Abstract
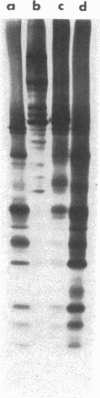
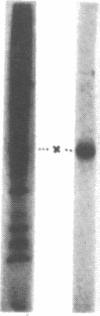
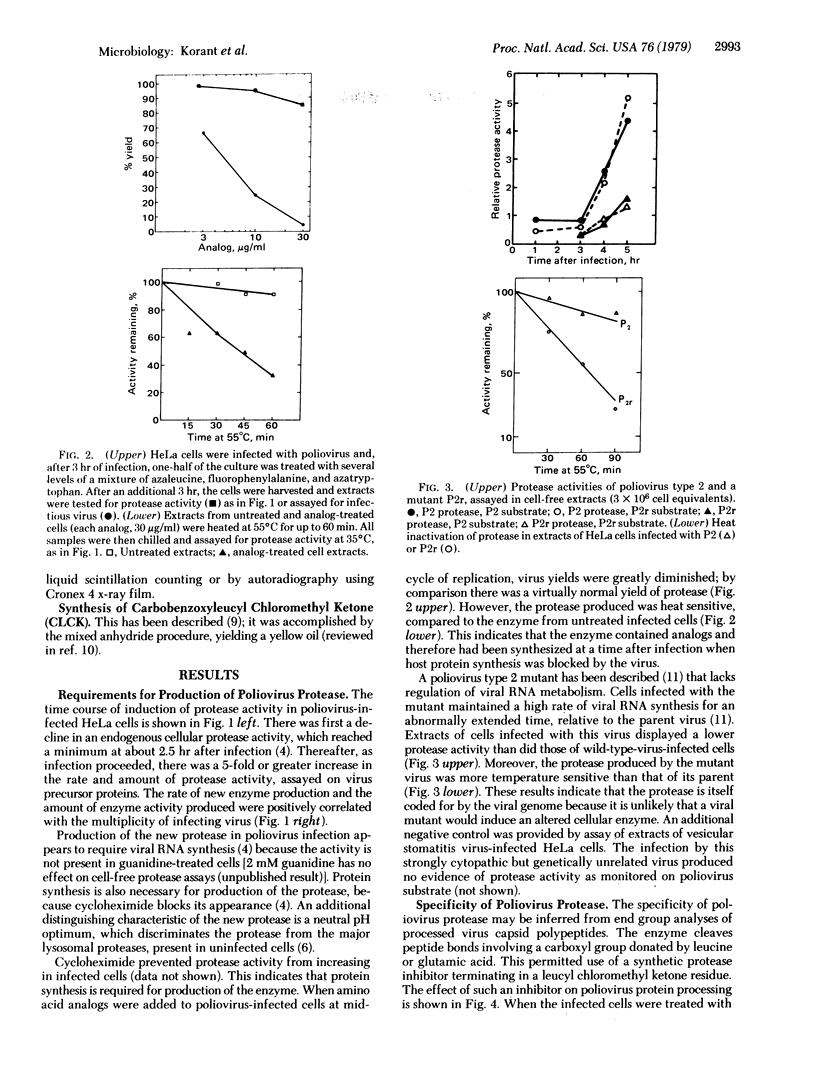
Previous studies have shown that primary cleavages in nascent picornavirus precursors are accomplished by cellular proteases. This study has characterized the enzyme in infected cells that produces the capsid polypeptides by secondary cleavages of viral precursors. The kinetics of the production of protease activity correlate with the time course of virus protein synthesis, and the new enzyme has characteristic pH and temperature optima. Guanidine and cycloheximide, which are inhibitors of virus RNA and protein synthesis, prevent production of the protease. As determined by introduction of amino acid analogs into the protease or inhibition by a leucyl chloromethyl ketone, the enzyme is synthesized at a time of infection when host cell proteins are not produced, and the enzyme copurified with a 40,000-dalton virus polypeptide present in the cytoplasm of infected cells. Wild-type levels of protease activity are produced by viral mutants that are defective in coat protein synthesis. The conclusion is that a non-structural poliovirus gene product participates in protein cleavages that produce the viral coat proteins.
Full text
PDF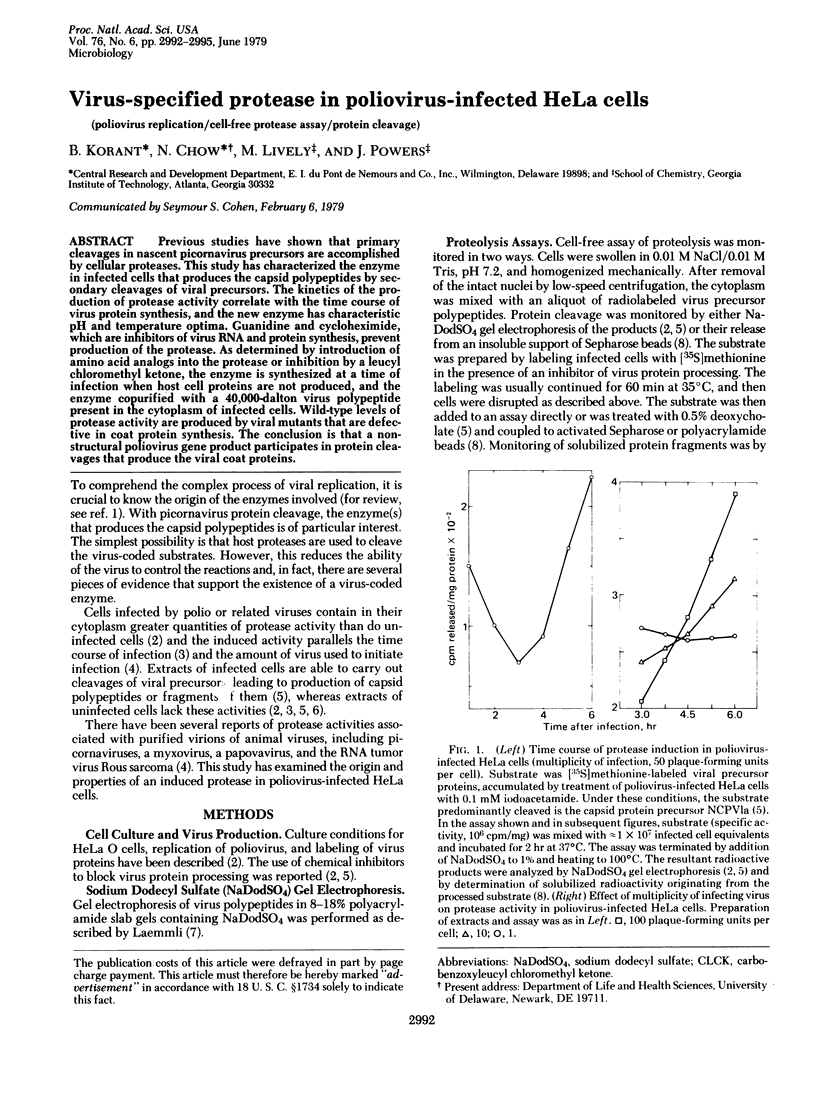
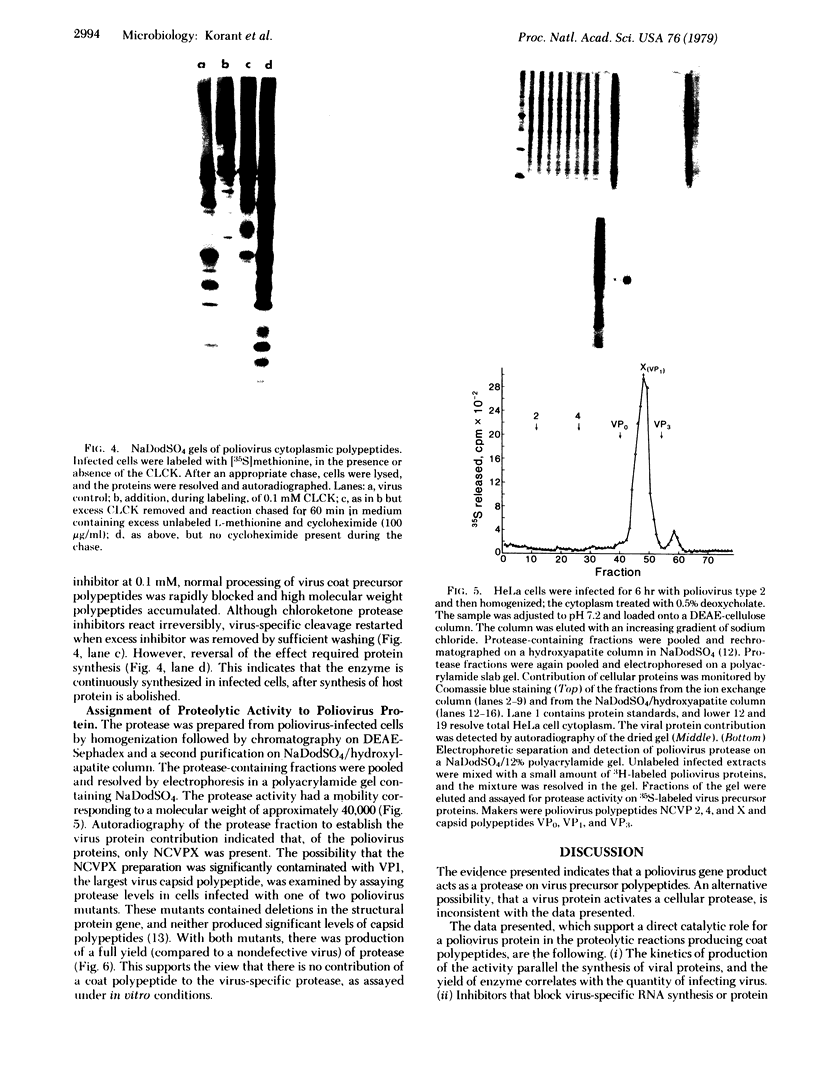
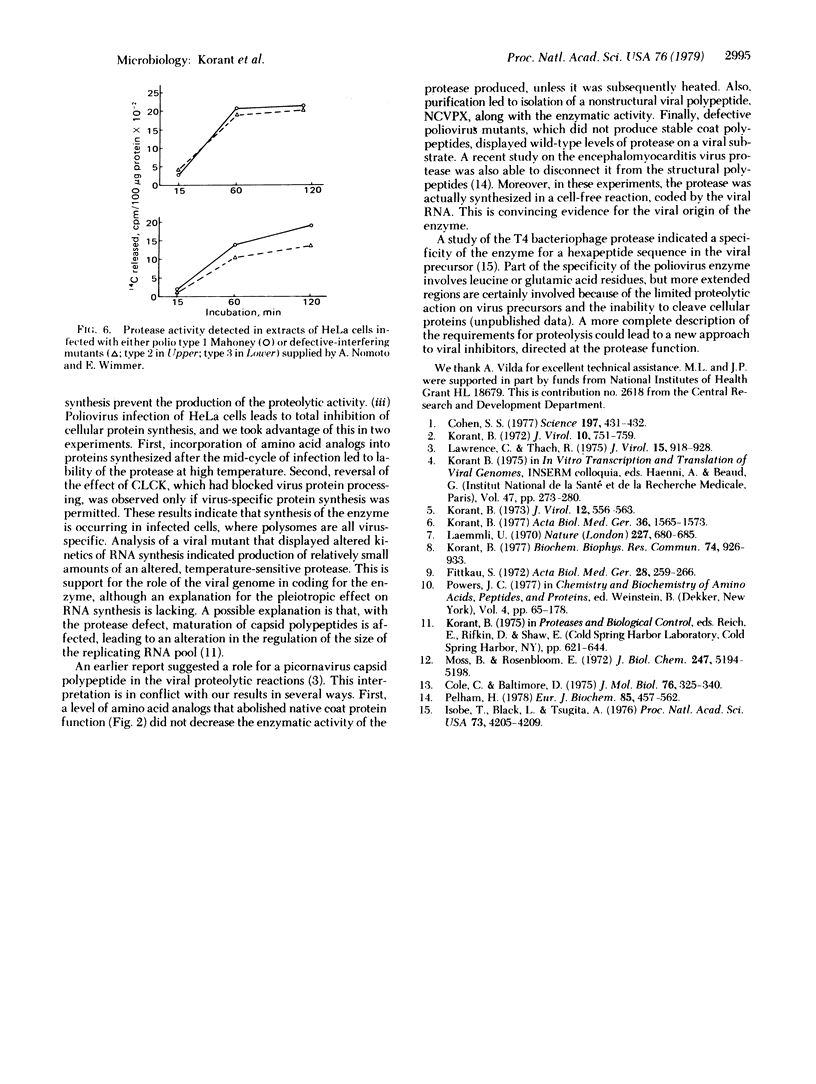
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cole C. N., Baltimore D. Defective interfering particles of poliovirus. II. Nature of the defect. J Mol Biol. 1973 May 25;76(3):325–343. doi: 10.1016/0022-2836(73)90508-1. [DOI] [PubMed] [Google Scholar]
- Fittkau S. Synthese und eigenschaften von 1-Chlor-3-amino-4-pphenylbutanon-2, einem Inhibitor der Leuzinaminopeptidase aus Rinderaugenlinsen. Acta Biol Med Ger. 1972;28(2):259–267. [PubMed] [Google Scholar]
- Isobe T., Black L. W., Tsugita A. Protein cleavage during virus assembly: a novel specificity of assembly dependent cleavage in bacteriophage T4. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4205–4209. doi: 10.1073/pnas.73.11.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korant B. D. Cleavage of poliovirus-specific polypeptide aggregates. J Virol. 1973 Sep;12(3):556–563. doi: 10.1128/jvi.12.3.556-563.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korant B. D. Cleavage of viral precursor proteins in vivo and in vitro. J Virol. 1972 Oct;10(4):751–759. doi: 10.1128/jvi.10.4.751-759.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korant B. D. Protease activity associated with HeLa cell ribosomes. Biochem Biophys Res Commun. 1977 Feb 7;74(3):926–933. doi: 10.1016/0006-291x(77)91607-2. [DOI] [PubMed] [Google Scholar]
- Korant B. D. Protein cleavage in virus-infected cells. Acta Biol Med Ger. 1977;36(11-12):1565–1573. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawrence C., Thach R. E. Identification of a viral protein involved in post-translational maturation of the encephalomyocarditis virus capsid precursor. J Virol. 1975 Apr;15(4):918–928. doi: 10.1128/jvi.15.4.918-928.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B., Rosenblum E. N. Hydroxylapatite chromatography of protein-sodium dodecyl sulfate complexes. A new method for the separation of polypeptide subunits. J Biol Chem. 1972 Aug 25;247(16):5194–5198. [PubMed] [Google Scholar]
- Pelham H. R. Translation of encephalomyocarditis virus RNA in vitro yields an active proteolytic processing enzyme. Eur J Biochem. 1978 Apr 17;85(2):457–462. doi: 10.1111/j.1432-1033.1978.tb12260.x. [DOI] [PubMed] [Google Scholar]