Abstract
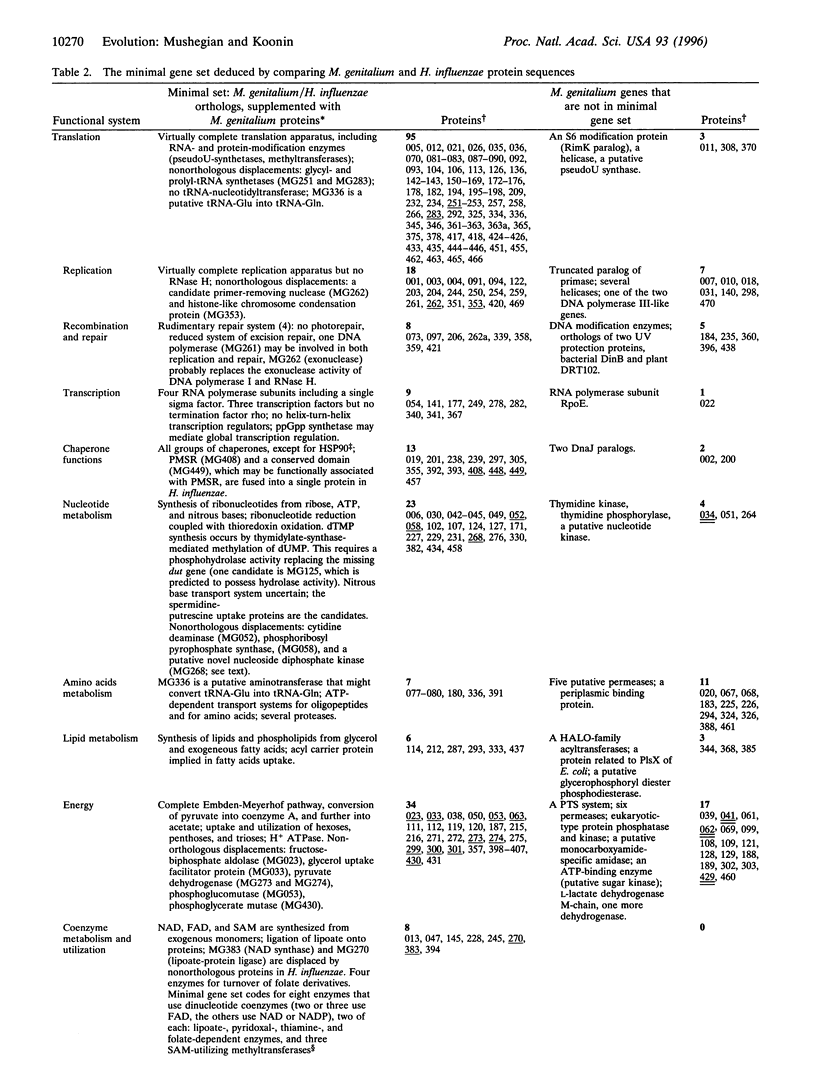
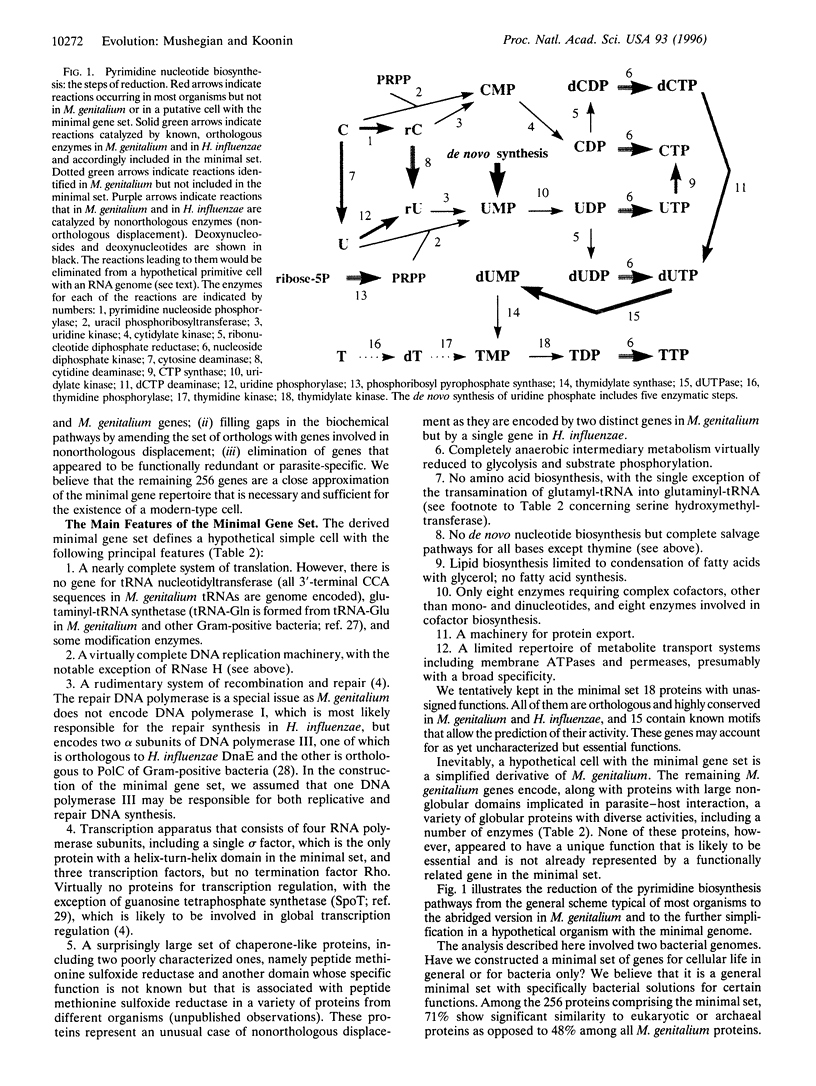
The recently sequenced genome of the parasitic bacterium Mycoplasma genitalium contains only 468 identified protein-coding genes that have been dubbed a minimal gene complement [Fraser, C.M., Gocayne, J.D., White, O., Adams, M.D., Clayton, R.A., et al. (1995) Science 270, 397-403]. Although the M. genitalium gene complement is indeed the smallest among known cellular life forms, there is no evidence that it is the minimal self-sufficient gene set. To derive such a set, we compared the 468 predicted M. genitalium protein sequences with the 1703 protein sequences encoded by the other completely sequenced small bacterial genome, that of Haemophilus influenzae. M. genitalium and H. influenzae belong to two ancient bacterial lineages, i.e., Gram-positive and Gram-negative bacteria, respectively. Therefore, the genes that are conserved in these two bacteria are almost certainly essential for cellular function. It is this category of genes that is most likely to approximate the minimal gene set. We found that 240 M. genitalium genes have orthologs among the genes of H. influenzae. This collection of genes falls short of comprising the minimal set as some enzymes responsible for intermediate steps in essential pathways are missing. The apparent reason for this is the phenomenon that we call nonorthologous gene displacement when the same function is fulfilled by nonorthologous proteins in two organisms. We identified 22 nonorthologous displacements and supplemented the set of orthologs with the respective M. genitalium genes. After examining the resulting list of 262 genes for possible functional redundancy and for the presence of apparently parasite-specific genes, 6 genes were removed. We suggest that the remaining 256 genes are close to the minimal gene set that is necessary and sufficient to sustain the existence of a modern-type cell. Most of the proteins encoded by the genes from the minimal set have eukaryotic or archaeal homologs but seven key proteins of DNA replication do not. We speculate that the last common ancestor of the three primary kingdoms had an RNA genome. Possibilities are explored to further reduce the minimal set to model a primitive cell that might have existed at a very early stage of life evolution.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Boguski M. S., Gish W., Wootton J. C. Issues in searching molecular sequence databases. Nat Genet. 1994 Feb;6(2):119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Benner S. A., Ellington A. D., Tauer A. Modern metabolism as a palimpsest of the RNA world. Proc Natl Acad Sci U S A. 1989 Sep;86(18):7054–7058. doi: 10.1073/pnas.86.18.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Ouzounis C., Casari G., Schneider R., Sander C., Dolan M., Gilbert W., Gillevet P. M. Exploring the Mycoplasma capricolum genome: a minimal cell reveals its physiology. Mol Microbiol. 1995 Jun;16(5):955–967. doi: 10.1111/j.1365-2958.1995.tb02321.x. [DOI] [PubMed] [Google Scholar]
- Bork P., Sander C., Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci U S A. 1992 Aug 15;89(16):7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C., Squires C., Squires C. L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995 Dec;59(4):623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danson M. J., Hough D. W. The enzymology of archaebacterial pathways of central metabolism. Biochem Soc Symp. 1992;58:7–21. [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Tsang S., Cho G., Little E. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science. 1996 Jan 26;271(5248):470–477. doi: 10.1126/science.271.5248.470. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Fitch W. M. Distinguishing homologous from analogous proteins. Syst Zool. 1970 Jun;19(2):99–113. [PubMed] [Google Scholar]
- Fleischmann R. D., Adams M. D., White O., Clayton R. A., Kirkness E. F., Kerlavage A. R., Bult C. J., Tomb J. F., Dougherty B. A., Merrick J. M. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995 Jul 28;269(5223):496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Fraser C. M., Gocayne J. D., White O., Adams M. D., Clayton R. A., Fleischmann R. D., Bult C. J., Kerlavage A. R., Sutton G., Kelley J. M. The minimal gene complement of Mycoplasma genitalium. Science. 1995 Oct 20;270(5235):397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- Green P., Lipman D., Hillier L., Waterston R., States D., Claverie J. M. Ancient conserved regions in new gene sequences and the protein databases. Science. 1993 Mar 19;259(5102):1711–1716. doi: 10.1126/science.8456298. [DOI] [PubMed] [Google Scholar]
- Itaya M. An estimation of minimal genome size required for life. FEBS Lett. 1995 Apr 10;362(3):257–260. doi: 10.1016/0014-5793(95)00233-y. [DOI] [PubMed] [Google Scholar]
- Kahane I., Horowitz S. Adherence of mycoplasma to cell surfaces. Subcell Biochem. 1993;20:225–241. doi: 10.1007/978-1-4615-2924-8_8. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Bork P. Ancient duplication of DNA polymerase inferred from analysis of complete bacterial genomes. Trends Biochem Sci. 1996 Apr;21(4):128–129. [PubMed] [Google Scholar]
- Koonin E. V., Mushegian A. R., Rudd K. E. Sequencing and analysis of bacterial genomes. Curr Biol. 1996 Apr 1;6(4):404–416. doi: 10.1016/s0960-9822(02)00508-0. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Tatusov R. L., Rudd K. E. Protein sequence comparison at genome scale. Methods Enzymol. 1996;266:295–322. doi: 10.1016/s0076-6879(96)66020-0. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Tatusov R. L., Rudd K. E. Sequence similarity analysis of Escherichia coli proteins: functional and evolutionary implications. Proc Natl Acad Sci U S A. 1995 Dec 5;92(25):11921–11925. doi: 10.1073/pnas.92.25.11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q., Zhang X., Almaula N., Mathews C. K., Inouye M. The gene for nucleoside diphosphate kinase functions as a mutator gene in Escherichia coli. J Mol Biol. 1995 Dec 1;254(3):337–341. doi: 10.1006/jmbi.1995.0620. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Woese C. R., Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994 Jan;176(1):1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R. Effective protein sequence comparison. Methods Enzymol. 1996;266:227–258. doi: 10.1016/s0076-6879(96)66017-0. [DOI] [PubMed] [Google Scholar]
- Saraste M., Sibbald P. R., Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990 Nov;15(11):430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- Strauch M. A., Zalkin H., Aronson A. I. Characterization of the glutamyl-tRNA(Gln)-to-glutaminyl-tRNA(Gln) amidotransferase reaction of Bacillus subtilis. J Bacteriol. 1988 Feb;170(2):916–920. doi: 10.1128/jb.170.2.916-920.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R. L., Altschul S. F., Koonin E. V. Detection of conserved segments in proteins: iterative scanning of sequence databases with alignment blocks. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):12091–12095. doi: 10.1073/pnas.91.25.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R. L., Mushegian A. R., Bork P., Brown N. P., Hayes W. S., Borodovsky M., Rudd K. E., Koonin E. V. Metabolism and evolution of Haemophilus influenzae deduced from a whole-genome comparison with Escherichia coli. Curr Biol. 1996 Mar 1;6(3):279–291. doi: 10.1016/s0960-9822(02)00478-5. [DOI] [PubMed] [Google Scholar]
- Wootton J. C., Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]




