Abstract
Metastatic melanoma is characterized by a poor response to chemotherapy. Furthermore, there is a lack of established predictive and prognostic markers. In this single institution study, we correlated mutation status and expression levels of BRAF and NRAS to dacarbazine (DTIC) treatment response as well as progression-free and overall survival in a cohort of 85 patients diagnosed with advanced melanoma. Neither BRAF nor NRAS mutation status correlated to treatment response. However, patients with tumors harboring NRAS mutations had a shorter overall survival (p < 0.001) compared to patients with tumors wild-type for NRAS. Patients having a clinical benefit (objective response or stable disease at 3 months) on DTIC therapy had lower BRAF and NRAS expression levels compared to patients progressing on therapy (p = 0.037 and 0.003, respectively). For BRAF expression, this association was stronger among patients with tumors wild-type for BRAF (p = 0.005). Further, low BRAF as well as NRAS expression levels were associated with a longer progression-free survival in the total population (p = 0.004 and <0.001, respectively). Contrasting low NRAS expression levels, which were associated with improved overall survival in the total population (p = 0.01), low BRAF levels were associated with improved overall survival only among patients with tumors wild-type for BRAF (p = 0.013). These findings indicate that BRAF and NRAS expression levels may influence responses to DTIC as well as prognosis in patients with advanced melanoma.
Electronic supplementary material
The online version of this article (doi:10.1007/s10585-013-9587-4) contains supplementary material, which is available to authorized users.
Keywords: Melanoma, BRAF, NRAS, Chemoresistance, Dacarbazine
Introduction
The incidence of cutaneous malignant melanoma is increasing among light-skin Caucasians [1]. Although most patients are cured by surgical excision of the tumor, approximately 20 % will relapse [2]. Systemic treatment for metastatic malignant melanomas remains palliative. Dacarbazine (DTIC) is considered standard chemotherapy treatment, even though objective response rates as low as 10 % are recorded [3].
Activating mutations in the BRAF (V600E) and NRAS (Q61K) genes are found at a frequency of 40–60 and 15–30 % in metastatic melanomas, respectively [4–10]. In vitro studies have shown that the V600E mutation, which is located in the protein’s activation loop, causes a 500-fold increase in the enzymatic activity of BRAF, enhancing activation of its downstream target, ERK [11]. Thus, for tumors harboring NRAS or BRAF mutations, activation of this pathway is thought to play a key role in driving tumor growth. This is further underlined by recent studies showing that targeted inhibition of mutated BRAF may cause tumor regression in metastatic melanomas harboring BRAF V600E mutations [12, 13], as well as findings indicating that treatment of BRAF-mutated melanomas may benefit from inhibition of the downstream effector MEK [14].
While the effects of the V600E mutation of BRAF have been extensively studied in experimental systems, several aspects of BRAF function remain poorly understood. Interestingly, copy-number gains of the BRAF gene have been proposed as an alternative mechanism of activation in both melanoma and glioma [15, 16], as well as being a cause of resistance towards BRAF inhibitor treatment of advanced melanoma [17]. Further, BRAF mRNA has been found subject to alternative splicing, with different transcript variants identified in colorectal cancer as well as in melanoma [18, 19]. Interestingly, expression of some splice variants has been related to resistance toward the BRAF inhibitor vemurafenib [19].
Although overexpression of wild-type BRAF has been reported to be an underlying mechanism of pathway activation in experimental systems [15], to the best of our knowledge the level of BRAF expression in tumors wild-type for BRAF has not been investigated as a potential predictive and prognostic factor in melanoma patients.
The aim of this study was to evaluate the predictive and prognostic impact of genetic disturbances and expression levels of BRAF together with NRAS mutations in patients treated with DTIC monotherapy for advanced melanomas.
Materials and methods
Patients and treatment
A total of 85 patients were enrolled in this protocol between January 2000 and November 2007. All patients were referred to the Department of Oncology at Haukeland University Hospital for locally advanced or metastatic melanoma. The protocol was approved by the Regional Ethical Committee, and was conducted in adherence to the Declaration of Helsinki. Each patient signed a written consent form. Details regarding the patient population studied are described in Table 1 and Supplementary Information, Table S1. Chemotherapy consisted of DTIC monotherapy, administered at a dose of 800–1,000 mg/m2 on a 3-weekly basis. Out of the total number of 85 patients, 75 commenced on chemotherapy and were available for response evaluation (the reason for non-compliance from the additional 10 patients is shown in Table S1). Evaluation of response was done at 6-weekly intervals. As the protocol was implemented in year 2000, the UICC response criteria were used for the whole series.
Table 1.
Patient characteristics according to BRAF and NRAS mutation status
| Total patient population | BRAF | NRAS | |||
|---|---|---|---|---|---|
| WT | Mutant | WT | Mutant | ||
| Patient characteristics | |||||
| Median age (range) | 62 (25–86) | 62 (38–84) | 63 (25–86) | 63 (25–86) | 61 (43–80) |
| Male (%) | 56.5 | 60 | 50 | 57.4 | 52.9 |
| Previous primary melanoma | |||||
| Cutaneous | 57a | 34 | 22 | 44 | 12 |
| Breslow thickness | |||||
| <1 | 6 | 3 | 3 | 6 | 0 |
| 1–2 | 15 | 6 | 9 | 12 | 3 |
| 2–4 | 11 | 10 | 1 | 8 | 3 |
| >4 | 16 | 12 | 4 | 11 | 5 |
| Not available | 8 | 3 | 5 | 7 | 1 |
| Acral | 4 | 4 | 0 | 4 | 0 |
| Mucosal | 5 | 4 | 1 | 5 | 0 |
| Uveal | 5a | 5 | 0 | 4 | 1 |
| No primary | 15 | 8 | 7 | 11 | 4 |
| Serum LDH (% elevated) | 39.7 | 47.2 | 24.0 | 36.1 | 52.9 |
| Metastases at inclusionb | |||||
| Soft tissuec | 20 | 12 | 8 | 17 | 3 |
| Visceral/skeletal | 54 | 33 | 21 | 44 | 10 |
| Brain | 11 | 10 | 1 | 7 | 4 |
| Stage at inclusion | |||||
| III | 3 | 1 | 2 | 3 | 0 |
| IV | 82 | 54 | 28 | 65 | 17 |
| Clinical benefit (%) | 20 | 20.8 | 18.5 | 20.3 | 18.8 |
aOne patient presented with both uveal and cutaneous primary
bRefers to the metastatic site associated with the worst prognosis
cDefined here as subcutaneous, cutaneous or lymph node metastases
Tumor tissue collection and handling
Prior to chemotherapy, tumor samples were obtained through incisional biopsies or ultrasound-guided tru-cut needle samples from deep lesions (liver deposits). Tissue samples were snap-frozen in the operating theatre immediately upon removal and stored in liquid nitrogen until analysis. In addition, some of the excised material was formalin-fixed and paraffin-embedded for histological examination.
Isolation of nucleic acids and cDNA synthesis
Total RNA was extracted using Trizol® reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Some of the samples contained high levels of melanin after the RNA extraction; here, further steps were performed to separate RNA from melanin [20]. Following purification, RNA was dissolved in DEPC-treated dH2O and stored at −80 °C.
DNA was extracted from the biopsies using QIAamp DNA Mini Kit® (Qiagen, Chatsworth, CA, USA). Some of the DNA samples also had to undergo further steps to separate DNA from melanin [21, 22]. DNA was dissolved in dH2O and kept at −20 °C.
First strand complementary DNA (cDNA) was synthesized using the Transcriptor reverse transcriptase (Roche Diagnostics, Basel, Switzerland) following the manufacturer’s instructions, using oligo-dT as primer and 250 ng total RNA as template.
BRAF and NRAS mutation screening
Regions covering the open reading frame of NRAS and the 3′ half of the BRAF open reading frame was amplified from cDNA by PCR. In about half of the cases (n = 47), findings in BRAF were subjected to confirmatory analyses performed on genomic DNA. The PCR amplifications were performed using the DyNazyme EXT polymerase system (FINNZYMES, Espoo, Finland) according to the manufacturer’s instructions. Primers and annealing temperatures used are listed in Supplementary Information, Table S2. DNA sequencing was performed using BigDye v.1.1 and a capillary DNA sequencer (ABI 3700).
Quantification of BRAF and NRAS mRNA transcripts
The expression levels of BRAF and NRAS and the two splice variants BRAFdel14–15 and BRAFdel12–15 were assessed by qPCR using the LightCycler 480 system (Roche Diagnostics, Basel, Switzerland). Specific amplification of each splice variant was ensured by using unique forward primers encompassing the specific splice site in combination with hybridization probes and reverse primers that were targeted to shared regions (primers and probes are listed in Supplementary Information, Table S2). To determine the specificity of the reactions, all reaction conditions were tested against a purified full-length template as well as other non-specific splice variants. The relative non-specific amplification of BRAF full-length/splice variants was determined to be lower than 5 × 10−4 as compared to specific amplification in all assays. Expression levels were normalized to ribosomal protein, large, P2 (RPLP2) expression before relative comparison between samples. Three patients were excluded from correlational analyses of BRAF full-length and NRAS, and an additional seven from splice variant mRNA analyses, due to total RNA levels falling below the sensitivity threshold of each assay.
Statistical analysis
Due to the low rate of objective response (n = 4), analysis of potential predictive factors was carried out by categorizing patients as having either a clinical benefit (Complete response, CR; partial response, PR; and stable disease, SD; n = 15), or progressive disease (PD; n = 60), as previously described for this patient cohort [23]. Correlating BRAF and NRAS expression levels to therapy response, we compared relative expression levels among responders versus non-responders using the Mann–Whitney rank test. Relative expression levels among BRAF mutation and amplification carriers were compared using the Mann–Whitney and Kruskal–Wallis rank tests. For correlation analyses, Spearman’s rank correlation coefficients were calculated. Comparisons of categorical data were performed using Fischer’s exact test. Assessing the prognostic role of BRAF and NRAS mRNA expression levels, we used the median value as cut-off limit. Log rank tests and Cox regression analyses were used for survival analyses. All p values are given as two-sided, and the p values from Fischer exact tests present cumulative values. All analyses were performed using the SPSS 19.0.0.2 statistical software package (Chicago, Illinois, USA).
Results
BRAF and NRAS mutation status
The BRAF single nucleotide substitution c.1799T>A (p.V600E) was detected in 29, and c.1798_1799GT>AA (p.V600K) in 1 out of 85 patients (35.3 % harboring BRAF mutations in total). No other mutations were observed in the region covered by sequencing (codon 463–715).
Sequencing of the protein-coding region of the NRAS gene revealed mutations in 19 out of 85 (22.4 %) patients, all located in the codon 61 hot spot. Notably, BRAF and NRAS mutations were found to be mutually exclusive (p < 0.001, Supplementary Information, Table S1), with no tumor harboring mutations in both genes. A summary of patient characteristics in regards to BRAF and NRAS mutation status is presented in Table 1.
Interestingly, the incidence of BRAF, but not NRAS mutations varied according to the anatomical site from which the sample was derived. Thus, BRAF mutations were found in 15/42 (35.7 %) of subcutaneous metastases, 12/20 (60.0 %) of lymph node metastases, but only in 3/22 (13.6 %) of the visceral metastases examined (p = 0.007; details available in Supplementary Information, Table S1). Studies have shown that uveal and mucosal primary tumors only rarely carry mutations in BRAF and NRAS [24]. Excluding metastatic deposits with primary tumors at these sites, which represented a substantial fraction of the visceral metastases (n = 9), from the analysis, moderated the difference observed (leaving 3/13 visceral deposits from cutaneous melanomas harboring BRAF mutations; 23.1 %), still, a significant difference in mutation distribution between metastases located to different organ systems remained (p = 0.040).
In a previous paper [25] using CGH array, we reported BRAF copy-number gains in 12 out of a subgroup of 53 samples of the tumors studied here. Interestingly, 9 out of 12 tumors with increased BRAF copy number were found to harbor BRAF V600 mutations in concert. In contrast, BRAF mutations were recorded in 16 out of 41 melanomas without BRAF gains, revealing a statistically significant association between BRAF copy-number gains and V600 mutation status (p = 0.047).
BRAF mRNA expression levels are elevated in metastatic melanoma and are higher in tumors harboring BRAF copy-number gains or V600 mutations
Expression levels of full-length BRAF mRNA was significantly higher in the malignant melanoma samples as compared to non-malignant nevi (p = 0.007, Fig. 1a).
Fig. 1.
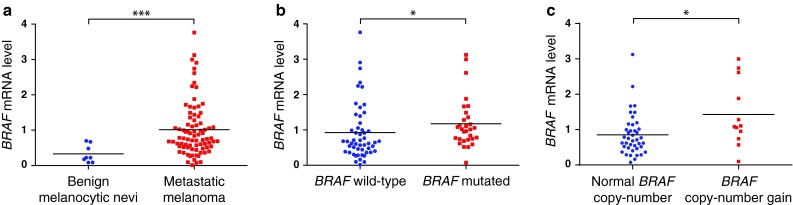
Expression characteristics of BRAF mRNA. Expression levels of BRAF mRNA in: a benign melanocytic nevi and in metastatic melanoma biopsies, b tumors with wild-type or mutated BRAF, and c tumors with or without BRAF copy-number gains. Asterisks denote significance levels: *p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.001
While analyzing for BRAF mutations, we observed two alternatively spliced variants, one lacking exons 14 and 15 (BRAFdel14–15); and one lacking exons 12–15 (BRAFdel12–15). Of these, the former has previously been reported in colorectal cancer [18], whereas the latter is novel.
We determined the mRNA expression levels of BRAF full-length and the two alternatively spliced variants of BRAF by qPCR. The expression levels of the alternatively spliced variants, BRAFdel14–15 and -12–15, were lower than the full-length transcript, and the expression levels of each correlated positively with the expression level of full-length BRAF (r s = 0.6609; p < 0.001 and r s = 0.4604; p < 0.001, respectively; Supplementary Information, Fig. S1).
Within the melanoma samples the BRAF expression levels varied between tumors harboring BRAF mutations and those wild-type for BRAF (p = 0.021; Fig. 1b). No difference in BRAF expression between NRAS mutated and NRAS wild-type was recorded (p > 0.1). Further, BRAF expression was higher in tumors with BRAF copy-number gains compared to those exhibiting a normal BRAF copy number (p = 0.028; Fig. 1c). Even though a low number of observations limits the strength of this analysis, comparing BRAF mRNA levels among tumors stratified for both BRAF mutation and copy-number status indicated that these two factors have an additive effect on BRAF expression (p = 0.005). Excluding tumors harboring BRAF copy-number gains from the analysis, BRAF expression levels remained higher among tumors harboring BRAF V600 mutations as compared to BRAF wild-type tumors (p = 0.014), suggesting an elevation of BRAF expression levels in tumors harboring the V600 mutations without additional gene copies in concert.
Low BRAF and NRAS expression levels but not BRAF or NRAS mutation status are associated with clinical benefit from DTIC chemotherapy
Only 4 out of a total of 75 tumors evaluated for response to DTIC treatment revealed an objective response. To determine potential correlations between BRAF (and, subsequently, NRAS) alterations and outcome, we therefore compared the BRAF status among patients having an objective response or stable disease recorded 3 months after commencing treatment (defined here as clinical benefit; n = 15) versus patients with PD (n = 60) within the same time interval [23]. No correlations between BRAF copy number (number compared = 45) or mutation status (number compared = 75) and response to chemotherapy were recorded (p > 0.1 for both).
Similar to what was observed for BRAF mutations, NRAS mutation status was not associated with clinical outcome after 3 months on therapy (number compared = 75; p > 0.5). Neither did we observe any difference in response to therapy between patients with tumors harboring either a BRAF or an NRAS mutation on the one side and those wild-type for both genes on the other (number compared = 75; p > 0.5).
In contrast, patients experiencing a clinical benefit to therapy after 3 months had lower BRAF mRNA expression levels compared to patients with PD (number compared = 72; p = 0.037; Fig. 2a). Limiting the analysis to patients with tumors expressing wild-type BRAF strengthened the association (number compared = 46; p = 0.005; Fig. 2b). The above comparisons were performed using rank tests, regarding BRAF expression level as a continuous variable. Assessing the robustness of the observed associations, we categorized BRAF expression as above or below the median, in the first quartile or in the first decile and compared the number of patients benefitting from therapy above or below these three cut-offs. For each analysis, we found low BRAF levels to be associated with clinical benefit to DTIC treatment in the total patient cohort as well as in the subgroup of patient with tumors wild-type for BRAF (p < 0.05 for all comparisons). In contrast, no association between BRAF expression level and benefit to therapy was observed among patients with tumors harboring BRAF mutations (number compared = 26; p > 0.5; Fig. 2c).
Fig. 2.

BRAF mRNA expression and response to DTIC treatment. BRAF mRNA expression among patients experiencing a clinical benefit or progressive disease (PD) 3 months after commencement of DTIC chemotherapy. a BRAF expression in the total patient group, b data as presented in a, but limited to patients with tumors harboring wild-type BRAF, c similar analysis performed for tumors harboring BRAF V600 mutations. Asterisks denote significance levels: *p ≤ 0.05, **p ≤ 0.005
Similar to what was recorded for full-length BRAF, low levels of BRAFdel14–15 were associated with a clinical benefit 3 months following commencement of DTIC therapy (number compared = 67; p = 0.045; Fig. S2a). In contrast, no correlation between BRAFdel12–15 levels and response to DTIC chemotherapy were observed (number compared = 67; Fig. S2b).
Based on the finding that BRAF expression levels were correlated to treatment response, we extended our analyses and determined NRAS mRNA levels in the same samples. Similar to what was seen for BRAF, NRAS expression levels were found to be significantly higher in malignant melanomas than in non-malignant nevi (p = 0.018; Fig. 3a). Moreover, there was a strong correlation between tumor NRAS and BRAF mRNA expression levels (r s = 0.627; p < 0.001; Fig. 3b). While 33 tumors revealed BRAF as well as NRAS levels both above median values, and 33 tumors had both values below median, only 16 tumors had one gene expressed above and the other expressed below median level. Further, we found that, similar to BRAF expression, low levels of NRAS expression were associated with a clinical benefit following DTIC treatment (number compared = 72; p = 0.003; Fig. 3c). This correlation was, however, not strengthened by restricting the analysis to patients with tumors wild-type for NRAS (number compared = 55; p > 0.05).
Fig. 3.
NRAS mRNA expression. a Expression levels of NRAS mRNA in benign melanocytic nevi (n = 8) and metastatic melanoma (n = 82). b Correlation plot showing the expression of NRAS in each tumor in relation to BRAF expression. c Expression levels of NRAS mRNA in patients experiencing a clinical benefit or progressive disease (PD) 3 months after commencement of chemotherapy. Asterisks denote significance levels: *p ≤ 0.05, **p ≤ 0.005
Low BRAF and NRAS expression is associated with overall and progression-free survival
Patients with clinical benefit from therapy revealed an improved progression-free and overall survival as compared to patients progressing at 3 months on therapy (number compared = 75; p < 0.001 for both; data not shown).
Next, we correlated progression-free and overall survival to BRAF and NRAS mutation status as well as BRAF and NRAS expression levels recorded as being below or above the median value.
BRAF mutation status was not associated with either overall or progression-free survival (number compared = 75; Fig. 4a, b). Conversely, while no correlation between NRAS mutation status and progression-free survival was recorded, patients harboring NRAS mutations had a significantly shorter overall survival as compared to patients with tumors wild-type for NRAS (median survival 3.2 and 8.2 months, respectively; p < 0.001; Fig. 4a).
Fig. 4.
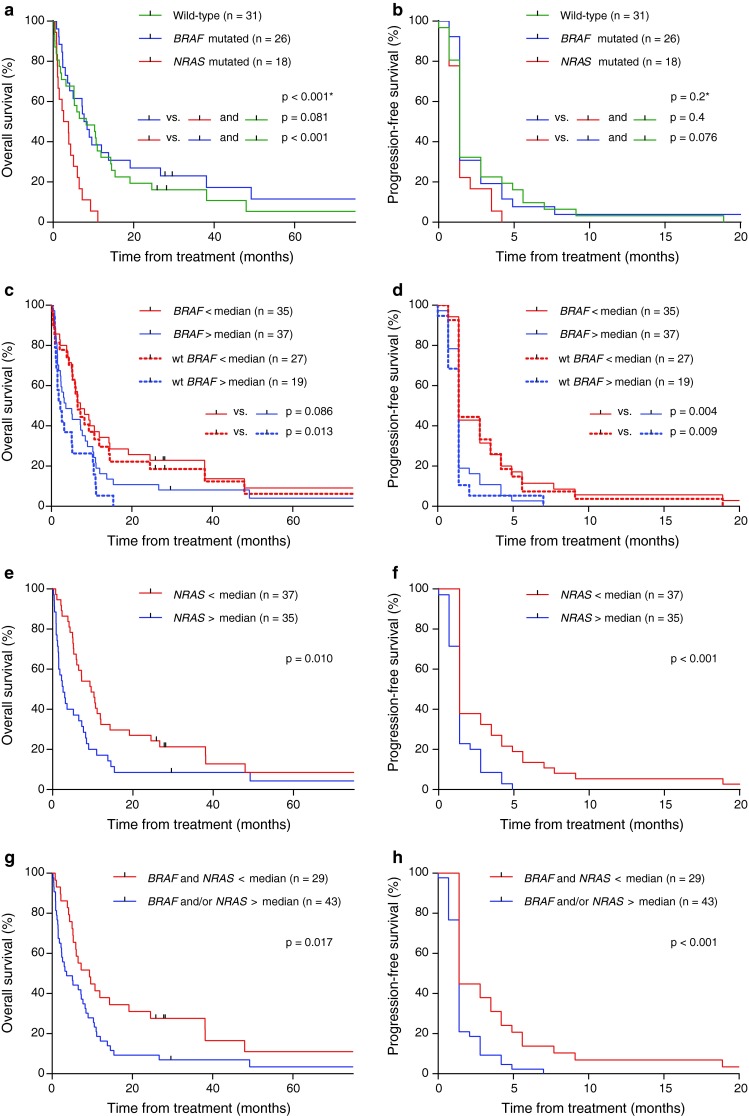
Progression-free and overall survival with respect to BRAF/NRAS status. Kaplan–Meier curves showing overall and progression-free survival: a, b patients with tumors harboring BRAF mutations or NRAS mutations, c, d effect of BRAF expression levels above or below the median value among patients evaluable for treatment response (median value determined among all patients). Solid curves represent all patients; dashed curves represent only patients with tumors harboring wild-type BRAF; e, f effect of NRAS expression levels above or below the median value among patients evaluable for treatment response (median value determined among all patients), g, h effect of combined BRAF/NRAS expression levels: one or both above compared to both below the median values. *All three groups included in the analysis
One potential confounding factor in assessing BRAF mutations as a prognostic factor could be lack of BRAF mutations in the visceral lesions, as well as a low prevalence of BRAF mutations in patients with brain metastases at inclusion, in as much as these patients may be expected to have a particularly poor prognosis. However, no effect of BRAF mutation status on survival was recorded after excluding patients with visceral biopsies or brain metastases from the analysis.
Analyzing patients with tumors mutated or wild-type for BRAF together (number compared = 72; Fig. 4c, d), BRAF expression below the median was associated with longer progression-free survival (p = 0.004). Excluding patients with tumors harboring V600 mutations from the analysis extended this association (number compared = 48; Fig. 4c, d); among patients with tumors wild-type for BRAF, low BRAF mRNA expression was associated with both a longer progression-free (p = 0.009) as well as an improved overall survival (median overall survival 6.5 months for low vs. 2.2 months for high BRAF mRNA expression; p = 0.013). In contrast, BRAF mRNA expression levels were not associated with overall or progression-free survival among patients with BRAF mutations (Fig. S3a, b). Comparing overall and progression-free survival among patients according to NRAS expression levels (number compared = 72; Fig. 4e, f) revealed expression levels below median value to be associated with an improved outcome in terms of progression-free (p < 0.001), as well as overall survival (median overall survival 9.6 months for low vs. 2.6 months for high NRAS expression; p = 0.01). Excluding patients with NRAS mutations did not strengthen this association (data not shown).
Combining BRAF and NRAS expression characteristics showed that patients with tumors expressing both genes below the median displayed an improved progression-free (p < 0.001) as well as overall (p = 0.017) survival as compared to patients with tumors in which either BRAF or NRAS or both were expressed above the median value (number compared = 72; Fig. 4g, h).
To assess the independent value of mutation status and expression of BRAF and NRAS, multivariate analysis was carried out using Cox regression entering serum LDH levels (available for 78 patients) and localization of most unfavorable metastasis present at treatment start (specified as locoregional, visceral or central nervous system metastases) together with BRAF and/or NRAS mutation/expression levels. Regarding progression-free survival, in general either NRAS expression levels or (when BRAF was added to the model) BRAF and NRAS expression levels combined (one or both elevated above median value) predicted a shorter time to progression (p < 0.05); neither serum LDH levels nor metastatic location were of significance. As for overall survival, both serum LDH and metastatic location consistently predicted survival (p values for both <0.005) together with either NRAS mutation status (p < 0.05) or NRAS/BRAF expression levels combined (p < 0.05).
Discussion
The RAF group of proto-oncogenes consists of three family members (A-, B- and CRAF). Among these, BRAF has been shown to play a key role through activating mutations in malignant melanomas. BRAF and NRAS proteins are both subject to activating mutations; with V600E (BRAF) and Q61K/R/L (NRAS) being the most frequent ones [7, 8]. Our detection of BRAF mutations in 41 % of patients with cutaneous primary melanomas but lack of mutations among uveal- and mucosa-derived tumors are in accordance with findings by others [24, 26–29]. Furthermore, our finding of NRAS mutations in 20 % of all tumors is consistent with previous reports [29]. In contrast to others reporting the BRAF V600K mutation to occur in about 6–20 % of BRAF-mutated melanomas [30], we only observed this mutation in one out of 30 metastases (3.3 %). Regarding the nature of BRAF mutations, in contrast to V600E, the V600K mutation has been reported to be associated with cumulative sun-induced damage (CSD) [31]. As CSD can be assumed to be relatively uncommon in Norway as compared to most other geographical areas, this could explain the low prevalence of BRAF V600K mutations observed in the current study.
While our finding of few BRAF mutations in visceral metastases from cutaneous melanomas contrasts the finding of others [32], due to a limited number of observation this discrepancy may be caused by chance only.
The association between BRAF mutation status and copy-number gains has been reported by others as well [33–35]. To the best of our knowledge, this study is the first to report elevated BRAF expression levels in human melanomas harboring BRAF mutations without elevated gene copy number. Notably, this finding contrasts data from cell lines [15].
In the current study, two products of alternative splicing of the BRAF pre-mRNA were observed, BRAFdel14–15 and BRAFdel12–15, of which the former has previously been reported [18]. While others have described BRAF splice variants promoting oncogenic activity [19], each of the alternative splices detected in the current study lacks the catalytic domain considered crucial to RAF activation [36], suggesting these splices not to be oncogenic.
As has been observed previously in the case of BRAF [9], we found no correlation between BRAF or NRAS mutations and response to DTIC treatment in our patients. In contrast, a low BRAF mRNA expression level was associated with benefit from DTIC treatment. Notably, this association was particularly strong in patients with tumors wild-type for BRAF. These findings are somewhat contradictory. The BRAF V600E mutation has been reported to enhance enzyme activity in vitro by a factor of about 500 [11]; thus, we may envision a poor drug response as well as a poor prognosis for tumors harboring the activating V600E mutation as well. However, the biological role of BRAF may be different between tumors with wild-type or V600E-mutated BRAF. In tumors harboring the V600E mutation, BRAF seems to be the key proliferation driver [37]; thus, vemurafenib causes dramatic tumor shrinkage in patients with V600E-mutated tumors but not in tumors wild-type for BRAF [38]. Notably, BRAF-mutated tumors in general reveal gene expression profiles different from those harboring wild-type BRAF [39], and V600E-mutated BRAF has been shown to interact with CRAF in a manner different from wild-type BRAF [40]. Taken together, these findings indicate effects of V600E-mutated BRAF protein not to be related to wild-type BRAF “dosing activity” only. These observations may have clinical implications; while vemurafenib administered as monotherapy is ineffective in BRAF wild-type tumors where it may even activate RAF signaling [41], future drugs suppressing BRAF levels may potentially sensitize wild-type tumors to chemotherapy if administered in concert.
Our finding that low NRAS expression levels was associated with improved treatment outcome indirectly supports the hypothesis that low BRAF activity is associated with improved treatment response. Moreover, this underlines the observation that activation of the RAS/RAF kinase pathway by increased expression levels may cause biological effects different from that of activation by mutations. Interestingly, the mRNA expression of BRAF and NRAS were positively correlated, in contrast to the mutation distribution of these genes, which were mutually exclusive.
Conflicting evidence has reported BRAF and NRAS mutations to be associated with a poor prognosis in melanoma [4–6, 9, 10, 42, 43]. Considering unresectable stages III and IV disease, our data are consistent with Jakob et al. [10] who reported shorter survival among patients with tumors harboring NRAS mutations. While our results do not support BRAF mutations to be associated with a poor prognosis in advanced melanoma, as suggested by Long et al. [9], their study reported survival for patients with BRAF-mutated tumors found ineligible for BRAF inhibitor trials who, as stated by the authors, are expected to be a poor-survival group due to intercurrent medical problems [9].
While the data currently presented should be interpreted carefully due to the limited number of patients studied, the association of low BRAF and NRAS expression with benefit from anti-tumor therapy as well as improved relapse-free and overall survival is interesting. Although the multivariate analyses carried out suggested an independent association between BRAF/NRAS expression levels and outcome, the results should be interpreted carefully due to the limited number of observations. While these findings may suggest that low BRAF/NRAS expression levels could be both a predictive as well as a prognostic factor [44], validation in independent studies are needed. For some patients, stable disease after 3 months on therapy could reflect slow tumor growth and may not necessarily signal response to DTIC therapy. As such, the effects of NRAS/BRAF status on survival may reflect an enhanced effect of treatment or, alternatively, tumor biology independent of drug therapy [44].
In conclusion, we present data linking BRAF, as well as NRAS, mRNA expression levels to outcome in advanced melanoma. Our data further indicate that low BRAF and NRAS expression levels may predict for benefit to DTIC chemotherapy. For patients with tumors harboring wild-type BRAF treated with DTIC chemotherapy, a low level of BRAF expression was associated with longer overall and progression-free survival, whereas low NRAS expression was associated with improved progression-free and overall survival irrespective of mutation status. Further studies are warranted to confirm a potential predictive and prognostic role of BRAF and NRAS expression levels in advanced melanomas.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was performed in Mohn Cancer Research Laboratory, Haukeland University Hospital and was supported by Grants from the Norwegian Cancer Society and the Western Norwegian Regional Health Authorities.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Einar Birkeland, Christian Busch have contributed equally.
References
- 1.Rigel DS. Cutaneous ultraviolet exposure and its relationship to the development of skin cancer. J Am Acad Dermatol. 2008;58(5 Suppl 2):S129–S132. doi: 10.1016/j.jaad.2007.04.034. [DOI] [PubMed] [Google Scholar]
- 2.Francken AB, Accortt NA, Shaw HM, et al. Follow-up schedules after treatment for malignant melanoma. Br J Surg. 2008;95(11):1401–1407. doi: 10.1002/bjs.6347. [DOI] [PubMed] [Google Scholar]
- 3.Eggermont AM. Reaching first base in the treatment of metastatic melanoma. J Clin Oncol. 2006;24(29):4673–4674. doi: 10.1200/JCO.2006.07.3593. [DOI] [PubMed] [Google Scholar]
- 4.Akslen LA, Angelini S, Straume O, et al. BRAF and NRAS mutations are frequent in nodular melanoma but are not associated with tumor cell proliferation or patient survival. J Investig Dermatol. 2005;125(2):312–317. doi: 10.1111/j.0022-202X.2005.23788.x. [DOI] [PubMed] [Google Scholar]
- 5.Ugurel S, Thirumaran RK, Bloethner S, et al. B-RAF and N-RAS mutations are preserved during short time in vitro propagation and differentially impact prognosis. PLoS ONE. 2007;2(2):e236. doi: 10.1371/journal.pone.0000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edlundh-Rose E, Egyhazi S, Omholt K, et al. NRAS and BRAF mutations in melanoma tumours in relation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Res. 2006;16(6):471–478. doi: 10.1097/01.cmr.0000232300.22032.86. [DOI] [PubMed] [Google Scholar]
- 7.Albino AP, Nanus DM, Mentle IR, et al. Analysis of ras oncogenes in malignant melanoma and precursor lesions: correlation of point mutations with differentiation phenotype. Oncogene. 1989;4(11):1363–1374. [PubMed] [Google Scholar]
- 8.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 9.Long GV, Menzies AM, Nagrial AM, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011;29(10):1239–1246. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 10.Jakob JA, Bassett RL, Jr, Ng CS, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer. 2012;118(16):4014–4023. doi: 10.1002/cncr.26724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF–ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–867. doi: 10.1016/S0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 12.Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366(8):707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauschild A, Grob JJ, Demidov LV, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380(9839):358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 14.Flaherty KT, Robert C, Hersey P, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367(2):107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 15.Tanami H, Imoto I, Hirasawa A, et al. Involvement of overexpressed wild-type BRAF in the growth of malignant melanoma cell lines. Oncogene. 2004;23(54):8796–8804. doi: 10.1038/sj.onc.1208152. [DOI] [PubMed] [Google Scholar]
- 16.Jeuken J, van den Broecke C, Gijsen S, et al. RAS/RAF pathway activation in gliomas: the result of copy number gains rather than activating mutations. Acta Neuropathol. 2007;114(2):121–133. doi: 10.1007/s00401-007-0239-0. [DOI] [PubMed] [Google Scholar]
- 17.Shi H, Moriceau G, Kong X, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seth R, Crook S, Ibrahem S, et al. Concomitant mutations and splice variants in KRAS and BRAF demonstrate complex perturbation of the RAS/RAF signalling pathway in advanced colorectal cancer. Gut. 2009;58(9):1234–1241. doi: 10.1136/gut.2008.159137. [DOI] [PubMed] [Google Scholar]
- 19.Poulikakos PI, Persaud Y, Janakiraman M, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480(7377):387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carninci P, Hayashizaki Y. High-efficiency full-length cDNA cloning. Methods Enzymol. 1999;303:19–44. doi: 10.1016/S0076-6879(99)03004-9. [DOI] [PubMed] [Google Scholar]
- 21.Lagonigro MS, De Cecco L, Carninci P, et al. CTAB–urea method purifies RNA from melanin for cDNA microarray analysis. Pigment Cell Res. 2004;17(3):312–315. doi: 10.1111/j.1600-0749.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- 22.Nozawa H, Yamamoto T, Uchihi R, et al. Purification of nuclear DNA from single hair shafts for DNA analysis in forensic sciences. Leg Med (Tokyo) 1999;1(2):61–67. doi: 10.1016/S1344-6223(99)80014-5. [DOI] [PubMed] [Google Scholar]
- 23.Busch C, Geisler J, Lillehaug JR, et al. MGMT expression levels predict disease stabilisation, progression-free and overall survival in patients with advanced melanomas treated with DTIC. Eur J Cancer. 2010;46(11):2127–2133. doi: 10.1016/j.ejca.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 24.Wong CW, Fan YS, Chan TL, et al. BRAF and NRAS mutations are uncommon in melanomas arising in diverse internal organs. J Clin Pathol. 2005;58(6):640–644. doi: 10.1136/jcp.2004.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonsson G, Busch C, Knappskog S, et al. Gene expression profiling-based identification of molecular subtypes in stage IV melanomas with different clinical outcome. Clin Cancer Res. 2010;16(13):3356–3367. doi: 10.1158/1078-0432.CCR-09-2509. [DOI] [PubMed] [Google Scholar]
- 26.Cruz F, III, Rubin BP, Wilson D, et al. Absence of BRAF and NRAS mutations in uveal melanoma. Cancer Res. 2003;63(18):5761–5766. [PubMed] [Google Scholar]
- 27.Shinozaki M, Fujimoto A, Morton DL, et al. Incidence of BRAF oncogene mutation and clinical relevance for primary cutaneous melanomas. Clin Cancer Res. 2004;10(5):1753–1757. doi: 10.1158/1078-0432.CCR-1169-3. [DOI] [PubMed] [Google Scholar]
- 28.Willmore-Payne C, Holden JA, Tripp S, et al. Human malignant melanoma: detection of BRAF- and c-kit-activating mutations by high-resolution amplicon melting analysis. Hum Pathol. 2005;36(5):486–493. doi: 10.1016/j.humpath.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 29.Platz A, Egyhazi S, Ringborg U, et al. Human cutaneous melanoma; a review of NRAS and BRAF mutation frequencies in relation to histogenetic subclass and body site. Mol Oncol. 2008;1(4):395–405. doi: 10.1016/j.molonc.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubinstein JC, Sznol M, Pavlick AC, et al. Incidence of the V600K mutation among melanoma patients with BRAF mutations, and potential therapeutic response to the specific BRAF inhibitor PLX4032. J Transl Med. 2010;8:67. doi: 10.1186/1479-5876-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Menzies AM, Haydu LE, Visintin L, et al. Distinguishing clinicopathologic features of patients with V600E and V600K BRAF-mutant metastatic melanoma. Clin Cancer Res. 2012;18(12):3242–3249. doi: 10.1158/1078-0432.CCR-12-0052. [DOI] [PubMed] [Google Scholar]
- 32.Colombino M, Capone M, Lissia A, et al. BRAF/NRAS mutation frequencies among primary tumors and metastases in patients with melanoma. J Clin Oncol. 2012;30(20):2522–2529. doi: 10.1200/JCO.2011.41.2452. [DOI] [PubMed] [Google Scholar]
- 33.Modrek B, Ge L, Pandita A, et al. Oncogenic activating mutations are associated with local copy gain. Mol Cancer Res. 2009;7(8):1244–1252. doi: 10.1158/1541-7786.MCR-08-0532. [DOI] [PubMed] [Google Scholar]
- 34.Maldonado JL, Fridlyand J, Patel H, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst. 2003;95(24):1878–1890. doi: 10.1093/jnci/djg123. [DOI] [PubMed] [Google Scholar]
- 35.Jonsson G, Dahl C, Staaf J, et al. Genomic profiling of malignant melanoma using tiling-resolution arrayCGH. Oncogene. 2007;26(32):4738–4748. doi: 10.1038/sj.onc.1210252. [DOI] [PubMed] [Google Scholar]
- 36.Rajakulendran T, Sahmi M, Lefrancois M, et al. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461(7263):542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 37.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pavey S, Johansson P, Packer L, et al. Microarray expression profiling in melanoma reveals a BRAF mutation signature. Oncogene. 2004;23(23):4060–4067. doi: 10.1038/sj.onc.1207563. [DOI] [PubMed] [Google Scholar]
- 40.Garnett MJ, Rana S, Paterson H, et al. Wild-type and mutant B-RAF activate C-RAF through distinct mechanisms involving heterodimerization. Mol Cell. 2005;20(6):963–969. doi: 10.1016/j.molcel.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 41.Poulikakos PI, Zhang C, Bollag G, et al. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreau S, Saiag P, Aegerter P, et al. Prognostic value of BRAF (V600) mutations in melanoma patients after resection of metastatic lymph nodes. Ann Surg Oncol. 2012;19(13):4314–4321. doi: 10.1245/s10434-012-2457-5. [DOI] [PubMed] [Google Scholar]
- 43.Mann GJ, Pupo GM, Campain AE, et al. BRAF mutation, NRAS mutation, and the absence of an immune-related expressed gene profile predict poor outcome in patients with stage III melanoma. J Investig Dermatol. 2013;133(2):509–517. doi: 10.1038/jid.2012.283. [DOI] [PubMed] [Google Scholar]
- 44.Lonning PE, Knappskog S, Staalesen V, et al. Breast cancer prognostication and prediction in the postgenomic era. Ann Oncol. 2007;18(8):1293–1306. doi: 10.1093/annonc/mdm013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



