Abstract
Nutrient richness, and specifically the abundance of mono- and disaccharides that characterize several food matrixes, such as milk and grape juice, has allowed the speciation of lactic acid bacteria and yeasts with a high fermentation capacity instead of energetically favorable respiratory metabolism. In these environmental contexts, rapid sugar consumption and lactic acid or ethanol production, accumulation, and tolerance, together with the ability to propagate in the absence of oxygen, are several of the “winning” traits that have apparently evolved and become specialized to perfection in these fermenting microorganisms. Here, we summarize and discuss the evolutionary context that has driven energetic metabolism in food-associated microorganisms, using the dairy species Lactococcus lactis and Streptococcus thermophilus among prokaryotes and the bakers’ yeast Saccharomyces cerevisiae among eukaryotes as model organisms.
Keywords: lactic acid bacteria, yeast, metabolism, evolution, food microbiology
INTRODUCTION
What is the role of environmental constraints in streamlining and shaping the energetic metabolism of food-adapted microorganisms? Here, we compare prokaryotic lactic acid bacteria (LAB) and the eukaryote Saccharomyces cerevisiae to draw bioenergetic analogies between these two classes of microorganisms that lead us to hypothesize a common evolutionary path. This paper will focus on the evolutionary forces that allow selection between selfish and cooperative energetic behaviors (Pfeiffer et al., 2001) in food-associated microorganisms. The analysis of energetic behaviors will be performed while considering the environmental context in which microorganisms evolve and the environmental changes that occur during microbial growth in a food matrix. Recently, the relevance of selfish and cooperative behaviors to the origins and the evolution of complex microbial communities in natural environments has been discussed (Sachs and Hollowell, 2012). These behaviors demonstrate how metabolic cooperation among microorganisms determines their genomic and metabolic streamlining. Here, we discuss the role of food environments generated by anthropic activities in shaping the energetic metabolism in food-associated microorganisms.
CHALLENGING METABOLISM WITH ENVIRONMENTAL STIMULI
During growth, every microorganism causes considerable changes in the environmental, chemical, and physical conditions by changing the concentrations of nutrients and organic acids and by other molecules generated through the organism’s catabolism. Certain habitats may fluctuate erratically, whereas others, which are more predictable, may offer the opportunity to prepare in advance for the next environmental change. In this context, microorganisms living in “predictable” fluctuating environments have evolved regulatory machinery to anticipate environmental perturbations by adapting to the perturbations’ temporal order of appearance (Mitchell et al., 2009; Zakrzewska et al., 2011). Food matrixes represent a good example of “predictable” fluctuating environments generated by anthropic activities. Nutrient richness, and specifically, the abundance of mono- and disaccharides that characterize several food matrixes, such as milk and grape juice (in which mono- and disaccharide resources are both large and dense), could have further allowed the speciation and/or domestication of LAB and yeasts with a high fermentation capacity instead of more energetically favorable respiratory metabolism (Bolotin et al., 2004; Fay and Benavides, 2005). S. cerevisiae has been exploited for several millennia throughout the world for its ability to produce beer, wine and bread. This widespread use is due to a robust fermentative metabolism. This organism’s ability to degrade sugars into ethanol and CO2, even under aerobic conditions, thus using fermentative metabolism instead of respiration, is at the basis of its popularity as “food” yeast and its benefit to human civilization for thousands of years (Piškur et al., 2006).
Among bacteria, the seemingly simplistic metabolism of LAB has been exploited throughout history for the preservation of foods and beverages in nearly all societies, dating back to the origins of agriculture (Dunne et al., 2012; Hayden et al., 2013). Certain LAB species have partially lost the genetic information required for respiratory metabolism, resulting in versatile fermentative metabolism. This metabolism involves a homofermentative pathway in which lactic acid is the primary product or a heterofermentative pathway in which lactic acid, CO2, acetic acid, and/or ethanol are produced (Figure 1). What were the natural habitats that drove the evolution of such peculiar metabolic traits in these microorganisms? The relevant type of environment appeared on Earth at the end of the Cretaceous age (between 145 and 65 mya), when an excess of fruits and therefore increased amounts of fermentable substrates became available to many microbial communities. Additionally, if we consider the availability of a high amount of lactose linked to the appearance of mammals on Earth, the adaptation of certain LAB species to the milk environment should have started in the late Paleocene (between 65 and 23 mya). However, the adaptation of LAB to food fermentations took place between 12000 and 6000 years ago (Dunne et al., 2012; Hayden et al., 2013) a period too short, in evolutionary terms, to justify that human activity directed evolution at the level of energetic metabolism, but sufficiently long to model regulatory mechanisms, and to drive genetic rearrangements through mobile genetic elements and horizontal gene transfer events.
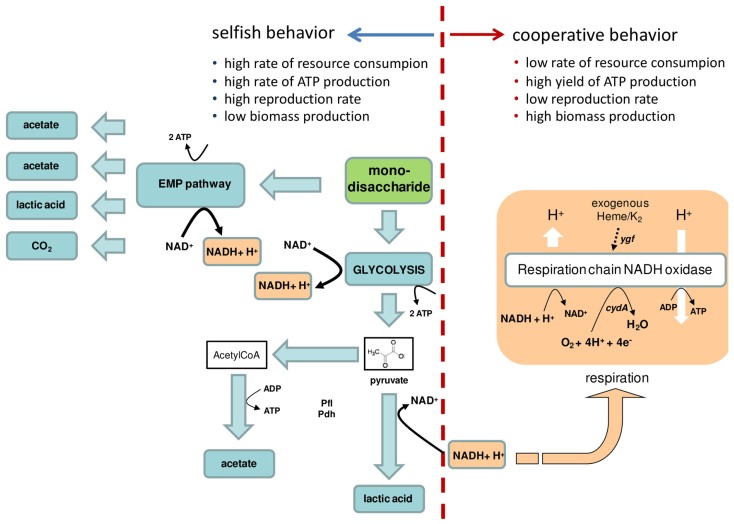
FIGURE 1.
Simplified representation of “selfish” and “cooperative” energetic behaviors in lactic acid bacteria. The key characteristics of “selfish” and “cooperative” energetic behaviors are indicated. The dotted red line separates the metabolic reactions that confer a “selfish” or a “cooperative” behavior. In lactic acid bacteria (LAB), mono- or disaccharides are catabolized: (i) to pyruvate via glycolysis, with the production of ATP and NADH, or ii) trough the exose monophosphate pathway (EMP), the heterofermentative pathway. The yellow arrow shows the regeneration of NAD+ that occurs during heme-dependent respiration. In Lactococcus lactis and in other LAB, the excess pyruvate can be metabolized by pyruvate formate lyase (Pfl), pyruvate dehydrogenase (Pdh) giving a mixed-acid fermentation with production of acetate. Ygf is the operon involved in heme homeostasis, and cydA is the gene for the cytochrome bd oxidase subunit I as described for L. lactis (Arioli et al., 2013). Arrows show the metabolic fluxes.
In those environmental contexts, LAB and yeasts could have evolved efficient fermentation metabolisms characterized by a high rate of sugar consumption associated with the accumulation of catabolic products, such as ethanol or lactic acid. It therefore appears that S. cerevisiae and LAB shared certain common constraints that have driven the evolution of their energetic metabolisms. What were the evolutionary driving forces that allowed this lifestyle to emerge? The answer depends on two very important parameters: gain and cost. The added value of the rapid utilization of available carbon sources and of the rapid accumulation of fermentation products is that these metabolic traits represent a strategy to efficiently outcompete other microbes in the same environmental context, thus becoming “winning” traits (MacLean and Gudelj, 2006; Piškur et al., 2006). Apparently, both S. cerevisiae and certain LAB species have evolved their energetic metabolism to reach maximum fitness in defined environmental niches characterized by high carbohydrate concentration.
“SELFISH” AND “COOPERATIVE” BEHAVIOR: THE POSSIBILITY OF CHOICE
In contrast to most LAB species, S. cerevisiae can use respiro-fermentative metabolism instead of only energetically more favorable respiratory metabolism (2 versus 32 mol of ATP per mol of glucose, respectively, for fermentation and respiration). This phenomenon is called the “Crabtree effect” (Crabtree, 1929). Extensive studies have been performed to study such complexity at the physiological, molecular, and structural levels (Fendt et al., 2010). It has been demonstrated that the Crabtree effect is mainly the result of a wide repression exerted by glucose on respiratory metabolism.
Respiration is characterized by a high yield of ATP production (the number of units of ATP per unit of resource consumed) associated with a low reproduction rate but high biomass production. This process is therefore considered as a “cooperative” behavior because the external resources are slowly consumed and are shared with other consumers that colonize the same environment. In contrast, fermentation is a “selfish” behavior characterized by a high rate of glucose consumption and low biomass production due to the use of carbon to produce ethanol (Pfeiffer et al., 2001; MacLean and Gudelj, 2006). In this context, it is interesting to note that in certain LAB species, such as Lactococcus lactis and Lactobacillus plantarum, heme (and menaquinone) can stimulate aerobic respiration (Figure 1), thereby increasing not only growth efficiency but also robustness (Brooijmans et al., 2009). In all of these examples, the shift in metabolic efficiency originates from a tradeoff between investments in enzyme synthesis and metabolic yields from alternative catabolic pathways (Molenaar et al., 2009).
From an ecological point of view, cellular decision-making is also based on the economics of nutrition and competition, and any organism has to find the most efficient way of colonizing its niche (Zakrzewska et al., 2011). In this context, growth rate and robustness are related properties that show a remarkable inverse correlation. Therefore, the net results of the evolutionary selection for microorganisms in constantly changing nutrient-abundant and stress-free environments automatically lead to selection for individuals with reduced robustness and rapid growth (Zakrzewska et al., 2011). More generally, the rate-yield tradeoff of metabolic pathways was experimentally shown to be an evolutionary constraint that reflects the evolutionary history of populations (MacLean, 2008). This tradeoff generates a fundamental social conflict (“the tragedy of the commons”) in microbial populations because the average fitness in a population is highest if all individuals exploit resources efficiently, but an individual’s reproductive rate is maximized by consuming common resources at the highest possible rate (MacLean and Gudelj, 2006; MacLean, 2008). This social and evolutionary dilemma arises only if there is competition for shared resources. Therefore, pathways with a high rate but low yield of ATP production should primarily be observed in association with the exploitation of external resources (Pfeiffer et al., 2001).
It can be assumed that microorganisms use energetically efficient pathways when growing at low concentrations of sugars and energy-wasting pathways at high sugar concentrations, as observed in S. cerevisiae (see Crabtree effect), Bacillus subtilis (Sonenshein, 2007), and LAB species, which have the ability to shift between low-efficiency homolactic fermentation and high-efficiency mixed acid fermentation (Teusink et al., 2006). In specific, if growing in a medium that contains an excess of glucose B. subtilis metabolizes a large proportion of the glucose only as far as pyruvate and acetyl CoA, and subsequently converts these compounds by-products including lactate, acetate, and acetoin (Sonenshein, 2007). For L. lactis, product formation at high dilution rates during continuous cultivations is similar to product formation during batch growth at high glucose concentrations, resulting in lactic acid as the sole metabolic product. On the other hand, growth at low dilution rates in continuous conditions or at low concentrations of glucose in batch conditions results in a mixed-acid fermentation (Thomas et al., 1979). On the other hand, L. plantarum that is strongly adapted to nutrient-rich environments, even at energy limitations uses a catabolic route that is less efficient in ATP production and uses futile cycles and parallel pathways to uncouple ATP production and anabolic ATP consumption especially under energy excess (Teusink et al., 2006).
The possibility of regulation of the ATP yield and the stoichiometry of oxidative phosphorylation may gain a selective ecological advantage during competition for energy resources. Among the yeasts belonging to the genus Saccharomyces, those yeasts that underwent whole-genome duplication exhibit a strong fermentative lifestyle due to the Crabtree effect and the ability to grow under strictly anaerobic conditions. However, only in S. cerevisiae have these traits been combined and developed to perfection (Merico et al., 2007). The “make-accumulate-consume ethanol” strategy not only acts to eliminate competitors but also becomes a newly available carbon source for the survivors when glucose is exhausted.
The preferred fermentative mode of energy production are also remarkably evident at the transcriptional regulation level. In this context, it was shown that single transcription-factor deletions can essentially abolish the tricarboxylic acid cycle flux and thus respiration, whereas only the orchestrated action of several glucose signaling pathways shift metabolism toward respiration, thereby inhibiting fermentation (Fendt et al., 2010).
DID HUMAN SELECTION BASED ON FOOD PROCESSING DRIVE “SELFISH” STRATEGY IN LAB AND YEAST METABOLIC BEHAVIORS?
A fundamental goal in evolutionary biology is to achieve an in-depth understanding of how human activities favor selection for specific metabolic traits in microbial species, guiding these species’ “domestication” or evolution toward new taxa. The question arising here is which type of energetic strategy, whether “selfish” or “cooperative” (Figure 1) has also been selected during the domestication of microbial species in environments showing predictable fluctuations, such as food niches. If we consider the energetic strategies adopted by a domesticated strain of S. cerevisiae and by the LAB species Streptococcus thermophilus and Lactobacillus delbrueckii subsp. bulgaricus, we can speculate that a “selfish” strategy has been evolutionarily selected in order to adapt to grape juice and milk environments, respectively. Thus, the role of human activities related to food forced the domestication of wild strains toward the selection of an energetic tradeoff between rate and yield on behalf of the rate of ATP production per unit of time. In contrast, the absence of close adaptation to a defined environmental niche should lead to an oscillation between selfish and cooperative behaviors that depends on environmental stimuli. In dairy LAB, the selective forces shaping microbial genotypes and phenotypes in L. lactis during evolutionary adaptation from the plant to the milk have been corroborated at the kinetic level based on the differences observed between plant and dairy L. lactis lactate dehydrogenase regulation. Dairy strains use fructose-1,6-biphosphate and Pi, whereas the lactate dehydrogenase of plant isolates is regulated by the NADH/NAD+ ratio (Levering et al., 2012). As a plant environmental niche is relatively devoid of phosphate, these observations underline a strict link between the organism’s natural environment and its metabolic regulation. More recently (Bachmann et al., 2012), in a plant-derived L. lactis strain, adaptation to a new environment, milk, was achieved in only the 1000th generation and resulted in an increased growth rate, yield, and fitness, together with a loss of mobile elements. More interestingly, it was observed that the transcriptome of milk-adapted strains converged toward the transcriptome of a dairy environment-derived reference strain (Bachmann et al., 2012), indicating an extremely rapid evolution of the first layer of regulatory mechanisms in response to the appearance of new environmental constraints. In particular, the new milk-adapted strains were characterized by the downregulation of transport systems dedicated to plant sugars and branched-chain amino acid biosynthesis, thus reflecting the ongoing process of genomic decay during adaptation to the milk environment. This genomic decay or regressive evolution linked to milk adaptation was already described as a key feature of the dairy species S. thermophilus and L. delbrueckii subsp. bulgaricus, the two species involved in yogurt production (Bolotin et al., 2004; van de Guchte et al., 2006). Genomic analysis of the latter species indicated its adaptation from a plant-associated habitat to the stable protein- and lactose-rich milk environment through the loss of superfluous functions and protocooperation with S. thermophilus (van de Guchte et al., 2006). Both the S. thermophilus and L. delbrueckii species are characterized by homofermentative lactic acid metabolism and do not have any heme-dependent respiratory metabolism, as observed for other LAB that are “still” able to colonize different ecological niches, as demonstrated by their metabolic and genetic flexibility (Brooijmans et al., 2009). It can therefore be hypothesized that adaptation to a dairy environment was generated by human activities that selected for microorganisms with “selfish metabolic behavior. Moreover, the phenotypic characteristics of S. salivarius, the closed phylogenetic neighbor of S. thermophilus, are further evidences of adaptation to milk environment by the latter species. S. salivarius is inhabitant of the oral cavity of mammals and despite the high phylogenetic relationship with S. thermophilus, these two species show an extremely different carbohydrate utilization pattern (Schleifer et al., 1991; Mora et al., 2002) and are characterized by differences in the transcriptional regulation of urease operon (Chen et al., 1998; Mora et al., 2005) that reflects the fluctuations of nutrients availability in milk and in the oral cavity of mammals.
The description and comparative genomic analysis of the LAB S. thermophilus (Bolotin et al., 2004) represented the first report providing insights into adaptive evolutionary mechanisms that led to the assembly of a “generally recognized as safe” (GRAS) species from closely related deadly human pathogens, such as S. pneumoniae, S. pyogenes, and S. agalactiae. Moreover, Bolotin et al. (2004) argued that the species S. thermophilus evolved from closely phylogenetically related pathogenic streptococci through loss-of-function events counterbalanced by the acquisition of relevant traits, such as lactose and urea utilization (Bolotin et al., 2004; Arioli et al., 2010), that have allowed the assembly of new genomic organization suitable for the colonization of the dairy niche.
In a more general view we can assume that the adaptation process to food environments it is far to be known because several food fermenting bacteria are clearly not adapted to food fermentation because distinct food-derived lineages do not exist. This is particularly true for L. plantarum and Lactobacillus reuteri that are involved in cereal fermentations since several millennia (Hayden et al., 2013). For L. plantarum the absence of the adaptation to a unique food-niche it is witnessed by an extremely high genome plasticity where the main features appears to be genomic life-style islands consisting of numerous functional gene cassettes, in particular for carbohydrates utilization, which can be acquired, shuffled, substituted, or deleted in response to niche requirements (Siezen and van Hylckama Vlieg, 2011). L. reuteri is both a gut symbiont and a stable member of sourdough microbiota (Walter et al., 2011; Su et al., 2012). Interestingly, it was recently demonstrated that sourdough isolates of L. reuteri emerge from the same phylogenetic line as rodent strains (Su et al., 2012) and it was excluded that the stable microbiota of sourdough could be subjected by recurrent fecal contamination of raw material. Additionally, it was reported that rodent L. reuteri isolates are capable of long-term persistence in food fermentation and that sourdough isolates were able to colonize Lactobacillus-free mouse model (Walter et al., 2008). It therefore appeared that L. reuteri can stably colonize two environmental niches, the upper intestine of mammals that consume cereal-based foods and sourdough, due to the similarities between the two habitats, i.e., availability of sucrose and maltose as the major carbon source (Su et al., 2012).
Has the domestication of S. cerevisiae caused similar metabolic selection? S. cerevisiae has been used for millennia in baking, brewing, and winemaking, and more recently, for ethanol production as a biofuel. All of these biotechnological applications are based on the very efficient fermentative metabolism of the yeast. Piškur et al. (2006) speculated that efficient metabolic regulation was the most unique “invention” of Saccharomyces yeast used in the industrial fermentations of breweries and wineries. The ability of S. cerevisiae to use respiro-fermentative metabolism has been an important characteristic in the evolutionary and ecological contexts and in many of the yeast’s industrial applications.
Progress in genomic sequencing has elucidated the phylogenetic relationships among yeasts and has revealed the importance of genomic events, such as whole-genome duplication (Wolfe and Shields, 1997), in the evolutionary history of the yeast, which occurred in the S. cerevisiae lineage approximately 100 mya, and the rewiring of the expression of hundreds of genes (Ihmels et al., 2005). Whole-genome duplication in yeast determined an increased glycolytic flux and therefore an increased fermentative capacity, which is a great advantage when glucose resources are both large and dense (Conant and Wolfe, 2007). Nevertheless, population genetic studies showed that S. cerevisiae strains associated with human activities are differentiated from the natural wild populations (Fay and Benavides, 2005; Liti et al., 2009). The recent analysis of wild S. cerevisiae population collected from primeval and rainforests across China (Wang et al., 2012) is now opening a new “path” to expand the knowledge about yeast’s history. The study of genetic, as well as of metabolic traits of these new and wild yeast lineages, could in fact keep many surprises and furthermore help to elucidate yeast genetic and metabolic evolution. It is noteworthy that so far most of what we know about S. cerevisiae is mainly based on studies carried out on laboratory strains.
Likewise on yeast, also in LAB genomic events resulted in new metabolic traits. A high relative activity of several glycolytic enzymes per cell, an outcome of diploidy, was measured in dairy strains of L. lactis (Michelsen et al., 2010). Although bacteria are normally haploid, maintaining one copy of their genome, several dairy strains of L. lactis fulfill the criterion for diploidy. The diploid dairy strains show higher sensitivity to UV light, increased cell size and higher glycolytic activity compared with haploid non-dairy strains. The authors concluded their study by proposing that diploidy has been selected during the 5,000–10,000 years that lactococci have been used in cheese production (Michelsen et al., 2010). Whole-genome duplication in yeast and diploidy in L. lactis were most likely events in a long process of adaptation that led to modern baker’s yeast and dairy lactococci, under the selective pressure of human food processing.
CONCLUSION
Food matrixes represent natural environments colonized by microorganisms, and fermentation is one of the oldest forms of food preparation and preservation. Here, we discussed how these environments have shaped the primary energetic metabolism of food-associated microorganisms, using the yeast S. cerevisiae and species of LAB known to be strictly adapted or “domesticated” to a defined food niche as examples. These microorganisms are characterized by a high phylogenetic distance but share similar cellular bioenergetics due to their evolution in food environments rich in easily assimilable carbon and nitrogen sources.
The adaptation to these environmental contexts caused genomic and metabolic streamlining in certain cases, resulting in a “reductive evolution” in which simple organisms derive from more complex ancestors (Bolotin et al., 2004; van de Guchte et al., 2006). Moreover, in such microorganisms, adaptation to food matrixes selected for “selfish” energetic behavior and for bioenergetic regulatory mechanisms that remarkably mirror the predictable environmental changes occurring during growth (i.e., during the fermentation process; Mitchell et al., 2009). The ecological forces and molecular mechanisms that govern this ability are not clear, but it is evident that the regulatory networks that link environmental stimuli to microbial responses are complex and can evolve rapidly. In conclusion, the bioenergetics of food-associated bacteria should be analyzed, always considering the environmental context, nutrient richness and availability, and the “predictable” succession of environmental stimuli that have driven the organisms’ domesticated speciation and evolution.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
- Arioli S., Zambelli D., Guglielmetti S., De Noni I., Pedersen M. B., Pedersen P. D., et al. (2013). Increasing the heme-dependent respiratory efficiency of Lactococcus lactis by inhibition of lactate dehydrogenase. Appl. Environ. Microbiol. 79 376–380 10.1128/AEM.02734-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli S., Ragg E., Scaglioni L., Fessas D., Signorelli M., Karp M., et al. (2010). Alkalizing reactions streamline cellular metabolism in acidogenic microorganisms. PLoS ONE 5:e15520. 10.1371/journal.pone.0015520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann H., Starrenburg M. J. C., Molenaar D., Kleerebezem M, van Hylckama Vlieg J. E. T. (2012). Microbial domestication signatures of Lactococcus lactis can be reproduced by experimental evolution. Genome Res. 22 115–124 10.1101/gr.121285.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A., Quinquis B., Renault P., Sorokin A., Ehrlich S. D., Kulakauskas S., et al. (2004). Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22 1554–1558 10.1038/nbt1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooijmans R., Smit B., Santos F., van Riel J., de Vos W. M. M, Hugenholtz J. (2009). Heme and menaquinone induced electron transport in lactic acid bacteria. Microb. Cell Fact. 8 28 10.1186/1475-2859-8-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. Y., Weaver C. A., Mendelsohn D. R., Burne R. A. (1998). Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J. Bacteriol. 180 5769–5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant G. C., Wolfe K. H. (2007). Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol. Syst. Biol. 3 129 10.1038/msb4100170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree H. G. (1929). Observations on the carbohydrate metabolism of tumors. Biochem. J. 23 536–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne J., Evershed R. P., Salque M., Cramp L., Bruni S., Ryan K., et al. (2012). First dairying in green Saharan Africa in the fifth millennium BC. Nature 486 390–394 10.1038/nature11186 [DOI] [PubMed] [Google Scholar]
- Fay J. C., Benavides J. A. (2005). Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 1:e5. 10.1371/journal.pgen.0010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt S.-M., Oliveira A. P., Christen S., Picotti P., Dechant R. C., Sauer U. (2010). Unravelling condition-dependent networks of transcription factors that control metabolic pathway activity in yeast. Mol. Syst. Biol. 6 432 10.1038/msb.2010.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J., Bergmann S., Gerami-Nejad M., Yanai I., McClellan M., Berman J., et al. (2005). Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309 938–940 10.1126/science.1113833 [DOI] [PubMed] [Google Scholar]
- Hayden B., Canuel N., Shance J. (2013). What was brewing in the Natufian? An archeological assessment of brewing technology in the epipaleolithic. J. Archaeol. Method Ther. 20 102–150 10.1007/s10816-011-9127-y [DOI] [Google Scholar]
- Levering J., Musters M. W. J. M., Bekker M., Bellomo D., Fiedler T., de Vos W. M., et al. (2012). Role of phosphate in the central metabolism of two lactic acid bacteria – a comparative system biology approach. FEBS J. 279 1274–1290 10.1111/j.1742-4658.2012.08523.x [DOI] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., James S. A., et al. (2009). Population genomics of domestic and wild yeasts. Nature 458 337–341 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean R. C. (2008). The tragedy of the commons in microbial populations: insights from theoretical, comparative and experimental studies. Heredity 100 471–477 10.1038/sj.hdy.6801073 [DOI] [PubMed] [Google Scholar]
- MacLean R. C., Gudelj I. (2006). Resource competition and social conflict in experimental populations of yeast. Nature 441 498–501 10.1038/nature04624 [DOI] [PubMed] [Google Scholar]
- Merico A., Sulo P., Piskur J., Compagno C. (2007). Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. FEBS J. 274 976–989 10.1111/j.1742-4658.2007.05645.x [DOI] [PubMed] [Google Scholar]
- Michelsen O., Hansen F. G., Albrechtsen B., Jensen P. R. (2010). The MG1363 and IL1403 laboratory strains of Lactococcus lactis and several dairy strains are diploid. J. Bacteriol. 192 1058–1065 10.1128/JB.00900-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, Romano G. H., Groisman B., Yona A., Dekel E., Kupiec M, et al. (2009). Adaptive prediction of environmental changes by microorganisms. Nature 460 220–224 10.1038/nature08112 [DOI] [PubMed] [Google Scholar]
- Molenaar D., van Berlo R, de Ridder D., Teusink B. (2009). Shifts in growth strategies reflect tradeoffs in cellular economics. Mol. Syst. Biol. 5 323 10.1038/msb.2009.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora D., Fortina M. G., Parini C., Ricci G., Gatti M., Giraffa G., et al. (2002). Genetic diversity and technological properties of Streptococcus thermophilus strains isolated from dairy products. J. Appl. Microbiol. 93 278–287 10.1046/j.1365-2672.2002.01696.x [DOI] [PubMed] [Google Scholar]
- Mora D., Monnet C., Parini C., Guglielmetti S., Mariani A., Pintus P., et al. (2005). Urease biogenesis in Streptococcus thermophilus. Res. Microbiol. 156 897–903 10.1016/j.resmic.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Pfeiffer T., Schuster S., Bonhoeffer S. (2001). Cooperation and competition in the evolution of ATP-producing pathways. Science 292 504–507 10.1126/science.1058079 [DOI] [PubMed] [Google Scholar]
- Piškur J., Rozpedowska E., Polakova S., Merico A., Compagno C. (2006). How did Saccharomyces evolve to become a good brewer. Trends Genet. 22 183–186 10.1016/j.tig.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Sachs J. L., Hollowell C. (2012). The origins of cooperative bacterial communities. MBio 3 e00099–e000112 10.1128/mBio.00099-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein A. L. (2007). Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5 917–927 10.1038/nrmicro1772 [DOI] [PubMed] [Google Scholar]
- Siezen R. J, van Hylckama Vlieg J. E. T. (2011). Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb. Cell Fact. 10(Suppl. 1) S3 10.1186/1475-2859-10-S1-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M. S.-W., Oh P. L., Walter J, Gänzle M. G. (2012). Intestinal origin of sourdough Lactobacillus reuteri isolates as revealed by phylogenetic, genetic, and physiological analysis. Appl. Environ. Microbiol. 78 6777–6780 10.1128/AEM.01678-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer K. H., Ehrmann M., Krusch U., Neve H. (1991). Revival of the species Streptococcus thermophilus (ex. Orla-Jenson, 1919) nom. Rev. Syst. Appl. Microbiol. 14 386–388 10.1016/S0723-2020(11)80314-0 [DOI] [Google Scholar]
- Teusink B., Wiersma A., Molenaar D., Francke C., de Vos W. M., Siezen R. J., et al. (2006). Analysis of growth of Lactobacillus plantarum WCFS1 on a complex medium using a genome-scale metabolic model. J. Biol. Chem. 281 40041–40048 10.1074/jbc.M606263200 [DOI] [PubMed] [Google Scholar]
- Thomas T. D., Ellwood D. C., Longyear V. M. (1979). Change from homo- to heterolactic fermentation by Streptococcus lactis resulting from glucose limitation in anaerobic chemostat cultures. J. Bacteriol. 138 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Guchte M., Pernaud S., Grimaldi C., Barbe V., Bryson K., Nicolas P., et al. (2006). The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc. Natl. Acad. Sci. U.S.A. 24 9274–9279 10.1073/pnas.0603024103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J., Britton R. A., Ross S. (2011). Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc. Natl. Acad. Sci. U.S.A. 15 4645–4652 10.1073/pnas.1000099107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J., Schwab C., Loach D. M., Gänzle M. G., Tannock G. W. (2008). Glucosyltransferase A (GtfA) and inulosucrase (Inu) of Lactobacillus reuteri TMW1.106 contribute to cell aggregation, in vitro biofilm formation, and colonization of the mouse gastrointestinal tract. Microbiology 154 72–80 10.1099/mic.0.2007/010637-0 [DOI] [PubMed] [Google Scholar]
- Wang Q.-M., Liu W.-Q., Liti G., Wang S.-A., Bay S.-W. (2012). Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol. Ecol. 21 5404–5417 10.1111/j.1365-294X.2012.05732.x [DOI] [PubMed] [Google Scholar]
- Wolfe K. H., Shields D. C. (1997). Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387 708–713 10.1038/42711 [DOI] [PubMed] [Google Scholar]
- Zakrzewska A., van Eikenhorst G., Burggraaff J. E. C., Vis D. J., Hoefsloot H., Delneri D., et al. (2011). Genome-wide analysis of yeast stress survival and tolerance acquisition to analyze the central trade-off between growth rate and cellular robustness. Mol. Biol. Cell 15 4435–4446 10.1091/mbc.E10-08-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]