Abstract
Translation, that is biosynthesis of polypeptides in accordance with information encoded in the genome, is one of the most important processes in the living cell, and it has been in the spotlight of international research for many years. The mechanisms of protein biosynthesis in bacteria and in the eukaryotic cytoplasm are now understood in great detail. However, significantly less is known about translation in eukaryotic mitochondria, which is characterized by a number of unusual features. In this review, we summarize current knowledge about mitochondrial translation in different organisms while paying special attention to the aspects of this process that differ from cytoplasmic protein biosynthesis.
Keywords: mitochondria, translation, ribosomes, translation factors
Almost all eukaryotic cells contain mitochondria. These organelles provide energy via the process of oxidative phosphorylation and are involved in fatty acid, heme, and iron-sulfur cluster biosynthesis, as well as programmed cell death. According to the generally accepted concept of endosymbiosis, mitochondria originated in the early stages of eukaryotic evolution from an endosym-biotic α-proteobacterium closely related to the modern Rickettsia prowazekii [1]. Over time, as a result of the tight association with the nuclear genome, many predecessors of mitochondrial genes have been transferred to the nucleus, but a few are retained in the mitochondrial DNA. Mitochondrial messenger RNAs (mRNAs) in the mitochondrion are translated by the organelle’s own apparatus of protein biosynthesis, and fine coordination between the two genomes is required.
Many aspects of mitochondrial translation remain poorly studied. This is largely due to the fact that mitochondrial translation systems capable of synthesis on natural matrices have still not been reconstructed in vitro. On the basis of early studies of sedimentation characteristics and RNA composition of mitochondrial ribosomes, as well as their inhibition by antibiotics, it became apparent that mitochondrial translation is similar to that of bacteria [2-4]. Recently, it has become clear that translation in the mitochondria has a number of distinctive features [5, 6].
A number of high-resolution structural studies have aided our understanding of translation at the molecular level. The majority of these were performed on bacterial and eukaryotic cytosolic ribosomes [7]. However, considerable success has also been achieved in the study of structural and functional properties of the mitochondrial translational system. In this review, we summarize the current understanding of protein synthesis in mitochondria.
GENERAL FEATURES OF PROTEIN SYNTHESIS IN MITOCHONDRIA
The mitochondrial genome, despite the universality of its function in energy generation, shows a high degree of variation in composition, size, and gene content across eukaryotes [8]. For example, the smallest mitochondrial genome described to date is the linear mitochondrial DNA (mtDNA) of Plasmodium falciparum (5966 bp). This encodes, besides the large and small ribosomal RNA (rRNA), only three proteins [9]. The human circular mtDNA (16,569 bp) contains 22 genes of transfer RNA (tRNA), 13 protein coding genes, and 12S and 16S rRNA genes. Exceeding this 22-fold in size, the Arabidopsis thaliana mitochondrial genome (366,924 bp) contains information for only 32 proteins and an incomplete set of tRNAs [10, 11]. Such disparity is explained by an unusually large number (>80%) of noncoding sequences in the mtDNA of A. thaliana compared with Leishmania (<10%) and mammals, whose genes directly follow each other or are separated by only a few nucleotides [12]. The rate of evolution of the mitochondrial genome is extremely high, partly due to an increased frequency of mutations due to the lack of an effective repair system and its proximity to reactive forms of oxygen produced near the inner mitochondrial membrane [13].
An important feature of mitochondrial translation is a deviation from the standard genetic code. For example, in vertebrate mitochondria one of the three stop codons – UGA – corresponds to tryptophan, and AUA encodes methionine instead of isoleucine. Mitochondria often use alternative triplets as start codons: AUA (Homo, Bos), AUU (Homo, Mus), AUC (Mus), and GUG (Coturnix, Gallus). The number of stop codons is reduced in mitochondrial systems: usually one or two, but 7 of 12 open reading frames of the parasitic nematode Radopholus similis mitochondria are deprived of canonical stop codons [14]. Additionally, in yeasts Saccharomyces cerevisiae and Torulopsis glabrata codons starting with CU have been reassigned from leucine to threonine [15].
In almost all eukaryotic mitochondria genes are transcribed by an RNA polymerase homologous to that of T3 and T7 phages [16]. The product of this transcription is a polycistronic primary transcript [16-18]. According to the “tRNA punctuation” model, it is further processed by an endonucleolytic excision of tRNA sequences generating the individual protein-encoding mRNAs [19]. There are 11 human mitochondrial mRNAs: nine monocistronic and two bicistronic with overlapping reading frames. In S. cerevisiae mitochondria, there are six monocistronic mRNAs and one bicistronic mRNA coding for subunits 6 and 8 of the FoF1-type ATP synthase [20]. Mitochondrial mRNAs lack the cap structure characteristic for the cytoplasmic translation, and they are characterized by dramatic lineage-specific differences. Human mitochondrial mRNAs have very short 5′- and 3′-untranslated regions (UTR) and are polyadenylated [20]. On the other hand, in S. cerevisiae mature mitochondrial transcripts have long UTRs that are important for translation regulation, and they are not polyadenylated [21-23]. Apparently, this reflects a fundamental difference in the mechanisms of recognition of the start codon and the initiator complex assembly in the mitochondria of yeast and mammals. Mitochondrial mRNAs, unlike those of bacteria, lack Shine–Dalgarno sequences and, accordingly, the complementary segment of the small subunit rRNA. The octanucleotide sequence UAUAAAUA was suggested to play the role of a common ribosome recognition site in yeast, but this hypothesis has not yet been experimentally confirmed [24]. The degradation of mitochondrial mRNA is controlled by a poly-subunit protein complex – the degradosome – possessing an RNA helicase as well as hydrolytic or phospholytic exonuclease activities [25]. In yeast, mitochondrial polynucleotide phosphorylase has not been found, while in mammals this enzyme is found in the intermembrane space, where, in addition to its main function, it participates in the regulation of RNA import into mitochondria [26].
The number of different tRNA species in mitochondria is usually significantly reduced in comparison to the cytoplasmic systems (22 and 24 in yeast and human, respectively), which is compensated by each of the tRNAs recognizing several codons. Mitochondrial tRNAs are usually shorter (59-75 nucleotides length in animals) than those of bacteria and the eukaryotic cytosol [27]. They often lack the conservative elements of the tertiary structure such as long-range interactions between dihydrouridyl (D) and thymidylpseudouridyl (T) arms, and binding sites of magnesium cations, with the latter feature leading to their low thermal stability [28, 29]. Despite the absence of high-resolution structural data, the results of chemical modification, nuclease protection, and NMR spectroscopy of T7 transcripts of bovine and nematode mitochondrial tRNA suggest that they assume a structure similar to the canonical L-form of the molecule [5]. This is confirmed by the results of cryoelectron microscopy of Bos taurus mitochondrial ribosomes: tRNA firmly bound in the peptidyl transferase center has a typical L-conformation with a cavity in the “elbow” [30]. Despite the presence of a complete set of tRNAs in the mitochondrial genome of S. cerevisiae, one of the two cytoplasmic isoacceptor lysine tRNAs with CUU anticodon is also imported into the organelle and is involved in translation [31]. In contrast to animals and yeasts, the majority or even all types of mitochondrial tRNAs in plants and trypanosomatids are imported from the cytoplasm [32, 33].
Mitochondrial ribosomes (mitoribosomes) differ from 70S bacterial and 80S eukaryotic particles in several ways. Budding yeast mitoribosomes have a sedimentation coefficient of 74S, mammals – 55S, and Leishmania tarentolae – just 50S [34 36]. Bos taurus and L. tarentolae mitoribosome structures resolved by cryoelectron microscopy to 9-14 Å are currently available [30, 35]. As in all ribosomes, mitoribosomes have large and small subunits consisting of ribosomal RNA (rRNA) and proteins (see table). The most significant difference between mitoribosomes and their cytoplasmic analogs is in the RNA/protein ratio, which is 2 : 1 in bacteria, while in B. taurus and L. tarentolae mitochondria it is 1 : 2 and 1 : 3, respectively. At the same time, mammalian mitoribosomes (2.71 MDa in mass and ~320 Å in diameter) are larger, but Leishmania mitoribosomes (2.2 MDa and ~245 Å) are smaller than the E. coli ribosome [30, 35]. Low values of sedimentation coefficients of mitoribosomes are due to their porous structure, which is a consequence of the absence or severe reduction in some segments of the rRNA [37]. For example, the small 12S rRNA subunit is completely missing the anti-Shine–Dalgarno sequence as well as helices 6, 8-10, 12, 13, 16, 17, 21, 33, 37, and 39-41 in the mammalian mitoribosome. Some of the lost elements are compensated for by new mitoribosome-specific proteins or by increased protein length relative to their bacterial counterparts. However, in mammals only 19% of missing sequences in small subunit rRNA and 28% in large subunit rRNA are replaced with proteins, while a similar compensation in L. tarentolae mitoribosomes reaches 50%. This has led to a number of distinctive morphological features while maintaining the general organization of the subunits. Such features include, for example, tunnel-like structures that link the intersubunit space with the external environment. Shielding of rRNA by proteins and the prevalence of protein–protein intersubunit bridges (9 of 15 in animals) are especially the case in mitoribosomes [30]. A unique feature of the small subunit of the mammalian mitoribosome is an extremely dynamic trihedral gate-shaped structure surrounding the mRNA entry site [30]. This is thought to help in attracting mRNAs lacking leader sequences to the small ribosomal subunit (SRS). The large ribosomal subunit (LRS) is characterized by a massive central protuberance and a lack of electron density corresponding to 5S rRNA. However, biochemical studies have shown that 5S rRNA is imported into human mitochondria from the cytosol, and it is associated with mitoribosomes [38, 39]. The absence of the core 16S rRNA elements constituting the classical E-site – helices 11 and 68 and the loop portion lying between the helices 76 and 77 – leads to very weak binding of tRNA to this region [40]. Another distinctive feature of the LRS is the presence of finger-like structures in the P-site formed by the extension of a portion of the mitochondria-specific proteins of the central protuberance. This finger interacts with the T-loop of the tRNA molecule located in the P-site, and it presumably exerts a stabilizing effect on the binding. Apparently, this is the reason that mitoribosomes, unlike bacterial ribosomes, are purified with tRNA firmly bound to the P-site. The organization of the tunnel through which the nascent peptide leaves the ribosome deserves special attention [41]. In the case of ribosomes of bacteria and eukaryotic cytoplasm, it leads to an opening referred to as the Polypeptide Exit Site (PES). This is a flat structure located in the lower part of the large subunit formed by elements of domains I and III of 23S rRNA and ribosomal proteins L22, L23, L24, and L29. The situation is different in the case of L. tarentolae and B. taurus mitoribosomes, where one more hole was discovered at some distance from the PES (~25 Å in the case of L. tarentolae), called PAS (Polypeptide-Accessible Site) [37, 42]. The formation of this structure results from the absence of most of domains I and III in the large subunit rRNA and its incomplete compensation by proteins. Thus, the exit of the nascent polypeptide chain from mitoribosome apparently can occur in two different ways.
Table 1. General features of mitochondrial and bacterial ribosomes.
| Bacteria | Bovine mitochondria | Leishmania mitochondria | |
|---|---|---|---|
|
| |||
| Molecular weight, MDa | 2.3 | 2.7 | 2.2 |
| Diameter, Å | ≈260 | ≈320 | ≈245 |
| Molar ratio RNA/protein | ≈2 : 1 | ≈1 : 2 | ≈1 : 3 |
| Small subunit rRNA and its size | 16S (1542 nt) | 12S (950 nt) | 9S (610 nt) |
| Number of proteins in small subunit | 21 | ≈29 | ≈56 |
| Large subunit rRNA and its size | 23S rRNA (2904 nt) 5S rRNA (120 nt) |
16S rRNA (1560 nt) |
12S rRNA (1173 nt) |
| Number of proteins in large subunit | 34 | ≈50 | ≈77 |
Mitochondrial ribosomes almost exclusively synthesize hydrophobic polypeptides that form the functional centers of electron transport chain complexes. To achieve efficient integration of nascent polypeptides into the inner mitochondrial membrane, translation and, most likely, assembly of the mitoribosomes occur directly on the membrane [43, 44]. The process of the anchoring the mitoribosome on the mitochondrial membrane in S. cerevisiae is fairly well understood. It involves a number of proteins (Oxa1p, Mba1p, and Mdm38p) as well as mRNA molecules associated with the membrane through translational activators – a group of proteins involved in the organization and regulation of the translational machinery. Oxa1p interacts with the PES via the C-terminal domain of the mitochondrial homolog of bacterial L23 [45, 46], Mba1p plays the role of mitoribosome receptor [47], and Mdm38p together with Mba1p bind the large subunit. Deletion of all genes encoding these proteins does not lead to complete dissociation of the ribosome from the membrane, indicating the existence of as yet undiscovered components involved in the process [48, 49].
Several recent studies have described unexpected and intimate functional links between the mitochondrial translation and transcription. In human mitochondria, there is a free pool of the mitoribosomal large subunit protein MRPL12, which selectively binds to mitochondrial RNA polymerase in vivo and directly stimulates a transcription system in vitro [50]. This is considered to be one of the mechanisms coordinating the assembly of mitochondrial ribosomes. On the other hand, human mitochondrial RNA polymerase (POLRMT) has a role in mitoribosomal biogenesis and/or translation that is completely independent of its function in transcription. Co-immunoprecipitation and fractionation of mitochondrial lysates indicate that POLRMT forms a complex with the 28S ribosomal subunit. This complex also includes a paralog of the mitochondrial transcription factor h-mtTFB2, protein h-mtTFB1, which acts as a 12S rRNA methyl transferase [51]. According to one hypothesis, by stimulating the methyltransferase activity of h-mtTFB1 and preventing the association of immature small subunits with 39S subunits, formation of the POLRMT·mitoribosome complex acts as a control point in the assembly path of the small subunit [51].
Below we describe translation in mammalian mitochondria, with differences in the yeast system noted when necessary.
TRANSLATION INITIATION IN MITOCHONDRIA
Common features
During the initiation of protein synthesis, scanning and positioning of the mRNA start codon occurs in the P-site of the small subunit. This is followed by binding of the initiator tRNA followed by association of the large subunit (subunit joining).
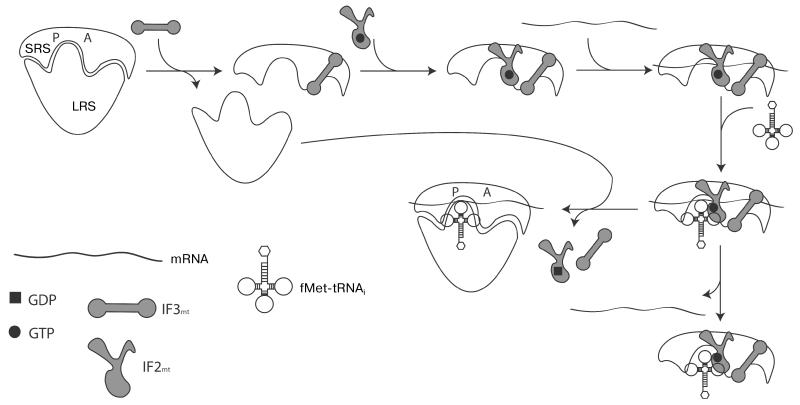
Like all stages of translation, initiation is assisted by specific protein factors. There are three universal initiation factors in bacteria, namely IF1, IF2, and IF3. In mitochondria, IF2mt is universally present and IF3mt has been identified in the vast majority of eukaryotes [52]. Both human factors have been characterized in detail in in vitro translation systems [53]. Using in vitro experiments with initiator complexes formed with 55S mitoribosomes from bovine liver on in vitro synthesized 5′- terminal fragments of cytochrome oxidase subunit I (COI) and subunit II of NADH dehydrogenase (ND2) mRNAs in the presence of yeast mitochondrial initiator tRNA (fMet-tRNAi), the following model of mitochondrial translation initiation was proposed (Fig. 1) [54]. First, IF3mt catalyzes the dissociation of 55S ribosomes, resulting in formation of free 39S subunits and 28S·IF3mt complexes. For convenience, binding of translational GTPase IF2mt·GTP is shown in the next stage, although the exact timing of its attachment to the complex is not known. Next, mRNA is threaded through the unique gate-shaped structure in the 28S subunit, followed by the binding of fMet-tRNAi. This order of events is dictated by IF3mt, which, unlike its bacterial homolog, destabilizes initiator tRNA binding in the absence of mRNA [55]. Primer extension reaction inhibition assays have shown that upon initiation of complex formation, the ribosome is stalled after the first 17 nucleotides of mammalian mitochondrial mRNAs. From this it was postulated that the mitoribosome scans the 5′-UTR of mRNA in a search for the start codon, and when it is located in the P-site, translational GTPase IF2mt in a GTP-bound form promotes docking of fMet-tRNAi, with resultant codon–anticodon interactions stabilizing the initiation complex. If the interactions between the start codon and anticodon are not formed, the 28S subunit continues sliding along the mRNA and eventually dissociates. After initiator tRNA binding, the 39S subunit joins the complex, IF2mt hydrolyzes bound GTP to GDP, and the initiator factors leave the mature 55S initiation complex, which is now ready for the elongation stage of polypeptide synthesis.
Fig. 1. Schematic representation of translation initiation in mitochondria.
SRS, small ribosomal subunit; LRS, large ribosomal subunit. See text for details.
The scheme of translation initiation described above is applicable only to mammalian mitochondria. Animal mitochondrial mRNAs lack long 5′ UTRs (up to 3 nt in case of COI mRNA). The presence of only three additional nucleotides upstream of the start codon in COI mRNA reduces the efficiency of initiation complex formation by 40%, and 12 additional nucleotides at the 5′- side of the AUG start codon lead to an 80% decrease [54]. A very different situation is observed in yeast, where the long 5′-UTRs precede the start codons and interact with specific activators of translation (see below). Also in yeast mitochondria, unlike mammals, there are two methionine tRNAs: initiator and elongater. It was originally assumed that similarly to the bacterial case, formylation of methionine of the acylated initiator tRNA is strictly necessary for the initiation of translation. Surprisingly, it was shown that in S. cerevisiae translation initiation can occur in the absence of a formyl residue of fMet-tRNAi. An auxiliary factor Aep3p, which promotes the binding of non formylated tRNAi to IF2mt, was discovered recently [56]. This protein is essential when only non-formylated initiator tRNA is available. Aep3p is a peripheral protein of the mitochondrial inner membrane and contains four RNA-binding PPR (pentatricopeptide) motifs, and in addition to its role in translation initiation, it stabilizes the bicistronic mitochondrial mRNA encoding subunits 6 and 8 of ATP synthase [57]. In mammals, the participation of a single methionyl-tRNA in initiation or elongation is determined by competition between transformylase and elongation factor EF-Tumt; affinity of IF2mt to the formylated form is 50 times higher than to non formylated, while EF-Tumt does not show detectable affinity to fMet-tRNAi [58].
The mitochondria of Saccharomyces fungi contain special translation activators, most of which are not found in mammalian mitochondria. In S. cerevisiae such specific protein factors are involved in translation of each of the eight polypeptides encoded in the mitochondrial genome [59]. Translation activators are encoded in the nuclear genome, and the proteins are transported to the mitochondrial inner membrane. Activators also coordinate the rate of synthesis of proteins encoded in the mitochondria with their subsequent assembly into the respiratory chain complexes. A detailed description of the translation activators can be found in a recent comprehensive review [60].
Mitochondrial translation initiation factors
As mentioned above, no mitochondrial ortholog of bacterial IF1 has been found in eukaryotes [61]. In bacteria, this small protein (71 amino acid residues in E. coli) together with IF3 is involved in fMet-tRNAi selection on the 30S subunit, blocking binding of the tRNA to the A-site of the ribosome and inhibiting 50S subunit joining in the absence of fMet-tRNAi. Until recently, it was thought that the absence of IF1 in mitochondria is generally compensated by a short insertion between domains V and VI (in accordance with the six-domain scheme of IF2) in IF2mt [62]. This hypothesis is supported by the fact that the expression of bovine IF2mt allows E. coli cells to grow with a deletion of the gene encoding IF1 [62]. Cryoelectron microscopy reconstruction of 70S ribosomes complexed with bovine IF2mt and fMet-tRNAi in the presence of mRNA and nonhydrolyzable analog of GTP, GDPNP, suggested that the insertion in IF2mt occupies the same area on the intersubunit surface of the 30S particle as IF1 in a bacterial system [61]. However, bioinformatic analysis showed that this amino acid sequence is only conserved in vertebrates [52]. Therefore, it appears that this phenomenon is not universal and the insertion has evolved secondarily to the loss of IF1mt.
IF2mt is a highly conserved protein, homologous to IF2 of bacteria and archaea and to the corresponding eukaryotic cytosolic factor eIF5B. By analogy with the six-domain organization of IF2 in E. coli, four domains are identified in IF2mt: III, IV, V, and VI; the latter is composed of two subdomains (CI and CII) [63]. Domain IV is a classic G-domain – the binding site of guanyl nucleotides [53]. In the presence of GDPNP, IF2mt binds to the 28S subunit three times more efficiently than in the presence of GDP and 10 times more efficiently than in the absence of nucleotides, whereas the dissociation constant for the complex of 28S·IF2mt, measured in experiments with biotinylated mitoribosomes in the presence of GDPNP, was found to be 10-20 nM [64]. Domain III forms contacts with the mitoribosome small subunit, and the VICII domain is involved in binding of the CCA-end of fMet tRNAi. The animal-specific insertion between domains V and VICI, which was suggested to play the role of a lost homolog of IF1, contacts helices h18 and h44 of rRNA and the protein S12.
The situation with the third mitochondrial translation initiation factor IF3mt is much more complicated. The low degree of homology with the bacterial factor (the human factor shares 20.8% identity with IF3 of E. coli) was for a long time an obstacle to detection of the respective factor in the mitochondria of eukaryotes. An initial search for IF3mt orthologs in the Homo sapiens EST data base using E. coli IF3 as the query was not successful [65]. However, candidate genes were successfully identified both in human and mouse using Mycoplasma sp. and Euglena gracilis chloroplast IF3s as queries [65]. Candidates for the role of this factor in the genomes of invertebrates and lower eukaryotes have proved harder to find. By using a sensitive PSI-Blast algorithm, an IF3mt ortholog in S. cerevisiae – Aim23p – was found [52]. The growth of yeast cells with AIM23 gene deletion on a medium with non-fermentable carbon sources was restored in the presence of the gene encoding S. pombe IF3mt, which fully confirms the in silico results. The mammalian IF3mt is 278 amino acid residues long and comprises two domains connected by a linker, as does its bacterial ortholog. According to computer modeling, the N-terminal domain of IF3mt (hereinafter the N-domain) is composed of an α-helical region located opposite a β-layer composed of four chains and a second α-helix joining the linker region [66]. The C-terminal domain (here inafter the C-domain) contains two α-helices (H3 and H4) lying on the β-layer consisting of four chains. One of the chains is divided into two segments by a proline residue. IF3mt has N- and C-terminal “tails” that are 31 and 33 amino acid residues long, respectively. It also has an N-terminal signal peptide that directs import into the mitochondrion and is cleaved to give the mature form upon the protein reaching the matrix. Neither tail segment can be modeled on the basis of currently available structural data; they presumably acquire a certain conformation only upon binding to the small subunit of the mitoribosome. The linker region is partially α-helical, and biochemical studies of the bacterial homolog indicate that the linker is highly flexible [67].
As is the case with the bacterial factor, IF3mt promotes the assembly of the initiator complex on the 28S subunit if the system contains mRNA and IF2mt·GTP. This is achieved through dissociation of active 55S ribosomes to large and small subunits [65]. Unlike its bacterial counterpart, IF3mt does not promote dissociation of noncognate initiation complexes, and the residues critical for this activity are not conserved in the mitochondrial factor [55].
There are functional differences between the N- and C-domains of bacterial and mitochondrial factors. In the case of the bacterial protein, the N-domain serves as an additional anchoring point for binding to the ribosome, while the functional activities reside in the C-domain. The two domains are connected by a linker region. The roles of the individual domains of IF3mt in the interactions of the factor with the ribosome were elucidated through a series of mutational experiments [66]. The C-domain plus linker region combined has the greatest affinity to the small subunit (Kd of 60 nM vs. 30 nM for the full-length protein). The binding of the N-domain to the 28S subunit is characterized by a larger Kd value of 240 nM. Removal of the linker leads to a decrease in affinity (Kd is 90 and 390 nM, in the case of the full-length protein and N-domain, respectively). Therefore, it was proposed that the linker region modulates IF3mt-mediated dissociation of the ribosome into subunits. Replacements of C-terminal domain amino acid residues at positions 170, 171, and 175 to alanine lead to a virtually complete loss of the activity of the factor in dissociation of the mitoribosome and stimulation of the initiation complex formation while having little effect the affinity of the protein to the small subunit. Most likely, these residues play an important role in blocking the intersubunit bridge B2b.
The roles of N- and C-terminal “tails” in the IF3mt functional cycle was analyzed in another series of mutational experiments [68]. Surprisingly, removal of these regions leads to a tenfold increase in the affinity of the factor to the large subunit, with the majority of this effect determined by the N-terminal extension. As mentioned in the previous section, IF3mt has another property, which differs from its bacterial homologs, namely dissociation of fMet-tRNAi from the complex with the 28S subunit in the absence of mRNA. This activity requires the presence of the C-terminal “tail” and the linker region, with the amino acid residues at positions 247 and 248 playing a key role. According to the available three-dimensional model, the C-terminal extension is directed toward the linker region, which allows these sections of the protein to act together.
There are considerable differences between the IF3 and the IF3mt binding sites on the small subunit as determined using chemical cross-linking followed by mass spectrometry [69]. The full-length IF3mt binds to ribosomal proteins MRPS5, MRPS9, MRPS10, and MRPS18, which have corresponding homologs in the 70S ribosome, as well as with a group of mitochondria-specific proteins of the small subunit: MRPS29, MRPS32, MRPS36, and PTCD3. The only ribosomal protein cross-linked to the N-domain of IF3mt is MRPS10, located in the head of 28S subunit, with the rest of the detected proteins being cross-linked to the C-domain and linker.
TRANSLATION ELONGATION IN MITOCHONDRIA
General features
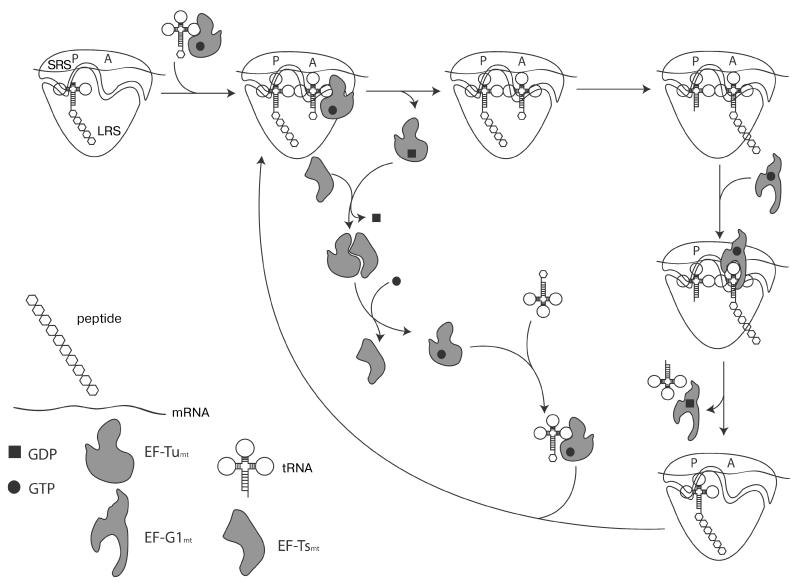
Comparison among all known translation systems shows that the elongation stage of translation has been particularly well conserved during evolution. The mitochondrial elongation factor GTPase EF-Tumt (Elongation Factor Thermo-unstable) forms a ternary complex with GTP and aminoacyl-tRNA and delivers the latter to the A-site of the mitoribosome [70]. Formation of the cognate codon–anticodon interaction promotes GTP hydrolysis by EF-Tu leading to the release of the factor from the ribosome. The guanine nucleotide exchange factor EF-Tsmt (Elongation Factor Thermostable) promotes replacement of GDP with GTP on EF-Tumt. Next, the ribosome catalyzes the transpeptidation reaction. This results in a pre-translocation complex with the P-site occupied by the deacylated tRNA and the A-site harboring the peptidyl-tRNA that has been extended by one amino acid residue. In the final step of elongation, the mitochondrial GTPase EF-G1 (Elongation Factor G 1) catalyzes the movement of mRNA by one codon in the 5′-3′ direction, resulting in deacylated tRNA leaving the ribosome, and the peptidyl-tRNA moving in the P-site, completing the cycle (Fig. 2). The E (Exit)-site that transiently binds the deacylated tRNA dissociating from the ribosome is universal for bacterial and eukaryotic cytosolic ribosomes. However, its existence is still being questioned in the case of the mitochondrial ribosomes [30].
Fig. 2. Schematic representation of translation elongation in mitochondria.
SRS, small ribosomal subunit; LRS, large ribosomal subunit. See text for detailed information.
Mitochondrial translation elongation factors
EF-Tumt is extracted from the mitochondrial fraction of bovine liver in a tight complex with EF-Tsmt (Elongation Factor Thermostable) and remains associated in the presence of GDP concentrations up to 1 mM [71]. This is due to the balance of EF-Tumt affinities to G nucleotides and EF-Tsmt; the Kd for binding of GDP and GTP is 1 and 18 μM, respectively, while binding to EF-Tsmt is consider ably tighter with Kd of 5.5 nM [72]. Both factors are present in mitochondria in vivo in a stoichiometric ratio of 1: 1, with around 400 copies per mitochondrion. This is different from the pattern in bacteria, where the ratio is 8: 1 and ≈200,000 molecules, respectively [73]. A mitochondrial localization signal was determined for both proteins by sequencing the N-terminus in purified preparations of isolated mitochondria [74].
X-Ray crystallography of EF-Tumt·GDP complex shows a three-domain organization scheme typical of bacterial homologs [75]. Domain I consists of eight α-helices and eight β-strands that form the binding pocket for guanyl nucleotides. A flexible linker of 11 amino acid residues joins the G-domain with domain II, which is formed by 11 β-strands folded in a “Greek key” motif. Domain III folds into a “jellyroll” structure, formed by a β-layer of six sheets. A unique feature of mitochondrial EF-Tu is the presence of a C-terminal extension. In humans and Bos taurus, its length is 11, and in C. elegans it is 57 amino acid residues. It is thought that this extension in nematodes is required for the formation of the ternary complex with tRNAs, which typically have unusually short T-stems, and a complete absence of the D-stem in these organisms. However, the function of the extension in mammals is associated with the binding of aminoacyl-tRNA to the small ribosomal subunit [75].
The binding pocket for the acceptor arm of amino-acyl-tRNA is formed at the junction between domains I and II/III of EF-Tumt. Interestingly, the mitochondrial factor is active in a system with bacterial tRNAs, while EF-Tu from E. coli is not able to participate in elongation using mitochondrial tRNAs [76, 77]. The formation of a correct codon–anticodon duplex in the A-site sends a signal through the tRNA molecule to the G-domain of EF Tu that leads to hydrolysis of GTP [78]. Presumably, the shorter mitochondrial tRNA molecules in complex with bacterial EF-Tu are not correctly positioned on the ribosome and cannot effectively stimulate nucleotide hydrolysis.
In addition to participating in translation elongation, EF-Tumt acts as a chaperone in the process of quality control of mitochondrial proteins [79]. EF-Tumt binds to unfolded or misfolded newly synthesized polypeptides and directs them to the protease complex, where they are degraded.
The low degree of sequence similarity of EF-Tsmt with its bacterial homologs (25-35%) has prevented reliable predictions from being made about its three-dimensional structure. Bacterial EF Ts consists of four domains: the N-terminal, a core domain (divided by N- and C-subdomains), a dimerization domain with a “coiled-coil” fold, and a C-terminal module [80]. The X ray structure of the EF-Tumt·EF Tsmt complex revealed dramatic differences from the organization outlined above [81]. The most conserved part of the protein – the N-domain – is assembled from three α-helices similarly to the bacterial homolog. The core domains of the two proteins share the “β-sandwich” fold, although the number and the organization of the β-strands varies. The most dramatic difference between the two proteins is a reduction of the dimerization domain to a large loop folded opposite to the C-subdomain and the complete absence of the C-terminal module in the mitochondrial counter part [81].
Interactions between EF-Tsmt and EF-Tumt·GDP lead to large-scale conformational changes in the latter, which is not the case for the corresponding E. coli and T. thermophilus complexes. GDP to GTP exchange is accompanied by significant conformational rearrangements in EF-Tumt leading to the disruption of the G-nucleotide binding site, which presumably occurs in three steps [82-84]. First, the Mg2+ ion-binding site is disrupted, followed by the destabilization of EF-Tumt interactions with the oxygen atoms of the GDP β-phosphate moiety, and, lastly, contacts with the ribose and purine ring are lost.
The equivalent of bacterial EF-Ts is absent in budding yeast S. cerevisiae, but it is found in the fission yeast Schizosaccharomyces pombe. In this respect at least, the fission yeast mitochondrial system is more similar to that of humans than is budding yeast. EF-Tumt in S. cerevisiae has profoundly different affinities to for GTP and GDP in comparison to that of E. coli EF-Tu (5 and 25 μM vs. 300 and 3 nM, respectively) and, therefore, does not require stabilization by GDP [85]. The functional link between the absence of EF Tsmt and low affinities to G nucleotides of S. cerevisiae EF-Tumt was demonstrated by mutational analysis [86]. Single point mutations in S. pombe EF-Tumt lowering its affinities to G nucleotides also lead to its independence from the EF Tsmt, rendering S. pombe EF-Tumt functionally similar to its S. cerevisiae counterpart.
EF G catalyzes the translocation of peptidyl-tRNA from the A to the P-site of the ribosome. In contrast to bacteria in which a single EF-G acts both during elongation and ribosomal recycling, in most eukaryotes the two functions are split between two mitochondrial factors, EF-G1mt and EF-G2mt [87, 88]. These two factors have amino acid sequence identity of 35% on average [87]. EF-G1mt has translocation activity with both 55S mitoribosomes and 70S bacterial monosomes. Bacterial EF-G, however, is incompatible with mitochondrial ribosomes. Experiments on hybrid ribosomes with heterologous ribosomal proteins L7/L12 demonstrated that the specificity lies in the interactions with these proteins [89].
Despite the fact that EF-G1mt was isolated from bovine liver many years ago, it is still one of the most poorly characterized proteins of the mitochondrial translation apparatus [90]. As with the bacterial protein, EF-G1mt can be structurally divided into five domains, and its shape is strikingly similar to that of the ternary complex EF-Tu·GDPNP·Phe tRNAPhe [91]. Despite the fact that the amino acid residues involved in the interaction of EF-G with the antibiotic fusidic acid are present in the mitochondrial factor, the antibiotic does not significantly inhibit EF-G1mt either in the presence of bacterial or mitochondrial ribosomes [90].
TRANSLATION TERMINATION AND RECYCLING IN MITOCHONDRIA
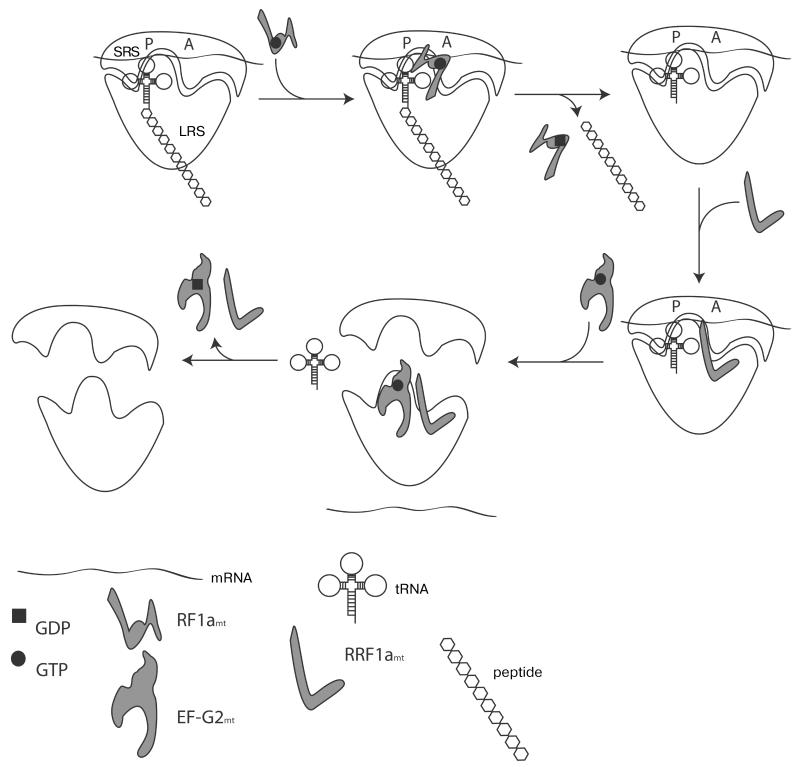
The third and fourth stages of protein synthesis in mitochondria occur when the stop codon of mRNA is present in the ribosomal A-site and is decoded by a termination, or Release Factor, RF. The stop codon is recognized by RF1amt (Release Factor 1), which promotes hydrolysis of the ester bond between P-site tRNA and the newly synthesized polypeptide, releasing the completed protein [92]. The cleavage occurs in the peptidyl transferase center of the large subunit. The mechanism of RF1amt release from its complex with mitoribosomes is still unknown. Next, two factors – RRF1mt (Ribosome Recycling Factor) and EF G2mt in complex with GTP – jointly promote the dissociation of tRNA and mRNA from the ribosome and split the ribosomal particles into subunits (Fig. 3), setting the stage for the reuse of the ribosome in the next round of translation. GTP hydrolysis by EF-G2mt is required for the release of RRF1mt and EF-G2mt from the ribosome [92].
Fig. 3. Schematic representation of translation termination in mitochondria.
SRS, small ribosomal subunit; LRS, large ribosomal subunit. See text for details.
Until recently, it was widely believed that human mitochondria have an extended repertoire of termination codons in comparison with the bacterial system [93]. Two open reading frames are terminated by nonstandard triplets: AGA and AGG in genes MTCOI and MTND6, respectively. However, recent investigations suggest that the 55S mitoribosome undergoes a −1 frameshift, which results in a standard UAG stop codon that is recognized by RF1mt in the A-site [94].
There are two classes of termination factors in bacteria, archaea, and the eukaryotic cytosol: class-I and class-II [95]. The class-I factors are involved in direct recognition of the stop codon and release of the peptide, where as class-II factors facilitate the release of the class-I factors from the ribosome. Class-II factors have not been found in mitochondria. In archaea and in the cytoplasm of eukaryotes all three stop codons are recognized by a single class-I release factor – aRF1 and eRF1, respectively; and in bacteria – by two proteins, RF1 and RF2. The UAA codon is recognized by both bacterial factors; RF1 specifically recognizes UAG, and RF2 recognizes UGA [96, 97]. Bioinformatic analyses have shown that the mitochondrial factors have greatest sequence similarity to the bacterial RF1 [98].
The three-dimensional structure of human RF1mt has been modeled using the available structure of Thermus thermophilus RF1 [92]. It was found that all protein domains and motifs typical for this class are present in the mitochondrial factor: the PXT motif of domain 2 (PKT in the case of the mitochondrial factor) participating in stop codon recognition, the 5 α-helix tip of domain 4, as well as the highly conserved GGQ motif of domain 3 that catalyzes the hydrolysis of an ester bond between the tRNA and the newly synthesized peptide [99]. Moreover, the arrangement of functional sites is compatible with the “open” conformation of the factor that is required for its functional activity on the ribosome [100]. Two additional proteins possessing the RF-specific GGQ motif are present in human mitochondria in addition to RF1mt, namely ICT1 and C12 [101, 102]. ICT1 has ribosome dependent, A-site codon independent peptidyl hydrolase activity. This protein is thought to promote the release of peptides from ribosomes “stuck” on mRNAs lacking a stop codon, thus performing a function analogous to tmRNA in bacteria [103]. Co-immunoprecipitation experiments show that ICT1 is tightly associated with mitoribosomal large subunits, and it is functionally active in a heterologous system with bacterial 70S ribosomes.
As mentioned in the previous chapter, there are two forms of the protein EF G, EF-G1mt and EF-G2mt, in the mitochondria of most eukaryotes. The former appears to be responsible for translocation, whereas the latter is involved in ribosomal recycling. This view is supported by recent in vivo results using an E. coli strain carrying a temperature-sensitive RRF variant. In this strain, EF-G2mt incombination with RRFmt is able to restore normal growth at non-permissive temperatures [103]. Molecular phylogenetic analysis of the EF-G protein family suggests an asymmetric separation of functional activities between the two EF-Gs: EF-G2mt is highly divergent, primarily conserving sites that maintain an EF-G like architecture and ribosome binding capabilities, while EF-G1mt has experienced substantial and well conserved changes in the G-domain that suggest a modified mechanism of interaction with nucleotides [104].
The ribosomal recycling stage in mitochondria differs from the bacterial system in another important aspect: GTP hydrolysis by EF-G2mt appears to occur after mitoribosome dissociation into subunits and is required for dissociation of EF-G2mt and RRFmt from the large subunit, allowing the subunits to associate again in the absence of IF3mt [103].
In this review we have attempted to compile the available data on protein synthesis in the mitochondria of eukaryotic cells. Our current knowledge of protein synthesis in mitochondria is still rather limited in comparison to that of bacterial and cytosolic translation in eukaryotes. Atomic resolution structures of the mitochondrial ribosome are lacking, and our biochemical understanding of the mitochondrial translation lacks kinetic details. Although mitochondrial translation is generally organized similarly to that in bacteria, it is char acterized by a number of significant differences. In recent years numerous scientific reports have elucidated these differences, reigniting the interest of the scientific community.
Acknowledgments
The authors are grateful to the members of P. Kamenski’s, V. Hauryliuk’s, and N. Zenkin’s laboratories for their assistance in writing this review.
Research in the P. Kamenski lab was supported by funding from the International Associated Laboratory “RNA mitocure”, the Russian Foundation for Basic Research, and the Russian Ministry of Education and Science (contract 8817); in the V. Hauryliuk lab by Estonian Science Foundation grants (ETF9012 and PUT37) and European Regional Development Fund (RLOTITIPP), and in the N. Zenkin lab by the UK Biotechnology and Biosciences Research Council and by European Research Council (ERC-2007 StG 202994-MTP). A. Kuzmenko is supported by the Archimedes Foundation and by the “U.M.N.I.K” program. G. C. Atkinson is supported by Estonian Science Foundation grant ETF9020 and European Social Fund grant “Mobilitas” MJD99.
Abbreviations
- LRS
large ribosomal subunit
- SRS
small ribosomal subunit
REFERENCES
- 1.Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR. Proc. Natl. Acad. Sci. USA. 1985;82:4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leon SA, Mahler HR. Arch. Biochem. Biophys. 1968;126:305–319. doi: 10.1016/0003-9861(68)90587-0. [DOI] [PubMed] [Google Scholar]
- 3.Linnane AW, Lamb AJ, Christodoulou C, Lukins HB. Proc. Natl. Acad. Sci. USA. 1968;59:1288–1293. doi: 10.1073/pnas.59.4.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner RP. Science. 1969;163:1026–1031. doi: 10.1126/science.163.3871.1026. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe K. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010;86:11–39. doi: 10.2183/pjab.86.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christian BE, Spremulli LL. Biochim. Biophys. Acta. 2012;1819:1035–1054. doi: 10.1016/j.bbagrm.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melnikov S, BenShem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. Nat. Struct. Mol. Biol. 2012;19:560–567. doi: 10.1038/nsmb.2313. [DOI] [PubMed] [Google Scholar]
- 8.Gray MW. Cold Spring Harb. Perspect. Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson RJ, Williamson DH. Microbiol. Mol. Biol. Rev. 1997;61:1–16. doi: 10.1128/mmbr.61.1.1-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJ, Staden R, Young IG. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 11.Unseld M, Marienfeld JR, Brandt P, Brennicke A. Nat. Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 12.Gray MW, Lang BF, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Brossard N, Delage E, Littlejohn TG, Plante I, Rioux P, SaintLouis D, Zhu Y, Burger G. Nucleic Acids Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown WM, George M, Jr., Wilson AC. Proc. Natl. Acad. Sci. USA. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob JE, Vanholme B, Van Leeuwen T, Gheysen G. BMC Res. Notes. 2009;2:192. doi: 10.1186/1756-0500-2-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Osawa S, Jukes TH, Watanabe K, Muto A. Microbiol. Rev. 1992;56:229–264. doi: 10.1128/mr.56.1.229-264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masters BS, Stohl LL, Clayton DA. Cell. 1987;51:89–99. doi: 10.1016/0092-8674(87)90013-4. [DOI] [PubMed] [Google Scholar]
- 17.Tiranti V, Savoia A, Forti F, D’Apolito MF, Centra M, Rocchi M, Zeviani M. Hum. Mol. Genet. 1997;6:615–625. doi: 10.1093/hmg/6.4.615. [DOI] [PubMed] [Google Scholar]
- 18.Ringel R, Sologub M, Morozov YI, Litonin D, Cramer P, Temiakov D. Nature. 2011;478:269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 19.Ojala D, Montoya J, Attardi G. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 20.Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM. Biochim. Biophys. Acta. 2010;1797:1081–1085. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montoya J, Ojala D, Attardi G. Nature. 1981;290:465–470. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- 22.Groot GS, Flavell RA, Van Ommen GJ, Grivell LA. Nature. 1974;252:167–169. doi: 10.1038/252167a0. [DOI] [PubMed] [Google Scholar]
- 23.Grohmann K, Amairic F, Crews S, Attardi G. Nucleic Acids Res. 1978;5:637–651. doi: 10.1093/nar/5.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunstan HM, GreenWillms NS, Fox TD. Genetics. 1997;147:87–100. doi: 10.1093/genetics/147.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP. J. Biol. Chem. 2003;278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]
- 26.Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, Lightowlers RN, Morse HC, 3rd, Koehler CM, Teitell MA. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suzuki T, Nagao A. Annu. Rev. Genet. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- 28.Helm M, Brule H, Friede D, Giege R, Putz D, Florentz C. RNA. 2000;6:1356–1379. doi: 10.1017/s1355838200001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yokogawa T, Watanabe Y, Kumazawa Y, Ueda T, Hirao I, Miura K, Watanabe K. Nucleic Acids Res. 1991;19:6101–6105. doi: 10.1093/nar/19.22.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 31.Kamenski P, Kolesnikova O, Jubenot V, Entelis N, Krasheninnikov IA, Martin RP, Tarassov I. Mol. Cell. 2007;26:625–637. doi: 10.1016/j.molcel.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 32.Hancock K, Hajduk SL. J. Biol. Chem. 1990;265:19208–19215. [PubMed] [Google Scholar]
- 33.Schneider A. Annu. Rev. Biochem. 2011;80:1033–1053. doi: 10.1146/annurev-biochem-060109-092838. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien TW. J. Biol. Chem. 1971;246:3409–3417. [PubMed] [Google Scholar]
- 35.Maslov DA, Sharma MR, Butler E, Falick AM, Gingery M, Agrawal RK, Spremulli LL, Simpson L. Mol. Biochem. Parasitol. 2006;148:69–78. doi: 10.1016/j.molbiopara.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 36.Borst P, Grivell LA. FEBS Lett. 1971;13:73–88. doi: 10.1016/0014-5793(71)80204-1. [DOI] [PubMed] [Google Scholar]
- 37.Agrawal RK, Sharma MR. Curr. Opin. Struct. Biol. 2012;22:797–803. doi: 10.1016/j.sbi.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smirnov A, Entelis N, Martin RP, Tarassov I. Genes Dev. 2011;25:1289–1305. doi: 10.1101/gad.624711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Entelis NS, Kolesnikova OA, Dogan S, Martin RP, Tarassov IA. J. Biol. Chem. 2001;276:45642–45653. doi: 10.1074/jbc.M103906200. [DOI] [PubMed] [Google Scholar]
- 40.Agrawal RK, Sharma MR, Yassin A, Lahiri I, Spremulli LL. Structure and Function of Organellar Ribosomes as Revealed by Cryo-EM. Springer; Wien-New York: 2011. [Google Scholar]
- 41.Gruschke S, Ott M. Bioessays. 2010;32:1050–1057. doi: 10.1002/bies.201000081. [DOI] [PubMed] [Google Scholar]
- 42.Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK. Proc. Natl. Acad. Sci. USA. 2009;106:9637–9642. doi: 10.1073/pnas.0901631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watson K. J. Cell Biol. 1972;55:721–726. doi: 10.1083/jcb.55.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fiori A, Mason TL, Fox TD. Eukaryot. Cell. 2003;2:651–653. doi: 10.1128/EC.2.3.651-653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM. EMBO J. 2003;22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia L, Dienhart M, Schramp M, McCauley M, Hell K, Stuart RA. EMBO J. 2003;22:6438–6447. doi: 10.1093/emboj/cdg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ott M, Prestele M, Bauerschmitt H, Funes S, Bonnefoy N, Herrmann JM. EMBO J. 2006;25:1603–1610. doi: 10.1038/sj.emboj.7601070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bauerschmitt H, Mick DU, Deckers M, Vollmer C, Funes S, Kehrein K, Ott M, Rehling P, Herrmann JM. Mol. Biol. Cell. 2010;21:1937–1944. doi: 10.1091/mbc.E10-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frazier AE, Taylor RD, Mick DU, Warscheid B, Stoepel N, Meyer HE, Ryan MT, Guiard B, Rehling P. J. Cell Biol. 2006;172:553–564. doi: 10.1083/jcb.200505060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Surovtseva YV, Shutt TE, Cotney J, Cimen H, Chen SY, Koc EC, Shadel GS. Proc. Natl. Acad. Sci. USA. 2011;108:17921–17926. doi: 10.1073/pnas.1108852108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Surovtseva YV, Shadel GS. Nucleic Acids Res. 2013;41:2479–2488. doi: 10.1093/nar/gks1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Atkinson GC, Kuzmenko A, Kamenski P, Vysokikh MY, Lakunina V, Tankov S, Smirnova E, Soosaar A, Tenson T, Hauryliuk V. Nucleic Acids Res. 2012;40:6122–6134. doi: 10.1093/nar/gks272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spremulli LL, Coursey A, Navratil T, Hunter SE. Prog. Nucleic Acid Res. Mol. Biol. 2004;77:211–261. doi: 10.1016/S0079-6603(04)77006-3. [DOI] [PubMed] [Google Scholar]
- 54.Christian BE, Spremulli LL. J. Biol. Chem. 2010;285:28379–28386. doi: 10.1074/jbc.M110.149054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bhargava K, Spremulli LL. Nucleic Acids Res. 2005;33:7011–7018. doi: 10.1093/nar/gki1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee C, Tibbetts AS, Kramer G, Appling DR. J. Biol. Chem. 2009;284:34116–34125. doi: 10.1074/jbc.M109.055350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ellis TP, Helfenbein KG, Tzagoloff A, Dieckmann CL. J. Biol. Chem. 2004;279:15728–15733. doi: 10.1074/jbc.M314162200. [DOI] [PubMed] [Google Scholar]
- 58.Spencer AC, Spremulli LL. Nucleic Acids Res. 2004;32:5464–5470. doi: 10.1093/nar/gkh886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Towpik J. Cell Mol. Biol. Lett. 2005;10:571–594. [PubMed] [Google Scholar]
- 60.Herrmann JM, Woellhaf MW, Bonnefoy N. Biochim. Biophys. Acta. 2013;1833:286–294. doi: 10.1016/j.bbamcr.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 61.Yassin AS, Haque ME, Datta PP, Elmore K, Banavali NK, Spremulli LL, Agrawal RK. Proc. Natl. Acad. Sci. USA. 2011;108:3918–3923. doi: 10.1073/pnas.1017425108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gaur R, Grasso D, Datta PP, Krishna PD, Das G, Spencer A, Agrawal RK, Spremulli L, Varshney U. Mol. Cell. 2008;29:180–190. doi: 10.1016/j.molcel.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laalami S, Sacerdot C, Vachon G, Mortensen K, Sperling-Petersen HU, Cenatiempo Y, Grunberg-Manago M. Biochimie. 1991;73:1557–1566. doi: 10.1016/0300-9084(91)90191-3. [DOI] [PubMed] [Google Scholar]
- 64.Spencer AC, Spremulli LL. Biochim. Biophys. Acta. 2005;1750:69–81. doi: 10.1016/j.bbapap.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 65.Koc EC, Spremulli LL. J. Biol. Chem. 2002;277:35541–35549. doi: 10.1074/jbc.M202498200. [DOI] [PubMed] [Google Scholar]
- 66.Christian BE, Spremulli LL. Biochemistry. 2009;48:3269–3278. doi: 10.1021/bi8023493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hua Y, Raleigh DP. J. Mol. Biol. 1998;278:871–878. doi: 10.1006/jmbi.1998.1736. [DOI] [PubMed] [Google Scholar]
- 68.Haque ME, Grasso D, Spremulli LL. Nucleic Acids Res. 2008;36:589–597. doi: 10.1093/nar/gkm1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haque ME, Koc H, Cimen H, Koc EC, Spremulli LL. Biochim. Biophys. Acta. 2011;1814:1779–1784. doi: 10.1016/j.bbapap.2011.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Smits P, Smeitink J, van den Heuvel L. J. Biomed. Biotechnol. 2010;2010:737385. doi: 10.1155/2010/737385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schwartzbach CJ, Spremulli LL. J. Biol. Chem. 1989;264:19125–19131. [PubMed] [Google Scholar]
- 72.Cai YC, Bullard JM, Thompson NL, Spremulli LL. Protein Sci. 2000;9:1791–1800. [PMC free article] [PubMed] [Google Scholar]
- 73.Woriax VL, Bullard JM, Ma L, Yokogawa T, Spremulli LL. Biochim. Biophys. Acta. 1997;1352:91–101. doi: 10.1016/s0167-4781(97)00002-x. [DOI] [PubMed] [Google Scholar]
- 74.Woriax VL, Burkhart W, Spremulli LL. Biochim. Biophys. Acta. 1995;1264:347–356. doi: 10.1016/0167-4781(95)00176-x. [DOI] [PubMed] [Google Scholar]
- 75.Andersen GR, Thirup S, Spremulli LL, Nyborg J. J. Mol. Biol. 2000;297:421–436. doi: 10.1006/jmbi.2000.3564. [DOI] [PubMed] [Google Scholar]
- 76.Hunter SE, Spremulli LL. Mitochondrion. 2004;4:21–29. doi: 10.1016/j.mito.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 77.Bullard JM, Cai YC, Zhang Y, Spremulli LL. Biochim. Biophys. Acta. 1999;1446:102–114. doi: 10.1016/s0167-4781(99)00077-9. [DOI] [PubMed] [Google Scholar]
- 78.Piepenburg O, Pape T, Pleiss JA, Wintermeyer W, Uhlenbeck OC, Rodnina MV. Biochemistry. 2000;39:1734–1738. doi: 10.1021/bi992331y. [DOI] [PubMed] [Google Scholar]
- 79.Suzuki H, Ueda T, Taguchi H, Takeuchi N. J. Biol. Chem. 2007;282:4076–4084. doi: 10.1074/jbc.M608187200. [DOI] [PubMed] [Google Scholar]
- 80.Kawashima T, Berthet-Colominas C, Wulff M, Cusack S, Leberman R. Nature. 1996;379:511–518. doi: 10.1038/379511a0. [DOI] [PubMed] [Google Scholar]
- 81.Jeppesen MG, Navratil T, Spremulli LL, Nyborg J. J. Biol. Chem. 2005;280:5071–5081. doi: 10.1074/jbc.M411782200. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Li X, Spremulli LL. FEBS Lett. 1996;391:330–332. doi: 10.1016/0014-5793(96)00789-2. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Y, Sun V, Spremulli LL. J. Biol. Chem. 1997;272:21956–21963. doi: 10.1074/jbc.272.35.21956. [DOI] [PubMed] [Google Scholar]
- 84.Zhang Y, Spremulli LL. J. Biol. Chem. 1998;273:28142–28148. doi: 10.1074/jbc.273.43.28142. [DOI] [PubMed] [Google Scholar]
- 85.Rosenthal LP, Bodley JW. J. Biol. Chem. 1987;262:10955–10959. [PubMed] [Google Scholar]
- 86.Chiron S, Suleau A, Bonnefoy N. Genetics. 2005;169:1891–1901. doi: 10.1534/genetics.104.037473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hammarsund M, Wilson W, Corcoran M, Merup M, Einhorn S, Grander D, Sangfelt O. Hum. Genet. 2001;109:542–550. doi: 10.1007/s00439-001-0610-5. [DOI] [PubMed] [Google Scholar]
- 88.Tsuboi M, Morita H, Nozaki Y, Akama K, Ueda T, Ito K, Nierhaus KH, Takeuchi N. Mol. Cell. 2009;35:502–510. doi: 10.1016/j.molcel.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 89.Terasaki M, Suzuki T, Hanada T, Watanabe K. J. Mol. Biol. 2004;336:331–342. doi: 10.1016/j.jmb.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 90.Chung HK, Spremulli LL. J. Biol. Chem. 1990;265:21000–21004. [PubMed] [Google Scholar]
- 91.Al Karadaghi S, Aevarsson A, Garber M, Zheltonosova J, Liljas A. Structure. 1996;4:555–565. doi: 10.1016/s0969-2126(96)00061-5. [DOI] [PubMed] [Google Scholar]
- 92.Chrzanowska-Lightowlers ZM, Pajak A, Lightowlers RN. J. Biol. Chem. 2011;286:34479–34485. doi: 10.1074/jbc.R111.290585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Richter R, Pajak A, Dennerlein S, Rozanska A, Lightowlers RN, Chrzanowska-Lightowlers ZM. Biochem. Soc. Trans. 2010;38:1523–1526. doi: 10.1042/BST0381523. [DOI] [PubMed] [Google Scholar]
- 94.Temperley R, Richter R, Dennerlein S, Lightowlers RN, Chrzanowska-Lightowlers ZM. Science. 2010;327:301. doi: 10.1126/science.1180674. [DOI] [PubMed] [Google Scholar]
- 95.Loh PG, Song H. Curr. Opin. Struct. Biol. 2010;20:98–103. doi: 10.1016/j.sbi.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 96.Dontsova M, Frolova L, Vassilieva J, Piendl W, Kisselev L, Garber M. FEBS Lett. 2000;472:213–216. doi: 10.1016/s0014-5793(00)01466-6. [DOI] [PubMed] [Google Scholar]
- 97.Ito K, Frolova L, Seit Nebi A, Karamyshev A, Kisselev L, Nakamura Y. Proc. Natl. Acad. Sci. USA. 2002;99:8494–8499. doi: 10.1073/pnas.142690099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Soleimanpour-Lichaei HR, Kuhl I, Gaisne M, Passos JF, Wydro M, Rorbach J, Temperley R, Bonnefoy N, Tate W, Lightowlers R, Chrzanowska-Lightowlers Z. Mol. Cell. 2007;27:745–757. doi: 10.1016/j.molcel.2007.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trobro S, Aqvist J. Biochemistry. 2009;48:11296–11303. doi: 10.1021/bi9017297. [DOI] [PubMed] [Google Scholar]
- 100.Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF. Nature. 2008;454:852–857. doi: 10.1038/nature07115. [DOI] [PubMed] [Google Scholar]
- 101.Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, Smeitink JA, Lightowlers RN, Chrzanowska-Lightowlers ZM. EMBO J. 2010;29:1116–1125. doi: 10.1038/emboj.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Antonicka H, Ostergaard E, Sasarman F, Weraarpachai W, Wibrand F, Pedersen AM, Rodenburg RJ, van der Knaap MS, Smeitink JA, Chrzanowska-Lightowlers ZM, Shoubridge EA. Am. J. Hum. Genet. 2010;87:115–122. doi: 10.1016/j.ajhg.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Haque ME, Spremulli LL. EMBO J. 2010;29:1019–1020. doi: 10.1038/emboj.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Atkinson GC, Baldauf SL. Mol. Biol. Evol. 2011;28:1281–1292. doi: 10.1093/molbev/msq316. [DOI] [PubMed] [Google Scholar]