Abstract
Although peroxisomes are ubiquitous organelles in all animal species, their importance for the functioning of tissues and organs remains largely unresolved. Because peroxins are essential for the biogenesis of peroxisomes, an obvious approach to investigate their physiological role is to inactivate a Pex gene or to suppress its translation. This has been performed in mice but also in more primitive organisms including D. melanogaster, C. elegans, and D. rerio, and the major findings and abnormalities in these models will be highlighted. Although peroxisomes are generally not essential for embryonic development and organogenesis, a generalized inactivity of peroxisomes affects lifespan and posthatching/postnatal growth, proving that peroxisomal metabolism is necessary for the normal maturation of these organisms. Strikingly, despite the wide variety of model organisms, corresponding tissues are affected including the central nervous system and the testis. By inactivating peroxisomes in a cell type selective way in the brain of mice, it was also demonstrated that peroxisomes are necessary to prevent neurodegeneration. As these peroxisome deficient model organisms recapitulate pathologies of patients affected with peroxisomal diseases, their further analysis will contribute to the elucidation of still elusive pathogenic mechanisms.
Keywords: inflammation, male fertility, phytanic acid, plasmalogens, PUFA, very long chain fatty acids, Zellweger syndrome
Introduction
Absence of peroxisomes in man leads to a devastating disease, clinically known as the hepato-renal syndrome of Zellweger. Affected baby's are born alive, but are severely hypotonic, mentally retarded with brain malformation, liver and kidney problems, and die generally with the first weeks of life (Wanders and Waterham, 2005). Understanding the anomalies at the cellular and organ level and the malformation during development in such patients with a peroxisome biogenesis disorder, requires access to suitable experimental material. Unfortunately, for man the sources are rather limited (fibroblasts, lymphoblasts, amniotic villi), and not representative for specialized cells/tissues. In addition, no natural occurring or inducible animal model is known. Hence, as soon as appropriate molecular techniques were established, animal models were created, starting of with PEX51 (Baes et al., 1997) and PEX2 (Faust and Hatten, 1997) deficient mice in 1997, followed later by inactivation of peroxins in other laboratory “pet-animals” like worms, fruitfly, or zebrafish.
In addition to these animal models, peroxisome deficient mutants were created in different yeasts, starting of with baker's yeast (Erdmann et al., 1989), followed by Hansenula polymorpha (Cregg et al., 1990) and Pichia pastoris (Gould et al., 1992); in filamentous fungi, Neurospora crassa (Sichting et al., 2003; Managadze et al., 2007), Magnaporthe grisea (Ramos-Pamplona and Naqvi, 2006), Aspergillus oryzae (Escano et al., 2009), in plants like Arabidopsis (Kaplan et al., 2001; Schumann et al., 2003; Fan et al., 2005), in trypanosomes (Banerjee et al., 2005; Galland et al., 2007). Some of the latter models are described elsewhere in this book, whereas for a treatise on human disorders linked to peroxisomes we refer to (Wanders and Waterham, 2005; Waterham and Ebberink, 2012).
Before discussing in more detail the different animal models, a general description of the metabolic functions of peroxisomes is given, followed by a short note about their biogenesis.
Peroxisomal metabolism
From a human pathological point of view, the main peroxisomal pathways are β-oxidation, α-oxidation, and ether lipid synthesis, and to a lesser extent glyoxylate metabolism and xanthine metabolism. Whereas peroxisomal β-oxidation seems universally present in all animals, although sometimes serving other purposes, some of the other pathways might be missing in lower vertebrates/invertebrates (e.g., etherlipid synthesis). In the following paragraphs the main pathways are briefly described, whereas their specific roles, if known, will be highlighted when discussing the different models (enzymes are named according to the mouse nomenclature).
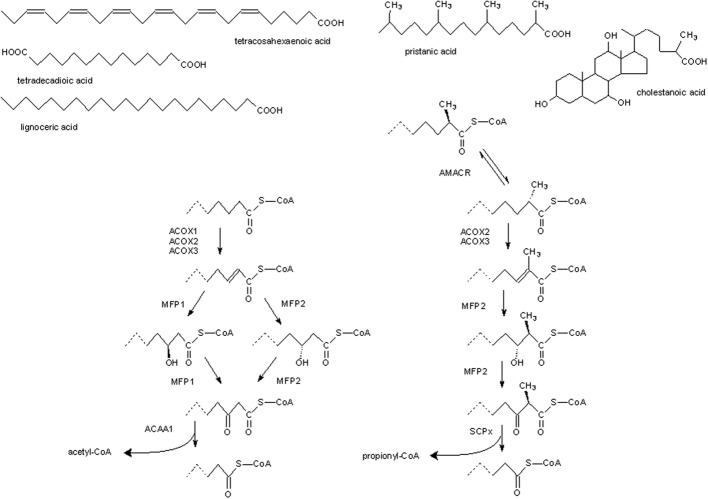
Typically, peroxisomes can β-oxidize a broad range of natural, often also xenobiotic, compounds containing a fatty acyl side chain with or without a methyl-branch, in α-position of the carboxy-group. This process consists of a sequence of four reactions, resulting in shortening of the main chain of an acyl-CoA by 2 carbons (see Figure 1) (Van Veldhoven, 2010). In a first step, acyl-CoA is converted into 2-trans-enoyl-CoA by an acyl-CoA oxidase (ACOX), thereby producing H2O2. The number of ACOXs varies between species and ACOXs acting on 2-methyl-acyl-CoAs (ACOX2 and ACOX3 in mammals) are stereospecific, only the 2S-isoform is desaturated, hence an additional peroxisomal enzyme, 2-methylacyl-CoA racemase (AMACR), is required to convert the 2R-isoforms. The oxidation is followed by a hydration of the double bond by a 2-enoyl-CoA hydratase, a dehydrogenation by 3-hydroxyacyl-CoA dehydrogenase, and finally a thiolytic cleavage, generating acetyl-CoA (or propionyl-CoA in case of 2-methylbranched acyl-CoA) and a shortened acyl-CoA. Generally more than one enzyme can catalyze each of these steps, either homologous proteins as is the case for ACOXs or totally different proteins, e.g., thiolases encoded by the Acaa1 or Scp2 genes, or activities can reside in multi-enzymes (e.g., EHHAHD, also called multifunctional protein 1 (MFP1), HSD17B4, often called MFP2), which catalyze the hydration and dehydrogenation steps in a stereoselective manner. In mammals, a well-characterized β-oxidation pathway is the formation of C24-bile acids, starting from C27-bile acids (cholestanoic acids). In lower vertebrates, such as reptiles, some amphibia, and lungfishes, however, no C24-bile acids are found (Hofmann et al., 2010). On the other hand, the genomes of amphibia, bony fishes and various invertebrates like insects, bivalves, and sea urchins (but not nematodes), encode a peroxisomal AMACR, suggestive for a role of peroxisomes in breakdown of other isoprenoid derived carboxylates in these species.
Figure 1.
Generalized scheme of peroxisomal β-oxidation in animals. On top, structures of some fatty carboxylates that, after activation (not shown), are degraded by peroxisomal β-oxidation. At the right, enzymatic reactions/enzymes involved in degradation of substrates containing a 2-methylbranch, based on the situation in mammals. Most of these enzymes can act on straight chain substrates, shown at the left, as well. The latter compounds are also recognized by more selective enzymes which do not tolerate a 2-methylbranch. ACAA1, 3-ketoacyl-CoA thiolase; ACOX, acyl-CoA oxidase; AMACR, 2-methylacyl-CoA racemase; MFP, multifunctional protein; SCPx, sterol carrier protein X-thiolase.
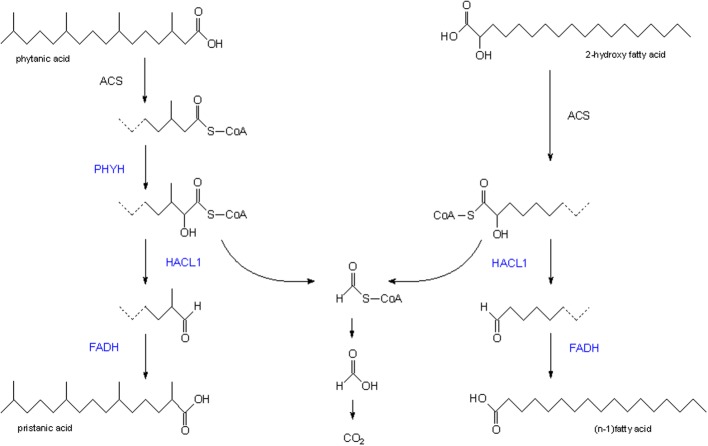
α-Oxidation is a process whereby fatty acids are shortened by one carbon atom, amply documented for phytanic acid in man, a diet derived 3-methylbranched fatty acid, and less well-known for long chain 2-hydroxy fatty acids (Van Veldhoven, 2010) (see Figure 2). For phytanic acid, the process starts with the hydroxylation of phytanoyl-CoA at position 2 (by phytanoyl-CoA hydroxylase, PHYH), followed by a cleavage into formyl-CoA and pristanal, catalyzed by 2-hydroxyacyl-CoA lyase (HACL1). 2-Hydroxy long chain fatty acids do not depend on PHYH and are, after activation, shortened into a (n-1)fatty aldehyde by HACL1. This pathway is present in all mammals, and representative species of birds, reptiles, amphibian, fish, insects, nematodes, echinoderms, cnidaria, ascidia.
Figure 2.
Peroxisomal α-oxidation. Scheme of the enzymatic reactions/enzymes involved in the peroxisomal breakdown of phytanic acid (left) and 2-hydroxy long chain fatty acids (right). Enzymes in blue are associated with peroxisomes. ACS, acyl-CoA synthetase; FADH, fatty aldehyde dehydrogenase; HACL1, 2-hydroxyacyl-CoA lyase; PHYH, phytanoyl-CoA α-hydroxylase.
In contrast to the bulk of glycerolipids containing ester-linked fatty acids, a small portion of glycerolipids contains an ether bond, the precursor of which is formed by peroxisomal enzymes (see Figure 3). A first one, dihydroxyacetone-phosphate acyltransferase (GNPAT) generates an obligate precursor, 1-acyl-dihydroxyacetone-phosphate, a second one catalyzes the exchange of the acyl for an alcohol (alkyl dihydroxyacetone-phosphate synthase, ADHAPS). After reduction, the generated 1-alkylglycerol-3-phosphate follows the same anabolic routes as 1-acylglycerol-3-phosphate in the ER, leading to neutral and phosphoglycerolipids with a 1-alkyl group. In mammals, 1-alkyl-2-acylglycerophosphoethanolamine is desaturated just adjacent to the ether linkage, generating plasmenylethanolamine which can be converted to the choline analogue. Phospholipids with this vinylether group are better known as plasmalogens. Based on genomic information, the key enzymes GNPAT and ADHAPS are expressed in nematodes, cnidaria, echinoderms, insects, fish, amphibia, reptiles and birds. The presence of plasmalogens will however, depend on the expression of plasmanylethanolamine desaturase, an orphan enzyme not yet cloned. Besides mammals, plasmalogens have been identified in various animals including birds, amphibia, fish, insects, molluscs, marine worms, jelly fish, echinodermata, slime mold (Horrocks and Sharma, 1982).
Figure 3.
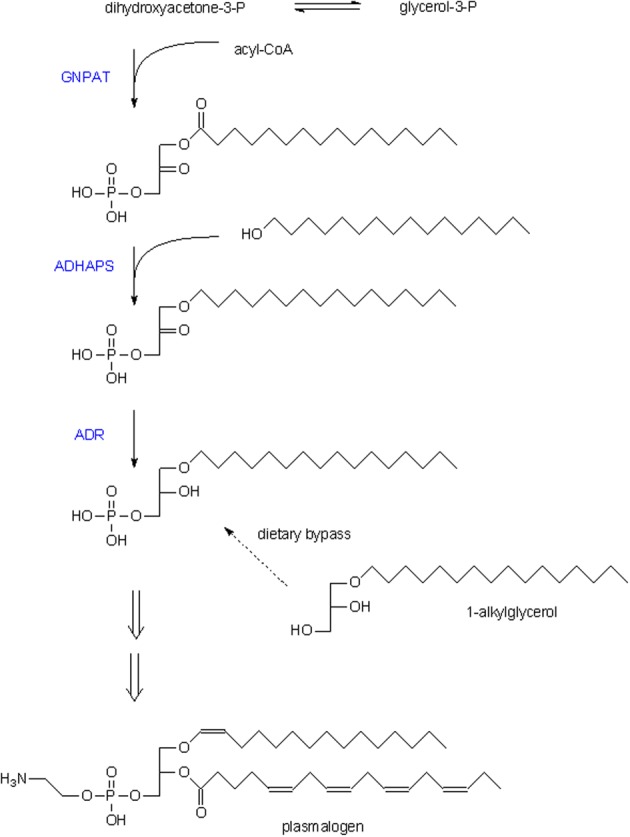
Etherlipid biosynthesis. Scheme of the reactions involved in formation of ether lipids, starting with dihydroxyacetone-phosphate. The long chain alcohol used by ADHAPS it generally 16–18 carbons, either saturated or with one double bond. Double arrows indicate multiple steps, catalyzed by ER-associated enzymes like headgroup addition and the introduction of the double bond in the 1-alkylchain. Generally, plasmalogens are enriched in PUFA at position 2 of the glycerol moiety (shown is 1-hexadecyl-2-arachidonoyl-plasmenylethanolamine). Dietary 1-alkylglycerol can bypass the peroxisomal steps in this pathway (dashed arrow). GNPAT, dihydroxyacetone-phosphate acyltransferase; ADHAPS, alkyl dihydroxyacetone-phosphate synthase; ADR, acyl/alkyl-dihydroxyacetone-phosphate reductase.
Depending on species, peroxisomes or related organelles (glyoxysomes) are more or less actively involved in glyoxylate metabolism and in the degradation of purines (purine oxidation pathway or ureide pathway). Depending on the phylogenetic position of the species, purines are degraded till the level of ureum (amphibian, fish) or only till uric acid (man).
Peroxisome biogenesis
Proteins involved in the formation (biogenesis) of peroxisomes were first identified in yeast (Erdmann et al., 1989), and the major players in this process are rather well-conserved throughout the different kingdoms. In yeast and lower eukaryotes, however, more peroxins are found that are involved in fission/fusion processes and regulation of the number of peroxisomes2, which is related to the fact that these species must be able to adapt their intracellular organelles quickly to changes in their environment. Briefly for animals, peroxisomal matrix proteins, synthesized in the cytosol, are captured by binding partners that recognize a specific motif within their primary sequence, either a C-terminal tripeptide, better known as Peroxisome Targeting Signal 1 (PTS1) which is recognized by PEX5, or an N-terminal nonapeptide (PTS2), bound by PEX7 (see Figure 4). Upstream residues of PTS1, often referred to as SKL-sequence, influence binding to Pex5, hence a more in depth analysis of the interaction has broadened PTS1 to a dodecamer (Brocard and Hartig, 2006). In all species investigated, only a minority of matrix proteins contain PTS2, and in certain species, such as nematodes (C. elegans; Motley et al., 2000), diatoms (Phaeodactylum tricornutum; Gonzalez et al., 2011), and insects (Drosophila; Faust et al., 2012), PEX7 is even missing. In those organisms, the classical PTS2 proteins are still associated with peroxisomes, but are decorated with PTS1 (de Vet et al., 1998; Motley et al., 2000; Faust et al., 2012).
Figure 4.
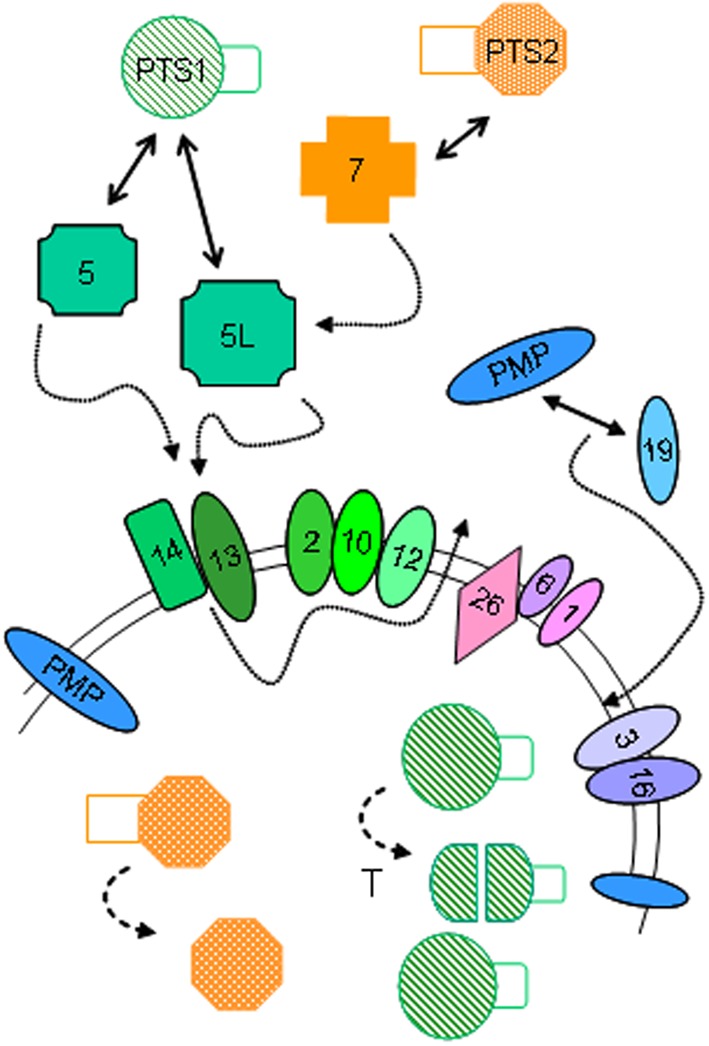
Peroxisome biogenesis in animals. Simplified scheme for the import of PTS1-proteins, PTS2-proteins and integral peroxisomal membrane proteins (PMP) in animals and the involved peroxins, indicated by their PEX number. Role of ubiquitination of PEX5 and farnesylation of PEX19 is not shown. After import, some PTS1-proteins are processed by TYSND1, a peroxisomal protease (T), while the signal peptide of PTS2-proteins is cleaved off. Adapted from Van Veldhoven (2010).
PEX5-PTS1 cargoes dock to the peroxisomal membrane via PEX14/PEX13, and are subsequently translocated through the bilayer. At the matrical side of the membrane, the cargo is released and PEX5 recycles to the membrane where it will undergo ubiquitination mediated by the RING-finger proteins PEX2, PEX10, and PEX12, and extracted back to the cytosol via PEX1/PEX6 in an ATP-dependent manner.
PEX7-PTS2 cargo also binds to PEX14, not directly but mediated via a long isoform of PEX5 (PEX5L) in mammals and other vertebrates (Figure 4). In fungi, the function of the latter is taken over by PEX18/PEX21 (Dodt et al., 2001).
Membrane biogenesis is depending on PEX19, PEX3, and (in animals) PEX16 (Fujiki et al., 2006). PEX19, a mainly cytosolic protein, plays a chaperone like function and binds most newly synthesized integral peroxisomal membrane proteins (PMP), and docks to PEX3, an integral peroxisomal membrane protein.
Finally, size and abundance of peroxisomes are regulated by PEX11 proteins, which play also a role in elongation of the organelles (Thoms and Erdmann, 2005; Koch et al., 2010). In mammals three isoforms are known, in lower animals only one (Table 1).
Table 1.
Overview of peroxins in animals used for peroxisomal research and in man.
| Peroxin2 | Description | Domain | Caenorhabditis elegans1 | Drosophila melanogaster | Danio rerio | Mus musculus | Homo sapiens | |
|---|---|---|---|---|---|---|---|---|
| PEX1 | AAA-ATPase | gi | 25153574 | 21355121 | 283046720 | 61657895 | 4505725 | |
| NP | NP_510386.2 | NP_652016.1I | NP_001164306.1 | NP_082053.1 | NP_000457.1 | |||
| alias | prx-1 (isoform a); C11H1.4a | FBgn0013563 | 793906; ZDB-GENE-070530-1 | ZWS1; 5430414H02Rik; E330005K07Rik | PBD1A; PBD1B; ZWS; ZWS1 | |||
| aa (MW) | 996 (111997) | 1006 (113739) | 1237 (136665) | 1244 (136613) | 1283 (142737) | |||
| PEX2 | E3 ligase | Zinc RING finger | gi | 133931002 | 21355975 | 189536742 | 254028168 | 4506343 |
| NP | NP_502201.2 | NP_648210.1 | XP_684073.2 | NP_001156773.1 | NP_000309.1 | |||
| alias | prx-2; ZK809.7 | CG7081; DmelPex2; Dmel; CG7081 | ZDB-GENE-070530-2 | D3Ertd138e; PAF-1; PMP35; Pxmp3 | PAF1; PBD5A; PBD5B; PMP3; PMP35; PXMP3; RNF72; ZWS3 | |||
| aa (MW) | 273 (31194) | 281 (32239) | 312 (35000) | 305 (34813) | 305 (34765) | |||
| PEX3 | PEX9 docking factor | gi | 193209553 | 21357431 | 41055494 | 9910484 | 4505727 | |
| NP | NP_001123111.1 | NP_648753.1 | NP_956522.1 | NP_064345.1 | NP_003621 | |||
| alias | prx-3; C15H9.8 | CG6859; DmelPex3; Dmel\CG6859; FBgn0036484 | zgc:56313; fd60g05.y1; ZDB-GENE-040426-979 | 1700014F15Rik; 2810027F19Rik | PBD10A; TRG18 | |||
| aa (MW) | 353 (39754) | 385 (43562) | 364 (41427) | 372 (42093) | 373 (42009) | |||
| PEX5 | PTS1-receptor | TPR | gi | 71983707 | 24639189 | 41055947 | 472339081 | 196259772 |
| NP | NP_001022019.1 | NP_569949.3 | NP_957450.1 | NP_001264259.1 | NP_001124496.1 | |||
| alias | prx-5; C34C6.6 | CG14815; DmelPex5; Dmel\CG14815; EG:63B12.5 | PXR1; zgc:56318; ZDB-GENE-040426-981 | AW212715; ESTM1; PTS1R; Pxr1; X83306 | PBD2A; PBD2B; PTS1-BP; PTS1R; PXR1 | |||
| aa (MW) | 502 (55344) | 559 (62994) | 600 (67012) | 602 (66675) | 602 (66699) | |||
| PEX5L | PTS1-receptor | TPR | gi | absent | absent | absent | 113930737 | 196259774 |
| NP | NP_033021.2 | NP_001124497.1 | ||||||
| alias | AW212715; ESTM1; PTS1R; Pxr1; X83306 | PBD2A; PBD2B; PTS1-BP; PTS1R; PXR1 | ||||||
| aa (MW) | 639 (70625) | (639) 70734 | ||||||
| PEX6 | AAA-ATPase | gi | 17562804 | 78707192 | 326678870 | 21703962 | 194018488 | |
| NP | NP_504268.1 | NP_001027403.1 | XP_001332652.4 | NP_663463.1 | NP_000278.3 | |||
| alias | prx-6; CELE_F39G3.7 | CG11919; CG30019; DmCG11919; DmelPex6; Dmel\CG11919 | ZDB-GENE-081104-252 | AI132582; mKIAA4177; PAF-2; D130055I09Rik; peroxisomal-type ATPase 1 | PAF-2; PAF2; PBD4A; PDB4B; PXAAA1 | |||
| aa (MW) | 720 (81223) | 897 (100990) | 865 (94312) | 981 (104418) | 980 (103930) | |||
| PEX7 | PTS2-receptor | WD40 | gi | absent | 24661084* | 61806636 | 6679283 | 4505731 |
| NP | NP_648251.1 | NP_001013550.1 | NP_032848.1 | NP_000279.1 | ||||
| alias | CG6486; DmelPex7; Dmel\CG6486 | zgc:103552; ZDB-GENE-050320-105 | MmPEX7 | PBD9B; PTS2R; RCDP1; RD | ||||
| aa (MW) | 339 (37486) | 314 (34818) | 318 (35371) | 323 (35761) | ||||
| PEX10 | Zinc RING-finger | gi | 392894943 | 54400490 | 109150414 | 24797089 | ||
| NP | NP_001021200.2 | NP_001005994.1 | NP_001035866.1 | NP_722540.1 | ||||
| alias | C34E10.4a3 | zgc:103520 | AV128229; Gm142 | NALD; PBD6A; PBD6B; RNF69 | ||||
| aa (MW) | 314 (35871) | 318 (37181) | 346 (39083) | |||||
| PEX11A | Peroxisome elongation Peroxisome constriction | gi | 17506083 | ?? | 156739285 | 6755034 | 4505717 | |
| NP | NP_493273.1 | NP_001096590.1 | NP_035198.1 | NP_003838.1 | ||||
| alias | prx-11; CELE_C47B2.8 | si:dkeyp-84g1.1; 565760; ZDB-GENE-050419-121 | PEX11alpha | hsPEX11p; PEX11-ALPHA; PMP28 | ||||
| aa (MW) | 214 (23780) | 246 (27867) | 246 (28022) | 247 (28222) | ||||
| PEX11B | Peroxisome elongation Peroxisome constriction | gi | ?? | 19922346 | 113951761 | 241666483 | 4505719 | |
| NP | NP_611071.1 | NP_001039319.1 | NP_001155859.1 | NP_003837.1 | ||||
| alias | CG8315; DmelPex11; Dmel\CG8315 | zgc:153402; 566742; ZDB-GENE-060825-289 | PEX11beta; Pex11pbeta | PEX11-BETA; PEX14B | ||||
| aa (MW) | 241 (27007) | 266 (29496) | 259 (28579) | 259 (28300) | ||||
| PEX11G | Peroxisome elongation Peroxisome constriction | gi | ?? | 285718374 | 71834488 | 21735445 | 18087833 | |
| NP | NP_651137.3 | NP_001025343.1 | NP_081227.1 | NP_542393.1 | ||||
| alias | BcDNA:RE30473; Dmel\CG13827 | 63203; ZDB-GENE-050913-79 | 1810022F11Rik; 1810049N02Rik; Pex11g; Pex11gamma | |||||
| aa (MW) | 233 (26208) | 240 (26210) | 241 (27021) | 241 (26505) | ||||
| PEX12 | RING Zinc finger, C3HC4 type | gi | 17551466 | 24580706 | 41055606 | 19527244 | 4505721 | |
| NP | NP_509908.1 | NP_608546.1 | NP_956499.1 | NP_598786.1 | NP_000277.1 | |||
| alias | prx-12; F08B12.2 | CG3639; DmelPex12; Dmel\CG3639 | zgc:56182; 393174; ZDB-GENE-040426-929 | AI451906 | PAF-3; PBD3A | |||
| aa (MW) | 359 (41158) | 297 (34413) | 303 (33979) | 359 (40502) | 359 (40666) | |||
| PEX13 | Docking PTS-cargo complex | SH3 | gi | 17533615 | 20129941 | 41055287 | 31543471 | 4505723 |
| NP | NP_495513.1 | NP_610850.1 | NP_956939.1 | NP_076140.2 | NP_002609.1 | |||
| alias | prx-13; F32A5.6 | CG4663; Dmel\Pex13; Dmel\CG4663; FBgn0033812 | zgc:66124; ZDB-GENE-040426-1544 | 2610008O20Rik | NALD; PBD11A; PBD11B; ZWS | |||
| aa (MW) | 330 (35635) | 440 (46529) | 416 (45338) | 405 (44479) | 403 (43999) | |||
| PEX14 | Docking PTS-cargo complex | gi | 17541806 | 21355205 | 292627105 | 9790153 | 4758896 | |
| NP | NP_502097.1 | NP_649253.1 | XP_688421.4 | NP_062755.1 | NP_004556.1 | |||
| alias | prx-14; R07H5.1 | CG4289; DmelPex14; Dmel\CG4289 | 559933; ZDB-GENE-060130-169 | R75137 | dJ734G22.2; NAPP2; PBD13A; Pex14p | |||
| aa (MW) | 258 (28025) | 280 (30673) | 422 (46234) | 376 (41077) | 377 (41106) | |||
| PEX16 | gi | Absent | 21355481 | 68448487 | 254750742 | 254750742 | ||
| NP | NP_649252.1 | NP_001020340.1 | NP_660104.2 | NP_660104.2 | ||||
| alias | CG3947; DmelPex16; Dmel\CG3947 | im:6894523; zgc:112248 | PBD8A; PBD8B | |||||
| aa (MW) | 341 (39228) | 335 (38424) | 336 (38546) | 336 (38546) (splice form; only on EST!) | ||||
| PEX19 | Cytosolic chaperone; PMP import receptor | CAAX-box | gi | 17553610 | 24583827 | 62899043 | 226958490 | 4506339 |
| NP | NP_498947.1 | NP_609547.2 | NP_001017399.1 | NP_075528.3 | NP_002848.1 | |||
| alias | F54F2.8 | BEST:GH03076; CG5325; DmelPex19; Dmel\CG5325 | wu:fb40d12; wu:fc41h09; zgc:110675 | Pxf | D1S2223E; HK33; PBD12A; PMP1; PMPI; PXF; PXMP1 | |||
| aa (MW) | 282 (30857) | 292 (31175) | 288 (31412) | 299 (32602) | 299 (32676) | |||
| PEX20 | Cytosolic chaperone | gi | ?? | 386768875 | Absent | Absent | Absent | |
| NP alias | NP_001245818.15 | |||||||
| CG3696; DmelPex20; Dmel\CG3696; EK2-4; kis; Su(Pc)21AB | ||||||||
| aa (MW) | 5343 (575803) | |||||||
| PEX23 | Peroxisome proliferation; peroxisomal growth regulation | gi | Absent | 24667330 | Absent | Absent | Absent | |
| NP | NP_730508.16 | |||||||
| alias | CG18565; CG32226; CG6468; DmelPex23; Dmel\CG32226 | |||||||
| aa (MW) | 1350 (149356) | |||||||
| PEX26 | Anchor for PEX1 and PEX6 to peroxisome membrane | gi | ?? | ?? | 41053983 | 21311973 | 189083737 | |
| NP | ?? | ?? | NP_956214.1 | NP_083006.1 | NP_001121121.1 | |||
| alias | fk41g06; wu:fk41g06; zgc:64014 | 4632428M11Rik; AI853212 | PBD7A; PBD7B; PEX26M1T; Pex26pM1T; FLJ20695 | |||||
| aa (MW) | 313 (34257) | 305 (33885) | 305 (33767) |
It should be noted that in the Worm database, prx-number has been proposed as acronym for peroxisome assembly factors given confusion with pex (pachytene exit defect). However, various entries related to prx are linked to both PeroxidoRedoXins and PeRoXisome assembly factors, given use of similar acronym.
Peroxins, not present in animals, include PEX4, PEX8, PEX15, PEX17, PEX18, PEX21, PEX22, PEX25, PEX27, PEX28, PEX29, PEX30, PEX31, PEX32, PEX34 (all present in yeasts), PEX9, PEX20, PEX23 (Yarrowia sp.), PEX24 (yeasts, plants), PEX33 (Neurospora sp.).
The C34E10.4 locus produces a primary transcript coding for two different proteins, PEX10 (at the 5′; C34E10.4a) or WARS-2 (at the 3′; C34E10.4b).
In addition to this entry, another PEX11 like protein (201 amino acids, MW 22671) is encoded by the fly genome. It concerns NP_995800.1 (gi 45552555), also named CG33474; Dmel\CG33474, which is most similar to PEX11G.
One of the six different isoforms encoded by CG3696, nowadays referred to as kismet; homologous to the human CHD7 (chromodomain-helicase-DNA-binding protein 7).
Whether NP-730508.1 represents the counterpart of Yarrowia PEX23 or yeast PEX31 is questionable; they all contain a Dysferlin domain, but likely this entry is the counterpart of TECP1 (tectonin beta-propeller repeat-containing protein 1).
not present in database, likely absent.
homologue, but functionality not proven.
Models
In the following sections, laboratory animals in which peroxisome biogenesis has been studied will be described. In Table 1, gene symbols and alternative names for peroxins in these animals are listed. Given differences in life cycle and organogenesis, the development of these animals will be shortly described, and specific metabolic roles of peroxisomes, if documented, will be highlighted.
Nematodes
A fertilized Caenorhabditis elegans egg develops into a small worm within the shell, in about 10–12 h. In the preceding 6 h (organogenesis/morphogenesis stage), the spheroid embryo started to elongate while its three germ layers differentiate into organs. After hatching, the post-embryonic development will start and the animal will pass, if food is present, through four larval stages (L1–L4, separated by 7–9 h) to reach sexual maturation, generally as a hermaphrodite, about 1.2 mm long, and will start to produce eggs. The cycle from egg to egg is therefore about 3 days; life span of the worm is 2 weeks.
In the adult nematode, peroxisomes are mainly present in the epithelial cells of the digestive tract, one of the largest organs, and in the pharyngeal gland (Yokota et al., 2002). In the gut, their volume density is 1.86/100 μm2 cytoplasm, similar to that in rat liver. Similar to rodents, fibrates increase the number of peroxisomes (Yokota et al., 2002). Based on the fluorescence pattern of animals expressing CFP-SKL, larvae contain more and larger peroxisomes than adult worms (Petriv et al., 2002).
In C. elegans, peroxisomal β-oxidation serves to generate acyl-CoAs used for the synthesis of dauer pheromone, also called daumone, a mixture of ascarosides which are excreted when the larvae are exposed to a hostile environment to block further development. Ascarosides are glycolipids and, in the case of dauer pheromone, consist of a hydroxylated medium chain fatty acid such as 6-hydroxyheptanoic acid or 8-hydroxy-2-nonenoic acid, O-glycosidically linked to ascarylose (3,6-didesoxymannose). Particularly daf-22 and dhs-28 (Butcher et al., 2009; Joo et al., 2009), the nematode counterparts of SCPX and the dehydrogenase moiety3 of D-specific MFP2, respectively, and acox-1 (Joo et al., 2010), one the seven nematodal ACOX proteins, are required for dauer pheromone production.
Regarding peroxisome biogenesis, it should be mentioned that the genome of C. elegans (and other nematodes) does not encode PEX7 (Motley et al., 2000). According to Thieringer et al. (2003) PEX16 is also missing and only one PEX11 isoform is present (Table 1).
During various large screenings by RNAi soaking, feeding or injection experiments, different peroxins were hit, however, the phenotype of the offspring was only minimally scored and the efficacy of silencing not investigated (see Table 2 and associated references). Moreover, these screens display some variability between approaches, are known to give rise to false negatives, and silencing is less effective in the nervous system. Efficacy can be increased by performing screens in the rrf-3 mutant, a strain being hypersensitive to RNAi, likely due to longer half life of RNAi (Simmer et al., 2003). Overall these screens, certainly those by Simmer et al. (2003) and Sonnichsen et al. (2005), indicate that normal larval development depends on functional peroxisomes (Table 2).
Table 2.
Overview of large scale silencing screens in C. elegans affecting peroxins.
| Pex1 | Pex2 | Pex3 | Pex5 | Pex6 | Pex10 | Pex11 | Pex12 | Pex13 | Pex14 | Pex19 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| References | Method | |||||||||||
| Gonczy et al., 2000 | Injection in gonads of dsRNA targeting genes of chromosome III | Slow growth | Larval arrest | |||||||||
| Maeda et al., 2001 | dsRNA soaking | Sick | ||||||||||
| Kamath et al., 2003 | Feeding E. coli expressing dsRNA | N | Slow growth | N | Slow growth; clear | N | Slow growth | Slow growth; clear | Slow growth | Slow growth | Slow growth | |
| Simmer et al., 2003 | Feeding E. coli expressing dsRNA in rrf-3 strain | Slow growth | Slow growth | Slow growth | Larval arrest | Slow growth | Slow growth | Slow growth | Larval arrest | Larval arrest | Larval arrest | |
| Ashrafi et al., 2003 | Bacterial feeding of L1 larvae; analysis of fat content | Reduced fat content | ||||||||||
| Rual et al., 2004 | Feeding at the first larval stage (L1) E. coli expressing inducible hairpin RNAi | No | No | None | Long | No | No | Embryonic lethal | No | No | No | |
| Sonnichsen et al., 2005 | Injection dsRNA into young adult worms; examination phenotype of F1 progeny | No | Early larval arrest; defective early embryogenesis | Embryonic lethal | No | Early larval lethal | No | No | Embryonic lethal | Early larval arrest | Early larval lethal | |
| Sieburth et al., 2005 | Feeding bacteria expressing dsRNA to larvae; screening adults for decreased acetylcholine secretion (aldicarb resistance) | Aldicarb resistant | ||||||||||
| Fernandez et al., 2005 | Soaking of L4 stage larvae; with dsRNA corresponding to ovary expressed genes; progeny | Embryonic lethal | N | Embryonic lethal | Embryonic lethal | |||||||
| Curran and Ruvkun, 2007 | Bacterial dsRNA feeding of L4-stage larvae (eri-1(mg366) strain); Screening life span of adult | Extended life span; Fat content reduced | ||||||||||
| Byrne et al., 2007 | Bacterial dsRNA feeding to L3-L4 stage worms; progeny and growth | Organism development variant | ||||||||||
| Ceron et al., 2007 | Bacterial dsRNA feeding to L1 larvae (lin-35(n2239) strain); progeny | Larval arrest; reduced brood size |
N, no abnormalities reported; clear, animals appear unusually transparent when compared to control; long, animals are longer and thinner than control animals at the same developmental stage; sick, animals exhibit some combination of abnormal features relating to size, movement, body integrity, pigmentation, viability, fertility; larval arrest: development halted at any larval stage, failure to reach adulthood; slow growth, any variation that causes a reduction in growth rate compared to control.
In more in depth investigations silencing dsRNAs were injected into the gonads of young adult hermaphrodites, followed by scoring of their effect on the progeny. Rachubinski and coworkers found that RNAi inactivation of Pex5, Pex12, Pex13, and Pex19 greatly reduced the percentage of adult progeny, at 3 days following injection of dsRNA, most progeny being developmentally delayed and still at the L1, L2, or L3 larval stage (Petriv et al., 2002) Targeting of Pex6, Pex1, or Pex2 was without effect, but the employed dsRNAs did also not affect the peroxisomal import of a fluorescent PTS1-protein (CFP-SKL). In contrast, injection of dsRNA targeting Pex5, Pex13, or Pex19, caused a cytosolic fluorescence of the reporter. Silencing of Pex12 resulted in fewer but larger peroxisomes.
Thieringer et al. (2003) reported similar experiments. Blocking either Pex5, Pex6, Pex12, Pex13, or Pex19 caused an arrest of the growth of their progeny at the L1 larval stage (Figure 5A). The arrested worms were viable, and resumed growth after 2–8 days, likely depending on quantity and stability of injected DNA, and developed into normal worms.
Figure 5.
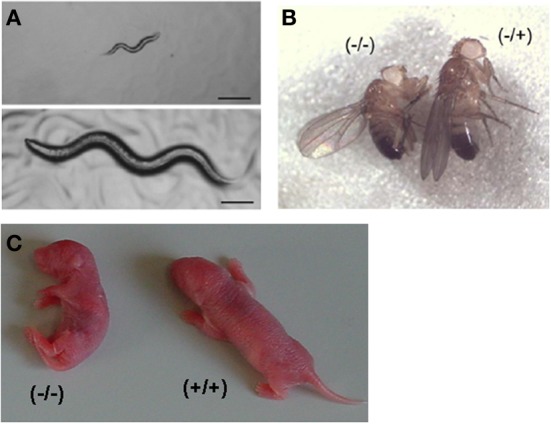
Developmental delay in peroxisome deficient animals. (A) Developmental arrest of C. elegans at the L1/L2 stage by Pex5 RNAi (top panel), compared to wild type nematode (bottom panel), being photographed 3 days after being laid. Bar, 10 μm. Reproduced/adapted with permission from Thieringer et al. (2003). (B) Reduced body size and weight of an adult male homozygous pex161 fruitfly compared to heterozygous animal. Taken from Nakayama et al. (2011). (C) Appearance of newborn mice pups, showing severe hypotonia and growth delay in a Pex5−/− pup compared to a wild type littermate.
Development seems less dependent on Pex10. During an ethyl methanesulfonate mutagenesis screen for genes affecting lipid droplets, Zhang et al. (2012) could classify surviving mutants having enlarged lipid droplets into four complementation groups, one group being linked to Pex10. In the mutant strain (prx-10(hj21)), PTS1 import was affected. Although not discussed in their paper, development and morphology of the worm appeared normal (based on pictures of 1 day adult). Given that the three other groups were linked to peroxisomal β-oxidation enzymes (maoc-1, dhs-28, daf-22, respectively, corresponding to an enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase and SCP2-containing thiolase), one could wonder why only one peroxin was hit in this screen or whether the others remained undetected due to lethality or slower development.
The mechanism underlying the developmental problems was not addressed, but might indeed be caused by peroxisomal metabolic inactivity. Pex5(RNAi) prevents initiation of post-embryonic cell divisions, and normal cell migrations including those of neuronal cells, are blocked (Thieringer et al., 2003). This phenotype resembles that of starved larvae, therefore division might require a peroxisomal metabolite. Furthermore, larval development of nematodes seems to be dependent on etherlipid synthesis. Eight days after injecting gonads of adults with dsRNA directed toward ADHAPS, their offspring was still in the larval stage, whereas those injected with non-specific dsRNA produced mature adults (Motley et al., 2000). Similar findings were reported by Petriv et al. (2002). Also β-oxidation might play a role in development. Upon silencing of Δ3,5-Δ2,4-dienoyl-CoA isomerase (encoded by Y25C1A.13), an enzyme required for degradation of polyunsaturated fatty acids, or silencing of the three ABC half-transporters (encoded by T02D1.5, C44B7.9, and C44B7.8), implicated in peroxisomal membrane translocation of fatty acids/acyl-CoAs, a similar phenotype is seen: no adult offspring three days after injection (Petriv et al., 2002). It should be noted, however, that silencing of peroxisomal thiolases, either the classical ones (encoded by T02G5.4 and T02G5.8) or SCP2-containing thiolase (encoded by daf-22), had no effect (Petriv et al., 2002). Also important to mention is that the C. elegans dienoyl-CoA isomerase, thought to be the counterpart of mammalian ECH1 (Petriv et al., 2002), which is targeted to both mitochondria and peroxisomes, does not have a PTS1. This complicates the interpretation of these silencing experiments.
Whereas a defective peroxisome biogenesis affects early larval development, silencing of Pex genes at a later stage seems beneficial. An extended life span, 22.7 days compared to 16.22, was seen upon silencing of Pex5 in L4 larvae in the eri-1(mg366) strain, a strain more sensitive to RNAi (Curran and Ruvkun, 2007). Similarly, Zhou et al. reported a 17% increase for Pex13 and 8% for Pex5 (15% for PMX4, a peroxisomal membrane protein), when silenced in 1 day old adult (Zhou et al., 2012). It is suggested that the longer survival is related to reduced generation of reactive oxygen species (ROS) when peroxisomes are less or not functional (see Fransen et al., 2012). The amount of measurable ROS is lowered in the Pex5, Pex11, or Pex13 silenced animals. Strangely, and in contrast to most other screens, silencing of these peroxins in L1 larvae had no effect, but controls on RNAi efficacy are missing.
Summarizing, RNAi based data suggest that peroxins play a critical role in nematode development, but are less important in the adult stage. A drawback of this technique is, however, the variability. In the near future, more solid data on the role of peroxins in nematodes are expected, given the increasing availability of deletion mutants (C.elegans mutation consortium. 2012): a Pex5 mutant (tm4948) with a 439 bp deletion is sterile4; a Pex1 mutant (tm0392) with a 681 bp deletion is classified as lethal or sterile5.
Fruitfly
About 21–22 h after fertilization (hpf), Drosophila larvae will hatch from the eggs. One distinguishes 17 steps during this period, known as Bownes stage numbers. In stage 6 (180–195 min), gastrulation starts, whereas formation of the Malphigian tubes (counterpart of kidney in mammals) starts in stage 10. In the late stage 11, the stomatogastric nervous system develops. During the subsequent larval stages, three in total, most of the organs/structures of the adult fly will develop, starting from imaginal discs. At the end of the third larval stage (120 hpf), metamorphosis starts, divided in a prepupal period and a pupal period, in total 4 days. Finally, the flies emerge from the pupal case (eclosion). They start mating 12 h after emergence and will live for about a month.
Based on fatty acid analysis of certain Pex mutants, very long chain fatty acids (VLCFA) are degraded via peroxisomal β-oxidation in fruitflies (Chen et al., 2010). Related to purine/xanthine metabolism, it should be noted that the eye pigment formation is dependent on peroxisomes. The rosy-506 eye-color mutant lacks xanthine dehydrogenase/oxidase, which is targeted to peroxisomes (Beard and Holtzman, 1987).
The genome of Drosophila encodes at least 15 peroxins (Chen et al., 2010; Mast et al., 2011), being homologous to mammalian peroxins. Whether orthologous of the fungal Pex20 and Pex23 are expressed (Mast et al., 2011), is questionable (see comments to the related entries in Table 1).
Based on RNAi in Drosophila S2 cells expressing GFP-SKL, silencing of Pex1, Pex5, Pex13, Pex16 results in import deficiency, silencing of Pex2, Pex3, Pex6, Pex12, Pex14 in impaired import. Interfering with Pex11 or Pex19 affects peroxisome number (reduced) and size (larger), whereas RNAi of the putative Pex20 or Pex23 has an opposite effect, more peroxisomes of smaller size (Mast et al., 2011).
Although for most of these peroxins, mutants with transposon P1-insertions were present in the repositories (see Table 3), these were not studied in depth. According to Spradling et al. (Spradling et al., 1999), the Pex2f018 allele was lethal, but this was later shown to be due to a second mutation (Chen et al., 2010). More recently, a library of RNAi transgenes, expressing inverted repeats causing conditional gene inactivation, became available, covering 88% of the predicted protein coding genes (Dietzl et al., 2007). For all fly peroxins, transgenic lines are available (unpublished data), but as far as known, not evaluated.
Table 3.
Overview of classical peroxin alleles in Drosophila melanogaster.
Related to fly development, and as far as studied in detail, PEX1, PEX3, and PEX13 appear critical. P-element insertion in Pex1 (pex1s4868) (Chen et al., 2010; Mast et al., 2011) or in Pex13 (pex13KG04339) (Chen et al., 2010), X-ray mutagenized Pex11 (Mast et al., 2011) or a deletion in Pex3, generated by P-element imprecise excision of pex3CG6859 (Nakayama et al., 2011), are lethal6 at the larval stage when homozygous. Expression of a wild type PEX1 rescues the pex1s4868 or pex11 mutants to survive past the second larval instar (Mast et al., 2011). Pex1 mutant larvae displayed a delay in development, little coordinated locomotion, poor feeding, and died at the L1–L2 stage (Mast et al., 2011). Some larvae even died a few hours after hatching, being unable to crawl out of the eggshells. In the peripheral and central nervous system various abnormalities were documented. These include malformation of the ventral nerve cord (lack of or underdeveloped commissures, breaks in longitudinal connectivities), reduced number of motor neurons, disorganization of glia cells, loss and hypoplasia of peripheral neurons, malformation of eye discs. In the malphigian tubules, structural abnormalities were noticed.
A dsRNA screen was conducted in preblastoderm embryos to detect genes that affect embryonic nervous system development. Although 50% of the Drosophila genes were covered, only one peroxin was hit, i.e., PEX19. Silencing of Pex19 resulted in disruption of the ventral nerve cord, misrouting of axons and disorganization of dorsal clusters of cells in the peripheral nervous system in stage 15–16 embryos (Koizumi et al., 2007).
Flies with insertional mutations in Pex2(pex2f0189 and pex2HP35039) (Chen et al., 2010), Pex12 (pex12f01300) (Chen et al., 2010), Pex1 (pex1S4868) (Zhou et al., 2012) or Pex13 (pex13KG04339) (Zhou et al., 2012) or a deletion in Pex10 (excision of P-element in pex10EY23523) (Chen et al., 2010) or Pex16 (excision of pex16CG3947) (Nakayama et al., 2011) are viable. Fertility, however, was reduced in Pex2, Pex10, or Pex12 female mutants and males were sterile (Chen et al., 2010). The latter phenotype was due to an arrest in the germ cell development at the level of the spermatocyte growth stage. Similarly, male fertility was compromised in the Pex16 mutant (Nakayama et al., 2011). Testes of this mutant were smaller and did not contain mature sperm cells, although early spermatocyte cysts were still present, due to an arrest in the maturation of spermatocytes at the young apolar stage (Figures 6A,B). This arrest and the fertility could be rescued by overexpression of PEX16 in the cyst cells, although germ cells still lacked peroxisomes. Expression of PEX16 in the germline cells, however, did not rescue the spermatogenesis, indicating that peroxisomes in the somatic cysts cells play an important role in spermatogenesis (Nakayama et al., 2011). This is, however, in contrast to the Pex2 mutant in which rescue of the germ cells normalized the phenotype (Chen et al., 2010). It is suggested that VLCFA, which show an age-dependent increase in Pex10 mutants (2.9- and 3.9-fold for whole body C26:0 at 2 and 15 days, respectively), play a critical role in spermatogenesis in insects (Chen et al., 2010).
Figure 6.
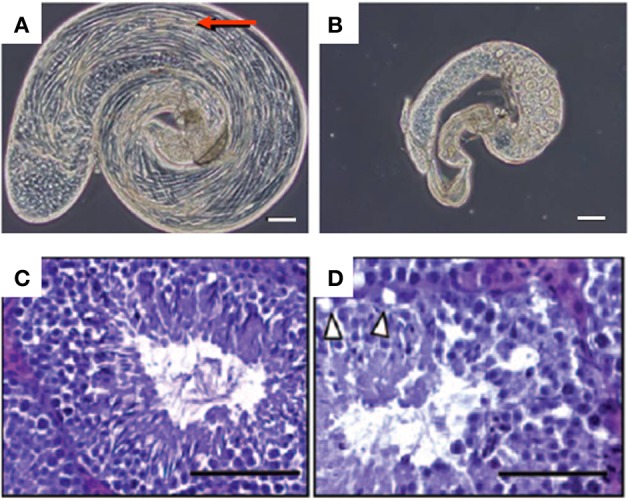
Male fertility problems in peroxisome deficient animals. (A,B) Phase-contrast micrographs of testes of fruitflies, with bundles of elongated spermatids (arrow) in a wild type (A) and arrest of germline cell maturation in a pex161 homozygous fruitfly (B). Bar, 100 μm. Taken from Nakayama et al. (2011). (C,D) Hematoxylin-eosin staining of 7 weeks old testis of wild type (C) and Sertoli PEX5 knockout mice (D), the latter showing lipid droplets that were emptied during the embedding procedure in the outer layer of the seminiferous epithelium (arrowheads) and reduced numbers of spermatozoa in the lumen of the tubuli. Bar 100 μm. Taken from Huyghe et al. (2006b).
Although viable, the Pex16 mutant adult flies were considerably smaller (30% females; 15% males) (Figure 5B) and their locomotion was affected, the latter likely being responsible for a severe reduction of their lifespan (to one third for females; to one-fourth for males) (Nakayama et al., 2011). Peroxisomes were still present in the malpighian tubule cells of Pex16 mutant flies, but their number is greatly reduced. Not unexpectedly, the eye pigmentation was affected in Pex16 mutants, resembling the rosy phenotype, and biochemically, an increase in (whole body) VLCFA levels was seen (2-fold for C24:0 in males, 3-fold in females). Histology of brain revealed a low density of dendritic trees in the lobula plate of the optic lobe; other cells in the optic lobe and other parts of the brain were unaffected. The dendritic reduction was already visible at the pupal stage and did not aggravate with age, suggesting a developmental problem, not a degeneration. Interestingly this defect can be rescued by expression of PEX16 in the fat body or in differentiated neurons (Nakayama et al., 2011).
The viability of the above mentioned Pex1 and Pex13 mutants can be explained by the nature of the mutation, affecting the promoter and resulting in lower expression (~20% of wild type) (Zhou et al., 2012). Interestingly, life span of these flies increased (16% in males; 13% in females), whereas their hydrogen peroxide levels were decreased. This is similar to findings reported in nematodes (see Nematodes). Along the same lines, Pex19 expression was reported to be repressed (1.8-fold) when feeding flies 4-phenylbutyrate, a diet which extends their lifespan by 36% (Kang et al., 2002). The latter compound is known in the peroxisomal field by its ability to induce the expression of ABCD2 (Kemp et al., 1998), an ABC-transporter functionally related to ABCD1 which is mutated in X-ALD.
Zebrafish
Given the translucency of the embryo and the short developmental period, zebrafish (Danio rerio) is an organism of choice for dynamic developmental studies. Gastrulation starts around 6 h post-fertilization (hpf), first somites are formed at 11 hpf, and at 24 hpf the embryo, surrounding the yolk sac, shows already the typical fish-like shape and tail and primary organs have been formed. In the subsequent day, the circulatory system and fins are formed. Cartilage development starts at 48 hpf, and at 3 days, fishes are self-supporting, first as larvae till 1 month of age, then juveniles till adulthood, around 90 days. Total life span is around 2 years.
Transcripts for peroxisomal matrix and membrane proteins can be detected starting at 24 hpf in the head region, whereas catalase-positive peroxisomes become visible in the liver and the pronephric duct in 4 days old fishes (Krysko et al., 2010). In adult fish, peroxisomes are most prominent in liver (Braunbeck et al., 1990; Krysko et al., 2010), renal proximal tubules (Krysko et al., 2010) and the intestinal epithelium (Krysko et al., 2010). For more information on expression in zebrafish during embryogenesis, the reader is referred to a large scale in situ hybridization screen (Thisse et al., 2004).
Similarly to rodents, zebrafish hepatic peroxisomes respond to peroxisome proliferators and an increased number is observed in liver when fishes are exposed to clofibrate (Venkatachalam et al., 2012) or phthalate esters (Ortiz-Zarragoitia et al., 2006).
Based on scattered information, the organelles are active in β-oxidation. Presence of ACOX1 was demonstrated (Ibabe et al., 2005; Morais et al., 2007) and the enzymes able to act on branched fatty acids, such as MFP2 (encoded by hsd17b4) and SCPX (encoded by scp2a), are expressed (Thisse et al., 2004), but apparently C24-bile acids are not formed in zebrafish, in contrast to other teleost fish (Hofmann et al., 2010). Based on genomic information, fish peroxisomes can synthesize etherlipids and contain an α-oxidation pathway.
Regarding peroxisome biogenesis, all classical peroxins are expressed in D. rerio (see Table 1), and based on high throughput analysis, Pex3, Pex5, Pex7, Pex10, Pex14, Pex19 are ubiquitously expressed from 24 hpf on, with higher expression in the head region (Thisse et al., 2004).
Despite the wide spread use of morpholinos to interfere with expression in zebrafish, in only few reports, as far as documented, this technique was applied to peroxisome biogenesis. Injection of morpholinos, intended to block the splice sites in Pex3 or Pex13, into one-cell embryos did not affect peroxisomal import. Subsequent RNA analysis revealed that these morphilinos did not eliminate exons, instead produced a short in frame insertion (Pex3) or deletion (Pex13) (Krysko et al., 2010). Blocking of the translation of Pex13 was more effective to reduce the number of hepatic peroxisomes, but high doses were needed and not all of the injected embryos showed such response. A Pex5 blocking morpholino had no effect at low dose, and caused embryonal death at higher dose. Finally, overexpression via mRNA injection of an N-terminal domain of (human) PEX3, having a dominant negative effect in human fibroblasts (Soukupova et al., 1999), did not affect biogenesis (Krysko et al., 2010). Coutinho et al. (2004) did not observe any abnormalities at 32 hpf when one cell stage embryos were injected with morpholinos directed against the 5′-end of Pex19 (notochord differentiation or pigmentation were normal), the efficacy of the morpholino was, however, not controlled.
Although technically easy, the dilution of morpholinos or mRNA upon subsequent cell divisions, combined with the turnover of peroxisomes, half life estimated at 2 days in cultured mammalian cells (Huybrechts et al., 2009), is a major obstacle in the embryonic injection approach. In the near future, more solid data might emerge from analysis of insertional zebrafish mutants. Although tools to carry out large scale insertional mutagenesis and positional cloning in zebrafish were developed several years ago using mouse retroviral vectors (Gaiano et al., 1996; Golling et al., 2002) the number of created, annotated and available mutants, however, remains low. For a more targeted approach, engineered Zn-finger nucleases are a promising tool to create zebrafish knockouts (Foley et al., 2009).
Mice
The intra-uterine development of mice takes 20–21 days. During this period, embryos are depending on the maternal circulation with regards to most nutrients. Examples of exceptions are brain poly-unsaturated fatty acids (PUFA) that are partly dependent on local synthesis (Janssen et al., 2000). At birth, organogenesis of most organs has been completed, except formation of the cerebellum which extends into the postnatal period and maturation of gonads before adulthood. After birth, pups are nursed and milk-fed till weaning, about 3 weeks later. At 6 weeks (females) or eight (males) of age, animals become sexually active and start to breed. Lifespan, under laboratory conditions, is 18–30 months.
In mammals, peroxisomal β-oxidation serves to generate PUFA and C24 bile acids. The first are implicated in many brain processes such as learning, memory, behavior; the latter are required for efficient uptake of lipophilic nutrients in the intestines. This pathway also shortens VLCFA, pristanic acid and dicarboxylic fatty acids (Van Veldhoven, 2010). Removal of the toxic phytanic acid requires an active α-oxidation. Plasmalogen deficiency in mammals is linked to a specific bone developmental problem, in man known as rhizomelic chondrodysplasia punctata (RCDP), and RCDP type I is linked to PEX7 deficiency.
Currently, the following peroxins have been inactivated in mice: PEX5 (Baes et al., 1997), PEX2 (Faust and Hatten, 1997), PEX11A (Li et al., 2002a; Weng et al., 2013), PEX11B (Li et al., 2002b), PEX13 (Maxwell et al., 2003), and PEX7 (Brites et al., 2003; Braverman et al., 2010). Mice lacking both PEX11A and PEX11B were also created (Li et al., 2002b), or lacking a peroxin together with another peroxisomal protein such as Pex7−/−:Abcd1−/− mice (Brites et al., 2009).
Given obvious similarities, PEX5, PEX2, and PEX13 deficiencies can be treated together, separately from the PEX7 knockout model. Considering that PEX11 proteins are not involved in peroxisome biogenesis per se and that this process is not affected in the Pex11a−/− and Pex11b−/− mice, but mainly their elongation and abundance, these models will not be discussed further in this chapter. Below we will summarize the main findings in the other mouse models [see also recent reviews by Baes and Van Veldhoven (2006, 2012)].
Related to PEX5, PEX2, and PEX13 deficient models, knockouts pups are born alive in the expected Mendelian ratio and without major deformities or skeletal malformations, suggesting a normal intra-uterine development (Baes et al., 1997; Maxwell et al., 2003). However, in case of PEX2 deficiency in an inbred 129 background, embryonic lethality was reported and only 20% of the pups are born (Faust and Hatten, 1997). In these three models, newborn pups are, however, growth retarded and severely hypotonic (Figure 5C), hence they do not feed and die 6–24 h after birth. Some Pex2−/− pups (20–30%), in a mixed Swiss Webster × 129SvEv background, survive for about 1–2 weeks (Faust and Hatten, 1997) and the postnatal survival can be improved by oral bile acid therapy (9% alive after 30 days) (Keane et al., 2007). The reason for this strain-dependent differences, although often seen in other mouse models, is not clear.
At closer inspection, there are some developmental problems, especially in the brain. Lamination of the cerebral cortex is affected due abnormal and delayed neuron migration (Baes et al., 1997; Faust and Hatten, 1997; Gressens et al., 2000). In the longer surviving Pex2−/− pups, dendritic arborization of the Purkinje cells in the cerebellum is reduced and their axons are dystrophic (Faust, 2003). Similar findings were seen in a Pex5 and Pex13 brain knockout (see further).
Finally, at the subcellular level, mitochondrial abnormalities were documented in liver (Baumgart et al., 2001; Keane et al., 2007) and lamellar lipid deposits were evident in the adrenocortical cells (Faust and Hatten, 1997).
Biochemically, various peroxisome dependent parameters are abnormal in pups with these Pex gene inactivations [accumulation of VLCFA, lack of plasmalogen, abnormal bile acids, shortage of docosahexaenoic acid (DHA)]. Changes in brain PUFA composition have been proposed to modify α-synuclein (Yakunin et al., 2010), which could contribute to the neuropathology. In whole brain extracts of these three models, Yakunin et al. (2010) showed increasing oligomerization and phosphorylation of α-synuclein. Such changes trigger intraneuronal deposition of α-synuclein (Lewy bodies), being a hallmark of synucleopathies such as Parkinson disease.
A different phenotype is seen in PEX7 deficient mice (Brites et al., 2003). Embryonic lethality is not seen, but these pups are also hypotonic and growth impaired (15–30% lower body weight at birth), and the majority (70%) dies before weaning (50% after 1 day, likely due to the hypotonia). The surviving animals do live till adulthood and longer, but males are infertile, the seminiferous epithelium being devoid of spermatogonia and spermatocytes. In brain, a delay in neuronal migration is seen, and ossification of distal bone elements of the limbs, skull and vertebrae, is defective. The amount of white, but not brown, adipose tissue is reduced (Brites et al., 2011). Bilateral cataracts develop 2 weeks after birth (Brites et al., 2011), the time pups open their eye lids. Biochemically, plasmalogens are depleted, phytanic acid cannot be degraded, and VLCFA oxidation is impaired in fibroblasts, but increased VLCFA levels are only found in spleen, spinal cord and neonatal brain (Brites et al., 2009).
In Pex7 hypomorphic mice, in which Pex7 transcripts are reduced to 5%, lifespan is normal (Braverman et al., 2010). The mice are still smaller, but are fertile. Their tissue content of plasmalogens is low but not absent, DHA in RBC is lowered and phytanic acid accumulates. Pathological findings include endochondral ossification defects, abnormalities in lens fibers and eye cataract (Braverman et al., 2010).
Feeding 6-weeks old Pex7−/− mice with 1-O-octadecylglycerol, an etherlipid which is bypassing the peroxisomal biosynthetic steps (see Figure 3), reveals that several phenotypic abnormalities are related to plasmalogen deficiency. The diet restores plasmalogen levels in non-nervous tissues. In parallel, testicular pathology is ameliorated (spermatogenesis was restored, although mature spermatozoa were still not detectable), and adipocytes displayed a normal size and fat content. When giving 1-O-octadecylglycerol to newborn pups, via supplementing it to the diet of the mother, testicular degeneration was prevented and cataract formation was absent or only unilateral and reduced to a small nuclear cataract (Brites et al., 2011). In the hypomorphic Pex7−/− mice, such treatment did not affect the cataracts (Braverman et al., 2010).
Severe bone abnormalities, a major hallmark in patients with PEX7 deficiency as reflected in their name (RCDP), are not observed in mice. Upon closer investigation, a delay, however, in endochondral bone formation was reported in both complete (Brites et al., 2003) and hypomorphic PEX7 (Braverman et al., 2010) deficient mice, likely due to a delayed maturation of chondrocytes at the pre-hypertrophic state, but further mechanistic insights were not generated.
Given the lethality of peroxin knockouts, especially of those with affected PTS1-import, developmental and behavioral studies are limited. This can be circumvented by conditional knock-out whereby peroxisomes are removed in specific tissues and/or at a certain stage. Tissue-specific removal of peroxisomes can be established by crossing mice containing a floxed Pex gene (Baes et al., 2002) with mice expressing cre in a promoter-specific manner. The promoter also determines the time point from when on the Pex gene is irreversibly inactivated in the targeted cells and their descendents. This technology was applied for Pex5 creating mice lacking peroxisomes in the central nervous system (CNS) [nestin-Cre, in neural precursors from embryonic (E) day 11 (Hulshagen et al., 2008)], hepatocytes [alfafoetoprotein-Cre, from E10 (Krysko et al., 2007) and albumin-Cre, from birth (Peeters et al., 2011)]. By using a similar approach, brain specific PEX13 knockouts were obtained (nestin-Cre) (Müller et al., 2011).
Pex5 was further inactivated in specific cell types by using appropriate Cre-expressing mice: Sertoli cells (Amh-Cre, from E14) (Huyghe et al., 2006b), oligodendrocytes (Cnp-Cre, from E14) (Kassmann et al., 2007), principal neurons in the forebrain (Nex-Cre, from E12) (Bottelbergs et al., 2010), and astrocytes (Gfap-Cre from E13) (Bottelbergs et al., 2010). The specific inactivation of PEX5 in adipocytes failed due to the non-selectivity of the aP2 promoter driving Cre expression (Martens et al., 2012).
Overall, these studies indicate that absence of peroxisomes in adipose (Martens et al., 2012), neurons (Bottelbergs et al., 2010), astrocytes (Bottelbergs et al., 2010), or Sertoli cells (Huyghe et al., 2006b) does not compromise life span. Postnatal thriving, however, requires functional liver and brain peroxisomes. Moreover, absence in liver results in life threatening development of hepatocarcinomas (Dirkx et al., 2005), absence in brain shortens life span considerably to 6 months with 20% dead before 3 weeks for Pex5-loxP:nestin-cre (Hulshagen et al., 2008) or 35 days for Pex13-loxP:nestin-cre (Müller et al., 2011) mice. Of the different models with specific brain cell inactivation, the oligodendrocyte knockout represents the worst outcome: almost none of the affected animals survive 1 year of age (Kassmann et al., 2007). Its phenotype resembles that of a total deficiency of peroxisomes in the brain, but with delayed onset of demyelination, axonal loss and neuroinflammation. The latter encompasses a strong activation of the innate immune system with microglia reactivity and increased expression of pro-inflammatory markers (Kassmann et al., 2007; Bottelbergs et al., 2012). The biochemical factor(s) contributing to or causing this phenotype remain unclear. To which extent peroxisomal metabolites can be transferred from one cell type to another in brain, or from the body to the brain, is not fully established, but an important role of peroxisomes in neurons or astrocytes in pre- and postnatal life can be excluded.
For more information about these models, and how peroxin deficiencies affect brain, liver and testis, we refer to recent reviews (Baes and Van Veldhoven, 2006, 2012; Baes and Aubourg, 2009). It should be stressed that part of the pathology seen in these mouse models might be related to the, not yet completely understood, interplay between peroxisomes, their metabolites and other organelles. As initially observed in PEX5 (Baumgart et al., 2001) and PEX2 knockouts (Keane et al., 2007), and further documented in the albumin-Cre/Pex5-loxP mice (Dirkx et al., 2005), absence of peroxisomes in hepatocytes affects their mitochondria severely. Structural alterations are seen in the inner mitochondrial membrane, and its potential is collapsed. Activities of complex I, III, and V are reduced. In addition, lipid droplets and ER stress are noticed. Based on the upregulation of ATF3, ATF4, ATF6, and CHOP, the unfolded protein response pathway is activated in absence of peroxisomes (Dirkx et al., 2005). Similar findings were seen in liver of surviving PEX2 pups, the integrated stress response mediated by PERK and ATF4 signaling being activated (Kovacs et al., 2009). It is postulated that perturbed peroxisomal β-oxidation metabolites (e.g., bile acid (intermediates), dicarboxylic acids), are causative factors given the fact that ER stress is also seen in mice with β-oxidation defects (Huang et al., 2011).
Conclusion
Although peroxisomes are not essential for cell functioning and survival, at the multicellular level they are indispensable as demonstrated by the different animal models treated in this chapter.
A common feature in animals with peroxisome biogenesis defects is a developmental delay, smaller size at hatching/birth and limited to very short lifespan (Figure 5). The reason for the delay is not clear. In most models, organogenesis seems to proceed normal, but the central nervous system appears sensitive to absence of peroxisomes (abnormal cerebellar lamination and delayed neuron migration in mice; malformation of the ventral nerve cord in fruitfly; block of neuron cell migration in L1 stage in nematodes). In a later stage of life, neuronal problems are manifested in reduced locomotion (larvae of insects) or coordination and motor skills (mice). In nematodes, normal larval development is dependent on ether lipids. In mice (and man), plasmalogen deficiency is compatible with prenatal development but the newborns exhibit already several abnormalities.
With regard to the nervous system, an intriguing question is to which extent myelinization/demyelinization and axonal integrity are linked to peroxisomes. Myelin, formed by the oligodendrocytes, is indeed enriched in metabolites related to peroxisomes (plasmalogens, VLCFA). It is therefore surprising that myelination is initially normal when peroxisomes are ablated from oligodendroglia and that in adulthood myelin becomes destabilized. Importantly, as both in the total brain and the oligodendrocyte knockout, degenerated axons are observed surrounded with a normal myelin sheet, it was postulated that oligodendroglial peroxisomes serve to support axons independent of myelination. This is further endorsed by the finding that peroxisomes are abundant in paranodes (Kassmann et al., 2011), sites where glia and axons interact. In this context, one should recall that in species in which axons are not myelinated such as fruitfly, neuronal abnormalities are seen when peroxisomes are ablated (Nakayama et al., 2011).
Another remarkable finding, although not studied in all models, is the male sterility, documented at least in Drosophila and in mice knockouts (Figure 6). In fly, peroxisomes of the cysts cell appear to be important for spermatogenesis (Nakayama et al., 2011), which is mirrored in mice where peroxisomes are necessary in the Sertoli cells (Huyghe et al., 2006b). In insects, the infertility was linked to accumulation of VLCFA, in mice experimental evidence points toward both an accumulation of VLCFA and VLCFA-PUFA (Huyghe et al., 2006b). The importance of normal peroxisomal β-oxidation for male fertility was further confirmed in ACOX1 (Fan et al., 1996) and MFP2 knockout mice (Huyghe et al., 2006b). In addition, a depletion of ether lipids also causes male infertility in PEX7 (Brites et al., 2011) and GNPAT (Rodemer et al., 2003) knockout mice.
Finally, related to aging and neurodegenerative diseases, and the emerging role of peroxisomes in ROS signaling (Titorenko and Terlecky, 2011; Fransen et al., 2012) scattered information derived from the animal models discussed above, suggest that less active peroxisomes in adulthood could positively contribute to longevity. This seems, however, in conflict with the general concept that the metabolic activity of these organelles becomes compromised during aging. On the other hand, it would be consistent with studies on the importance of catalase in aging. Improving the removal of peroxide in peroxisomes, by expressing an engineered catalase with a higher affinity for PEX5, delays the appearance of senescence markers in human fibroblasts (Koepke et al., 2007). Hence, not the peroxisomal metabolic activity, but the ratio of ROS-generation/removal (Fransen et al., 2013), might be a determining factor in aging.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research by the authors is supported by grants from the Fonds voor Wetenschappelijk Vlaanderen (FWO) (G.0760.09 and G.0675.12 to Myriam Baes; G.0721.10N to Paul P. Van Veldhoven) and of the KU Leuven (OT/08/40 and OT12/79 to Myriam Baes; OT/09/045 to Paul P. Van Veldhoven).
Footnotes
1For the sake of consistency, the nomenclature guidelines as formulated for rodents (http://www.informatics.jax.org/mgihome/nomen/gene.shtml) are followed throughout this text, gene symbols being italicized, first letter capitalized, whereas the protein is referred to by the corresponding gene symbol in standard capitalized font. Guidelines related to nomenclature in other species can be found at following URLs: nematodes (http://www.wormbase.org/about/userguide/nomenclature#fda31g748bjh9c650ie2—10); fruitflies (http://flybase.org/static_pages/docs/nomenclature/nomenclature3.html#1.2.3.); zebrafish (https://wiki.zfin.org/display/general/ZFIN+Zebrafish+Nomenclature+Guidelines#ZFINZebrafishNomenclatureGuidelines-1.1); man (http://www.genenames.org).
2In sensu stricto of the original definition (Distel et al., 1996), these proteins should not be called peroxins.
3In contrast to most other higher eukaryotes, the two catalytic domains of MFP2 are expressed as separate proteins in C. elegans (Huyghe et al., 2006a).
6The pex1S084807 and Pex102402 alleles were reported to be semi-lethal and in homozygous third instar larvae necrosis was observed in salivary gland cells (Burmester et al., 2000).
References
- Ashrafi K., Chang F. Y., Watts J. L., Fraser A. G., Kamath R. S., Ahringer J., et al. (2003). Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268–272 10.1038/nature01279 [DOI] [PubMed] [Google Scholar]
- Baes M., Aubourg P. (2009). Peroxisomes, myelination, and axonal integrity in the, CNS. Neuroscientist 15, 367–379 10.1177/1073858409336297 [DOI] [PubMed] [Google Scholar]
- Baes M., Dewerchin M., Janssen A., Collen D., Carmeliet P. (2002). Generation of Pex5-loxP mice allowing the conditional elimination of peroxisomes. Genesis 32, 177–178 10.1002/gene.10047 [DOI] [PubMed] [Google Scholar]
- Baes M., Gressens P., Baumgart E., Carmeliet P., Casteels M., Fransen M., et al. (1997). A mouse model for Zellweger syndrome. Nat. Genet. 17, 49–57 10.1038/ng0997-49 [DOI] [PubMed] [Google Scholar]
- Baes M., Van Veldhoven P. P. (2006). Generalised and conditional inactivation of Pex genes in mice. Biochim. Biophys. Acta 1763, 1785–1793 10.1016/j.bbamcr.2006.08.018 [DOI] [PubMed] [Google Scholar]
- Baes M., Van Veldhoven P. P. (2012). Mouse models for peroxisome biogenesis defects and beta-oxidation enzyme deficiencies. Biochim. Biophys. Acta 1822, 1489–1500 10.1016/j.bbadis.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Banerjee S. K., Kessler P. S., Saveria T., Parsons M. (2005). Identification of trypanosomatid PEX19: functional characterization reveals impact on cell growth and glycosome size and number BANERJEE2005. Mol. Biochem. Parasitol. 142, 47–55 10.1016/j.molbiopara.2005.03.008 [DOI] [PubMed] [Google Scholar]
- Baumgart E., Vanhorebeek I., Grabenbauer M., Borgers M., Declercq P. E., Fahimi H. D., et al. (2001). Mitochondrial alterations caused by defective peroxisomal biogenesis in a mouse model for Zellweger syndrome (PEX5 knockout mouse). Am. J. Pathol. 159, 1477–1494 10.1016/S0002-9440(10)62534-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard M. E., Holtzman E. (1987). Peroxisomes in wild-type and rosy mutant Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 84, 7433–7437 10.1073/pnas.84.21.7433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottelbergs A., Verheijden S., Hulshagen L., Gutmann D. H., Goebbels S., Nave K. A., et al. (2010). Axonal integrity in the absence of functional peroxisomes from projection neurons and astrocytes. Glia 58, 1532–1543 10.1002/glia.21027 [DOI] [PubMed] [Google Scholar]
- Bottelbergs A., Verheijden S., Van Veldhoven P. P., Just W., Devos R., Baes M. (2012). Peroxisome deficiency but not the defect in ether lipid synthesis causes activation of the innate immune system and axonal loss in the central nervous system. J. Neuroinflam. 9, 61 10.1186/1742-2094-9-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunbeck T., Gorge G., Storch V., Nagel R. (1990). Hepatic steatosis in zebra fish (Brachydanio rerio) induced by long-term exposure to gamma-hexachlorocyclohexane. Ecotoxicol. Environ. Saf. 19, 355–374 10.1016/0147-6513(90)90036-5 [DOI] [PubMed] [Google Scholar]
- Braverman N., Zhang R., Chen L., Nimmo G., Scheper S., Tran T., et al. (2010). A Pex7 hypomorphic mouse model for plasmalogen deficiency affecting the lens and skeleton. Mol. Genet. Metab. 99, 408–416 10.1016/j.ymgme.2009.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brites P., Ferreira A. S., da Silva T. F., Sousa V. F., Malheiro A. R., Duran M., et al. (2011). Alkyl-glycerol rescues plasmalogen levels and pathology of ether-phospholipid deficient mice. PLoS ONE 6:e28539 10.1371/journal.pone.0028539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brites P., Mooyer P. A., el Mrabet L., Waterham H. R., Wanders R. J. (2009). Plasmalogens participate in very-long-chain fatty acid-induced pathology. Brain 132, 482–492 10.1093/brain/awn295 [DOI] [PubMed] [Google Scholar]
- Brites P., Motley A. M., Gressens P., Mooyer P. A., Ploegaert I., Everts V., et al. (2003). Impaired neuronal migration and endochondral ossification in Pex7 knockout mice: a model for rhizomelic chondrodysplasia punctata. Hum. Mol. Genet. 12, 2255–2267 10.1093/hmg/ddg236 [DOI] [PubMed] [Google Scholar]
- Brocard C., Hartig A. (2006). Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta 1763, 1565–1573 10.1016/j.bbamcr.2006.08.022 [DOI] [PubMed] [Google Scholar]
- Burmester T., Mink M., Pal M., Laszloffy Z., Lepesant J., Maroy P. (2000). Genetic and molecular analysis in the 70CD region of the third chromosome of Drosophila melanogaster. Gene 246, 157–167 10.1016/S0378-1119(00)00066-4 [DOI] [PubMed] [Google Scholar]
- Butcher R. A., Ragains J. R., Li W., Ruvkun G., Clardy J., Mak H. Y. (2009). Biosynthesis of the Caenorhabditis elegans dauer pheromone. Proc. Natl. Acad. Sci. U.S.A. 106, 1875–1879 10.1073/pnas.0810338106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne A. B., Weirauch M. T., Wong V., Koeva M., Dixon S. J., Stuart J. M., et al. (2007). A global analysis of genetic interactions in Caenorhabditis elegans. J. Biol. 6:8 10.1186/jbiol58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- C.elegans mutation consortium (2012). Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 2, 1415–1425 10.1534/g3.112.003830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceron J., Rual J. F., Chandra A., Dupuy D., Vidal M., van den Heuvel S. (2007). Large-scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity. BMC Dev. Biol. 7:30 10.1186/1471-213X-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Liu Z., Huang X. (2010). Drosophila models of peroxisomal biogenesis disorder: peroxins are required for spermatogenesis and very-long-chain fatty acid metabolism. Hum. Mol. Genet. 19, 494–505 10.1093/hmg/ddp518 [DOI] [PubMed] [Google Scholar]
- Coutinho P., Parsons M. J., Thomas K. A., Hirst E. M., Saude L., Campos I., et al. (2004). Differential requirements for COPI transport during vertebrate early development. Dev. Cell 7, 547–558 10.1016/j.devcel.2004.07.020 [DOI] [PubMed] [Google Scholar]
- Cregg J. M., van der Klei I. J., Sulter G. J., Veenhuis M., Harder W. (1990). Peroxisome-deficient mutants of Hansenula polymorpha. Yeast 6, 87–97 10.1002/yea.320060202 [DOI] [PubMed] [Google Scholar]
- Curran S. P., Ruvkun G. (2007). Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 3:e56 10.1371/journal.pgen.0030056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vet E. C., Prinsen H. C., van den Bosch H. (1998). Nucleotide sequence of a cDNA clone encoding a Caenorhabditis elegans homolog of mammalian alkyl-dihydroxyacetonephosphate synthase: evolutionary switching of peroxisomal targeting signals. Biochem. Biophys. Res. Commun. 242, 277–281 10.1006/bbrc.1997.7950 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Dirkx R., Vanhorebeek I., Martens K., Schad A., Grabenbauer M., Fahimi D., et al. (2005). Absence of peroxisomes in mouse hepatocytes causes mitochondrial and ER abnormalities. Hepatology 41, 868–878 10.1002/hep.20628 [DOI] [PubMed] [Google Scholar]
- Distel B., Erdmann R., Gould S. J., Blobel G., Crane D. I., Cregg J. M., et al. (1996). A unified nomenclature for peroxisome biogenesis factors. J. Cell Biol. 135, 1–3 10.1083/jcb.135.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt G., Warren D., Becker E., Rehling P., Gould S. J. (2001). Domain mapping of human PEX5 reveals functional and structural similarities to Saccharomyces cerevisiae Pex18p and Pex21p. J. Biol. Chem. 276, 41769–41781 10.1074/jbc.M106932200 [DOI] [PubMed] [Google Scholar]
- Erdmann R., Veenhuis M., Mertens D., Kunau W. H. (1989). Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 86, 5419–5423 10.1073/pnas.86.14.5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escano C. S., Juvvadi P. R., Jin F. J., Takahashi T., Koyama Y., Yamashita S., et al. (2009). Disruption of the Aopex11-1 gene involved in peroxisome proliferation leads to impaired Woronin body formation in Aspergillus oryzae. Eukaryotic Cell 8, 296–305 10.1128/EC.00197-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C. Y., Pan J., Chu R., Lee D., Kluckman K. D., Usuda N., et al. (1996). Hepatocellular and hepatic peroxisomal alterations in mice with a disrupted peroxisomal fatty acyl-coenzyme A oxidase gene. J. Biol. Chem. 271, 24698–24710 10.1074/jbc.271.40.24698 [DOI] [PubMed] [Google Scholar]
- Fan J., Quan S., Orth T., Awai C., Chory J., Hu J. (2005). The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol. 139, 231–239 10.1104/pp.105.066811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust J. E., Verma A., Peng C., McNew J. A. (2012). An inventory of peroxisomal proteins and pathways in Drosophila melanogaster. Traffic 13, 1378–1392 10.1111/j.1600-0854.2012.01393.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust P. L. (2003). Abnormal cerebellar histogenesis in PEX2 Zellweger mice reflects multiple neuronal defects induced by peroxisome deficiency. J. Comp. Neurol. 461, 394–413 10.1002/cne.10699 [DOI] [PubMed] [Google Scholar]
- Faust P. L., Hatten M. E. (1997). Targeted deletion of the PEX2 peroxisome assembly gene in mice provides a model for Zellweger syndrome, a human neuronal migration disorder. J. Cell Biol. 139, 1293–1305 10.1083/jcb.139.5.1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A. G., Gunsalus K. C., Huang J., Chuang L. S., Ying N., Liang H. L., et al. (2005). New genes with roles in the C. elegans embryo revealed using RNAi of ovary-enriched ORFeome clones. Genome Res. 15, 250–259 10.1101/gr.3194805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J. E., Maeder M. L., Pearlberg J., Joung J. K., Peterson R. T., Yeh J. R. (2009). Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat. Protoc. 4, 1855–1867 10.1038/nprot.2009.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen M., Nordgren M., Wang B., Apanasets O. (2012). Role of peroxisomes in ROS/RNS-metabolism: implications for human disease. Biochim. Biophys. Acta 1822, 1363–1373 10.1016/j.bbadis.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Fransen M., Nordgren M., Wang B., Apanasets O., Van Veldhoven P. P. (2013). Aging, age-related diseases and peroxisomes. Subcell. Biochem. 69, 45–65 10.1007/978-94-007-6889-5_3 [DOI] [PubMed] [Google Scholar]
- Fujiki Y., Matsuzono Y., Matsuzaki T., Fransen M. (2006). Import of peroxisomal membrane proteins: the interplay of Pex3p- and Pex19p-mediated interactions. Biochim. Biophys. Acta 1763, 1639–1646 10.1016/j.bbamcr.2006.09.030 [DOI] [PubMed] [Google Scholar]
- Gaiano N., Amsterdam A., Kawakami K., Allende M., Becker T., Hopkins N. (1996). Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature 383, 829–832 10.1038/383829a0 [DOI] [PubMed] [Google Scholar]
- Galland N., Demeure F., Hannaert V., Verplaetse E., Vertommen D., Van der Smissen P., et al. (2007). Characterization of the role of the receptors PEX5 and PEX7 in the import of proteins into glycosomes of Trypanosoma brucei. Biochim. Biophys. Acta 1773, 521–535 10.1016/j.bbamcr.2007.01.006 [DOI] [PubMed] [Google Scholar]
- Golling G., Amsterdam A., Sun Z., Antonelli M., Maldonado E., Chen W., et al. (2002). Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 31, 135–140 10.1038/ng896 [DOI] [PubMed] [Google Scholar]
- Gonczy P., Echeverri C., Oegema K., Coulson A., Jones S. J., Copley R. R., et al. (2000). Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408, 331–336 10.1038/35042526 [DOI] [PubMed] [Google Scholar]
- Gonzalez N. H., Felsner G., Schramm F. D., Klingl A., Maier U. G., Bolte K. (2011). A single peroxisomal targeting signal mediates matrix protein import in diatoms. PLoS ONE 6:e25316 10.1371/journal.pone.0025316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould S. J., McCollum D., Spong A. P., Heyman J. A., Subramani S. (1992). Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast 8, 613–628 10.1002/yea.320080805 [DOI] [PubMed] [Google Scholar]
- Gressens P., Baes M., Leroux P., Lombet A., Van Veldhoven P. P., Janssen A., et al. (2000). Neuronal migration disorder in Zellweger mice is secondary to glutamate receptor dysfunction. Ann. Neurol. 48, 336–343 [DOI] [PubMed] [Google Scholar]
- Hofmann A. F., Hagey L. R., Krasowski M. D. (2010). Bile salts of vertebrates: structural variation and possible evolutionary significance. J. Lipid Res. 51, 226–246 10.1194/jlr.R000042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horrocks L. A., Sharma M. (1982). Plasmalogens and O-alkyl glycerophospholipids, in Phospholipids, eds Hawthorne J. N., Anselleditors G. B. (Amsterdam: Elsevier; ), 51–93 [Google Scholar]
- Huang J., Viswakarma N., Yu S., Jia Y., Bai L., Vluggens A., et al. (2011). Progressive endoplasmic reticulum stress contributes to Hepatocarcinogenesis in fatty acyl-CoA oxidase 1-deficient mice. Am. J. Pathol. 179, 703–713 10.1016/j.ajpath.2011.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulshagen L., Krysko O., Bottelbergs A., Huyghe S., Klein R., Van Veldhoven P. P., et al. (2008). Absence of functional peroxisomes from mouse CNS causes dysmyelination and axon degeneration. J. Neurosci. 28, 4015–4027 10.1523/JNEUROSCI.4968-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huybrechts S. J., Van Veldhoven P. P., Brees C., Mannaerts G. P., Los G. V., Fransen M. (2009). Peroxisome dynamics in cultured mammalian cells. Traffic 10, 1722–1733 10.1111/j.1600-0854.2009.00970.x [DOI] [PubMed] [Google Scholar]
- Huyghe S., Mannaerts G. P., Baes M., Van Veldhoven P. P. (2006a). Peroxisomal multifunctional protein-2: the enzyme, the patients and the knockout mouse model. Biochim. Biophys. Acta 1761, 973–994 10.1016/j.bbalip.2006.04.006 [DOI] [PubMed] [Google Scholar]
- Huyghe S., Schmalbruch H., De Gendt K., Verhoeven G., Guillou F., Van Veldhoven P. P., et al. (2006b). Peroxisomal multifunctional protein 2 is essential for lipid homeostasis in Sertoli cells and male fertility in mice. Endocrinology 147, 2228–2236 10.1210/en.2005-1571 [DOI] [PubMed] [Google Scholar]
- Ibabe A., Bilbao E., Cajaraville M. P. (2005). Expression of peroxisome proliferator-activated receptors in zebrafish (Danio rerio) depending on gender and developmental stage. Histochem. Cell Biol. 123, 75–87 10.1007/s00418-004-0737-2 [DOI] [PubMed] [Google Scholar]
- Janssen A., Baes M., Gressens P., Mannaerts G. P., Declercq P., Van Veldhoven P. P. (2000). Docosahexaenoic acid deficit is not a major pathogenic factor in peroxisome-deficient mice. Lab. Invest. 80, 31–35 10.1038/labinvest.3780005 [DOI] [PubMed] [Google Scholar]
- Joo H. J., Kim K. Y., Yim Y. H., Jin Y. X., Kim H., Kim M. Y., et al. (2010). Contribution of the peroxisomal acox gene to the dynamic balance of daumone production in Caenorhabditis elegans. J. Biol. Chem. 285, 29319–29325 10.1074/jbc.M110.122663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo H. J., Yim Y. H., Jeong P. Y., Jin Y. X., Lee J. E., Kim H., et al. (2009). Caenorhabditis elegans utilizes dauer pheromone biosynthesis to dispose of toxic peroxisomal fatty acids for cellular homoeostasis. Biochem. J. 422, 61–71 10.1042/BJ20090513 [DOI] [PubMed] [Google Scholar]
- Kamath R. S., Fraser A. G., Dong Y., Poulin G., Durbin R., Gotta M., et al. (2003). Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Kang H. L., Benzer S., Min K. T. (2002). Life extension in Drosophila by feeding a drug. Proc. Natl. Acad. Sci. U.S.A. 99, 838–843 10.1073/pnas.022631999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C. P., Thomas J. E., Charlton W. L., Baker A. (2001). Identification and characterisation of PEX6 orthologues from plants KAPLAN2001. Biochim. Biophys. Acta 1539, 173–180 10.1016/S0167-4889(01)00091-X [DOI] [PubMed] [Google Scholar]
- Kassmann C. M., Lappe-Siefke C., Baes M., Brugger B., Mildner A., Werner H. B., et al. (2007). Axonal loss and neuroinflammation caused by peroxisome-deficient oligodendrocytes. Nat. Genet. 39, 969–976 10.1038/ng2070 [DOI] [PubMed] [Google Scholar]
- Kassmann C. M., Quintes S., Rietdorf J., Mobius W., Sereda M. W., Nientiedt T., et al. (2011). A role for myelin-associated peroxisomes in maintaining paranodal loops and axonal integrity. FEBS Lett. 585, 2205–2211 10.1016/j.febslet.2011.05.032 [DOI] [PubMed] [Google Scholar]
- Keane M. H., Overmars H., Wikander T. M., Ferdinandusse S., Duran M., Wanders R. J., et al. (2007). Bile acid treatment alters hepatic disease and bile acid transport in peroxisome-deficient PEX2 Zellweger mice. Hepatology 45, 982–997 10.1002/hep.21532 [DOI] [PubMed] [Google Scholar]
- Kemp S., Wei H. M., Lu J. F., Braiterman L. T., McGuinness M. C., Moser A. B., et al. (1998). Gene redundancy and pharmacological gene therapy: implications for X-linked adrenoleukodystrophy. Nat. Med. 4, 1261–1268 10.1038/3242 [DOI] [PubMed] [Google Scholar]
- Koch J., Pranjic K., Huber A., Ellinger A., Hartig A., Kragler F., et al. (2010). PEX11 family members are membrane elongation factors that coordinate peroxisome proliferation and maintenance. J. Cell Sci. 123, 3389–3400 10.1242/jcs.064907 [DOI] [PubMed] [Google Scholar]
- Koepke J. I., Nakrieko K. A., Wood C. S., Boucher K. K., Terlecky L. J., Walton P. A., et al. (2007). Restoration of peroxisomal catalase import in a model of human cellular aging. Traffic 8, 1590–1600 10.1111/j.1600-0854.2007.00633.x [DOI] [PubMed] [Google Scholar]
- Koizumi K., Higashida H., Yoo S., Islam M. S., Ivanov A. I., Guo V., et al. (2007). RNA interference screen to identify genes required for Drosophila embryonic nervous system development. Proc. Natl. Acad. Sci. U.S.A. 104, 5626–5631 10.1073/pnas.0611687104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs W. J., Tape K. N., Shackelford J. E., Wikander T. M., Richards M. J., Fliesler S. J., et al. (2009). Peroxisome deficiency causes a complex phenotype because of hepatic SREBP/Insig dysregulation associated with endoplasmic reticulum stress. J. Biol. Chem. 284, 7232–7245 10.1074/jbc.M809064200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysko O., Hulshagen L., Janssen A., Schutz G., Klein R., De B. M., et al. (2007). Neocortical and cerebellar developmental abnormalities in conditions of selective elimination of peroxisomes from brain or from liver. J. Neurosci. Res. 85, 58–72 10.1002/jnr.21097 [DOI] [PubMed] [Google Scholar]
- Krysko O., Stevens M., Langenberg T., Fransen M., Espeel M., Baes M. (2010). Peroxisomes in zebrafish: distribution pattern and knockdown studies. Histochem. Cell Biol. 134, 39–51 10.1007/s00418-010-0712-z [DOI] [PubMed] [Google Scholar]
- Li X., Baumgart E., Dong G. X., Morrell J. C., Jimenez-Sanchez G., Valle D., et al. (2002a). PEX11alpha is required for peroxisome proliferation in response to 4-phenylbutyrate but is dispensable for peroxisome proliferator-activated receptor alpha-mediated peroxisome proliferation. Mol. Cell. Biol. 22, 8226–8240 10.1128/MCB.22.23.8226-8240.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Baumgart E., Morrell J. C., Jimenez-Sanchez G., Valle D., Gould S. J. (2002b). PEX11 beta deficiency is lethal and impairs neuronal migration but does not abrogate peroxisome function. Mol. Cell. Biol. 22, 4358–4365 10.1128/MCB.22.12.4358-4365.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda I., Kohara Y., Yamamoto M., Sugimoto A. (2001). Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr. Biol. 11, 171–176 10.1016/S0960-9822(01)00052-5 [DOI] [PubMed] [Google Scholar]
- Managadze D., Wurtz C., Sichting M., Niehaus G., Veenhuis M., Rottensteiner H. (2007). The peroxin PEX14 of Neurospora crassa is essential for the biogenesis of both glyoxysomes and Woronin bodies. Traffic 8, 687–68701 10.1111/j.1600-0854.2007.00560.x [DOI] [PubMed] [Google Scholar]
- Martens K., Bottelbergs A., Peeters A., Jacobs F., Espeel M., Carmeliet P., et al. (2012). Peroxisome deficient aP2-Pex5 knockout mice display impaired white adipocyte and muscle function concomitant with reduced adrenergic tone. Mol. Genet. Metab. 107, 735–747 10.1016/j.ymgme.2012.10.015 [DOI] [PubMed] [Google Scholar]
- Mast F. D., Li J., Virk M. K., Hughes S. C., Simmonds A. J., Rachubinski R. A. (2011). A Drosophila model for the Zellweger spectrum of peroxisome biogenesis disorders. Dis. Model. Mech. 4, 659–672 10.1242/dmm.007419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell M., Bjorkman J., Nguyen T., Sharp P., Finnie J., Paterson C., et al. (2003). Pex13 inactivation in the mouse disrupts peroxisome biogenesis and leads to a Zellweger syndrome phenotype. Mol. Cell. Biol. 23, 5947–5957 10.1128/MCB.23.16.5947-5957.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais S., Knoll-Gellida A., Andre M., Barthe C., Babin P. J. (2007). Conserved expression of alternative splicing variants of peroxisomal acyl-CoA oxidase 1 in vertebrates and developmental and nutritional regulation in fish. Physiol. Genomics 28, 239–252 10.1152/physiolgenomics.00136.2006 [DOI] [PubMed] [Google Scholar]
- Motley A. M., Hettema E. H., Ketting R., Plasterk R., Tabak H. F. (2000). Caenorhabditis elegans has a single pathway to target matrix proteins to peroxisomes. EMBO Rep. 1, 40–46 10.1093/embo-reports/kvd010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C. C., Nguyen T. H., Ahlemeyer B., Meshram M., Santrampurwala N., Cao S., et al. (2011). PEX13 deficiency in mouse brain as a model of Zellweger syndrome: abnormal cerebellum formation, reactive gliosis and oxidative stress. Dis. Model. Mech. 4, 104–119 10.1242/dmm.004622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama M., Sato H., Okuda T., Fujisawa N., Kono N., Arai H., et al. (2011). Drosophila carrying pex3 or pex16 mutations are models of Zellweger syndrome that reflect its symptoms associated with the absence of peroxisomes. PLoS ONE 6:e22984 10.1371/journal.pone.0022984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Zarragoitia M., Trant J. M., Cajaravillet M. P. (2006). Effects of dibutylphthalate and ethynylestradiol on liver peroxisomes, reproduction, and development of zebrafish (Danio rerio). Environ. Toxicol. Chem. 25, 2394–2404 10.1897/05-456R.1 [DOI] [PubMed] [Google Scholar]
- Peeters A., Swinnen J. V., Van Veldhoven P. P., Baes M. (2011). Hepatosteatosis in peroxisome deficient liver despite increased beta-oxidation capacity and impaired lipogenesis. Biochimie 93, 1828–1838 10.1016/j.biochi.2011.06.034 [DOI] [PubMed] [Google Scholar]
- Petriv O. I., Pilgrim D. B., Rachubinski R. A., Titorenko V. I. (2002). RNA interference of peroxisome-related genes in, C. elegans: a new model for human peroxisomal disorders. Physiol Genomics 10, 79–91 10.1152/physiolgenomics.00044.2002 [DOI] [PubMed] [Google Scholar]
- Ramos-Pamplona M., Naqvi N. I. (2006). Host invasion during rice-blast disease requires carnitine-dependent transport of peroxisomal acetyl-CoA. Mol. Microbiol. 61, 61–75 10.1111/j.1365-2958.2006.05194.x [DOI] [PubMed] [Google Scholar]
- Rodemer C., Thai T. P., Brugger B., Kaercher T., Werner H., Nave K. A., et al. (2003). Inactivation of ether lipid biosynthesis causes male infertility, defects in eye development and optic nerve hypoplasia in mice. Hum. Mol. Genet. 12, 1881–1895 10.1093/hmg/ddg191 [DOI] [PubMed] [Google Scholar]
- Rual J. F., Ceron J., Koreth J., Hao T., Nicot A. S., Hirozane-Kishikawa T., et al. (2004). Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14, 2162–2168 10.1101/gr.2505604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U., Wanner G., Veenhuis M., Schmid M., Gietl C. (2003). AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. U.S.A. 100, 9626–9631 10.1073/pnas.1633697100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichting M., Schell-Steven A., Prokisch H., Erdmann R., Rottensteiner H. (2003). Pex7p and Pex20p of Neurospora crassa function together in PTS2-dependent protein import into peroxisomes. Mol. Biol. Cell 14, 810–821 10.1091/mbc.E02-08-0539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth D., Ch'ng Q., Dybbs M., Tavazoie M., Kennedy S., Wang D., et al. (2005). Systematic analysis of genes required for synapse structure and function. Nature 436, 510–517 10.1038/nature03809 [DOI] [PubMed] [Google Scholar]
- Simmer F., Moorman C., van der Linden A. M., Kuijk E., van den Berghe P. V., Kamath R. S., et al. (2003). Genome-wide RNAi of, C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1:E12 10.1371/journal.pbio.0000012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B., Koski L. B., Walsh A., Marschall P., Neumann B., Brehm M., et al. (2005). Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434, 462–469 10.1038/nature03353 [DOI] [PubMed] [Google Scholar]
- Soukupova M., Sprenger C., Gorgas K., Kunau W. H., Dodt G. (1999). Identification and characterization of the human peroxin PEX3. Eur. J. Cell Biol. 78, 357–374 10.1016/S0171-9335(99)80078-8 [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Stern D., Beaton A., Rhem E. J., Laverty T., Mozden N., et al. (1999). The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153, 135–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieringer H., Moellers B., Dodt G., Kunau W. H., Driscoll M. (2003). Modeling human peroxisome biogenesis disorders in the nematode Caenorhabditis elegans. J. Cell Sci. 116, 1797–1804 10.1242/jcs.00380 [DOI] [PubMed] [Google Scholar]
- Thisse B., Heyer V., Lux A., Alunni V., Degrave A., Seiliez I., et al. (2004). Spatial and temporal expression of the zebrafish genome by large-scale in situ hybridization screening. Methods Cell Biol. 77, 505–519 10.1016/S0091-679X(04)77027-2 [DOI] [PubMed] [Google Scholar]
- Thoms S., Erdmann R. (2005). Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J. 272, 5169–5181 10.1111/j.1742-4658.2005.04939.x [DOI] [PubMed] [Google Scholar]
- Titorenko V. I., Terlecky S. R. (2011). Peroxisome metabolism and cellular aging. Traffic 12, 252–259 10.1111/j.1600-0854.2010.01144.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Veldhoven P. P. (2010). Biochemistry and genetics of inherited disorders of peroxisomal fatty acid metabolism. J. Lipid Res. 51, 2863–2895 10.1194/jlr.R005959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam A. B., Lall S. P., Denovan-Wright E. M., Wright J. M. (2012). Tissue-specific differential induction of duplicated fatty acid-binding protein genes by the peroxisome proliferator, clofibrate, in zebrafish (Danio rerio). BMC Evol. Biol. 12:112 10.1186/1471-2148-12-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders R. J., Waterham H. R. (2005). Peroxisomal disorders I: biochemistry and genetics of peroxisome biogenesis disorders. Clin. Genet. 67, 107–133 10.1111/j.1399-0004.2004.00329.x [DOI] [PubMed] [Google Scholar]
- Waterham H. R., Ebberink M. S. (2012). Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim. Biophys. Acta 1822, 1430–1441 10.1016/j.bbadis.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Weng H., Ji X., Naito Y., Endo K., Ma X., Takahashi R., et al. (2013). Pex11alpha deficiency impairs peroxisome elongation and division and contributes to nonalcoholic fatty liver in mice. Am. J. Physiol. Endocrinol. Metab. 304, E187–E196 10.1152/ajpendo.00425.2012 [DOI] [PubMed] [Google Scholar]
- Yakunin E., Moser A., Loeb V., Saada A., Faust P., Crane D. I., et al. (2010). alpha-Synuclein abnormalities in mouse models of peroxisome biogenesis disorders. J. Neurosci. Res. 88, 866–876 10.1002/jnr.22246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota S., Togo S. H., Maebuchi M., Bun-Ya M., Haraguchi C. M., Kamiryo T. (2002). Peroxisomes of the nematode Caenorhabditis elegans: distribution and morphological characteristics. Histochem. Cell Biol. 118, 329–336 10.1007/s00418-002-0450-y [DOI] [PubMed] [Google Scholar]
- Zhang G. F., Sadhukhan S., Ibarra R. A., Lauden S. M., Chuang C. Y., Sushailo S., et al. (2012). Metabolism of gamma-hydroxybutyrate in perfused rat livers. Biochem. J. 444, 333–341 10.1042/BJ20112046 [DOI] [PubMed] [Google Scholar]
- Zhou B., Yang L., Li S., Huang J., Chen H., Hou L., et al. (2012). Midlife gene expressions identify modulators of aging through dietary interventions. Proc. Natl. Acad. Sci. U.S.A. 109, E1201–E1209 10.1073/pnas.1119304109 [DOI] [PMC free article] [PubMed] [Google Scholar]