Background: Polyphosphate (polyP) accumulates in an acidic calcium store named the acidocalcisome.
Results: TbVtc4 is an acidocalcisome polyP kinase required for osmoregulation and virulence.
Conclusion: TbVtc4 is a potential drug target in T. brucei
Significance: This is the first demonstration of an essential polyP kinase in trypanosomes.
Keywords: Enzyme Catalysis, Parasite, Parasite Metabolism, Trypanosoma brucei, Vacuolar Acidification, Acidocalcisome, Osmoregulation, Polyphosphate, Vacuolar Transporter Chaperone
Abstract
Polyphosphate (polyP) is an anionic polymer of orthophosphate groups linked by high energy bonds that typically accumulates in acidic, calcium-rich organelles known as acidocalcisomes. PolyP synthesis in eukaryotes was unclear until it was demonstrated that the protein named Vtc4p (vacuolar transporter chaperone 4) is a long chain polyP kinase that localizes to the yeast vacuole. Here, we report that TbVtc4 (Vtc4 ortholog of Trypanosoma brucei) encodes, in contrast, a short chain polyP kinase that localizes to acidocalcisomes. The subcellular localization of TbVtc4 was demonstrated by fluorescence and electron microscopy of cell lines expressing TbVtc4 in its endogenous locus fused to an epitope tag and by purified polyclonal antibodies against TbVtc4. Recombinant TbVtc4 was expressed in bacteria, and polyP kinase activity was assayed in vitro. The in vitro growth of conditional knock-out bloodstream form trypanosomes (TbVtc4-KO) was significantly affected relative to the parental cell line. This mutant had reduced polyP kinase activity and short chain polyP content and was considerably less virulent in mice. The wild-type phenotype was recovered when an ectopic copy of the TbVtc4 gene was expressed in the presence of doxycycline. The mutant also exhibited a defect in volume recovery under osmotic stress conditions in vitro, underscoring the relevance of polyP in osmoregulation.
Introduction
Polyphosphate (polyP)3 is an inorganic, linear polymer of orthophosphate (Pi) units linked by phosphoanhydride bonds. PolyP can exist as short (3 to ∼300 Pi) or long chain (∼300 to ∼1,000 Pi) polymers, is abundant in nature, and has been conserved during evolution (1, 2). PolyP has been extensively studied in bacteria, where it is involved in several essential functions, such as DNA replication, sporulation, germination, motility, and pathogenesis. Much less is known of the functions of polyP in eukaryotes (1, 2). The recent discoveries that polyP can be released from some mammalian cells, such as blood platelets (3) and mast cells (4), and has potent modulatory activity on blood coagulation (5) and inflammation (6) have renewed interest in this polymer. Interestingly, polyP with chain lengths characteristic of microorganisms modulates coagulation and inflammation differently than polyP with chain lengths typically found in mammalian cells (7).
In many organisms, polyP is mobilized primarily by the synthetic activity of polyP kinases and degradation by endo- and exopolyphosphatases, respectively. A few genes encoding exopolyphosphatases (8–11) and endopolyphosphatases (12) have been described in eukaryotes. Recently, the first eukaryotic enzyme involved in synthesis and translocation of polyP, ScVtc4p (Saccharomyces cerevisiae vacuolar transporter chaperone 4), was identified (13). The Vtc complex consists of four proteins (Vtc1–4) that form hetero-oligomeric complexes and are able to synthesize and transfer polyP into the vacuole as well as impacting membrane trafficking and vacuole fusion (14–16). Vtc4 forms the catalytic core of the complex, although null mutations of each of the Vtc proteins result in reduced accumulation of polyP. Vtc proteins are present in fungi, algae, trypanosomatids, and Apicomplexan parasites but are absent in mammalian cells.
In many cells, short and long chain polyP accumulate in acidocalcisomes, acidic calcium stores (17) where polyP is complexed with several cations (18, 19). These organelles were first described in Trypanosoma brucei (20) but later identified in a broad range of organisms from bacterial to human cells (18) and are involved in Ca2+ signaling, as inferred from the presence in them of an inositol 1,4,5-trisphosphate receptor (21). T. brucei belongs to the group of trypanosomes that causes human African trypanosomiasis (also known as sleeping sickness), an endemic disease of Sub-Saharan Africa. There is no vaccine available for this disease, and chemotherapy also remains unsatisfactory, especially for advanced cases when a neurological phase has been reached and the disease becomes potentially fatal.
Previous work has shown that polyP has a critical role in survival of trypanosomes under sharp environmental changes, including osmotic stress (22–24). This resistance to osmotic stress is essential for digenetic trypanosomatids as they encounter drastic osmotic changes in both the insect vectors and vertebrate hosts (23, 25, 26). Regulation of cell volume is, in addition, a homeostatic process needed at all times by all cells. PolyP hydrolysis occurs during hyposmotic stress of trypanosomes (22), probably increasing the osmotic pressure of the acidocalcisomes and facilitating water movement. On the other hand, an increase in long chain polyP levels has been observed in T. cruzi during hyperosmotic stress (22, 23). This latter work suggested that polyP could play an important role at the early stages of hyperosmotic stress response by sequestering ions into the acidocalcisomes to reduce the ionic strength of the cytosol (23).
Homologs of S. cerevisiae Vtc1 and Vtc4 genes are present in the genome of T. brucei. TbVtc1, a protein present in T. brucei acidocalcisomes (27), is essential for polyP synthesis and acidocalcisome biogenesis. However, this protein does not have a polyP kinase domain or PPK activity. An ScVtc4p homolog (TbVtc4) was detected in a proteomic analysis of T. brucei acidocalcisomes.4 In the present study, we investigated the role of this enzyme by biochemical and genetic approaches, elucidating important aspects of its physiological role in T. brucei, where polyP seems to be essential for parasite survival in the mammalian host. Because Vtc4 proteins are absent in vertebrates, we propose this enzyme as a potential target for drug development and disease control.
EXPERIMENTAL PROCEDURES
Culture Methods
Cultivation of 29-13 procyclic form (PCF) (28) and single marker bloodstream form (BSF) (29) trypanosomes derived from T. brucei Lister strain 427 was carried out as described previously (30). Cell growth was followed using a Beckman® Coulter Z1 dual cell and particle counter.
Chemicals and Reagents
TRIzol reagent, MagicMedia, Taq polymerase, BenchMark protein ladder, Alexa-conjugated secondary antibodies, and Escherichia coli BL21 Codon Plus (DE3)-RIPL were purchased from Invitrogen. Vector pET32 Ek/LIC, Benzonase® nuclease, anti-histidine tag antibodies, and S-protein HRP conjugate were from Novagen (EMD Millipore, Billerica, MA). [α-32P]dCTP (3,000 Ci/mmol) and [γ-32P]ATP (3,000 Ci/mmol) were from PerkinElmer Life Sciences. Rabbit and mouse antibodies against T. brucei vacuolar H+-pyrophosphatase (TbVP1) (31) were a gift from Dr. Norbert Bakalara (Ecole Nationale Supérieure de Chimie de Montpellier, Montpellier, France). Anti-HA high affinity rat monoclonal antibody (clone 3F10) was purchased from Roche Applied Science. The pMOTag4H vector (32) was a gift from Dr. Thomas Seebeck (University of Bern, Bern, Switzerland). PD-10 desalting columns were from GE Healthcare. Pierce ECL Western blotting substrate and Pierce BCA protein assay reagent were from Thermo Fisher Scientific Inc. Zeta-Probe GT genomic testing blotting and nitrocellulose membranes were from Bio-Rad. The AMAXA human T-cell Nucleofector kit was purchased from Lonza (Germany). The Prime-a-Gene labeling system was from Promega (Madison, WI). QIAprep Spin Miniprep and Midiprep kits, the QIAquick gel extraction kit, and the MinElute PCR purification kit were from Qiagen (Valencia, CA). The fluorimetric ADP assay kit was from PhosphoWorks (AAT Bioquest, Inc., Sunnyvale, CA). The primers were purchased from Integrated DNA Technologies (Coralville, IA). Antibiotics and all other reagents of analytical grade were from Sigma.
Sequence Analysis
The analysis of TbVtc4 sequence (gene ID Tb11.01.4040) was performed using DNAMAN software (version 7.212, Lynnon Corp., Quebec, Canada) for alignments, BLAST for searching homologous sequences, the Motif Scan algorithm for prediction of functional domains (33), and the TopPred algorithm for the prediction of transmembrane domains (34). General information available for this sequence was obtained from TriTrypDB (35).
Gene Cloning and Protein Heterologous Expression
DNA sequence corresponding to the TbVtc4 catalytic core (nucleotides 595–1518 of the TbVtc4 open reading frame, amino acids 199–506 of the full-length protein) was PCR-amplified from T. brucei 29-13 strain gDNA (forward primer, 5′-GACGACGACAAGATACCTTGTGGTACCGTTGG-3′; reverse primer, 5′-GAGGAGAAGCCCGGTGTGGAAGCGCGAATGTCAA-3′) and ligation-independently cloned into vector pET32 Ek/LIC for heterologous expression in bacteria. The sequence of several recombinant clones was verified, and they were transformed by heat shock into E. coli BL21 Codon Plus (DE3)-RIPL chemically competent cells. Induction of TbVtc4(199–506) expression was performed with MagicMedia following the manufacturer's dual temperature protocol to avoid aggregation of protein in inclusion bodies for purification under native conditions. Protein expression was alternatively induced with 1 mm isopropyl-β-d-thiogalactopyranoside in LB broth for 3 h at 37 °C for purification under denaturing conditions.
The catalytic domain of ScVtc4p (aa 189–480 (13)) was amplified from yeast gDNA using standard PCR protocols (forward primer, 5′-GAGCTCAAGGGAAGCAACAAAATTTC-3′; reverse primer, 5′-CACGTGTCATTGAGGTAACCAAAATG-3′) and Pfu Ultra HF (Stratagene). The fragment was cloned with a TOPO-TA cloning kit, verified by sequencing, and ligated into the expression vector pQE-2 (Qiagen) using SacI and PmlI sites that were added to the 5′- and 3′-primers, respectively. E. coli (BL21 Codon Plus DE3 RIPL, Stratagene) were transformed with ScVtc4p(189–480)/pQE-2. Histidine-tagged ScVtc4p(189–480) was induced overnight at 25 °C with 0.5 m isopropyl-β-d-thiogalactopyranoside, isolated by metal ion affinity chromatography, and desalted on a HiTrap column (GE Healthcare) with 25 mm Tris, 200 mm NaCl, 2 mm DTT (13).
Purification of Recombinant TbVtc4 Catalytic Core under Native Conditions
Cell pellets from a 200-ml culture of recombinant E. coli BL21 expressing TbVtc4(199–506) grown in MagicMedia were resuspended and incubated for 30 min on ice in 20 ml of cold lysis buffer: 50 mm sodium phosphate, pH 8.0, 0.3 m sodium chloride, 10 mm imidazole, 0.1% Triton X-100, 0.1 mg/ml lysozyme, 25 units/ml Benzonase® nuclease and protease inhibitor mixture for purification of histidine-tagged proteins (Sigma; 50 μl/g of cell paste). Then three sonication pulses (40% amplitude, 30 s, on ice) were applied to ensure the complete disruption of cells. After centrifugation at 20,000 × g for 30 min at 4 °C, supernatant was passed through a 0.8-μm pore nitrocellulose filter in order to obtain a clarified crude protein extract that was kept on ice and used for immediate purification of recombinant TbVtc4(199–506). Protein purification was performed at 4 °C using HIS-Select® Cartridge (Sigma), in an immobilized nickel-ion affinity chromatography system, following the manufacturer's protocol for histidine-tagged protein purification under native conditions. One-ml fractions were eluted (elution buffer: 50 mm sodium phosphate, pH 8.0, 0.3 m sodium chloride, 250 mm imidazole), and buffer exchange was performed immediately using PD-10 desalting columns to finally obtain the protein in assay buffer (20 mm Hepes, pH 6.5). All purification steps were verified by SDS-PAGE and Western blot analyses using anti-histidine tag commercial antibodies or S-protein HRP conjugate. For antibody production, recombinant TbVtc4(199–506) was purified under denaturing conditions using HIS-Select® Cartridge (Sigma).
Antibody Production
Polyclonal anti-TbVtc4 antibodies were generated in a guinea pig by Covance (Princeton, NJ) against a synthetic peptide (CSRSRRVYARRKIRYDDRRG) that corresponds to a conserved hydrophilic region located between the second and the third predicted transmembrane domains of the TbVtc4 amino acid sequence. In addition, polyclonal anti-TbVtc4 antibodies were produced in mice using recombinant TbVtc4(199–506) as an antigen. These antibodies were generated at the Monoclonal Antibody Facility of the College of Veterinary Medicine, University of Georgia (Athens, GA). Final bleeds from five inoculated mice were affinity-purified by immunoadsorption to the recombinant protein immobilized on nitrocellulose strips. The adsorbed antibodies were eluted with 0.1 m glycine, pH 2.5, and neutral pH was restored immediately by adding 1 m Tris-HCl buffer, pH 8.0.
Fluorescence Microscopy
For immunofluorescence assays, T. brucei PCF trypanosomes were centrifuged at 1,000 × g for 10 min at 25 °C; washed twice with PBS, pH 7.4; and fixed with 4% paraformaldehyde in PBS for 1 h on ice. Afterward, cells were adhered to poly-l-lysine-coated coverslips; permeabilized with 0.3% Triton X-100 for 3 min; washed three times; and blocked with PBS containing 3% BSA, 1% fish gelatin, 50 mm NH4Cl, and 5% goat serum for 1 h. Next, cells were incubated for 1 h at room temperature with primary antibodies: polyclonal guinea pig anti-TbVtc4 (1:50) or rat anti-HA tag high affinity mAb (Roche Applied Science; diluted 1:10) and polyclonal rabbit anti-TbVP1 (1:250), as acidocalcisome marker. After washing three times with 3% BSA in PBS (pH 8.0), cells were incubated for 45 min at room temperature in the dark with secondary antibodies: Alexa Fluor 488-conjugated goat anti-guinea pig or anti-rabbit (1:1,000) and Alexa Fluor 546-conjugated goat anti-rabbit or anti-mouse (1:1,500). Then, cells were counterstained with 5 μg/ml DAPI to label nuclei and kinetoplast (mitochondrial DNA). Finally, all preparations were washed again three times with 3% BSA in PBS (pH 8.0) and mounted on glass slides with Fluoromount-G (Southern Biotechnology). Differential interference contrast and fluorescence optical images were captured under non-saturating conditions and identical exposure times using an Olympus IX-71 inverted fluorescence microscope with a Photometrix CoolSnapHQ charge-coupled device (CCD) camera driven by DeltaVision software (Applied Precision).
Electron Microscopy
BSF trypanosomes were washed twice in 0.1 m sodium cacodylate buffer, pH 7.4, and fixed for 1 h on ice with 0.1% glutaraldehyde, 4% paraformaldehyde, and 0.1 m sodium cacodylate buffer, pH 7.4. Samples were processed for cryo-immunoelectron microscopy at the Molecular Microbiology Imaging Facility, Washington University School of Medicine. HA fusion protein localization was detected with a polyclonal antibody against HA and anti-rabbit gold-conjugated as a secondary antibody. Mouse anti-TbVP1 polyclonal antibodies and anti-mouse gold-conjugated secondary antibodies were used.
Enzymatic Assays for PolyP Synthesis-ADP Assay
For determination of TbVtc4 and ScVtc4p kinetic parameters, the specific activity of the enzyme was assayed using an ADP determination kit (PhosphoWorksTM fluorimetric ADP assay kit, AAT Bioquest, Inc.) to quantify the amount of ADP or GDP synthesized during polyP polymerization at different ATP or GTP concentrations. Analysis of ATP conversion by the recombinant catalytic domain of ScVtc4p was carried out in buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm MnCl2, and 1 mm ATP, at room temperature using 1 μm ScVtc4p. TbVtc4 polyP kinase activity was assayed in buffer containing 50 mm Hepes (pH 6.5), 150 mm NaCl, and 1 mm MnCl2, at room temperature using 1 μm TbVtc4 and 1 mm ATP or different concentrations of ATP or GTP. When indicated, 1 mm PPi was included in the reactions. Twenty-μl reactions were incubated 1 h at room temperature. Components A and B of the ADP/GDP determination kit were added immediately, and after a 30-min incubation at room temperature, fluorescence was detected at a 540/590-nm excitation/emission ratio in a Molecular Devices plate reader. An ADP (or GDP) standard curve was obtained for quantification purposes. The data were fit to a Michaelis-Menten equation, and GraphPad Prism software version 5.0 was used for data analysis and determination of Km, Vmax, and kcat.
Coupled Assay
ATP hydrolysis was also monitored via NADH oxidation enzymatically coupled to the rephosphorylation of produced ADP. NADH concentration was measured optically at 340 nm in buffer containing 150 μm NADH, 0.15 mm phosphoenolpyruvate, 0.1 mg/ml pyruvate kinase, and 0.1 mg/ml lactate dehydrogenase. The reaction was initiated by the addition of 1 mm ATP, and the incubations were done under the same conditions described above.
Radioactive Assay
To visualize TbVtc4 and ScVtc4p reaction products, newly synthesized polyP chains were detected by autoradiography using [γ-32P]ATP as substrate. PolyP was separated by electrophoresis on Tris borate-EDTA (TBE)-polyacrylamide gels. Reactions were carried out as described above using 1 mm [γ-32P]ATP (20 Ci/mmol) in a final volume of 50 μl for 8 h at room temperature and stopped by adding EDTA (final concentration = 1 mm) and 10× sample buffer (2 mg/ml Orange G, 30% glycerol). Before loading the samples, TBE-polyacrylamide gels (0.1 × 16 × 20 cm, 1× TBE, 10% polyacrylamide (19:1 acrylamide/bisacrylamide), 0.05% tetramethylenediamine, 0.05% (w/w) ammonium persulfate) were prerun at 200 V for 30 min in a cold room using a PROTEAN® II xi Cell (Bio-Rad). Thirty μl of sample and commercial polyP markers were loaded per well as indicated. Gels were run with a 4-mA constant current for 20 h at 4 °C. Different pH and cation requirements were assayed. Dried gels were exposed to films for at least 48 h at −80 °C for autoradiography.
Molecular Constructs for TbVtc4 Mutant Cell Lines
For TbVtc4 knock-out construction in BSF trypanosomes, one TbVtc4 allele was knocked out by replacement with a puromycin selectable marker in the single marker line that expresses T7 polymerase and tetracycline repressor maintained by a single G418 resistance marker (29). This puromycin cassette was obtained by PCR using a set of long primers (ultramers) containing 100–120 nucleotides from the 5′- and 3′-UTRs flanking regions of the TbVtc4 ORF (forward primer, 5′-GCTGTTGTTGTTTTCTTTTTCATTATTGTTTACAAAGAAGTACGATAAGAGGAACATTAAGTGTTGAGAGGCAAGGAGAAGCAAACAAACAGAGTTATAACGTGGTACCGGGCCCCCCCTCGAG-3′; reverse primer, 5′-CGTTAAACATAGCAGAACATCAGCACATTACTGACAATCAACCAACATGTACACGTTCTTTTCCGTGAAAGCCAACATATTTCCTGCCCCTCCCTCAGTCTGGCGGCCGCTCTAGAACTAGTGGAT-3′) and pMOTag23M vector as template (32). To replace the second allele, we first introduced an ectopic copy of the gene (TbVtc4ec) under the control of the tetracycline-inducible promoter and selectable by blasticidin resistance inserted at the ribosomal non-transcribed spacer. This cassette was constructed, amplifying the TbVtc4 gene by PCR from T. brucei single marker strain gDNA (forward primer, 5′-CAGTGATCATATGCCCTTCAGCAAAGCATG-3′; reverse primer, 5′-AGATGATCATCAGAAGGTGTCGCTTCCGG-3′) to clone TbVtc4ec into pLEW100v5b1d-BSD expression vector (a gift from Dr. George Cross, The Rockefeller University, New York), a modified version of the original pLEW100 vector (29). The construct was linearized with NotI restriction enzyme before cell transfection. Finally, the second TbVtc4 allele was knocked out by replacement with a phleomycin selectable marker while keeping the ectopic copy “on” by the addition of tetracycline to the selection medium. The phleomycin cassette was also PCR-amplified using a primer set containing 100–120 nucleotides from TbVtc4 ORF 5′- and 3′-UTRs (forward primer, 5′-GCTGTTGTTGTTTTCTTTTTCATTATTGTTTAACAAAGAAGTACGATAAGAGGAACATTAAGTGTTGAGAGGCAAGGAGAAGCAAACAAACAGAGTTATAACGTATGGCCAAGTTGACCAGTGCCG-3′; reverse primer, 5′-CGTTAAACATAGCAGAACATCAGCACATTACTGACAATCAACCAACATGTACACGTTCTTTTCCGTGAAAGCCAACATATTTCCTGCCCCTCCCTCAGTCTCAGTCCTGCTCCTCGGCCA-3′) and pUB39 vector, a modified version of the original pLEW82 vector (29), as DNA template. The linear constructs were used for transfection of BSF trypanosomes (single marker strain) and selection of stable resistant clones. Finally, a C-terminal HA-tagged mutant (TbVtc4-HA) was generated using also ultramers (forward primer, 5′-GTAGCGTTAACATTTGTGATATTAGCCGTTATTCTTATAACTGTTATGATGCACGTTATGGTCCGGTACGGGCCTATGCTCACCGGAAGCGACACCTTCGGTACCGGGCCCCCCCTCGAG-3′; reverse primer, 5′-CGTTAAACATAGCAGAACATCAGCACATTACTGACAATCAACCAACATGTACACGTTCTTTTCCGTGAAAGCCAACATATTTCCTGCCCCTCCCTCAGTCTGGCGGCCGCTCTAGAACTAGTGGAT-3′) and pMOTag4H vector (32) as DNA template to generate a linear fragment that was used to transfect T. brucei PCF and BSF.
Cell Transfections
T. brucei BSF were transfected as described previously (30) with some modifications. Briefly, 3 × 107 mid-log phase cells (cell density in culture below 1 × 106 cells/ml) were harvested by centrifugation at 1,300 × g for 10 min and resuspended in 100 μl of AMAXA human T-cell Nucleofector solution. Then 10 μg of NotI-linearized plasmid DNA or purified PCR product (<10 μl) was added to cells in 2-mm gap cuvettes. Immediately, one electroporation pulse was applied using program X-001 of the AMAXA Nucleofector® II apparatus. Following each transfection, resistant cells were selected and cloned by limiting dilution in HMI-9 medium containing 20% tetracycline-free FBS with appropriate antibiotics (2.5 μg/ml G418, 0.1 μg/ml puromycin, 2.5 μg/ml blasticidin, 2.5 μg/ml phleomycin, or 5.0 μg/ml hygromycin) in 24-well plates. Integration of the constructs into genomic DNA of each transfectant cell line was verified by PCR and Southern blot analysis.
For PCF transfection, mid-log phase parasites (cell density around 5 × 106 cells/ml) were harvested by centrifugation at 1,000 × g for 7 min, washed with cold cytomix (2 mm EGTA, 5 mm MgCl2, 120 mm KCl, 0.5% glucose, 0.15 mm CaCl2, 0.01% BSA, 10 mm K2HPO4/KH2PO4, 1 mm hypoxanthine, 25 mm Hepes, pH 7.6), and resuspended in 0.5 ml of cytomix at a cell density of 5 × 107 cells/ml. Then cells were mixed with 10 μg of PCR product (<50 μl) in a 0.4-cm electroporation cuvette and subjected to two pulses (1,500 V, 25 microfarads) in a Bio-Rad Gene Pulser electroporator. The stable transfectants were obtained in SDM-79 medium supplemented with 15% FBS plus the appropriate antibiotic (50 μg/ml hygromycin).
Southern Blot Analysis
Genomic DNA from parental and TbVtc4 mutant cell lines was extracted as described (36). Two μg of gDNA were digested overnight with BamHI and HindIII restriction enzymes. Digestion products were resolved by electrophoresis on a 1% agarose gel in Tris-acetate EDTA buffer at 50 V. DNA was transferred from agarose gels onto Zeta-Probe blotting membranes (Bio-Rad) by capillarity overnight using 0.4 m NaOH as transfer solution. Membranes were hybridized with a radiolabeled TbVtc4 probe, generated by PCR (forward primer, 5′-GACGACGACAAGATACCTTGTGGTACCGTTGG-3′; reverse primer, 5′-GAGGAGAAGCCCGGTGTGGAAGCGCGAATGTCAA-3′), and labeled with [α-32P]dCTP using random hexanucleotide primers and the Klenow fragment of DNA polymerase I (Prime-a-Gene labeling system, Promega). Membranes were exposed to films for 24–72 h at −80 °C and developed in a dark room.
Northern Blot Analysis
Northern blot analysis was performed as described previously (23). Briefly, total RNA was isolated from BSF using TRI Reagent®. RNA samples were subjected to electrophoresis in 1% agarose gels containing 2.2 m formaldehyde, 20 mm MOPS, pH 7.0, 1 mm EDTA, and 8 mm sodium acetate, transferred to nylon membranes, and hybridized with radiolabeled probes for TbVtc4 and the Tb-β-tubulin gene (Tb927.1.2390). Tb-β-tubulin probe was also generated by PCR (forward primer, 5′-TGCGTGAGATTGTGTGCGTTCAGG-3′; reverse primer, 5′-AGTGCAGACGAGGGAACGGCACCA-3′) and labeled with [α-32P]dCTP using random hexanucleotide primers and the Klenow fragment of DNA polymerase I (Prime-a-Gene labeling system, Promega). The Tb-β-tubulin gene was used as a loading control, assuming a similar level of expression of this gene in all mutants at the same stage. Finally, membranes were exposed to films for 24–72 h at −80 °C and developed in a dark room.
Western Blot Analysis
Parental and mutant cell lines were separately harvested. Parasites were washed twice in PBS (PCF) or buffer A with glucose (BAG; 116 mm NaCl, 5.4 mm KCl, 0.8 mm MgSO4, 50 mm HEPES, and 5.5 mm glucose, pH 7.3) for BSF and resuspended in radioimmune precipitation assay buffer (150 mm NaCl, 20 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1% SDS, and 0.1% Triton X-100) plus protease inhibitors (mammalian cell protease inhibitor mixture (Sigma P8340, diluted 1:250), 1 mm EDTA, 1 mm PMSF, 2.5 mm tosylphenylalanyl chloromethyl ketone and 100 μm E64) and Benzonase® nuclease (25 units/ml of culture). Then cells were incubated for 30 min on ice, and five rounds of freeze-thaw were applied (5 min on dry ice/ethanol bath, 1 min on 37 °C water bath). Cell lysis was verified under a light microscope, and protein concentration was determined by a BCA protein assay (Pierce). Thirty μg of protein from each cell lysate were mixed with 4× Laemmli sample buffer and analyzed by SDS-PAGE in 10% gels. Separated proteins were transferred onto nitrocellulose membranes (Bio-Rad) using a Bio-Rad transblot apparatus. Membranes were blocked with 5% nonfat dried skim milk in PBST (PBS containing 0.1% (v/v) Tween 20) overnight at 4 °C. Next, blots were incubated for 1 h at room temperature with different primary antibodies: polyclonal mouse anti-TbVtc4 (1:500), rat anti-HA tag mAb (1:100), or anti-tubulin mAb (1:50,000). After five washes with PBST, blots were incubated with the appropriate secondary antibody: HRP-conjugated goat anti-mouse or anti-rat IgG (1:15,000) for 1 h at room temperature. After washing five times with PBST, the immunoblots were visualized using ECL Western blotting substrate (Pierce) according to the manufacturer's instructions.
Short Chain and Long Chain Polyphosphate Quantification
Determination of polyP levels in BSF parental and mutant cell lines was performed as described previously by measuring Pi release by recombinant yeast expolyphosphatase (37).
Regulatory Volume Changes under Osmotic Stress Conditions
For osmotic stress under constant ionic strength, the following buffers described previously (37, 38) with some modifications were used: isotonic (64 mm NaCl, 4 mm KCl, 1.8 mm CaCl2, 0.53 mm MgCl2, 5.5 mm glucose, 5 mm Na-Hepes, pH 7.4, and 150 mm mannitol; 320 ± 5 mosm, as determined using an Advanced Instruments 3D3 osmometer), hypotonic (the same as isotonic but without mannitol; 160 ± 5 mosm), and hypertonic (the same as isotonic but with increased mannitol concentration to 1.2 m; 980 ± 5 mosm). Samples of 1 × 107 BSF (single marker strain and TbVtc4-KO with or without tetracycline) were collected, washed with isotonic buffer prewarmed at 37 °C, and resuspended in 100 μl of isotonic buffer. Next, cells were transferred to a 96-well plate, and changes in cell volume were followed monitoring absorbance at 550 nm in a plate reader with continuous agitation to avoid decantation of parasites. Osmotic stress was induced after 3 min of absorbance recording as follows. Hyposmotic stress was induced by the addition of 200 μl of hypotonic buffer to 100 μl of cells in isotonic buffer (final osmolarity, 213 mosm), hyperosmotic stress was induced by the addition of 100 μl of hypertonic buffer to 100 μl of cells in isotonic buffer (final osmolarity, 650 mosm), and controls adding 100 and 200 μl of isotonic buffer to the cells were carried out in parallel. After inducing osmotic stress, absorbance at 550 nm was recorded for an additional 10 min. Cell viability was verified in the microscope after 10 min under osmotic stress.
In Vivo Studies
To evaluate the infectivity of TbVtc4-KO BSF trypanosomes, the cells were cultivated for 14 days in the absence of tetracycline. Exponentially growing cells (single marker and TbVtc4-KO with or without tetracycline) were washed once in HMI-9 medium without selectable drugs and resuspended in the same medium. Eight-week-old BALB/c mice (5 mice/group) were infected with a single intraperitoneal injection of 2 × 104 BSF trypanosomes in 0.2 ml of HMI-9 medium. Mice were given either normal water or water containing 200 μg/ml doxycycline in a 5% sucrose solution (30, 31). The drinking water with or without doxycycline was provided 3 days before infection and exchanged every 2–3 days, continuing throughout the 30-day period. Animals were fed ad libitum on standard chow. Parasitemia levels were monitored 1–2 times/week during the whole experiment (39). Mice were euthanized upon attaining a parasite density over 1 × 108 cells/ml. This study was carried out in strict accordance with the recommendations in the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The animal protocol was approved by the Institutional Animal Care and Use Committee of the University of Georgia.
Statistical Analyses
For all experiments, results were expressed as mean values of three independent experiments ± S.D. Statistical analyses were performed using GraphPad Prism software version 5.0. Comparison of polyP kinase activity and short and long chain polyP content in different cell lines was performed by Student's t test with a significance level of 0.05. Comparison of changes in cell volume after osmotic stress in different cell lines was done by Bonferroni's multiple comparison a posteriori test of one-way analysis of variance at all time points after induction of osmotic stress (significance level = 0.05). In this way, the pattern of response to osmotic stress of all cell lines was analyzed during the entire period of observation.
RESULTS
TbVtc4 Sequence Analysis
The TbVtc4 amino acid sequence (NCBI Reference Sequence: XP_829284.1; TriTrypDB sequence: Tb11.01.4040) was aligned with orthologs from other kinetoplastids and also with ScVtc4p (not shown). TbVtc4 sequence shares 72.2, 66.9, and 23.3% amino acid identity with T. cruzi Vtc4 (TcCLB.511127.100), Leishmania major Vtc4 (LmjF09.0220), and ScVtc4p (AAP21767), respectively. The ORF of TbVtc4 encodes a protein of 793 amino acids with a predicted molecular mass of 91.3 kDa. TbVtc4 has a VTC domain at the hydrophilic region (aa 210–482) and three predicted transmembrane domains (TMDs), one putative (aa 476–496) and two certain (aa 716–738 and aa 759–781). The catalytic domain (VTC) is highly conserved (61.5% identity) among these four orthologs.
TbVtc4 Localizes to Acidocalcisomes
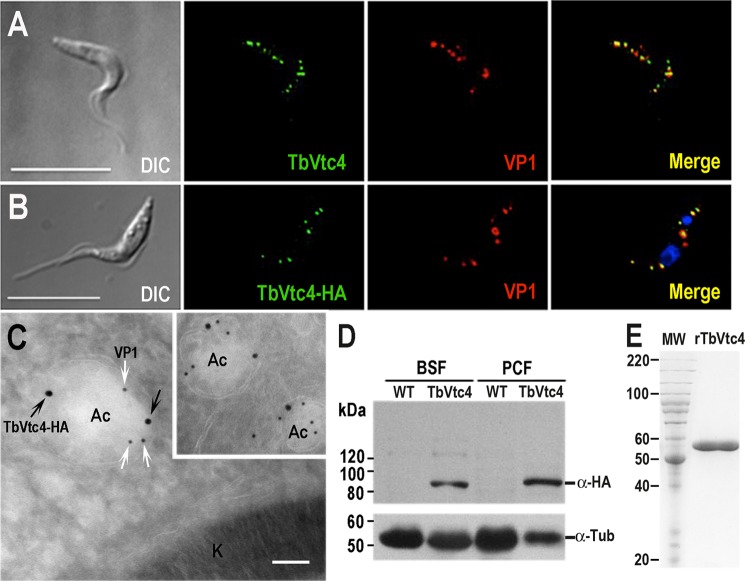
To establish the localization of TbVtc4, an in situ tagging technique was used. We designed the tagged gene to remain under wild-type regulation, avoiding the pitfall of overexpression and consequently increased risk of abnormal distribution of the tagged protein. The linear epitope-tagging cassette was transfected into PCF and BSF trypanosomes, where it integrated into the original TbVtc4 locus via homologous recombination. TbVtc4 co-localized with the vacuolar proton pyrophosphatase (TbVP1), an acidocalcisomal marker (31, 40), in both PCF (Fig. 1B) and BSF trypanosomes (Fig. 1C), as detected by immunofluorescence (Fig. 1B) and immunoelectron microscopy (Fig. 1C), respectively. Western blot analysis confirmed the expression of the tagged protein of the expected size and showed that TbVtc4 expression levels were higher in PCF (Fig. 1D). Similar co-localization results were observed in PCF using affinity-purified, polyclonal antibodies against synthetic peptide fragment from TbVtc4 (aa 740–759; Fig. 1A).
FIGURE 1.
Localization of TbVtc4 in PCF and BSF trypanosomes. TbVtc4 co-localizes with TbVP1 in acidocalcisomes of PCF (A and B). TbVtc4 was detected with monoclonal anti-HA antibodies in trypanosomes expressing TbVtc4-HA (B) or with polyclonal anti-TbVtc4 antibodies in wild type trypanosomes (A) (green), and co-localized with antibodies against TbVP1 (red). The merge shows co-localization in yellow. Bars, 10 μm. C, TbVtc4-HA fusion protein was also detected in BSF trypanosomes with anti-HA antibodies and gold-conjugated anti-mouse secondary antibody (18 nm) and co-localized with antibodies against TbVP1 and gold-conjugated anti-rabbit antibody (12 nm). Acidocalcisomes (Ac) and kinetoplast (k) are indicated. Bar, 100 nm. D, Western blot analysis in BSF and PCF WT and endogenously tagged parasites (TbVtc4) using monoclonal anti-HA antibodies. Anti-tubulin antibodies were used as a loading control. Molecular weights are shown on the left. E, recombinant catalytic region of TbVtc4 (aa 199–506) affinity-purified from E. coli (lane rTbVTC4). Lane MW, protein molecular mass standards (kDa). The 10% SDS-polyacrylamide gel was stained with Coomassie Brilliant Blue. A single protein band corresponds to rTbVtc4.
TbVtc4 Synthesizes Short Chain PolyP and Requires Divalent Cations but Not Pyrophosphate
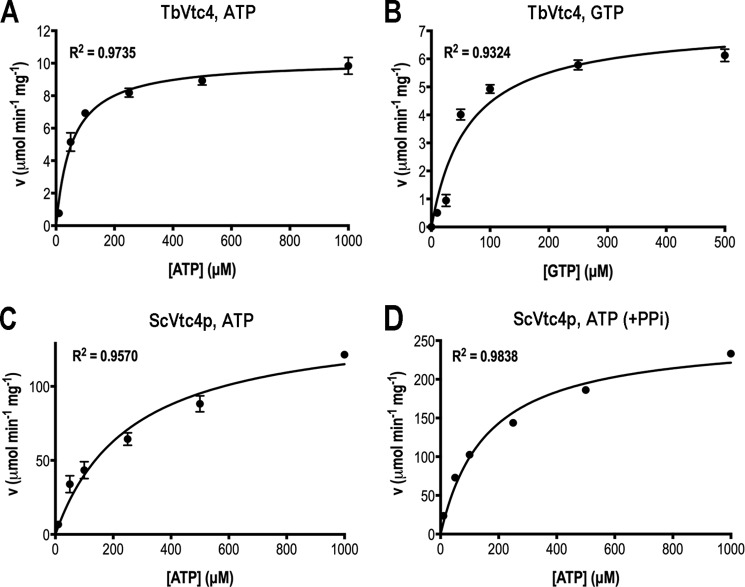
To characterize the enzymatic activity of TbVtc4, we expressed its catalytic domain (TbVtc4(199–506)) as a fusion protein with an N-terminal polyhistidine tag. In Fig. 1E, lane rTbVtc4 shows that the recombinant protein (including the His tag) appears as a strong single band with a molecular mass comparable with the predicted molecular mass (53.4 kDa). The catalytic core of the S. cerevisiae enzyme (ScVtc4p(189–480)) (13) was used as a control. We tested the activity of TbVtc4 with ATP or GTP (Fig. 2, A and B, respectively). ScVtc4p was assayed with ATP in the absence and presence of PPi (Fig. 2, C and D, respectively). TbVtc4 has a higher affinity for ATP than ScVtc4p (Table 1) in the absence of PPi (TbVtc4 Km = 54.8 ± 7.3 μm versus ScVtc4p Km = 261.2 ± 48.6 μm). However, the yeast enzyme is much more efficient than TbVtc4 (ScVtc4p kcat/Km = 9.3 × 103 s−1 m−1 versus TbVtc4 kcat/Km = 3.1 × 103 s−1 m−1). The presence of PPi did not “prime” or stimulate TbVtc4 activity (see also Fig. 3C), as was observed with the yeast enzyme (13). On the other hand, the reported priming effect of PPi on the polyP kinase activity of ScVtc4p was confirmed by the increase in its efficiency in the presence of PPi with a kcat/Km ratio of 1.7 × 104 s−1 m−1 (Table 1).
FIGURE 2.
Recombinant polyP kinase activity as a function of substrate concentration. TbVtc4polyP kinase activity was assayed in buffer containing 50 mm Hepes (pH 6.5), 150 mm NaCl, 1 mm MnCl2, and different concentrations of ATP (A) or GTP (B) at room temperature, using 1 μm TbVtc4. ScVtc4ppolyP kinase activity was assayed in buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm MnCl2, and different concentrations of ATP at room temperature, using 1 μm ScVtc4p in the absence (C) and presence (D) of 1 mm PPi. Other experimental conditions were as described under “Experimental Procedures.” Values are means ± S.D. (error bars) of three independent experiments. Error bars are smaller than the symbols used for some data points.
TABLE 1.
Kinetic parameters of purified recombinant Vtc4 catalytic regions from T. brucei and S. cerevisiae with different substrates
| Enzyme | Substrate | Vmax | Km | kcat/Km |
|---|---|---|---|---|
| μmol min−1 mg protein−1 | μm | s−1 m−1 | ||
| TbVtc4 | ATP | 10.2 ± 0.3 | 54.8 ± 7.3 | 3.1 × 103 |
| GTP | 7.2 ± 0.5 | 63.0 ± 13.0 | 1.9 × 103 | |
| ScVtc4 | ATP | 145.0 ± 10.2 | 261.2 ± 48.6 | 9.3 × 103 |
| ATP + PPi | 256.6 ± 8.6 | 158.5 ± 16.5 | 1.7 × 104 |
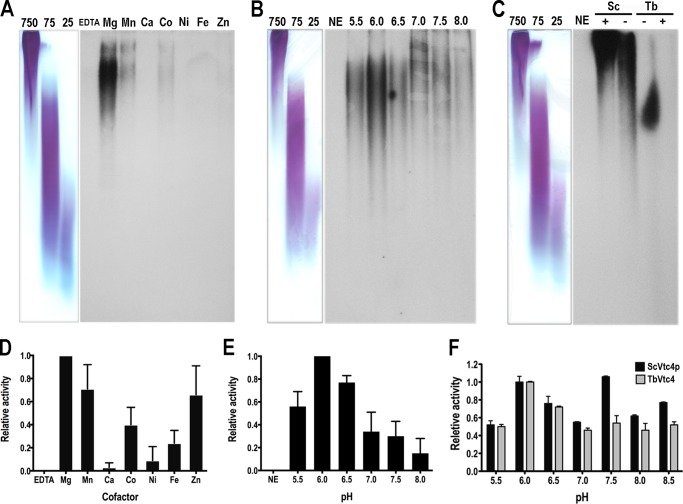
FIGURE 3.
TBE-PAGE analysis of polyP produced by TbVtc4 and ScVtc4p catalytic cores. PolyP synthesized by recombinant TbVtc4 and ScVtc4p using [γ-32P]ATP as substrate was analyzed by TBE-PAGE. A, autoradiography of TbVtc4 reaction products in the presence of different cations. B, autoradiography of TbVtc4 reaction products at different pH. C, autoradiography of TbVtc4 (Tb; in the presence of Mn2+, pH 6.0) and ScVtc4p (Sc; in the presence of Mn2+, pH 7.5) reaction products in the presence (+) or absence (−) of PPi. Lanes to the left of each autoradiograph in A–C show polyP of 25, 75, and 750 phosphate units loaded on the same gel and stained with toluidine blue. D and E, results from four (D) and three (E) independent experiments similar to those shown in A and B were quantified by densitometry using the SpotDenso tool of the AlphaImager® gel documentation system (ProteinSimple, Santa Clara, CA) and then normalized and plotted. F, optimal pH of recombinant TbVtc4 and ScVtc4p was confirmed by an alternative method. Coupling the reaction to lactate synthesis from phosphoenolpyruvate allows the regeneration of ATP to the Vtc4 reaction. Absorbance (340 nm) turnover indicated NADH consumption during the coupled reaction and was used for measuring polyP kinase activity of affinity-purified recombinant Vtc4s. Results are expressed in relative activity units at different pH values. Values are means ± S.D. (error bars) of three independent experiments.
PolyP produced by TbVtc4 reactions was visualized using [γ-32P]ATP as substrate followed by TBE-PAGE. TbVtc4 requires divalent cations, preferentially Mg2+, Mn2+, or Zn2+ (Fig. 3, A and D) and an acidic pH (Fig. 3, B and E) for optimal activity. The increased activity of TbVtc4 at acidic pH was corroborated using a different method based on a coupled assay that generates NADH upon polyP production. In contrast to TbVtc4 (optimal pH = 6.0), ScVtc4p exhibited pH optima at 6.0 and 7.5 (Fig. 3F). Finally, TBE-PAGE/autoradiography demonstrated that polyP chains synthesized by TbVtc4 catalytic core are much shorter (∼100–300 Pi residues) than those produced by ScVtc4p (∼750 Pi residues) (Fig. 3C), and TbVtc4 activity was inhibited in the presence of PPi (Fig. 3C).
Reduced Expression of TbVtc4 in BSF Trypanosomes Results in Decreased PolyP Kinase Activity and Short Chain PolyP Content
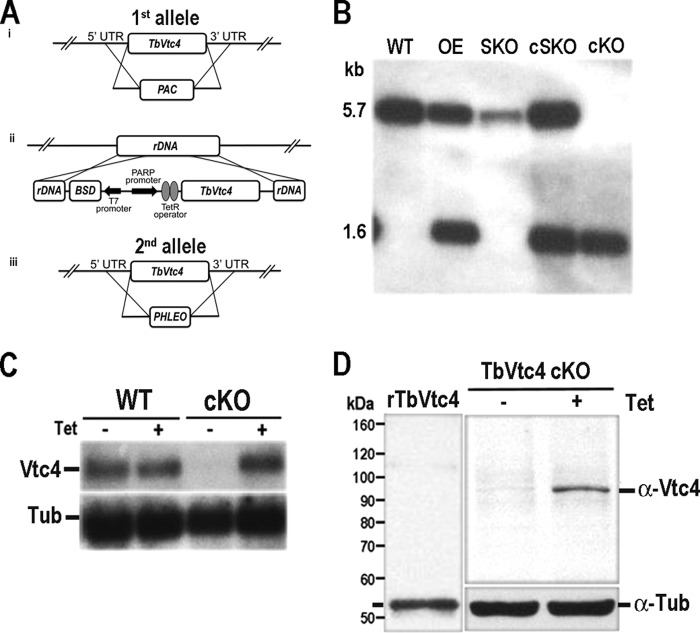
Previous studies demonstrated that polyP is important for trypanosome growth and osmoregulation in trypanosomes (22, 23, 27, 30, 31, 37), but the length of the polyP responsible for these roles was not investigated. To investigate whether the short chain polyP synthesized by TbVtc4 is involved in these processes, we analyzed the phenotypic changes of BSF trypanosomes with a conditional KO of TbVtc4. In these cells, we replaced both TbVtc4 alleles with drug resistance genes, but, because TbVtc4 could be required for growth, we introduced an ectopic copy of the TbVtc4 gene whose expression depended on the presence of tetracycline or doxycycline in the culture medium (Fig. 4A). The genotype of the mutant cell line was verified by Southern blot analysis (Fig. 4B). Levels of mRNA in the presence or absence of tetracycline were analyzed by Northern blot (Fig. 4C). As expected, there was a decrease in TbVtc4 mRNA levels in the absence of tetracycline for the KO cell line. In the presence of tetracycline, TbVtc4 mRNA levels of the KO mutant were normal because of the ectopic gene expression. The expression level of TbVtc4 in these mutants was confirmed by Western blot analysis using mouse polyclonal antibodies against TbVtc4 (Fig. 4D).
FIGURE 4.
Generation of TbVtc4 cKO cell line. A, schematic representation of the strategy used for the generation of a stable TbVtc4 conditional knock-out mutant in T. brucei BSF. i, one allele of TbVtc4 was replaced with the puromycin resistance gene (PAC) by homologous recombination, generating the TbVtc4 SKO cell line. ii, an ectopic TbVtc4 cassette under the control of the tetracycline-inducible PARP promoter and selectable by blasticidin resistance (BSD) was inserted at the ribosomal non-transcribed spacer (rDNA), generating the TbVtc4 cSKO cell line. This cassette was constructed using pLEW100v5b1d-BSD expression vector. iii, while keeping induced the expression of the ectopic TbVtc4, the second allele of the gene was replaced with a phleomycin resistance gene (PHLEO) by homologous recombination, resulting in conditional knock-out cell line TbVtc4 cKO. B, Southern blot analysis of parental cell line (single marker; WT), overexpressing (OE), single knock-out (SKO), complemented single knock-out (cSKO), and complemented double KO (cKO). C, Northern blot analysis of WT and TbVtc4-KO mutant (cKO) in the absence or presence of tetracycline (± Tet), using a Vtc4 probe. A tubulin probe was used as a loading control. D, Western blot analysis of TbVtc4 cKO mutant cell line in the absence or presence of tetracycline, using polyclonal antibodies anti-TbVtc4 and anti-tubulin antibodies as a loading control. Affinity-purified recombinant TbVtc4(199–506) (rTbVtc4) was included as control. Molecular weights are shown to the left of the blot.
The in vitro growth rate of the mutant cell line in the absence of tetracycline was monitored during 2 weeks and compared with that of the parental single marker strain (WT). The growth rate of TbVtc4-KO BSF progressively decreased relative to the parental cell line. TbVtc4-KO BSF partially recovered as escape mutants arose after 14 days (Fig. 5A). PolyP kinase activity after 2 weeks of withdrawal of tetracycline was significantly decreased in total cell lysates (Fig. 5B). Reduced activity was accompanied by a 35% decrease in short chain polyP (Fig. 5C) but no significant changes in long chain polyP levels (Fig. 5D). In summary, our results indicate that disruption of TbVtc4 in BSF trypanosomes decreases polyP kinase activity and, as a consequence, results in significantly lower levels of short chain polyP.
FIGURE 5.
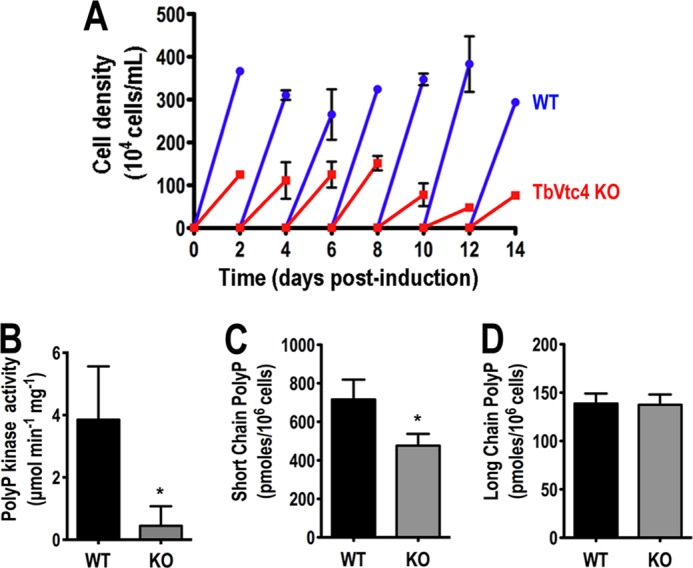
Effect of inhibition of TbVtc4 expression on cell growth, polyP kinase activity, and short and long chain polyP levels. A, in vitro growth of single marker BSF (WT, full circles, blue) and TbVtc4 conditional knock-out parasites (TbVtc4 KO, full squares, red). B, lysates from TbVtc4-KO BSF showed an ∼8-fold lower polyP kinase activity than those from single marker BSF trypanosomes (WT). C and D, extracts from TbVtc4-KO BSF trypanosomes showed a ∼35% reduction in short chain polyP content (C) with no significant changes in long chain polyP content (D), as compared with the parental cell line (WT). Values are means ± S.D. (error bars) of three different experiments. *, differences are statistically significant as compared with respective controls, p < 0.05 (Student's t test).
TbVtc4-KO Mutant Parasites Display an Osmoregulatory Defect
To investigate the role of TbVtc4 in osmoregulation, we exposed TbVtc4-deficient BSF trypanosomes to hyposmotic and hyperosmotic conditions and evaluated changes of cell volume with time. These parasites showed a defect in the ability to recover cell volume (a process known as regulatory volume decrease (41)) during hyposmotic stress when compared with parental (WT) and complemented (+tet) cell lines (Fig. 6A). Loss of water was also more pronounced in these parasites during hyperosmotic treatment compared with the single marker cell line. This defect in hyperosmotic response was overcome when we induced the expression of an ectopic copy of the gene (Fig. 6B).
FIGURE 6.
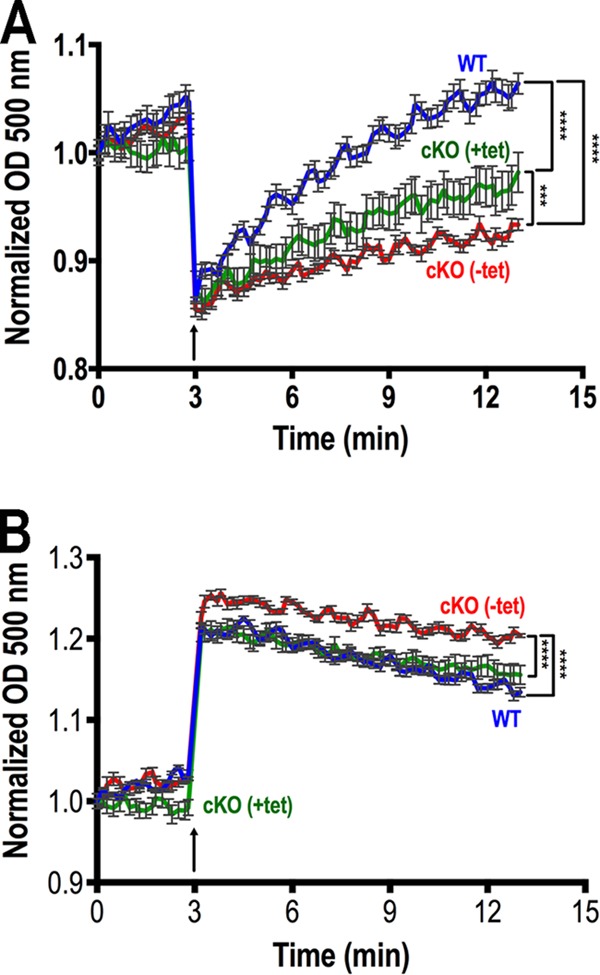
Effect of inhibition of TbVtc4 expression on the response of BSF trypanosomes to hyposmotic and hyperosmotic stresses. The same amounts of single marker (WT; blue), TbVtc4 knock-out (cKO (−Tet); red), and TbVtc4 complemented knock-out (cKO (+Tet); green) BSF trypanosomes were suspended in isotonic buffer. TbVtc4-KO BSF trypanosomes (cKO (−Tet)) were cultured in the absence of tetracycline for 14 days before the experiment. The cells were then treated as described under “Experimental Procedures,” and relative changes in cell volume were followed by monitoring the absorbance at 550 nm. A, changes in cell volume after hyposmotic stress (213 mosm). B, changes in cell volume after hyperosmotic stress (650 mosm). Arrows indicate the time point (3 min) when osmotic stress was induced. A decrease in absorbance corresponds to an increase in cell volume and vice versa. Values are means ± S.D. (error bars) of three different experiments. Asterisks indicate statistically significant differences between cell line patterns, p < 0.05 (Bonferroni's multiple comparison “a posteriori” test of one-way analysis of variance at all time points after induction of osmotic stress).
TbVtc4 Is Required for Effective in Vivo Infection
We tested the infectivity of the TbVtc4-KO mutants in vivo using a mouse model, and we found that the mutant cells were considerably less virulent in mice. Once again, the phenotype reverted when an ectopic copy of TbVtc4 gene was induced by doxycycline (Fig. 7A). It is important to mention that TbVtc4-KO (−dox) BSF trypanosomes were able to infect mice, as verified by detection of parasites in blood at day 3–4 postinfection. However, the amount of parasites was significantly lower than that observed in the control groups, and, 1 week postinfection, TbVtc4-KO (−dox) BSF trypanosomes were no longer detected in the blood of surviving animals (Fig. 7B). Thus, these parasites were unable to persist in the mammalian host. Taken together, our data indicate that TbVtc4 plays an important role in the infectivity of BSF trypanosomes, and this role is possibly related to their reduced ability to survive under the osmotic stress conditions of the host.
FIGURE 7.
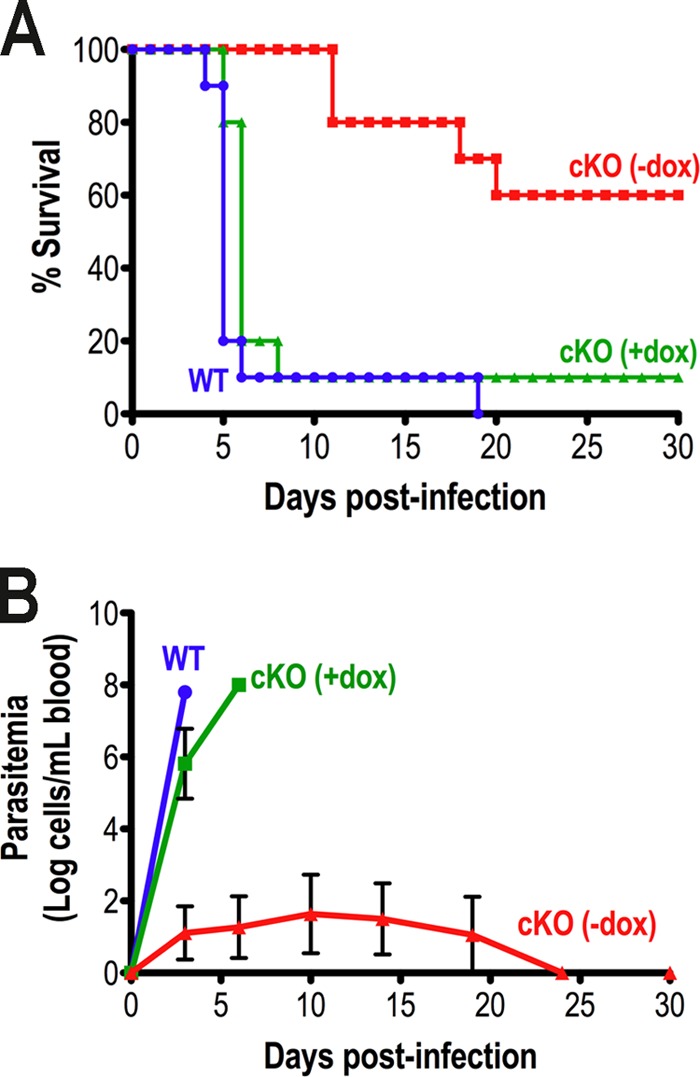
Effect of inhibition of TbVtc4 expression on virulence in mice. Two experiments involving three groups of five mice that were infected with single marker (WT) and TbVtc4-KO mutant (cKO) BSF trypanosomes (−dox,+dox), were performed. 200 μg/ml doxycycline was supplied in the drinking water of one group of mice for the induction of an ectopic copy of TbVtc4 gene in the cKO cell line (complemented knock-out control). The percentage of mouse survival (A) and parasitemia levels in the blood of infected mice (B) were monitored during 30 days postinfection. The charts combine results from two independent experiments. Error bars, S.D.
DISCUSSION
We report here that TbVtc4 encodes a short chain polyP kinase that localizes to the acidocalcisomes of T. brucei. The enzyme is important for osmoregulation, in vitro growth, and infectivity of BSF trypanosomes in vivo.
In contrast to the S. cerevisiae enzyme, which is a very long chain polyP kinase, TbVtc4 catalyzes production of polyP of about 100–300 Pi units and was not activated but rather inhibited by PPi. Despite these differences, we found some characteristics of the enzyme similar to those of ScVtc4p (13). Bivalent cation requirements for both enzymes are slightly different (TbVtc4 metal ion specificity is Mg2+ = Mn2+ > Zn2+ > Co2+ > Fe2+ > Ni2+ > Ca2+, and ScVtc4p metal ion specificity is Mn2+ > Zn2+ > Co2+ > Mg2+ > Fe2+ > Ni2+). It is important to mention that the acidocalcisome environment of TbVtc4 is rich in zinc, magnesium, and calcium (42). Both enzymes can catalyze polyP synthesis at acidic pH. TbVtc4 activity is highest at a pH of 6.0, but ScVtc4p exhibits two optimal pH values (6.0 and 7.5). Although these enzymes are located in the membrane of acidic calcium stores (the acidocalcisome and the yeast vacuole, respectively), their catalytic domains are facing the cytosol. It is possible that the microenvironment close to the outer leaflet of the acidocalcisome and vacuolar membranes has a lower pH due to the presence of Na+/H+ exchangers that move protons out from the acidocalcisome (40) and vacuole lumen (43).
A significant decrease in the level of short chain polyP was observed in the knock-out BSF trypanosomes. However, a large reservoir was still present after parasites had been cultured for 2 weeks without tetracycline. The persistence of short chain polyP could be due to the known slow turnover of polyP (44) but more likely arises from the methodological limitations in distinguishing between abundant forms of very short polyP (polyP3, polyP4, and polyP5) (45) and medium size polyP (100–300 Pi), which are the main product of TbVtc4. The synthetic mechanism of very short polyP is unknown. Nevertheless, our results suggest that medium size polyP is important for osmoregulation and viability. Although previous reports have shown that polyP has important roles in growth and osmoregulation in trypanosomes (22, 23, 27, 30, 31, 37), the length of the polyP responsible for these roles was not investigated. In this work, we report that polyP of 100–300 Pi units, which is synthesized by TbVtc4, is required for regulatory volume decrease during hyposmotic stress and also relevant for the response of the parasites to hyperosmotic stress conditions. The ability of these parasites to overcome such drastic changes in osmolarity is critical from their survival in the mammalian host. BSF trypanosomes in humans must be able to resist osmolality as high as 1,400 mosm when passing through the renal medulla and rapidly accommodate a return to the isosmotic environment (300 mosm) of the general circulation (46). Our results demonstrate that depletion of TbVtc4 in BSF trypanosomes leads to defective osmoregulation and infectivity. Both phenotypes are rescued with expression of an ectopic copy of the gene. A possible explanation for these observations is that mutant parasites, with lower short chain polyP levels, are not able to overcome the dramatic cell volume stresses of mammalian renal circulation and consequently cannot establish an infection. Alternatively, the increased osmotic sensitivity of TbVtc4-deficient trypanosomes weakens the parasites and makes them unable to evade the host immune response.
Long chain polyP is present in trypanosomes (22), but TbVtc4 is not able to synthesize these polymers; therefore, other enzyme(s) could be involved in this process. We measured the activity of the catalytic region of TbVtc4 because our attempts to recombinantly express the full-length protein in soluble form were unsuccessful. It is possible that the native enzyme can synthesize long chain polyP. However, this is not likely because only short chain polyP synthesis was affected in knock-out parasites.
The palmitoyl proteome of T. brucei has recently been reported (47). Both TbVtc1 and TbVtc4 were reported to be palmitoylated. However, our attempts to demonstrate palmitoylation of TbVtc4 in PCF following established protocols (48, 49) were unsuccessful (data not shown). In summary, the essentiality of TbVtc4 for growth and establishment of an efficient infection suggests that this enzyme is a potential drug target and that it would be possible to develop inhibitors.
Acknowledgments
We thank George A. M. Cross for providing strains 29-13 (PCF) and single marker (BSF) and vectors, Norbert Bakalara for the TbVP1 antibody, Thomas Seebeck for the pMOTag4H vector, and Wandy Beatty for help with the immunogold electron microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grant AI077538 (to R. D.). This work was also supported by an American Heart Association predoctoral fellowship (to N. L.) and postdoctoral fellowship (to P. N. U.).
G. Huang, P. N. Ulrich, D. Johnson, S. N. J. Moreno, R. Orlando, and R. Docampo, unpublished data.
- polyP
- polyphosphate
- PCF
- procyclic form
- BSF
- bloodstream form
- gDNA
- genomic DNA
- aa
- amino acids
- TBE
- Tris borate-EDTA.
REFERENCES
- 1. Rao N. N., Gómez-Garcia M. R., Kornberg A. (2009) Inorganic polyphosphate. Essential for growth and survival. Annu. Rev. Biochem. 78, 605–647 [DOI] [PubMed] [Google Scholar]
- 2. Moreno S. N., Docampo R. (2013) Polyphosphate and its diverse functions in host cells and pathogens. PLoS Pathog. 9, e1003230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ruiz F. A., Lea C. R., Oldfield E., Docampo R. (2004) Human platelet dense granules contain polyphosphate and are similar to acidocalcisomes of bacteria and unicellular eukaryotes. J. Biol. Chem. 279, 44250–44257 [DOI] [PubMed] [Google Scholar]
- 4. Moreno-Sanchez D., Hernandez-Ruiz L., Ruiz F. A., Docampo R. (2012) Polyphosphate is a novel pro-inflammatory regulator of mast cells and is located in acidocalcisomes. J. Biol. Chem. 287, 28435–28444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Smith S. A., Mutch N. J., Baskar D., Rohloff P., Docampo R., Morrissey J. H. (2006) Polyphosphate modulates blood coagulation and fibrinolysis. Proc. Natl. Acad. Sci. U.S.A. 103, 903–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Müller F., Mutch N. J., Schenk W. A., Smith S. A., Esterl L., Spronk H. M., Schmidbauer S., Gahl W. A., Morrissey J. H., Renné T. (2009) Platelet polyphosphates are proinflammatory and procoagulant mediators in vivo. Cell 139, 1143–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morrissey J. H., Choi S. H., Smith S. A. (2012) Polyphosphate. An ancient molecule that links platelets, coagulation, and inflammation. Blood 119, 5972–5979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wurst H., Shiba T., Kornberg A. (1995) The gene for a major exopolyphosphatase of Saccharomyces cerevisiae. J. Bacteriol. 177, 898–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rodrigues C. O., Ruiz F. A., Vieira M., Hill J. E., Docampo R. (2002) An acidocalcisomal exopolyphosphatase from Leishmania major with high affinity for short chain polyphosphate. J. Biol. Chem. 277, 50899–50906 [DOI] [PubMed] [Google Scholar]
- 10. Fang J., Ruiz F. A., Docampo M., Luo S., Rodrigues J. C., Motta L. S., Rohloff P., Docampo R. (2007) Overexpression of a Zn2+-sensitive soluble exopolyphosphatase from Trypanosoma cruzi depletes polyphosphate and affects osmoregulation. J. Biol. Chem. 282, 32501–32510 [DOI] [PubMed] [Google Scholar]
- 11. Tammenkoski M., Koivula K., Cusanelli E., Zollo M., Steegborn C., Baykov A. A., Lahti R. (2008) Human metastasis regulator protein H-prune is a short-chain exopolyphosphatase. Biochemistry 47, 9707–9713 [DOI] [PubMed] [Google Scholar]
- 12. Sethuraman A., Rao N. N., Kornberg A. (2001) The endopolyphosphatase gene. Essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 98, 8542–8547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hothorn M., Neumann H., Lenherr E. D., Wehner M., Rybin V., Hassa P. O., Uttenweiler A., Reinhardt M., Schmidt A., Seiler J., Ladurner A. G., Herrmann C., Scheffzek K., Mayer A. (2009) Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 324, 513–516 [DOI] [PubMed] [Google Scholar]
- 14. Cohen A., Perzov N., Nelson H., Nelson N. (1999) A novel family of yeast chaperons involved in the distribution of V-ATPase and other membrane proteins. J. Biol. Chem. 274, 26885–26893 [DOI] [PubMed] [Google Scholar]
- 15. Müller O., Bayer M. J., Peters C., Andersen J. S., Mann M., Mayer A. (2002) The Vtc proteins in vacuole fusion. Coupling NSF activity to V(0) trans-complex formation. EMBO J. 21, 259–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Müller O., Neumann H., Bayer M. J., Mayer A. (2003) Role of the Vtc proteins in V-ATPase stability and membrane trafficking. J. Cell Sci. 116, 1107–1115 [DOI] [PubMed] [Google Scholar]
- 17. Patel S., Docampo R. (2010) Acidic calcium stores open for business. Expanding the potential for intracellular Ca2+ signaling. Trends Cell Biol. 20, 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Docampo R., de Souza W., Miranda K., Rohloff P., Moreno S. N. (2005) Acidocalcisomes. Conserved from bacteria to man. Nat. Rev. Microbiol. 3, 251–261 [DOI] [PubMed] [Google Scholar]
- 19. Docampo R., Moreno S. N. (2011) Acidocalcisomes. Cell Calcium 50, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vercesi A. E., Moreno S. N., Docampo R. (1994) Ca2+/H+ exchange in acidic vacuoles of Trypanosoma brucei. Biochem. J. 304, 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang G., Bartlett P. J., Thomas A. P., Moreno S. N., Docampo R. (2013) Acidocalcisomes of Trypanosoma brucei have an inositol 1,4,5-trisphosphate receptor that is required for growth and infectivity. Proc. Natl. Acad. Sci. U.S.A. 110, 1887–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ruiz F. A., Rodrigues C. O., Docampo R. (2001) Rapid changes in polyphosphate content within acidocalcisomes in response to cell growth, differentiation, and environmental stress in Trypanosoma cruzi. J. Biol. Chem. 276, 26114–26121 [DOI] [PubMed] [Google Scholar]
- 23. Li Z. H., Alvarez V. E., De Gaudenzi J. G., Sant'Anna C., Frasch A. C., Cazzulo J. J., Docampo R. (2011) Hyperosmotic stress induces aquaporin-dependent cell shrinkage, polyphosphate synthesis, amino acid accumulation, and global gene expression changes in Trypanosoma cruzi. J. Biol. Chem. 286, 43959–43971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Docampo R., Jimenez V., King-Keller S., Li Z. H., Moreno S. N. (2011) The role of acidocalcisomes in the stress response of Trypanosoma cruzi. Adv. Parasitol. 75, 307–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rohloff P., Docampo R. (2008) A contractile vacuole complex is involved in osmoregulation in Trypanosoma cruzi. Exp. Parasitol. 118, 17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ulrich P. N., Jimenez V., Park M., Martins V. P., Atwood J., 3rd, Moles K., Collins D., Rohloff P., Tarleton R., Moreno S. N., Orlando R., Docampo R. (2011) Identification of contractile vacuole proteins in Trypanosoma cruzi. PLoS One 6, e18013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fang J., Rohloff P., Miranda K., Docampo R. (2007) Ablation of a small transmembrane protein of Trypanosoma brucei (TbVTC1) involved in the synthesis of polyphosphate alters acidocalcisome biogenesis and function and leads to a cytokinesis defect. Biochem. J. 407, 161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wirtz E., Clayton C. (1995) Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science 268, 1179–1183 [DOI] [PubMed] [Google Scholar]
- 29. Wirtz E., Leal S., Ochatt C., Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 30. Huang G., Fang J., Sant'Anna C., Li Z. H., Wellems D. L., Rohloff P., Docampo R. (2011) Adaptor protein-3 (AP-3) complex mediates the biogenesis of acidocalcisomes and is essential for growth and virulence of Trypanosoma brucei. J. Biol. Chem. 286, 36619–36630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lemercier G., Dutoya S., Luo S., Ruiz F. A., Rodrigues C. O., Baltz T., Docampo R., Bakalara N. (2002) A vacuolar-type H+-pyrophosphatase governs maintenance of functional acidocalcisomes and growth of the insect and mammalian forms of Trypanosoma brucei. J. Biol. Chem. 277, 37369–37376 [DOI] [PubMed] [Google Scholar]
- 32. Oberholzer M., Morand S., Kunz S., Seebeck T. (2006) A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 145, 117–120 [DOI] [PubMed] [Google Scholar]
- 33. Falquet L., Pagni M., Bucher P., Hulo N., Sigrist C. J., Hofmann K., Bairoch A. (2002) The PROSITE database, its status in 2002. Nucleic Acids Res. 30, 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Claros M. G., von Heijne G. (1994) TopPred II. An improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10, 685–686 [DOI] [PubMed] [Google Scholar]
- 35. Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B. P., Carrington M., Depledge D. P., Fischer S., Gajria B., Gao X., Gardner M. J., Gingle A., Grant G., Harb O. S., Heiges M., Hertz-Fowler C., Houston R., Innamorato F., Iodice J., Kissinger J. C., Kraemer E., Li W., Logan F. J., Miller J. A., Mitra S., Myler P. J., Nayak V., Pennington C., Phan I., Pinney D. F., Ramasamy G., Rogers M. B., Roos D. S., Ross C., Sivam D., Smith D. F., Srinivasamoorthy G., Stoeckert C. J., Jr., Subramanian S., Thibodeau R., Tivey A., Treatman C., Velarde G., Wang H. (2010) TriTrypDB. A functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 38, D457–D462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Medina-Acosta E., Cross G. A. (1993) Rapid isolation of DNA from trypanosomatid protozoa using a simple “mini-prep” procedure. Mol. Biochem. Parasitol. 59, 327–329 [DOI] [PubMed] [Google Scholar]
- 37. de Jesus T. C., Tonelli R. R., Nardelli S. C., da Silva Augusto L., Motta M. C., Girard Dias W., Miranda K., Ulrich P., Jimenez V., Barquilla A., Navarro M., Docampo R., Schenkman S. (2010) Target of rapamycin (TOR)-like 1 kinase is involved in the control of polyphosphate levels and acidocalcisome maintenance in Trypanosoma brucei. J. Biol. Chem. 285, 24131–24140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kocic I., Hirano Y., Hiraoka M. (2001) Ionic basis for membrane potential changes induced by hypoosmotic stress in guinea-pig ventricular myocytes. Cardiovasc. Res. 51, 59–70 [DOI] [PubMed] [Google Scholar]
- 39. Herbert W. J., Lumsden W. H. (1976) Trypanosoma brucei: a rapid “matching” method for estimating the host's parasitemia. Exp. Parasitol. 40, 427–431 [DOI] [PubMed] [Google Scholar]
- 40. Rodrigues C. O., Scott D. A., Docampo R. (1999) Characterization of a vacuolar pyrophosphatase in Trypanosoma brucei and its localization to acidocalcisomes. Mol. Cell. Biol. 19, 7712–7723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rohloff P., Rodrigues C. O., Docampo R. (2003) Regulatory volume decrease in Trypanosoma cruzi involves amino acid efflux and changes in intracellular calcium. Mol. Biochem. Parasitol. 126, 219–230 [DOI] [PubMed] [Google Scholar]
- 42. Scott D. A., Docampo R., Dvorak J. A., Shi S., Leapman R. D. (1997) In situ compositional analysis of acidocalcisomes in Trypanosoma cruzi. J. Biol. Chem. 272, 28020–28029 [DOI] [PubMed] [Google Scholar]
- 43. Cagnac O., Leterrier M., Yeager M., Blumwald E. (2007) Identification and characterization of Vnx1p, a novel type of vacuolar monovalent cation/H+ antiporter of Saccharomyces cerevisiae. J. Biol. Chem. 282, 24284–24293 [DOI] [PubMed] [Google Scholar]
- 44. Rao N. N., Kornberg A. (1996) Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178, 1394–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moreno B., Urbina J. A., Oldfield E., Bailey B. N., Rodrigues C. O., Docampo R. (2000) 31P NMR spectroscopy of Trypanosoma brucei, Trypanosoma cruzi, and Leishmania major. Evidence for high levels of condensed inorganic phosphates. J. Biol. Chem. 275, 28356–28362 [DOI] [PubMed] [Google Scholar]
- 46. Lang F., Busch G. L., Ritter M., Völkl H., Waldegger S., Gulbins E., Häussinger D. (1998) Functional significance of cell volume regulatory mechanisms. Physiol. Rev. 78, 247–306 [DOI] [PubMed] [Google Scholar]
- 47. Emmer B. T., Nakayasu E. S., Souther C., Choi H., Sobreira T. J., Epting C. L., Nesvizhskii A. I., Almeida I. C., Engman D. M. (2011) Global analysis of protein palmitoylation in African trypanosomes. Eukaryot. Cell 10, 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Furuya T., Kashuba C., Docampo R., Moreno S. N. (2000) A novel phosphatidylinositol-phospholipase C of Trypanosoma cruzi that is lipid modified and activated during trypomastigote to amastigote differentiation. J. Biol. Chem. 275, 6428–6438 [DOI] [PubMed] [Google Scholar]
- 49. de Paulo Martins V., Okura M., Maric D., Engman D. M., Vieira M., Docampo R., Moreno S. N. (2010) Acylation-dependent export of Trypanosoma cruzi phosphoinositide-specific phospholipase C to the outer surface of amastigotes. J. Biol. Chem. 285, 30906–30917 [DOI] [PMC free article] [PubMed] [Google Scholar]