Background: Polyglutamine expansion of the ataxin-7 protein, a subunit of the SAGA (Spt-Ada-Gcn5-acetyltransferase) complex, causes spinocerebellar ataxia type 7.
Results: Polyglutamine-expanded ataxin-7 aberrantly binds to SAGA, affecting Gcn5 acetyltransferase activity at SAGA-regulated genes.
Conclusion: Ataxin-7 exhibits a dominant-negative effect on Gcn5 catalytic activity in vivo.
Significance: We show a direct dominant effect of polyglutamine-expanded ataxin-7 on SAGA acetyltransferase function.
Keywords: Gene Expression, Histone Acetylase, Histone Modification, Neurodegeneration, Polyglutamine Disease, Protein Complexes, Spinocerebellar Ataxia Type 7
Abstract
Spinocerebellar ataxia type 7 (SCA7) is a neurodegenerative disease caused by polyglutamine (polyQ) expansion within the N-terminal region of the ataxin-7 protein, a known subunit of the SAGA complex. Although the mechanisms of SCA7 pathogenesis remain poorly understood, previous studies have shown perturbations in SAGA histone acetyltransferase function and transcriptional alterations. We sought to determine whether and how polyQ-expanded ataxin-7 affects SAGA catalytic activity. Here, we determined that polyQ-expanded ataxin-7 directly bound the Gcn5 catalytic core of SAGA while in association with its regulatory proteins, Ada2 and Ada3. This caused a significant decrease in Gcn5 histone acetyltransferase activity in vitro and in vivo at two SAGA-regulated galactose genes, GAL1 and GAL7. However, Gcn5 occupancy at the GAL1 and GAL7 promoters was increased in these cells, revealing a dominant-negative phenotype of the polyQ-expanded ataxin-7-incorporated, catalytically inactive SAGA. These findings suggest a dominant mechanism of polyQ-mediated SAGA inhibition that potentially contributes to SCA7 disease pathogenesis.
Introduction
Dense chromatin structure poses a problem for nuclear machinery that requires access to DNA (1). For efficient transcription to occur in cells, the obstacle imposed by chromatin must be overcome. To achieve this, distinct chromatin-modifying enzymes exist that help alter the structure of chromatin to provide for correct gene transcriptional regulation. Chromatin regulatory proteins, including most histone acetyltransferases (HATs),2 frequently function in the context of larger macromolecular complexes (2, 3). SAGA (Spt-Ada-Gcn5-acetyltransferase) is one such transcriptional coactivator complex with 20+ subunits. SAGA has been shown to be required for expression of a subset of genes, mainly stress-induced genes, and serves multiple transcriptional coactivator functions, which vary depending on the gene in context. Its transcriptional coactivator functions include TATA-binding protein recruitment; preinitiation complex formation; and two enzymatic activities, histone acetylation and deubiquitination (DUB) (4). SAGA subunit composition and organization are conserved from yeast to humans, and proper arrangement of its subunits is important for SAGA function (4). Thus, multisubunit protein complexes such as SAGA require correct assembly and synergistic effects among their subunits to fulfill their multiple cellular duties.
Ataxin-7 and its yeast homolog Sca7 (spinocerebellar ataxia type 7)/Sgf73 are subunits of the conserved SAGA family of transcriptional complexes (5–7). Polyglutamine (polyQ) expansion toward the N terminus of ataxin-7 is known to cause the autosomal dominant neurodegenerative disorder spinocerebellar ataxia type 7 (SCA7). The tract is polymorphic, ranging from 4 to 35 repeats, with expansion generally above 35 glutamines resulting in disease pathogenesis (8). The majority of normal alleles contain 10 glutamines, and pathogenic forms have reached upwards of 250 repeats (8). SCA7 disease manifests as retinal and cerebellar degeneration and pigmentary macular dystrophy, eventually leading to blindness (8). Patients display the greatest effect of neuronal loss and degeneration in the Purkinje cell layers of the cerebellum and in the inferior olivary complex. Both the normal and pathogenic forms of ataxin-7 are known to associate with the SAGA complex (6, 9). Human ataxin-7 (normal and polyQ-expanded) is able to complement the loss of Sgf73 (yeast ataxin-7) and restores subunit composition (6, 7, 9). Within SAGA, ataxin-7 is known to bridge the DUB and HAT modules (10–12). Ataxin-7 is a physical member of the DUB module in SAGA and is responsible for tethering the DUB module to the rest of the complex.
A complete understanding of precisely how SAGA subunits are arranged and assembled is no easy feat, much less understanding how these interactions and functions may go awry when a pathogenic protein is incorporated. Additionally, whereas it is clear that polyQ expansion of the SCA7 gene product, ataxin-7, is at the root of the disease, to date, little is known about the specific mechanism of pathology. Previous studies have attempted to answer how the ataxin-7 polyQ tract alters the activity of SAGA. Our previous work and work by Roeder and co-workers have shown that polyQ-expanded ataxin-7 disrupts SAGA integrity and lowers HAT activity (6, 9). Also, reducing Gcn5 expression accelerates disease phenotypes in a SCA7 mouse model (13). However, a separate study suggested transcriptional defects in a SCA7 mouse model, without loss of histone acetylation (14). To resolve this issue, we decided to focus on a specific SAGA function and sought to determine whether and how the polyQ expansion of ataxin-7 directly affects the Gcn5 catalytic core. We employed in vitro biochemical methods to assess, directly, if polyQ-expanded ataxin-7 associates with the Gcn5 module of SAGA. Then, using yeast as a model system, we assessed if these biochemical alterations affect SAGA function, particularly HAT function in vivo.
In this study, we were able to determine that polyQ-expanded ataxin-7 bound the Gcn5 catalytic core of SAGA while in association with its regulatory proteins, Ada2 and Ada3. However, wild-type ataxin-7 did not incorporate into this trimeric complex. When comparing the HAT activities of the trimeric complex versus a tetrameric complex containing polyQ-expanded ataxin-7, we found that the addition of ataxin-7-60Q abrogated the in vitro HAT activity on core histones, nucleosomes, and histone H3 N-terminal peptides. Noticing that yeast cells expressing the 60Q form of ataxin-7 exhibited growth defects when grown on galactose medium, we utilized chromatin immunoprecipitation methods to show that H3 acetylation at the GAL1 and GAL7 promoter nucleosomes in this strain was also significantly decreased. However, when Gcn5 occupancy at the upstream activating sequences (UASs) of GAL1 and GAL7 was measured, we found that the Gcn5 levels were above wild-type levels. These findings suggest that the SAGA complex containing the polyQ-expanded protein directly alters normal Gcn5 acetyltransferase function by inhibiting its activity and not by excluding it from the complex or chromatin per se.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
The WT, sgf73Δ, Δ+10Q, and Δ+60Q strains used for growth assays, mRNA expression, and the H3K9 acetylation ChIP experiments were derivatives of BY4741 and BY4742 (Research Genetics, Huntsville, AL). Gcn5-tandem affinity purification tag (TAP), Gcn5-TAP, and sgf73Δ (Δ; MATα Gcn5TAP:HIS3, BY4741) strains were obtained from Dr. Jerry Workman's laboratory for use in Gcn5 ChIP assays, and Δ strains were transformed with pRS416-His6-Atxn10Q or pRS416-His6-Atxn60Q (CEN, URA3) using the lithium acetate method. The yeast expression vector pRS416 (CEN, URA3) containing ataxin-7-10Q or ataxin-7-60Q was modified using the QuikChange II Site-directed mutagenesis kit (catalog no. 200523, Agilent Technologies) to add a His6 tag at the N terminus of ataxin-7. CAG repeat lengths and correct insertion of the His6 tag were verified by DNA sequencing after mutagenesis.
Expression and Purification of Recombinant Proteins
Yeast Gcn5 and human ataxin-7-10Q and ataxin-7-60Q were expressed in bacteria as fusion proteins with six histidine residues at the N terminus and purified under nondenaturing conditions on nickel-nitrilotriacetic acid (Ni2+-NTA)-agarose. Dimeric (Ada2 and Gcn5) and trimeric (Ada3, Ada2, and Gcn5) complexes were expressed in the polycistronic vector pST44 alone or with either C-terminal FLAG-tagged ataxin-7-10Q or ataxin-7-60Q added in the fourth coding region of the vector (15). Ada2 and Ada3 contain the N-terminal His6 tag in the dimer and trimer, respectively. These vectors were expressed in Escherichia coli, purified under nondenaturing conditions on Ni2+-NTA-agarose, and eluted with 0.3 m imidazole. Eluates were then run on a 10% SDS-polyacrylamide gel and immunoblotted for Gcn5, Ada2, and His6 (Ada3).
The tetramer was doubly purified to enrich for tetramers in the recombinant mixture. First, the tetramer was purified using Ni2+-NTA-agarose as described above for trimer purification. Second, eluates were then immunoprecipitated with anti-FLAG M2 affinity gel overnight at 4 °C and washed with 1× TBS. Proteins were eluted from the anti-FLAG affinity gel using 3×FLAG-peptide. Eluates were then run on a 10% SDS-polyacrylamide gel and immunoblotted for Gcn5, Ada2, His6 (Ada3), and FLAG (ataxin-7).
In Vitro Binding Assays
His6-tagged recombinant Gcn5, trimer, ataxin-7-10Q, and ataxin-7-60Q were purified using Ni2+-NTA-agarose and eluted with 0.3 m imidazole. Eluates were then subjected to size exclusion chromatography using a Superose 12 column on an ÄKTA purifier (GE Healthcare) for further purification. Fractions containing each protein were run on an SDS-polyacrylamide gel and immunoblotted for antibodies against ataxin-7-10Q (His6), ataxin-7-60Q (His6), Ada3 (His6), Ada2, and Gcn5. Equal amounts of ataxin-7-10Q and ataxin-7-60Q were mixed with either Gcn5 or trimer. Gcn5 or trimer + ataxin-7-10Q or ataxin-7-60Q were rotated for 30 min at 30 °C and then placed on ice. Mixtures were run over either a Superose 12 column (for recombinant Gcn5 binding assay) or a Superose 6 column (for trimer binding assay). Odd-numbered fractions were then run on a 10% SDS-polyacrylamide gel; transferred to a nitrocellulose membrane; and immunoblotted for antibodies against His6 (ataxin-7-10Q, ataxin-7-60Q, Ada3), Ada2, and Gcn5.
HAT Assays
Fluorography and liquid HAT assays were performed as described by McMahon et al. (16). Core histones were obtained from Millipore (catalog no. 13-107); recombinant reconstituted Xenopus nucleosome core particles were obtained from the laboratory of Dr. Song Tan. H3 N-terminal peptides were synthesized as described by Grant et al. (17). Wild-type peptides span N-terminal amino acids 5–28 of human histone H3, and derivatives contain either no or single non-acetylated lysines available at amino acid position 9, 14, 18, or 23. Assays were performed using 300 ng of peptides. Gcn5 levels between samples were normalized by Western blotting for Gcn5 (sc-9078, Santa Cruz Biotechnology).
Growth Assays
Yeast cultures were grown overnight in selective medium with 2% dextrose. The next day, the absorbance at 600 nm was measured using a spectrophotometer. Cells were spun down, washed twice with water, and then reseeded in selective medium with 2% galactose. All strains were normalized to an A600 of 0.2. Cells were placed back in the shaker at 30 °C, and the absorbance was measured in triplicate every 2 h for a total time of 14 h.
mRNA Expression
Yeast cultures (10 ml) were grown in selective synthetic complete (SC) medium containing 2% dextrose to an absorbance at 600 nm of 0.5, and cells were then reseeded in SC medium containing 2% galactose for 4 h. Total RNA was extracted from the cell pellets, and the RNA was then treated with DNase using the TURBO DNA-free kit from Ambion. RNA (1 μg) was reverse-transcribed using a SuperScript VILO cDNA synthesis kit (catalog no. 11750, Invitrogen). The cDNA generated was amplified in real time using iQ SYBR Green Supermix (catalog 2012-03, Bio-Rad) and the Bio-Rad MyiQ single color real-time PCR detection system. cDNA was amplified using primers specific for the GAL1 and ACT1 genes. Wild-type values were set to 1.
ChIP
Yeast cultures (50 ml) were grown in selective SC medium containing 2% dextrose to an absorbance at 600 nm of 0.5. The cells were then split; half were kept in 2% dextrose, and half were spun down, washed, and reseeded in SC medium containing 2% galactose. Cells were then placed back on a shaking platform at 30 °C for 2 h. ChIP assays were performed as described by Kuo and Allis (18) with a few modifications. Cells were sonicated using a of Bioruptor Twin system (UCD-400) on high 20× for 30 s. Equal amounts of yeast extracts (0.8 mg of total protein) were immunoprecipitated with 3 μl of antibody against Lys9-acetylated histone H3 (catalog no. 07-352, Millipore) or 8 μl of anti-H3 antibody (ab1791, Abcam) in a total reaction volume of 500 μl. Protein A-Sepharose CL-B4 beads (GE Healthcare) were added to the antibody/lysate mixture the following day and rotated for 2 h at 4 °C. IgG-Sepharose was used for TAP immunoprecipitation. DNA was isolated from the samples using a PCR purification kit (catalog no. K3100-02, Invitrogen). DNA was amplified using oligonucleotide primers (Invitrogen) specific for the GAL1 and GAL7 UASs or control primers for the ACT1 promoter or TEL (noncoding region of Saccharomyces cerevisiae chromosome 6R). The recovered DNA was quantified in triplicate by real-time PCR using Power SYBR Green PCR Master Mix (Applied Biosystems) and the Bio-Rad MyiQ single color real-time PCR detection system. ChIP values obtained are presented as relative enrichment/occupancy; relative enrichment = (2(ΔCTcontrol−ΔCTsample)). Wild-type enrichment values were set to 1, and values for samples grown in galactose were divided by values for samples grown in dextrose. For H3K9 acetylation ChIPs, H3K9 acetylation CT values for galactose were normalized to account for nucleosome eviction.
RESULTS
Polyglutamine-expanded Ataxin-7 Binds Gcn5 and the Gcn5 Trimeric Complex in Vitro
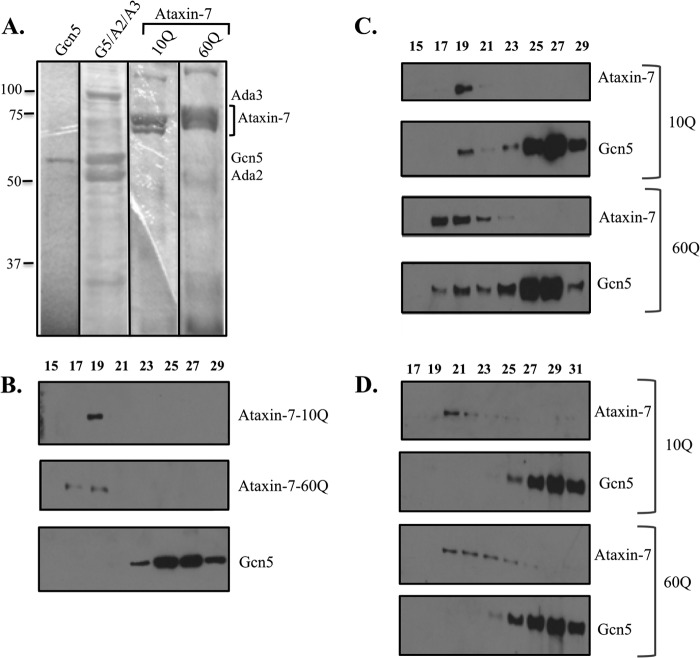
We set out to determine whether ataxin-7 binds directly to the catalytic core and if differential binding to Gcn5 occurs when ataxin-7 is polyQ-expanded. Previous studies have suggested that aberrant interactions may take place in SAGA complexes that contain polyQ-expanded ataxin-7, resulting in loss of some proteins from the complex (6, 9). However, to determine whether there is any direct binding with the catalytic core, we conducted in vitro binding assays with purified recombinant proteins. His6-Gcn5 and His6-ataxin-7 were independently expressed in and isolated from E. coli, purified using Ni2+-NTA-agarose, and eluted with 0.3 m imidazole (Fig. 1A). The proteins were first fractionated separately using a Superose 12 size exclusion column to determine where each protein ran individually (Fig. 1B). Ataxin-7 eluted from the column with a larger than expected molecular weight, suggesting either multimerization or a non-globular organization. Ataxin-7-10Q or ataxin-7-60Q was then mixed with Gcn5 in binding buffer and fractionated with a Superose 12 column. Under these conditions, a portion of Gcn5 eluted in an earlier fraction (from fractions 23–29 to 19), co-eluting with ataxin-7, consistent with a physical interaction between these proteins in a complex with a higher molecular weight than Gcn5 alone. A shift in Gcn5 to an earlier fraction was seen when Gcn5 was incubated with either ataxin-7-10Q or ataxin-7-60Q (Fig. 1C).
FIGURE 1.
Polyglutamine-expanded ataxin-7 binds Gcn5 in vitro. A, Coomassie Blue staining of recombinant proteins (eluates of individually purified Gcn5, Gcn5-Ada2-Ada3 (G5/A2/A3) trimeric complex, ataxin-7-10Q, and ataxin-7-60Q). Complexes were purified using Ni2+-NTA-agarose and then eluted with imidazole. B, Western blots of Superose 12 fractions of recombinant Gcn5, ataxin-7-10Q, and ataxin-7-60Q. C, Western blots of Superose 12 fractions from in vitro binding assays of ataxin-7-10Q or ataxin-7-60Q with Gcn5. D, Western blots of Superose 6 fractions from in vitro binding assays of ataxin-7-10Q or ataxin-7-60Q with the trimer (Ada2, Ada3, and Gcn5).
Our next step was to determine, using the binding assay described above, if either ataxin-7-10Q or ataxin-7-60Q binds to a purified recombinant Gcn5 catalytic core complex consisting of His6-Ada3, Ada2, and Gcn5. This association of Gcn5 with Ada3, via the bridging factor Ada2, has been found to increase the overall catalytic activity of Gcn5, to confer its ability to acetylate nucleosomes, and to increase its lysine site specificity across the histone H3 tail (19). Ataxin-7-10Q or ataxin-7-60Q was incubated with the purified recombinant Gcn5, Ada2, and Ada3 complex, and the binding mixture was then run over a Superose 6 column to observe any fractionation shifts in Gcn5, Ada2 or Ada3 that would be indicative of physical interaction and/or disruption of the trimeric complex. Interestingly, neither the 10Q or 60Q form of ataxin-7 bound to the trimer when mixed independently, as there was no shift in Gcn5 (Fig. 1D) or Ada2 or Ada3 (data not shown) to an earlier fraction. However, a possible explanation for this may be that the Gcn5 catalytic core already was formed, and it is possible that ataxin-7 may still bind the catalytic core during assembly of SAGA and/or the Gcn5 catalytic core when coexpressed.
To elucidate whether ataxin-7 binds the catalytic core during complex formation, we used a polycistronic vector expression system. This method, used mainly for expressing small protein complexes, allowed us to express all members of the catalytic core along with either the 10Q and 60Q form of ataxin-7. The polycistronic vector pST44 has four positions in which a gene can be easily inserted via traditional cloning methods. All genes cloned are then expressed from the same promoter (15). A gene encoding FLAG-tagged ataxin-7-10Q or ataxin-7-60Q was inserted into the fourth position for either the dimeric His6-Ada2 and Gcn5 constructs or the His6-Ada3-Ada2-Gcn5 trimer construct. These vectors were then expressed in E. coli, and translated proteins were purified by Ni2+-NTA-agarose chromatography (Fig. 2B). Next, the eluates were run on an SDS-polyacrylamide gel; transferred to nitrocellulose membrane; and blotted with antibodies against Gcn5, Ada2, Ada3, and ataxin-7 (Fig. 2A). The results obtained from these experiments revealed that polyQ-expanded ataxin-7 bound the Gcn5 catalytic core, but little to no ataxin-7-10Q bound. Interestingly, ataxin-7-10Q was able to bind the dimer (Gcn5 and Ada2) with similar efficiency to ataxin-7-60Q (Fig. 2A). Used as a negative control, recombinant ataxin-7-60Q was shown not to elute alone, without Gcn5 or one or both of its accessory proteins, from Ni2+-NTA-agarose resin (Fig. 2A, lower panel). This confirms that the alternative binding activity of ataxin-7-60Q is not a result of the polyQ expansion nonspecifically binding to Ni2+-NTA-agarose. These results suggest that ataxin-7-10Q and ataxin-7-60Q exhibit differential binding with the catalytic core containing Ada3.
FIGURE 2.
Coexpression of ataxin-7 with catalytic core proteins reveals different binding behaviors of ataxin-7-10Q and ataxin-7-60Q in vitro. A, ataxin-7-10Q or ataxin-7-60Q was coexpressed and purified with either the Gcn5-Ada2 dimer (G5/A2) or the Gcn5-Ada2-Ada3 trimer (G5/A2/A3). Ada2 and Ada3 are His6-tagged at their N termini in the dimer and trimer, respectively. Shown are Western blots from Ni2+-NTA-agarose purifications. The lower panel shows a negative control of unbound fractions and eluates of recombinant ataxin-7-60Q alone. B, eluates of ataxin-7-10Q or ataxin-7-60Q were coexpressed with either Gcn5-Ada2 or Gcn5-Ada2-Ada3 and purified using Ni2+-NTA-agarose and then eluted with imidazole. Ada2 and Ada3 are His6-tagged at their N termini in the dimer and trimer, respectively. C, a two-step purification of the tetramer (ataxin-7-60Q, Ada3, Ada2, and Gcn5) allows for enrichment of the complex. A schematic representation of purification of the tetrameric complex is shown. The His6 and FLAG affinity tags on Ada3 and ataxin-7, respectively, allowed for a two-step purification. FT, flow-through fraction. D, Coomassie Blue staining and Western analysis of eluates from a two-step purification of the tetrameric complex (Gcn5, Ada2, Ada3, and ataxin-7-60Q).
Polyglutamine-expanded Ataxin-7 Abrogates Gcn5 Catalytic Activity in Vitro
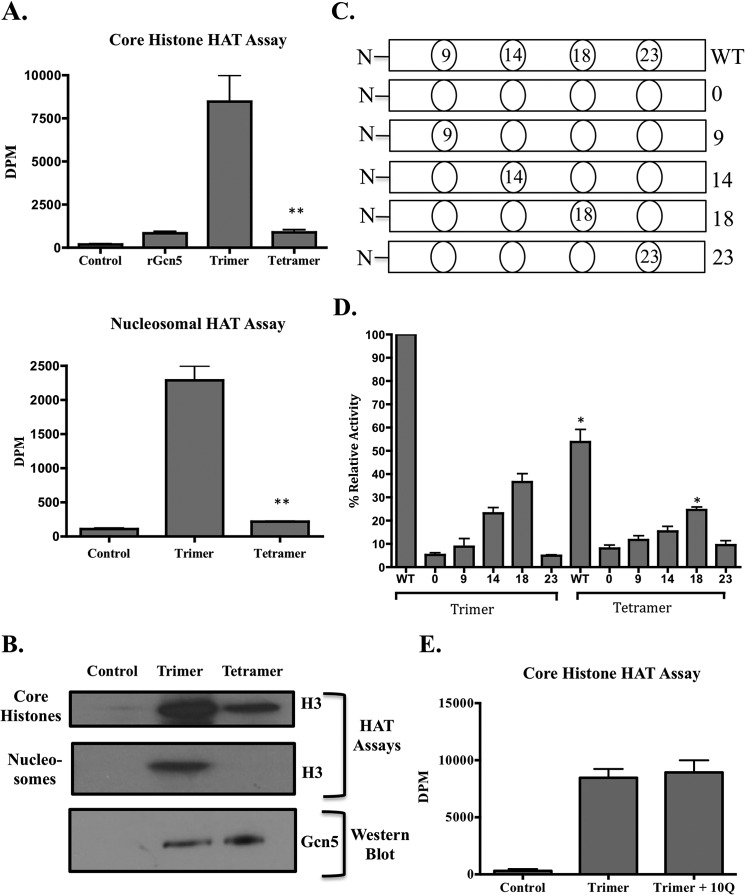
After observing different binding behavior of ataxin-7-60Q to the Gcn5 catalytic core as compared with a physiologically normal ataxin-7-10Q protein, our next step was to determine whether these differential interactions affect the functional properties of this core complex. Conducting in vitro HAT assays with our purified recombinant trimeric catalytic core complex or tetrameric complex (catalytic core with ataxin-7-60Q) allowed us to determine the differences between HAT activities of these complexes and to assess if the differences were due to direct binding of the ataxin-7-60Q protein to the catalytic core. For these functional assays, the tetrameric complex was doubly purified to enrich for complexes containing all four proteins (Fig. 2C). It is likely that purifying the complexes with just Ni2+-NTA-agarose would result in a very heterogeneous population, a milieu of complexes such as Ada3-Ada2-Gcn5 and Ada3-Ada2-Gcn5-ataxin-7-60Q resulting from the elution. Early attempts to conduct HAT assays with a singly purified complex resulted in erratic HAT assay results most likely due to eluates being a varied mixture of complexes. The two-step purification, taking advantage of affinity tags on both Ada3 (His6) and ataxin-7 (FLAG), allowed for a more homogeneous complex isolation (Fig. 2D). The trimeric complex was purified as described for Fig. 1A, and Gcn5 levels between the two complexes were equalized via Western analysis (Fig. 3B, lower panel). Attempts to doubly purify a tetrameric complex with ataxin-7-10Q were not successful as Gcn5, Ada2, and Ada3 fractionated away from ataxin-10Q (Fig. 2A and data not shown). Therefore, the proceeding HAT assays were conducted to compare the acetylation activities of trimer versus a tetramer containing ataxin-7-60Q.
FIGURE 3.
Ataxin-7 expansion abrogates Ada2- and Ada3-mediated enhancement of Gcn5 catalytic activity and substrate specificity in vitro. A, histograms displaying dpm of 3H incorporation into free histones or nucleosomes after liquid HAT assays using purified recombinant Gcn5, trimer (Gcn5, Ada2, and Ada3), or tetramer (ataxin-7-60Q, Gcn5, Ada2, and Ada3). Control samples contained no Gcn5. The trimer is a positive control for both histone and nucleosomal HAT assays. Error bars indicate S.E. Asterisks represent p values < 0.05 as determined using Student's t two-tailed test (16). B, fluorogram of HAT assays using core histone and nucleosomal substrates incubated with purified complexes of the trimer or tetramer. The lower panel is a Western blot showing equalized protein levels of Gcn5. C, diagram of the N-terminal peptides used in D. The positions of the non-acetylated lysines are marked in each peptide. Open ovals indicate acetylated lysines. D, the recombinant trimer or tetramer was incubated with the WT peptide, a peptide lacking any non-acetylated lysines (0), or peptides with a single non-acetylated lysine (position 9, 14, 18, or 23). Histograms displaying dpm of 3H incorporation from liquid assays are shown. Data are normalized to the relative activity of the WT peptide (17). Asterisks represent p values < 0.05 as determined using Student's two-tailed t test (**, values compared with the WT trimer). E, fluorogram of HAT assay using core histone substrates incubated without enzyme (Control) or with the purified trimer alone or the purified trimer and separately purified ataxin-7–10.
HAT assays conducted with core histones used as a substrate showed a sharp decrease in the amount of histone acetylation when the tetrameric complex was used (Fig. 3, A and B). These results show that the addition of ataxin-7-60Q to the catalytic core complex cancelled out or inhibited the effects of the adaptor protein Ada2 on core histone acetylation by Gcn5, returning acetylation levels to those with Gcn5 alone. Ada2 is known to increase the in vitro HAT activity of Gcn5 when core histones are used as a substrate; thus, the potentiation of Gcn5 activity by Ada2 is lost when ataxin-7-60Q is present (19). Although we were unable to purify a tetrameric complex using ataxin-7-10Q, we wanted to verify that the simple addition of separately purified ataxin-7-10Q to the trimer would not result in a change in its HAT activity. The addition of ataxin-7-10Q to the trimer resulted in little to no change in the HAT activity of the trimer (Fig. 3E). These results are also consistent with the reduction in HAT activity by the tetrameric complex being a specific effect of polyQ-expanded ataxin-7. Next, we conducted in vitro HAT assays using nucleosomes as a substrate. Ada3 is needed for efficient acetylation of nucleosomal substrates by Gcn5 (19). Again, the results show a sharp decline in the amount of lysine acetylation on nucleosomes when the tetramer was used relative to the trimer (Fig. 3A, lower panel). These results are consistent with the loss of SAGA-mediated acetylation of nucleosomes observed when polyQ-expanded ataxin-7 is incorporated into the entire yeast or human complexes (6, 9).
Ada3 is also important for proper lysine acetylation specificity. To determine whether lysine specificity changes when ataxin-7-60Q is assembled with the catalytic core, we conducted HAT assays with histone H3 N-terminal peptides (Fig. 3C). The peptides used have all sites open for acetylation (WT), no sites open (0), or one available non-acetylated site open (lysine position 9, 14, 18, or 23). As expected, the trimer exhibited an acetylation pattern of WT > 18 > 14 > 9 > 23 (17), whereas the tetramer exhibited a much lower overall level of acetylation, but with a similar pattern. The results show that peptide acetylation levels catalyzed by the tetramer were ∼50% less than those catalyzed by the trimer (Fig. 3D). These results suggest that Ada3 is bound, exerting its lysine specificity role in Gcn5, but Gcn5 is unable to perform effective nucleosome acetylation.
Yeast Cells Expressing Polyglutamine-expanded Ataxin-7 Exhibit Acetylation Deficiencies in Vivo
All data thus far have focused on in vitro studies showing the direct effects of polyQ-expanded ataxin-7 on the Gcn5 catalytic core, biochemically and functionally. Our next focus was to determine whether the influence of polyQ-expanded ataxin-7 on Gcn5 HAT activity exerts effects in vivo. Growth curve analysis allowed us to follow the division of these strains in galactose medium. We observed that yeast cells expressing ataxin-7-10Q were able to at least partially rescue the growth defect of sgf73Δ yeast on plates containing galactose medium, whereas ataxin-7-60Q was not. The sgf73Δ strain grew slower than the wild-type strain over a 14-h time course, whereas expression of ataxin-7-10Q partially rescued this defect (Fig. 4). However, we found that the Δ+60Q strain was the slowest growing strain, further exacerbating the growth defect found in the sgf73Δ strain (Fig. 4).
FIGURE 4.

Yeast cells expressing polyQ-expanded ataxin-7 exhibit growth defects on alternate carbon sources. Shown are the results from the growth curve analysis of sgf73Δ, Δ+10Q, and Δ+60Q relative to the parental WT strain in SC medium with 2% galactose. Error bars represent three separate spectrometer readings.
We decided to look more closely to determine a molecular explanation for the galactose phenotype. We asked whether the inhibited cell growth seen in the Δ+60Q cells corresponded to deficits in Gcn5-mediated histone H3 acetylation in vivo. We focused on two well studied galactose genes, GAL1 and GAL7, whose expression is induced in yeast grown in galactose medium, because SAGA is an important transcriptional regulator of this gene (20, 21). Although containing their own promoter regions, GAL1 and GAL7 are known to be co-regulated upon galactose induction and, along with GAL10, are contained on chromosome 2 (22), The GAL1 gene codes for the galactokinase protein, which is responsible for phosphorylating galactose, the first step in galactose conversion to glucose to enter the glycolytic pathway. The GAL7 gene codes for the protein galactose-1-phosphate uridylyltransferase, which catalyzes the second step in glucose-to-galactose conversion. SAGA directly binds the Gal4 protein at the GAL1 and GAL7 UASs and is important for complete assembly of the preinitiation complex at these promoters (20, 21). Yeast cells grown in SC/dextrose medium were shifted to SC/galactose medium for 2 h. Measurement of GAL1 mRNA abundance revealed a modest decline in GAL1 mRNA in the Δ+60Q strains compared with the Δ+10Q strains (Fig. 5A). This reduction mirrored the growth of the Δ+60Q strains shown in Fig. 4.
FIGURE 5.
Strains expressing polyQ-expanded ataxin-7 show increased recruitment of Gcn5 but decreased acetylation in vivo. A, ataxin-7-60Q showed a modest decrease in mRNA expression of the GAL1 gene. mRNA expression of the GAL1 gene in the WT, sgf73Δ, Δ+10Q, and Δ+60Q strains grown in galactose medium for 2 h was determined by RT-PCR in real time. Values are presented as relative mRNA abundance, with WT values set to 1, and normalized against the ACT1 gene. B, ChIP for H3K9 acetylation at the GAL1 UAS. Values are presented as relative enrichment of TEL, with WT values set to 1.0. C, ChIP for Gcn5 occupancy at the GAL1 UAS. Values are presented as relative occupancy of the ACT1 promoter, with WT values set to 1.0. D, ChIP for H3K9 acetylation at the GAL7 UAS. Values are presented as relative enrichment of TEL, with WT values set at 1.0. E, ChIP for Gcn5 occupancy at the GAL7 UAS. For B–E, total H3 values were normalized between cells grown in galactose and dextrose to correct for total H3 lost during nucleosome eviction. Asterisks represent p values <0.05 as determined using Student's two-tailed t test.
ChIP assays were performed in WT, sgf73Δ, Δ+10Q, and Δ+60Q strains using antibodies against histone H3, Lys9-acetylated histone H3, and Gcn5. ChIP results for H3K9 acetylation at the UASs of GAL1 and GAL7 showed a reduction in the sgf73Δ strain compared with the WT strain. However, when we compared the Δ+60Q and Δ+10Q strains, we saw a very drastic decline in acetylation levels (Fig. 5, B and D). These results mimic the in vitro HAT assay data in Fig. 3, where the addition of polyQ-expanded ataxin-7 to the catalytic core caused a sharp decrease in HAT activity. We also observed an increase in acetylation in the ataxin-7-10Q strain at the GAL1 locus and, to a lesser extent, at the GAL7 promoter, which is consistent with a somewhat higher nucleosomal catalytic activity of the purified ataxin-7-10Q-SAGA complex (6). ChIP assays for Gcn5 occupancy at the GAL1 and GAL7 UASs revealed a reduction in Gcn5 binding in the sgf73Δ strain compared with the WT strain (Fig. 5, C and E). This is consistent with a reduction in SAGA complex stability in sgf73Δ mutant yeast (6). Interestingly, when we performed ChIP assays looking at Gcn5 occupancy at the GAL1 and GAL7 UASs in the Δ+60Q strain, we observed more Gcn5 occupancy at the UAS compared with the Δ+10Q strain, even though acetylation values in the Δ+60Q strain were drastically decreased (Fig. 5, C and E). Therefore, we can conclude that Gcn5 is effectively recruited to the GAL1 and GAL7 UASs under inducing conditions in the Δ+60Q strain; however, it is unable to perform proper acetylation activity, leading to reduced mRNA production.
DISCUSSION
We have shown direct inhibition of Gcn5 catalytic activity by polyQ-expanded ataxin-7 and that inhibition contributes to loss of Gcn5 catalytic activity both in vitro and in vivo. Our in vitro biochemical results suggest that polyQ-expanded ataxin-7 exhibits a gain-of-function phenotype by increasing binding to a known binding partner, Gcn5, and/or improper binding to other SAGA complex subunits. Differential binding behavior is a hallmark of polyQ-expanded proteins (23). Studies on other polyQ diseases show that abnormal interactions may contribute to their pathogenesis as well. In SCA1, for example, polyQ expansion of ataxin-1 results in an increased interaction with one of the multisubunit complexes it is known to associate with. This complex, containing the protein RBM17, preferentially interacts with the polyQ-expanded version of ataxin-1 and participates in that complex's gain of function, which is a contributing factor to SCA1 pathogenesis (24). Additionally, the protein huntingtin, which is polyQ-expanded in Huntington disease, has a number of interactions that can either decrease or increase depending on whether its polyQ tract is expanded (25). The alternative and aberrant binding behavior of ataxin-7 is dependent upon polyQ stretches greater than 35 CAG-encoded repeats (26), which form a β-sheet amyloid plaque, putting strain on the normal secondary structure of the protein (26). Formation of the β-sheet by the protein monomer leads to assembly of β-sheet oligomers, and eventually, it is believed, these β-sheet oligomers of polyQ-expanded proteins accumulate to form amyloid fibrillar aggregates (26). During β-sheet formation and early oligomer formation, aberrant interactions with known binding partners and other proteins are most likely to take place. Therefore, it is possible that the increased binding of Gcn5 by ataxin-7-60Q seen in Fig. 1C could be the result of Gcn5 binding directly to the polyQ-expanded stretch in addition to/or instead of its binding seen with ataxin-7-10Q.
In vitro functional studies sought to determine whether the binding of polyQ-expanded ataxin-7 to the Gcn5 catalytic core affects the HAT activity of Gcn5. We found that this is indeed the case; polyQ-expanded ataxin-7 bound to the Gcn5 catalytic core, causing a severe decrease in HAT activity using both free histones and nucleosomes. Consistent with this, both yeast and mammalian SAGA complexes display decreased catalytic activity in the presence of polyQ-expanded ataxin-7 (6, 9).
Because Ada2 and Ada3 are both present in the recombinant tetramer used in this study, an inhibition by the polyQ stretch of Gcn5 or one of its adaptor proteins is a likely cause of the abrogated HAT activity seen on both free histones and nucleosomes. Previous reports have shown that incorporation of polyQ-expanded ataxin-7 into SAGA results in decreased association of some subunits of SAGA, including Ada2 and Ada3 (6, 9). Although it is difficult to infer loss of subunits by polyQ-expanded ataxin-7 in the experiments employed in these studies, that idea is not contradictory to our findings. Our results suggest that the activities of the catalytic core may be affected before complete SAGA destabilization happens.
In our in vivo experiments, the Δ+60Q strain exhibited a modest 30% decrease in GAL1 mRNA compared with the Δ+10Q strain (Fig. 5A). A total loss of acetylation but a somewhat modest decline in mRNA transcription are not a complete surprise because acetylation by Gcn5 is not required for full mRNA transcription at the GAL1 locus. The reduction in GAL1 mRNA transcription seen in the Δ+60Q strain is very similar to the mRNA reductions seen in gcn5Δ cells at the GAL1 locus (27). Therefore, these results show that the mRNA reduction seen in the Δ+60Q strain is consistent with an inhibition of Gcn5 acetylation at the GAL1 UAS rather than another SAGA-related function. What we have learned from studying SAGA at various loci is that many genes require the different activities of SAGA subunits for full transcriptional activity (28, 29). Recent studies suggest that SAGA may function at more genes than previously thought (30). Thus, when considering how an aberrant protein is incorporated into such a multisubunit, multifunctioning complex, it is highly possible that disruption by this protein will vary depending on the gene context and cellular environment. Indeed, ataxin-7 associates with the DUB module of SAGA, and histone H2B monoubiquitination accumulates at the promoter of the transcriptionally repressed Reelin gene in human astrocytes upon expression of polyQ-expanded ataxin-7 (31).
Upon investigation of the acetylation and occupancy of Gcn5 at the GAL1 and GAL7 UASs, we found that although acetylation was severely reduced in Δ+60Q cells compared with Δ+10Q cells, the occupancy of Gcn5 was increased in the Δ+60Q strains. These results follow previous findings indicating that although Gcn5 is present at a SAGA-regulated gene in cells expressing polyQ-expanded ataxin-7, less acetylation takes place (9). In studies conducted by Palhan et al. (9), retina cells from transgenic ataxin-7–92Q mice showed severely decreased acetylation of histone H3 in symptomatic 11-week-old mice. Although ChIPs for Gcn5 were not conducted in this study, immunohistochemistry on retinal sections was used to determine the levels of Gcn5 in these cells, demonstrating a dramatic increase in signal intensity. Our results point toward a dominant-negative effect of polyQ-expanded ataxin-7 at the GAL1 and GAL7 UASs, where SAGA containing polyQ-expanded ataxin-7 is properly recruited to the promoter but does not have a functional acetyltransferase. SCA7 is a dominant disorder and because polyQ-expanded ataxin-7 is known to accumulate in the nucleus, recruitment of an inactive or compromised complex may compete with the normal complex and sway the balance toward an increased mutant complex abundance with possibly an either greater promoter occupancy or residency time.
The results obtained show a possible two-pronged approach to how ataxin-7 may dominantly affect SAGA function and contribute to SCA7 pathogenesis. One way is a direct biochemical inhibition of the catalytic core, affecting the ability of the Gcn5 adaptor proteins Ada2 and Ada3 to potentiate HAT activity on nucleosomes. Then, this SAGA complex containing polyQ-expanded ataxin-7 is able to be properly recruited to promoters, but it is unable to efficiently acetylate nucleosomes. Increased Gcn5 (and presumably SAGA) levels measured at the GAL1 and GAL7 promoters by ChIP could represent either increased abundance of the mutant complex or a longer residency time. Theoretically, this could be part of a mechanism for cells to attempt to correct for low acetylation levels or a failure for SAGA to dissociate from target promoters following normal histone acetylation. In either scenario, these mutant complexes may occlude the recruitment of fully functional complexes or other compensatory acetyltransferases. This coupled with the increased abundance of polyQ-expanded ataxin-7 relative to normal ataxin-7 in heterozygous diseased cells (32) would significantly limit the ability of healthy SAGA complexes to function in promoter histone acetylation. Our in vitro and in vivo results suggest for the first time a direct molecular mechanism by which polyQ-expanded ataxin-7 dominantly inhibits the function of the SAGA complex in histone acetylation, helping to explain certain transcriptional alterations associated with SCA7 pathogenesis.
Acknowledgments
We thank Dr. Song Tan for providing the pST44 polycistronic vectors as well as recombinant nucleosomes and Drs. Kenneth Lee and Jerry Workman for Gcn5-TAP strains and technical help with ChIP assays.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 NS049065 (to P. A. G.).
- HAT
- histone acetyltransferase
- DUB
- deubiquitination
- polyQ
- polyglutamine
- SCA7
- spinocerebellar ataxia type 7
- UAS
- upstream activating sequence
- TAP
- tandem affinity purification tag
- Ni2+-NTA
- nickel-nitrilotriacetic acid
- SC
- synthetic complete.
REFERENCES
- 1. Peterson C. L., Laniel M. A. (2004) Histones and histone modifications. Curr. Biol. 14, R546–R551 [DOI] [PubMed] [Google Scholar]
- 2. Lee J., Thompson J. R., Botuyan M. V., Mer G. (2008) Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 15, 109–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burke T. L., Grant P. A. (2011) Histone Acetylation Complexes. in Regulation of Organelle and Cell Compartment Signaling: Cell Signaling Collection (Bradshaw R. A., Dennis E. A., eds) p. 109, Academic Press, New York [Google Scholar]
- 4. Baker S. P., Grant P. A. (2007) The SAGA continues: expanding the cellular role of a transcriptional co-activator complex. Oncogene 26, 5329–5340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sanders S. L., Jennings J., Canutescu A., Link A. J., Weil P. A. (2002) Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22, 4723–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McMahon S. J., Pray-Grant M. G., Schieltz D., Yates J. R., 3rd, Grant P. A. (2005) Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc. Natl. Acad. Sci. U.S.A. 102, 8478–8482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Helmlinger D., Hardy S., Sasorith S., Klein F., Robert F., Weber C., Miguet L., Potier N., Van-Dorsselaer A., Wurtz J. M., Mandel J. L., Tora L., Devys D. (2004) Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum. Mol. Genet. 13, 1257–1265 [DOI] [PubMed] [Google Scholar]
- 8. David G., Abbas N., Stevanin G., A., Yvert G., Cancel G., Weber C., Imbert G., Saudou F., Antoniou E., Drabkin H., Gemmill R., Giunti P., Benomar A., Wood N., Ruberg M., Agid Y., Mandel J. L., Brice A. (1997) Cloning of the SCA7 gene reveals a highly unstable CAG repeat expansion. Nat. Genet. 17, 65–70 [DOI] [PubMed] [Google Scholar]
- 9. Palhan V. B., Chen S., Peng G. H., Tjernberg A., Gamper A. M., Fan Y., Chait B. T., La Spada A. R., Roeder R. G. (2005) Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc. Natl. Acad. Sci. U.S.A. 102, 8472–8477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee K. K., Sardiu M. E., Swanson S. K., Gilmore J. M., Torok M., Grant P. A., Florens L., Workman J. L., Washburn M. P. (2011) Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol. Syst. Biol. 7, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Köhler A., Schneider M., Cabal G. G., Nehrbass U. (2008) Yeast ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 10, 707–715 [DOI] [PubMed] [Google Scholar]
- 12. Lee K. K., Swanson S. K., Florens L., Washburn M. P., Workman J. L. (2009) Yeast Sgf73/Ataxin-7 serves to anchor the deubiquitination module into both SAGA and Slik(SALSA) HAT complexes. Epigenetics Chromatin 21, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen Y. C., Gatchel J. R., Lewis R. W., Mao C. A., Grant P. A., Zoghbi H. Y., Dent S. Y. (2012) Gcn5 loss-of-function accelerates cerebellar and retinal degeneration in a SCA7 mouse model. Hum. Mol. Genet. 21, 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Helmlinger D., Hardy S., Abou-Sleymane G., Eberlin A., Bowman A. B., Gansmüller A., Picaud S., Zoghbi H. Y., Trottier Y., Tora L. (2006) Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 4, e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tan S., Kern R. C., Selleck W. (2005) The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr. Purif. 40, 385–395 [DOI] [PubMed] [Google Scholar]
- 16. McMahon S. J., Doyon Y., J., Grant P. A. (2004) Identification and analysis of native HAT complexes. Methods Enzymol. 377, 154–167 [DOI] [PubMed] [Google Scholar]
- 17. Grant P. A., Eberharter A., John S., Cook R. G., Turner B. M., Workman J. L. (1999) Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274, 5895–5900 [DOI] [PubMed] [Google Scholar]
- 18. Kuo M. H., Allis C. D. (1999) In vivo cross-linking and immunoprecipitation for studying dynamic protein: DNA associations in a chromatin environment. Methods 19, 425–433 [DOI] [PubMed] [Google Scholar]
- 19. Balasubramanian R., Pray-Grant M. G., Selleck W., Grant P. A., Tan S. (2002) Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277, 7989–7995 [DOI] [PubMed] [Google Scholar]
- 20. Bhaumik S. R., Green M. R. (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15, 1935–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bryant G. O., Ptashne M. (2003) Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol. Cell 11, 1301–1309 [DOI] [PubMed] [Google Scholar]
- 22. Lohr D., Venkov P., Zlatanova J. (1995) Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 9, 777–787 [DOI] [PubMed] [Google Scholar]
- 23. Gatchel J. R., Zoghbi H. Y. (2005) Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 6, 743–755 [DOI] [PubMed] [Google Scholar]
- 24. Lim J., Crespo-Barreto J., Jafar-Nejad P., Bowman A. B., Richman R., Hill D. E., Orr H. T., Zoghbi H. Y. (2008) Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature 452, 713–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li S. H., Li X. J. (2004) Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet. 20, 146–154 [DOI] [PubMed] [Google Scholar]
- 26. Nagai Y., Popiel H. A. (2008) Conformational changes and aggregation of expanded polyglutamine proteins as therapeutic targets of the polyglutamine diseases: exposed-sheet hypothesis. Curr. Pharm. Des. 14, 3267–3279 [DOI] [PubMed] [Google Scholar]
- 27. Govind C. K., Zhang F., Qiu H., Hofmeyer K., Hinnebusch A. G. (2007) Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25, 31–42 [DOI] [PubMed] [Google Scholar]
- 28. Bhaumik S. R., Green M. R. (2002) Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22, 7365–7371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Roberts S. M., Winston F. (1997) Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147, 451–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Venters B. J., Pugh B. F. (2009) A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 19, 360–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McCullough S. D., Xu X., Dent S. Y. R., Bekiranov S., Roeder R. G., Grant P. A. (2012) Reelin is a target of polyglutamine expanded ataxin-7 in human spinocerebellar ataxia type 7 (SCA7) astrocytes. Proc. Natl. Acad. Sci. U.S.A. 109, 21319–21324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCullough S. D., Grant P. A. (2010) Histone acetylation, acetyltransferases, and ataxia–alteration of histone acetylation and chromatin dynamics is implicated in the pathogenesis of polyglutamine-expansion disorders. Adv. Protein Chem. Struct. Biol. 79, 165–203 [DOI] [PMC free article] [PubMed] [Google Scholar]