizontal dashed line.
To generate a stable RAD51-ssDNA filament, we added a 500 nm concentration of the RAD51K133R protein, together with 4 mm ATP, to the flow chamber and incubated the reaction for 5 min before data acquisition. Formation of RAD51K133R-ssDNA filaments was observed as a dramatic decrease in EFRET (from ∼0.6 to ∼0.3 and ∼0.1 for the 30- and 20-nt substrates, respectively), resulting from the RAD51-dependent extension in ssDNA contour length in the filament (Fig. 6, C and D, gray histograms; illustrated in Fig. 6B). RAD51K133R was found to form stable filaments on the ssDNA, as evidenced by the narrow FRET distribution of smFRET histograms. This was also apparent in the majority of smFRET trajectories, which showed a steady low FRET state (EFRET ∼0.1 and 0.3 for the 20- and 30-nt substrates, respectively; Fig. 6, C and D), corresponding to stable RAD51-ssDNA filaments. There were occasional transitions to higher FRET states (EFRET ∼0.4–0.5), probably reflecting the dynamics of RAD51 filament rearrangement. Fig. 6E shows a representative single-molecule trajectory of a RAD51K133R-ssDNA filament, where the top panel displays the donor (green) and acceptor (red) intensities and the bottom panel shows the corresponding FRET efficiency (blue). The smFRET trajectory shows that the RAD51K133R protein forms a stable filament, corresponding to a lower FRET value (∼0.3; dashed line). Hence, we conclude that RAD51K133R protein can generate stable filaments under our experimental conditions.
To verify that spontaneous RAD51K133R filament disassembly would not mask FBH1-mediated disassembly, we monitored the stability of RAD51K133R-ssDNA over time. Following initial filament assembly, the flow chamber was washed with imaging buffer to remove excess RAD51K133R in solution, and DNA substrates were monitored again 30 and 60 min later. The smFRET histogram obtained 60 min after excess RAD51K133R removal was very similar to the histogram obtained immediately after filament formation, indicating that the RAD51K133R filament is stable on ssDNA for a prolonged time (data not shown). We also verified that the observed decrease in FRET was indeed due to RAD51K133R-ssDNA filament formation by using a high salt (500 mm NaCl) buffer to remove all bound proteins. As anticipated, the smFRET histograms obtained after high salt buffer wash were identical to the initial smFRET histograms obtained for DNA only (data not shown).
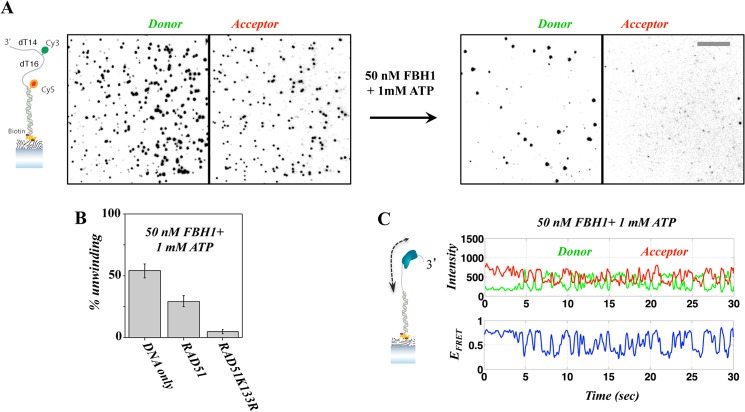
We next characterized the behavior of FBH1 on the tailed DNA substrate in the absence of RAD51. 50 nm FBH1 along with 1 mm ATP were added to the flow chamber containing surface-tethered DNA-FRET substrates, and data were recorded immediately thereafter. We found unwinding of the surface-tethered DNA by FBH1, as evidenced by the rapid reduction of the observed FRET molecules on the surface (Fig. 7, A and B). Quantification of FBH1 unwinding efficiency (Fig. 7B), calculated as the percentage of molecules removed within the first 2 min of the unwinding reaction, showed that FBH1 unwinds more than 50% of the surface-bound DNA molecules within this time period. We also tested FBH1 unwinding activity using DNA substrates on which WT RAD51 or RAD51K133R filaments were preformed. This was achieved by the addition of 500 nm WT RAD51 or RAD51K133R protein together with 4 mm ATP and incubation for 5 min prior to the addition of FBH1. Notably, the efficiency of FBH1-mediated unwinding of DNA containing preformed RAD51 filaments showed only a modest decrease in efficiency when using WT RAD51 but nearly complete suppression of unwinding when using RAD51K133R filaments (Fig. 2B). As expected, analysis of individual FRET trajectories acquired during FBH1 unwinding reactions displayed rapid, repetitive FRET fluctuations, indicative of ATP-dependent translocation activity, which has been reported previously for other helicases (35, 36). Fig. 7C shows a characteristic single-molecule FRET trajectory in the presence of FBH1 and ATP, where the donor-acceptor intensities and corresponding FRET efficiency are shown in the top and bottom panels, respectively. The smFRET trajectory shows a clear repetitive fluctuation induced by FBH1 translocation on ssDNA, observed as recurring transitions from high FRET base-line value to lower FRET values. This characteristic behavior was also observed with the 20-nt tailed DNA substrates (data not shown).
FIGURE 7.
Unwinding of a duplex DNA substrate by FBH1. A, unwinding of 30-nt tailed DNA substrate illustrated on the left. The images show spots corresponding to individual molecules observed in the donor (left) and acceptor (right) channels. Following the addition of FBH1 and ATP (right), there is a sharp reduction in the number of observed DNA molecules. B, unwinding yield calculated for the initial 2 min of the unwinding reaction, showing that rapid unwinding is dramatically reduced in the presence of a RAD51K133R filament. C, representative smFRET trajectory showing repetitive translocation of FBH1 on the DNA FRET substrate in the absence of a RAD51-ssDNA filament. Error bars, S.E.
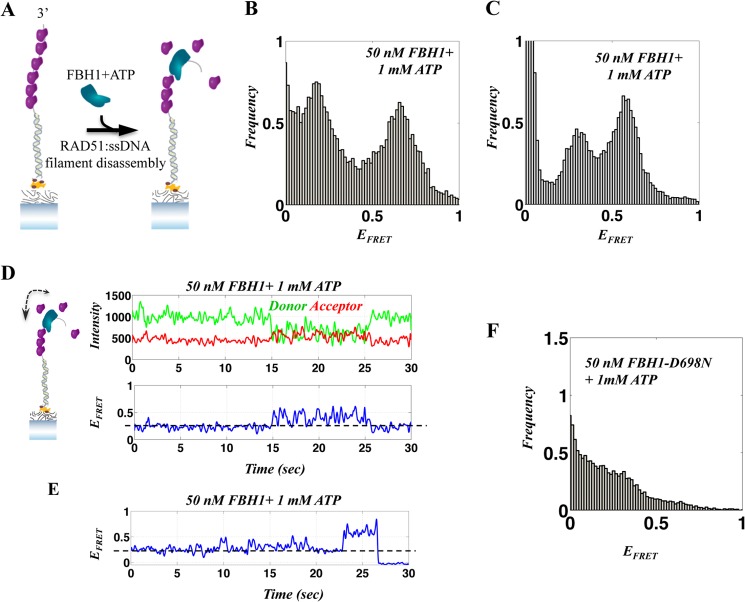
Next, we analyzed FBH1-mediated disruption of a preformed RAD51K133R-ssDNA filament. For this, a stable RAD51K133R-ssDNA filament was formed by incubating the surface tethered DNA substrate with RAD51K133R and 4 mm ATP for 5 min. Thereafter, excess RAD51K133R was removed, and FBH1 (50 nm) and ATP (1 mm) were added (illustrated in Fig. 8A). Data acquisition followed immediately. The resulting histograms (Fig. 8, B and C, for the 20- and 30-nt substrates, respectively) display a clear broadening of the peak corresponding to the RAD51K133R-ssDNA filament (∼0.3 and ∼0.1 for the 30- and 20-nt substrates, respectively) together with the appearance of another FRET population at ∼0.6, corresponding to removal of the RAD51K133R-ssDNA filament from ssDNA tail. The broadening of the low FRET RAD51K133R-ssDNA filament peak in the presence of FBH1 (Fig. 8, B and C; compared with the narrower peak (Fig. 6, C and D)) is indicative of the dynamics of the RAD51K133R-ssDNA filaments resulting from FBH1 translocation. The formation of two distinct FRET populations upon the addition of FBH1 strongly suggests that translocation-mediated directional removal of RAD51K133R from ssDNA is occurring. This is probably initiated at the 3′-end of the DNA. A broad, continuous FRET distribution would be expected if random binding and translocation were to occur along the ssDNA tail.
FIGURE 8.
Single-molecule FRET analysis of the FBH1-RAD51 interaction. A, illustration of the DNA FRET assay for single-molecule, FBH1-mediated, RAD51-ssDNA filament disassembly. The addition of FBH1 to a preformed RAD51 filament causes filament disassembly enabling relaxation (shortening) of ssDNA and an increase in FRET efficiency. B and C, FRET histograms after the addition of FBH1 and ATP to a preformed RAD51K133R-ssDNA filament for the 20-nt (B) and 30-nt (C) substrates. D and E, representative smFRET trajectories of the dynamics associated with repetitive translocation of FBH1 on ssDNA in the presence of a RAD51K133R filament and FBH1-mediated RAD51K133R-ssDNA filament disassembly. The FRET efficiency corresponding to a RAD51K133R-ssDNA filament (dashed line) displays changes due to repetitive FBH1 ssDNA translocation that fluctuate around the filament base line (D), indicating the co-existence of FBH1 and the RAD51K133R filament. E, fluctuations of FRET corresponding to FBH1 translocations in the presence of RAD51K133R-ssDNA filament, followed by an abrupt increase to higher FRET due to the removal of RAD51K133R filament. F, FRET histogram after the addition of FBH1-D698N mutant (ATPase-dead) and ATP to a preformed RAD51K133R-ssDNA filament for the 20-nt DNA, showing the persistence of the RAD51K133R filament.
Analysis of individual single-molecule FRET trajectories revealed direct filament disruption events, where the low FRET values corresponding to RAD51K133R-ssDNA base line (∼0.3) were now accompanied by low amplitude repetitive transitions to higher FRET. Fig. 8, D and E, shows three representative smFRET trajectories showing RAD51K133R-ssDNA filament disassembly mediated by FBH1. The trajectory shown in Fig. 8D displays repetitive FRET modulation occurring around the RAD51K133R-ssDNA filament FRET base line (as compared with the RAD51K133R-ssDNA filament shown in Fig. 6E). This may indicate that FBH1 first binds and clears RAD51K133R from ssDNA at the 3′-end and undergoes repetitive translocation, hence destabilizing the RAD51K133R-ssDNA filament that is still bound further down the ssDNA strand (34). Fig. 8E shows that the fluctuations in RAD51K133R-ssDNA FRET base line (t = 0–23 s) are followed by an abrupt transition to higher FRET, where the RAD51K133R protein is removed. These observations denote FBH1 translocation-dependent RAD51-ssDNA filament disassembly. Similar features were also observed in trajectories obtained from the 20-nt tailed FRET substrate (data not shown).
To further verify that the observed filament disruption is due to FBH1 ATPase activity, we repeated the RAD51K133R-ssDNA disruption experiment with the ATPase-defective mutant of FBH1, FBH1-D698N, and 1 mm ATP. This resulted in no substantial disruption of RAD51 filament, even for the shorter 20-nt substrate, as shown in Fig. 8F (as compared with wild-type FBH1; Fig. 8, B and C), indicating that the RAD51-ssDNA filament disruption is indeed due to FBH1 ATP-dependent ssDNA translocation, as has been observed previously for yeast Srs2 protein (34).
DISCUSSION
The FBH1 helicase has been implicated in the regulation of the RAD51 recombinase in a number of organisms, including fission yeast, as well as chicken and mammalian cells (14–19). Here, we have defined the molecular mechanism underlying the anti-recombinase activity of FBH1 in mammalian cells. Using biochemical and biophysical techniques, we have demonstrated that human FBH1 has the capacity to disrupt the ATP-bound form of the RAD51 presynaptic filament by acting as a ssDNA translocase. This serves to expel RAD51 monomers from the ssDNA. A similar activity has been shown previously for human RECQ5, which belongs to a different family of helicases from FBH1 (RecQ, not UvrD) (33). Thus, mammalian cells possess at least two evolutionarily divergent helicases that can regulate RAD51 filament assembly by a mechanism similar to that shown for the canonical enzyme in yeast, Srs2. It remains to be determined whether FBH1 and RECQ5 have redundant roles in HR regulation or whether they act in different HR pathways. Evidence in favor of the latter proposal comes from the finding that RecQ5-deficient mouse cells show increased levels of RAD51 foci that are accompanied by excessive SCEs and γ-H2AX foci (33), neither of which are seen in our mouse Fbh1helΔ/helΔ cells. Other putative regulators of RAD51 are expressed in human cells, including PARI, BLM, and HelQ, but none of these has properties identical to FBH1. PARI appears to be the closest mammalian homolog of yeast Srs2. However, PARI lacks key residues essential in other UvrD family proteins for DNA translocation (11), making it very unlikely that it disrupts RAD51 filaments via a similar, translocation-based, mechanism to that used by FBH1. In contrast to FBH1, BLM is incapable of disrupting the ATP-bound form of RAD51 but instead was reported to stimulate DNA strand exchange by ATP-bound RAD51 (37, 38). HelQ also behaves differently from FBH1, in that it seems to be specific for RAD51 bound to dsDNA and to utilize an ATP-independent disruption mechanism (39). Taken together, these data suggest that FBH1 has properties that distinguish it from any of the other known RAD51 regulators in mammalian cells.
A previous study (18) suggested that the primary role of FBH1 in mammalian cells is in the control of mitosis, more specifically in restoring normal progression through mitosis following exposure of cells to catalytic inhibitors of topoisomerase II that prevent normal sister chromatid disjunction. This conclusion was based on analysis of mouse ES cells lacking Fbh1 or expressing F-Box-deficient Fbh1. In contrast to those cell lines, we studied ES cells expressing helicase-deficient Fbh1. Our Fbh1helΔ/helΔ mutant cells are not hypersensitive to topoisomerase II catalytic inhibitors,6 unlike the ES mutants described by Laulier et al. (18). Other than the obvious difference in genotype of the ES cells characterized in the Laulier et al. (18) study and ours, we do not have a clear explanation for why we see hyper-recombination, but they did not, and why we do not see obvious mitotic defects. In Schizosaccharomyces pombe, Fbh1 appears to function as a dedicated HR factor, and moreover, many of the phenotypes seen in S. pombe fbh1 mutants are seen in our Fbh1helΔ/helΔ mutant cells. Hence, we believe that our data are consistent with the evolutionarily conserved role of Fbh1 being in the regulation of HR.
An unexpected feature of mouse ES cells deficient in FBH1 helicase function is hyper-recombination for some, but not all, forms of HR. It will be informative in future studies to define whether FBH1 possesses an inherent ability to regulate only some forms of HR or whether RAD51 itself is differentially sensitive to the action of FBH1, depending on the circumstances. One possibility would be that RAD51 post-translational modifications influence the ability of FBH1 to disrupt the nucleoprotein filament. Our data suggest that replication-associated HR that generates SCEs is insensitive to the effects of FBH1, whereas DSB-induced HR is far more effectively regulated by FBH1. Loss of FBH1 helicase function also does not impact strongly on the cell death response following DNA damage, at least not with the range of agents that we analyzed. This indicates that FBH1 helicase deficiency does not markedly impair HR repair functions required for viability in mouse cells. In the Fbh1helΔ/helΔ mutant cells, the frequency of DSB-induced HR was strongly elevated, as determined by the widely used DR-GFP assay. This assay is designed to measure non-crossover forms of HR that most likely derive from repair events mediated by the synthesis-dependent strand-annealing pathway (21). Taken together with the lack of altered SCE levels in Fbh1helΔ/helΔ cells, which reflect crossover recombination events, we propose a working model in which FBH1 acts as a negative regulator of the synthesis-dependent strand-annealing pathway, but only in cells that are not experiencing excessive DNA damage. This activity would have the effect of preventing spontaneous synthesis-dependent strand-annealing reactions being initiated inappropriately. However, when cells are damaged, FBH1 is apparently no longer functional in restraining RAD51. Hence, a mechanism must exist to suppress the action of FBH1 in cells experiencing DNA damage, thus derepressing RAD51 function to permit essential HR reactions to proceed. This would be logical and consistent with the notion that RAD51-dependent HR is essential for survival in cells exposed to DNA-damaging agents. Interestingly, and consistent with their hyperrecombination phenotype, the Fbh1helΔ/helΔ cells are more resistant than wild-type cells to the PARP inhibitor olaparib. It is known from previous studies that recombination-deficient cells are hypersensitive to PARP inhibitors (29–31). However, our data indicate that the reciprocal relationship is seen in Fbh1helΔ/helΔ cells, namely hyperrecombination accompanied by PARP inhibitor resistance. Hence, our data also suggest that the relative sensitivity of cells to PARP inhibitors correlates closely with their overall recombination efficiency and could have implications for anti-tumor therapies based on the use of PARP inhibitors.
Acknowledgments
We thank members of the Hickson, Janscak, and Rothenberg laboratories for helpful discussions; Roland Kanaar and Giordano Liberi for reagents; and Kamila Burdova for help with protein production and purification.
This work was supported by Cancer Research UK, the Nordea Foundation (Denmark), Czech Science Foundation Grant GAP305/10/0281, and Swiss National Science Foundation Grant 31003A_129747.
M. J. Payne, W. K. Chu, K. Hanada, and I. D. Hickson, unpublished data.
- HR
- homologous recombination
- LOH
- loss of heterozygosity
- DSB
- double strand break
- ES
- embryonic stem
- SCE
- sister chromatid exchange
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- smFRET
- single-molecule fluorescence resonance energy transfer
- PARP
- poly(ADP-ribose) polymerase
- nt
- nucleotide(s)
- DR
- direct repeat.
REFERENCES
- 1. Heyer W. D., Ehmsen K. T., Liu J. (2010) Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Luo G., Santoro I. M., McDaniel L. D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R. A., Bradley A. (2000) Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 26, 424–429 [DOI] [PubMed] [Google Scholar]
- 3. Benson F. E., Stasiak A., West S. C. (1994) Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 13, 5764–5771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu J., Doty T., Gibson B., Heyer W. D. (2010) Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tan T. L., Kanaar R., Wyman C. (2003) Rad54, a Jack of all trades in homologous recombination. DNA Repair 2, 787–794 [DOI] [PubMed] [Google Scholar]
- 6. Sigurdsson S., Van Komen S., Bussen W., Schild D., Albala J. S., Sung P. (2001) Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 15, 3308–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu L., Hickson I. D. (2006) DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 40, 279–306 [DOI] [PubMed] [Google Scholar]
- 8. Antony E., Tomko E. J., Xiao Q., Krejci L., Lohman T. M., Ellenberger T. (2009) Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell 35, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Veaute X., Jeusset J., Soustelle C., Kowalczykowski S. C., Le Cam E., Fabre F. (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 [DOI] [PubMed] [Google Scholar]
- 10. Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., Klein H., Ellenberger T., Sung P. (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 [DOI] [PubMed] [Google Scholar]
- 11. Moldovan G. L., Dejsuphong D., Petalcorin M. I., Hofmann K., Takeda S., Boulton S. J., D'Andrea A. D. (2012) Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell 45, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim J., Kim J. H., Lee S. H., Kim D. H., Kang H. Y., Bae S. H., Pan Z. Q., Seo Y. S. (2002) The novel human DNA helicase hFBH1 is an F-box protein. J. Biol. Chem. 277, 24530–24537 [DOI] [PubMed] [Google Scholar]
- 13. Kim J. H., Kim J., Kim D. H., Ryu G. H., Bae S. H., Seo Y. S. (2004) SCFhFBH1 can act as helicase and E3 ubiquitin ligase. Nucleic Acids Res. 32, 2287–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morishita T., Furukawa F., Sakaguchi C., Toda T., Carr A. M., Iwasaki H., Shinagawa H. (2005) Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates. Mol. Cell Biol. 25, 8074–8083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Osman F., Dixon J., Barr A. R., Whitby M. C. (2005) The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol. Cell Biol. 25, 8084–8096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lorenz A., Osman F., Folkyte V., Sofueva S., Whitby M. C. (2009) Fbh1 limits Rad51-dependent recombination at blocked replication forks. Mol. Cell Biol. 29, 4742–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kohzaki M., Hatanaka A., Sonoda E., Yamazoe M., Kikuchi K., Vu Trung N., Szüts D., Sale J. E., Shinagawa H., Watanabe M., Takeda S. (2007) Cooperative roles of vertebrate Fbh1 and Blm DNA helicases in avoidance of crossovers during recombination initiated by replication fork collapse. Mol. Cell Biol. 27, 2812–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Laulier C., Cheng A., Huang N., Stark J. M. (2010) Mammalian Fbh1 is important to restore normal mitotic progression following decatenation stress. DNA Repair 9, 708–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fugger K., Mistrik M., Danielsen J. R., Dinant C., Falck J., Bartek J., Lukas J., Mailand N. (2009) Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J. Cell Biol. 186, 655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yusa K., Horie K., Kondoh G., Kouno M., Maeda Y., Kinoshita T., Takeda J. (2004) Genome-wide phenotype analysis in ES cells by regulated disruption of Bloom's syndrome gene. Nature 429, 896–899 [DOI] [PubMed] [Google Scholar]
- 21. Pierce A. J., Jasin M. (2005) Measuring recombination proficiency in mouse embryonic stem cells. Methods Mol. Biol. 291, 373–384 [DOI] [PubMed] [Google Scholar]
- 22. Sigurdsson S., Van Komen S., Petukhova G., Sung P. (2002) Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 277, 42790–42794 [DOI] [PubMed] [Google Scholar]
- 23. Henricksen L. A., Umbricht C. B., Wold M. S. (1994) Recombinant replication protein A. Expression, complex formation, and functional characterization. J. Biol. Chem. 269, 11121–11132 [PubMed] [Google Scholar]
- 24. Garcia P. L., Liu Y., Jiricny J., West S. C., Janscak P. (2004) Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 23, 2882–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rothenberg E., Ha T. (2010) Single-molecule FRET analysis of helicase functions. Methods Mol. Biol. 587, 29–43 [DOI] [PubMed] [Google Scholar]
- 26. Ha T., Rasnik I., Cheng W., Babcock H. P., Gauss G. H., Lohman T. M., Chu S. (2002) Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature 419, 638–641 [DOI] [PubMed] [Google Scholar]
- 27. Fugger K., Chu W. K., Haahr P., Kousholt A. N., Beck H., Payne M. J., Hanada K., Hickson I. D., Sørensen C. S. (2013) FBH1 co-operates with MUS81 in inducing DNA double-strand breaks and cell death following replication stress. Nat. Commun. 4, 1423. [DOI] [PubMed] [Google Scholar]
- 28. Fong P. C., Boss D. S., Yap T. A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O'Connor M. J., Ashworth A., Carmichael J., Kaye S. B., Schellens J. H., de Bono J. S. (2009) Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 361, 123–134 [DOI] [PubMed] [Google Scholar]
- 29. Farmer H., McCabe N., Lord C. J., Tutt A. N., Johnson D. A., Richardson T. B., Santarosa M., Dillon K. J., Hickson I., Knights C., Martin N. M., Jackson S. P., Smith G. C., Ashworth A. (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 [DOI] [PubMed] [Google Scholar]
- 30. Bryant H. E., Schultz N., Thomas H. D., Parker K. M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N. J., Helleday T. (2005) Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 [DOI] [PubMed] [Google Scholar]
- 31. Chi P., Van Komen S., Sehorn M. G., Sigurdsson S., Sung P. (2006) Roles of ATP binding and ATP hydrolysis in human Rad51 recombinase function. DNA Repair 5, 381–391 [DOI] [PubMed] [Google Scholar]
- 32. Ogawa T., Yu X., Shinohara A., Egelman E. H. (1993) Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science 259, 1896–1899 [DOI] [PubMed] [Google Scholar]
- 33. Hu Y., Raynard S., Sehorn M. G., Lu X., Bussen W., Zheng L., Stark J. M., Barnes E. L., Chi P., Janscak P., Jasin M., Vogel H., Sung P., Luo G. (2007) RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 21, 3073–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qiu Y., Antony E., Doganay S., Ran Koh H., Lohman T. M., Myong S. (2013) Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat. Commun. 4, 2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yodh J. G., Stevens B. C., Kanagaraj R., Janscak P., Ha T. (2009) BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO J. 28, 405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Myong S., Rasnik I., Joo C., Lohman T. M., Ha T. (2005) Repetitive shuttling of a motor protein on DNA. Nature 437, 1321–1325 [DOI] [PubMed] [Google Scholar]
- 37. Bugreev D. V., Mazina O. M., Mazin A. V. (2009) Bloom syndrome helicase stimulates RAD51 DNA strand exchange activity through a novel mechanism. J. Biol. Chem. 284, 26349–26359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bugreev D. V., Yu X., Egelman E. H., Mazin A. V. (2007) Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 21, 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ward J. D., Muzzini D. M., Petalcorin M. I., Martinez-Perez E., Martin J. S., Plevani P., Cassata G., Marini F., Boulton S. J. (2010) Overlapping mechanisms promote postsynaptic RAD-51 filament disassembly during meiotic double-strand break repair. Mol. Cell 37, 259–272 [DOI] [PubMed] [Google Scholar]