Ryanodine Receptors as Regulated Guardians of Intracellular Ca2+ Stores
Ryanodine-sensitive Ca2+ release channels, also known as the ryanodine receptors (RyRs),2 are homotetramers of an ∼550-kDa subunit (Fig. 1) that are resident proteins of intracellular membranes such as the sarcoplasmic/endoplasmic reticulum. RyRs are responsible for the regulated release of Ca2+ from these lumenal stores.
FIGURE 1.
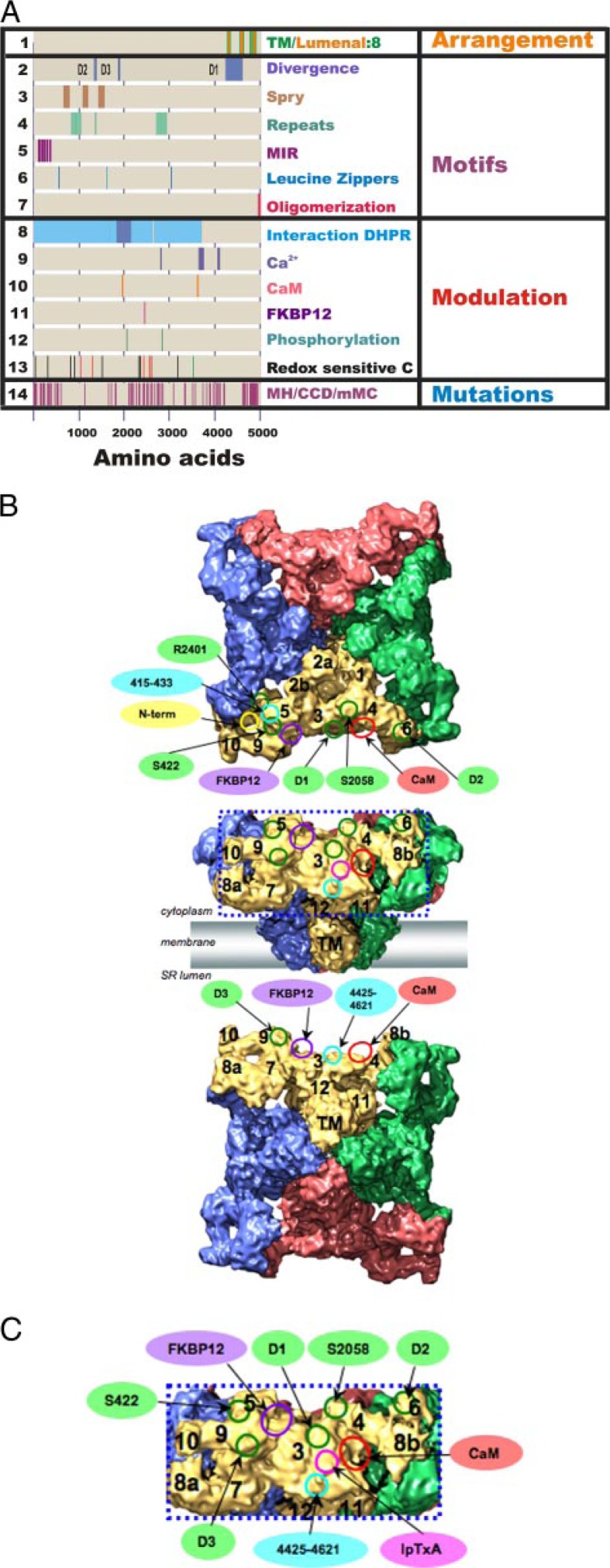
Correlating primary sequence motifs with subdomains in the 9.6-Å three-dimensional structure. A, sites identified in the primary sequence of RyR1. DHPR, dihydropyridine receptor; MH, malignant hyperthermia; CCD, central core disease; mMC, multiminicore disease. B, cryo-EM map viewed from the cytoplasm (upper panel) and from the sarcoplasmic reticulum (SR) lumen (lower panel). RyR1 subunits are color-coded; putative subregions are indicated. Circles indicate the ligand-binding sites and RyR sequences on the three-dimensional map of RyR by single-particle cryo-EM: red, CaM-binding site; purple, FKBP12-binding site; magenta, imperatoxin A (IpTxA)-binding site; green, sequences mapped by reconstructions of expressed RyR2-GFP fusion proteins; cyan, sequence-specific antibody-binding sites; yellow, the position of the glutathione S-transferase tag fused to the N terminus of RyR3. C, enlargement of the boxed region in B. Ligand-binding sites are labeled as described for B.
Over the last 10 years, single-particle cryoelectron microscopy (cryo-EM) has produced several low resolution (∼30 Å) structures of RyRs (for review, see Ref. 1). Recently, cryo-EM reconstructions of RyR1 have reached subnanometer resolution (2, 3), a breakthrough due to improvements in cryo-specimen preparation, instrumentation, image processing, and three-dimensional reconstruction techniques.
RyRs have a mushroom shape with 4-fold symmetry (Fig. 1B) (2). Most of the mass of RyR forms a large cytoplasmic (CY) assembly (280 × 280 × 120 Å) that is connected to the transmembrane (TM) region by a stalk-like structure. The CY region is strikingly empty with numerous distinctive structural domains and intervening cavities that appear suitable for interaction with modulators that bind within the N-terminal regions of RyR (Fig. 1). The clamp-shaped regions, located at the corners of the CY assembly, are likely regions for the interdigitation of neighboring RyRs seen in situ (5) or for interaction with modulators. The clamp-shaped regions are interconnected to form a continuous network between the central rim and the CY stalk-like structure via several bridging densities. The TM region (120 ×120 × 60 Å) is rotated by ∼40° with respect to the CY region.
Pore Region
The pore is thought to be composed of six to eight TM segments (RyR1 TM1–8, amino acids (aa) 4277–4323, 4343–4363, 4557–4576, 4637–4662, 4776–4800, 4803–4825, 4834–4854, and 4911–4935 with aa 4854–4911 forming the pore helix and selectivity filter) (Fig. 1A, line 1) (6). This arrangement places both the N and C termini of RyR in the cytoplasm. Sequence numbers used in this review refer to rabbit RyR1 (Swiss-Prot accession number P11716) and, where indicated, RyR2 (Swiss-Prot accession number P30957).
The best current maps of RyR1 have resolutions of ∼10 Å (2, 3), allowing delineation of secondary structure elements in the pore region (Fig. 2A): the helix running across the membrane (helix 1, possibly TM8), the short helix (helix 2, which is likely to be part of the pore helix) that forms the lumenal entrance of the channel, and helix 3 (located parallel to the membrane plane on the CY side of the TM region). Helices 1 and 2 lie near the 4-fold channel axis and resemble the inner (pore-lining) and pore helices in K+ channel structures (7, 8). The orientation of helix 3 is similar to the slide helix seen in the KirBac1.1 structure (9).
FIGURE 2.
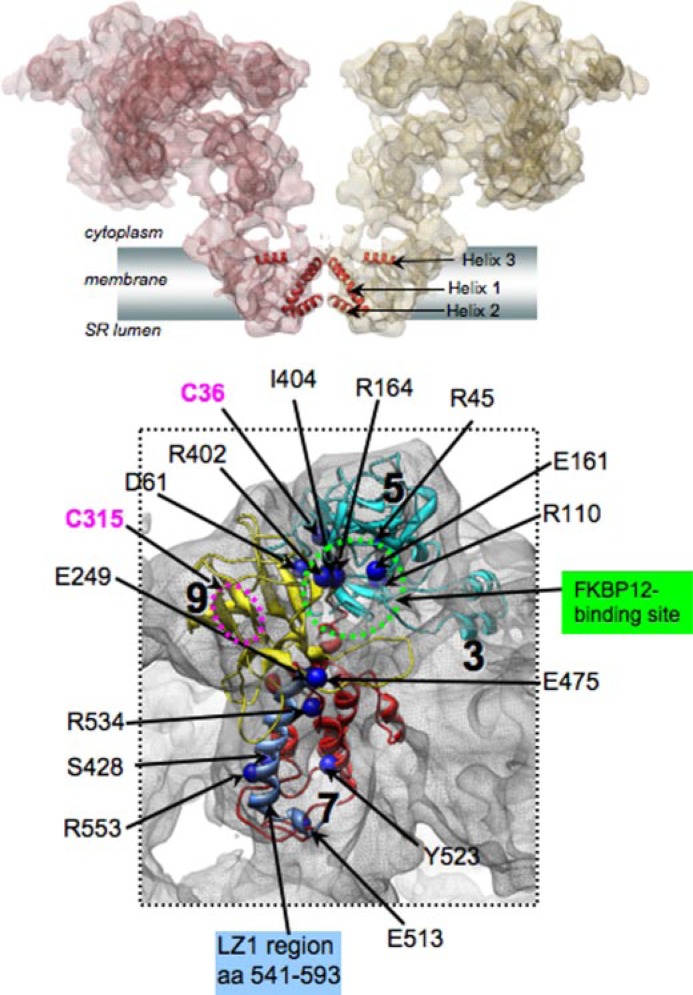
RyR1 Ca2+ release channel closed-state structure at 9.6-Å resolution (2, 4). A, two subunits of RyR1 are shown as semitransparent surfaces. α-Helices in the Ca2+ conduction pathway of the TM region are annotated. SR, sarcoplasmic reticulum. B, malignant hyperthermia/central core disease mutations are mapped to the homology models for the N-terminal region of RyR1 (aa 12–565), localized to the clamp-shaped region of the channel structure (aa numbers correspond to Swiss-Prot accession number P11716). The FKBP12-binding region is indicated in green; the LZ1 region is shown in blue; and hyper-reactive cysteines (Cys36 and Cys315) are indicated in magenta. The location of Cys315 (magenta circle) is tentatively assigned because sequence Ala311–Glu343 was excluded from the homology model due to lack of a structural template for this region.
Two recent RyR1 structures (2, 3) were determined under conditions favoring the closed channel, the arrangement of the membrane-spanning helix that, in these structures, is quite different. Ludtke et al. (2) found a bent pore-lining helix (Fig. 2A) and suggested that the structure is similar to the open MthK channel (8). This raises questions as to whether “kinking” of the inner helix alone, as proposed for K+ channels, is adequate to open RyRs. However, the conformation of the pore-lining helix in the other high resolution RyR1 cryo-EM structure (3) is straight and resembles the closed KcsA channel structure (7). These differences may due to resolution or to the conformational state of the channel during the imaging. At the current level of resolution in the three-dimensional structure of RyR, it is not possible to elucidate the number of TM helices.
Mass Movements Associated with Channel Opening
Single-particle cryo-EM has been used to generate structures of RyR1 in different conformational states to explore the structural transitions associated with the RyR gating. RyR1 was frozen under conditions determined from electrophysiological and biochemical studies (10–12) to drive the channel into either an opened or a closed state. Although the resolution of these structures was only 25–30 Å, global conformational changes associated with the closed-open transition of the RyR channel were detected in both the CY region (opening of the clamps) and the TM region (mass depletion from the center and a twisting motion of the TM region). Channel opening was proposed to be similar to the opening-closing of the iris in a camera diaphragm.
Four approaches are used to identify the locations of specific sequences in the three-dimensional structures: 1) difference mapping with and without bound modulators; 2) difference mapping with and without sequence-specific antibodies; 3) insertion of tags such as green fluorescent protein (GFP) into defined positions in the primary sequence, followed by difference mapping; and 4) docking of molecular models of domains derived from crystal structures into the three-dimensional maps.
RyR Isoforms and Divergent Regions
There are three known RyR isoforms encoded by three distinct genes: RyR1 (expressed primarily in skeletal muscle), RyR2 (expressed in cardiac muscle and brain), and RyR3 (expressed in diaphragm, smooth muscle, and brain). These isoforms share ∼65% sequence identity. Regions of divergence (D1, aa 4254–4631 in RyR1 and aa 4210–4562 in RyR2; D2, aa 1342–1403 in RyR1 and aa 1353–1397 in RyR2; and D3, aa 1872–1923 in RyR1 and aa 1852–1890 in RyR2) are indicated in Fig. 1A (line 2). These divergent regions could be responsible for important isoform-specific functions, or they could represent regions that are not evolutionarily conserved. Three-dimensional structures of all three RyR isoforms determined at ∼30 Å using single-particle cryo-EM are virtually identical (10–12).
Using GFP insertions, D1 was mapped to subregion 3, whereas both D2 and D3 were mapped to the clamp in subregions 6 and 9, respectively (Fig. 1B). The three divergent regions all appear to be located in the CY domains close to the top of the structure, as would be expected for regions of RyR1 involved in interactions with CaV1.1. However, whether they are involved remains to be determined.
Primary Sequence Motifs
Analysis of the primary sequence of RyR1 has revealed several motifs found in other proteins, but the functional roles of these motifs in RyRs are unknown. These motifs include 1) three SPRY domains (aa 582–798, 1014–1209, and 1358–1571) (Fig. 1A, line 3), which are domains found in members of the immunoglobulin superfamily; 2) a sequence that is repeated four times in two tandem domains (aa 842–955, 956–1069, 2726–2845, and 2846–2959) (Fig. 1A, line 4) with two additional truncated repeats located between the two domains (aa 1345–1360 and 1373–1388); 3) five MIR domains (aa 98–153, 160–205, 211–265, 271–334, and 336–394) (Fig. 1A, line 5), which are sequences found in RyRs, inositol trisphosphate receptors, and protein O-mannosyltransferases; and 4) two or more leucine zippers (aa 554–575 and 3039–3071 in RyR1 and aa 565–594, 1604–1632, and 3005–3036 in RyR2) (Fig. 1A, line 6), which are protein-protein interaction motifs and have been suggested to anchor PP1/spinophilin, PP2A/PR130, and protein kinase A/RII (protein kinase A regulatory subunit II)/ mAKAP (muscle A-kinase anchoring protein) to RyR2 (13). The MIR domains are all located close to the N terminus, which is part of the clamp domain, suggesting that these domains may contribute to the structure/function of the clamps (4). Computational placement of a homology model based on the crystal structure of the inositol trisphosphate (IP3)-binding and suppressor domains of the IP3 receptor (4) places the first leucine zipper (aa 554–574) in the clamp domain in subregion 7 (Figs. 1B and 2B). If this is indeed the site that anchors kinases or phosphatases to RyR1, the targets of the enzymes may be sequences in close proximity to this site.
Stewart et al. (15) proposed that the last 15 amino acids in the C-terminal tail are required for tetramer formation. Galvan and Mignery (16) also suggested a C-terminal tetramerization domain but suggested that this was between aa 4869 and 5019 in RyR1. These regions are indicated in Fig. 1A (line 7). Because the C termini of RyRs are cytoplasmic, the region of tetramerization must also be in CY domains.
Channel Modulators
The activities of all three RyR isoforms are modulated by a large number of agents, including ATP, Ca2+, Mg2+, calmodulin, FKBP12, junctin, triadin, calsequestrin, S100A, sorcin, and many others. Most of these modulators bind to the CY domains of the RyRs and allosterically regulate the opening of the Ca2+ conduction pathway in the TM region. RyR activities are also modulated by post-translation modifications such as phosphorylation, oxidation, and S-nitrosylation. Prerequisites for the delineation of the mechanisms of channel regulation by individual modulators and modifications are the identity of the sites of modulator interaction or modification in the primary sequence and the location of these sites in the three-dimensional structure. Single-particle cryo-EM has been used to analyze macromolecular interactions of RyR with the modulatory proteins calmodulin and FKBP12 (for original references on these studies, see reviews in Refs. 1 and 17). Because most channel modulators bind in the CY assembly, there must be mechanisms whereby changes in the conformation of specific domains in the CY assembly are transduced, first into changes in the column-like structure and then into changes in the TM helices of the pore.
Identification of Modulator-binding Sites in the Primary Sequence of RyRs
CaV1.1—In skeletal muscle, the interaction between CaV1.1 in the transverse tubules and RyR1 in the sarcoplasmic reticulum has been shown to involve an orthograde signal, whereby CaV1.1 signals RyR1 to open, and a retrograde signal, through which RyR1 increases the Ca2+ current through CaV1.1 (18). Multiple regions of RyR1 are likely to be involved in the coupling to CaV1.1 (Fig. 1A, line 8) (reviewed in Ref. 18), including aa 1635–2636, 2659–3720, and 1837–2154 and multiple regions between aa 1 and 1680 (19). The sites of interaction appear to be distributed over a rather large part of the CY domain of RyR1.
Based on freeze-fracture studies (20), the most likely regions of RyR1 to be in contact with CaV1.1 include subregions 4 and 6 (Fig. 1B). Several of the sequences suggested to contribute to mechanical coupling have been found within these two subdomains. Ser437 of RyR2 (Ser422 in RyR1) maps to a location between subregions 5 and 9. Ser2368 in RyR2 (Arg2400 in RyR1) maps to a location between subregions 5 and 6. An interaction of CaV1.1 with these regions of RyR1 could directly control the opening of the clamp and allosterically modulate the channel gating.
Ca2+—Regulation of RyRs by Ca2+ varies with the RyR isoform. The most important modulator of RyR2 activity in cardiac muscle is Ca2+, where Ca2+ enters through CaV1.2 and activates RyR2. Ca2+ also regulates RyR1 activity, but depending on the concentration, it can either activate or inhibit its activity. These findings suggest that RyR1 has at least two Ca2+-binding sites. Both RyR1 and RyR2 are activated at 0.1–10 μm Ca2+ (21), but RyR1 is more sensitive to inhibition by higher concentrations of Ca2+. Lumenal Ca2+ also regulates the activity of RyRs (22).
Possible binding sites for Ca2+ (Fig. 1A, line 9) are located between aa 1861 and 2094 (21), 3657 and 3776 (21), and 4381 and 4626 (23) and two putative EF-hand motifs located at aa 4079–4092 (EF1) and 4115–4126 (EF2) (24). Mutation E3986A (Glu4042 in rabbit RyR1) in mouse RyR2 reduced Ca2+ activation by >1000-fold (25). Fessenden et al. (26) scrambled the putative EF-hand motifs and found that these mutations did not alter the response to either depolarization or RyR agonists (caffeine, 4-chloro-m-cresol) in intact myotubes. Surprisingly, no high affinity [3H]ryanodine binding was observed in membranes expressing the EF2 mutation, but the channels in bilayers were activated by Ca2+ in the micromolar range. These findings suggest that the putative EF-hands contribute to Ca2+ modulation of the channel but are not sufficient for Ca2+ regulation of this channel.
Sites close to these putative Ca2+-binding sites have not yet been mapped in the three-dimensional structures of RyRs. However, Xiong et al. (27) suggested an interaction of the regions containing putative EF1 and EF2 with the calmodulin (CaM)-binding region (aa 3614–3643), suggesting that these two EF-hands are close to subdomain 3, where the CaM-binding site is located (see below).
Calmodulin—RyRs also bind CaM, which is itself a Ca2+-binding protein. RyR1 is the major CaM-binding protein of sarcoplasmic reticulum membranes (28). At micromolar Ca2+ concentrations, CaM inhibits both RyR1 and RyR2. However, at lower concentrations, CaM activates RyR1 but inhibits RyR2 (29). RyR3 also binds CaM, and the effects of CaM on the channel are redox-sensitive (30). The CaM-binding site on RyR1 involves aa 3614–3643 (Fig. 1A, line 10) (31, 32) and exists at an intersubunit boundary close to aa 1975–1999 (33).
By difference mapping, the CaM-binding site was found to be located in CY subregion 3 (Fig. 1B). Moreover, the location of the CaM-binding site is displaced to ∼33 Å in the presence of Ca2+ with respect to its position for Ca2+-free CaM (14). This displacement of CaM could be due to a movement of CaM upon binding Ca2+ and/or a movement of the CaM-binding site when RyR1 binds Ca2+. These findings suggest that subregion 3 includes a crucial regulatory site(s) for channel activation and inhibition, but additional studies are required to further elucidate the molecular mechanism of CaM regulation of RyR1.
Immunophilins—The immunophilins FKBP12 and FKBP12.6 bind to RyRs and regulate their activities (34). Although FKBP12 and FKBP12.6 are able to bind to all RyR isoforms, FKBP12 copurifies with RyR1, whereas FKBP12.6 copurifies with RyR2. The FKBP isoform normally bound to RyR3 is not known. FKBPs bind to RyR1 and RyR2 on one site per RyR1 subunit and stabilize a closed state of the channel (34). The binding site has been suggested to be between aa 2458 and 2468 in RyR1 (Fig. 1A, line 11). Mutation of this site abolishes FKBP12 binding (35), but this may be due to an allosteric regulation of the actual binding site (36). Using difference mapping of three-dimensional reconstructions of RyR1 with and without FKBP12, the binding site for FKBP12 is found at the interface between clamp subregions 5 and 9 and subregion 3, connecting neighboring clamps in the RyR CY region (Fig. 1B). Comparison of the location of FKBP12 by difference mapping (37) with the docking of the IP3 homology model (4) suggests that there is a binding pocket for FKBP12 created by Glu161, Arg164, Arg402, and Ile404 (Fig. 2B). Whether or not FKBP12 binds at this pocket remains to be tested.
Homer—The scaffolding protein Homer has been shown to interact with RyR1 and to regulate its response to both voltage and caffeine (38). A possible Homer-binding sequence is found between aa 1773 and 1783. Region 1727–2503 has been mapped between domains 5 and 6 (39), suggesting that Homer may also interact via the clamp domain of RyR1.
Phosphorylation—Marks and co-workers (40) have suggested that RyR hyperphosphorylation (aa 2843 in RyR1 and aa 2808 in RyR2) displaces FKBPs from RyRs, leading to Ca2+ leak. A number of other laboratories have failed, however, to reproduce these findings (41–43). Another potential site of phosphorylation is aa 2065 (aa 2031 in RyR2). Possible phosphorylation sites are indicated in Fig. 1A (line 12). Insertion of GFP at position 2801 in RyR2 (Ser2843 in RyR1) localized this site to domain 6, distal to the FKBP12.6-binding site (44). The second putative phosphorylation site was mapped by inserting GFP at position 2023 and was found to be located in subdomain 4, again distal to the FKBP-binding site (45).
Redox Modifications—RyRs are highly redox-sensitive. Cys3635, in the CaM-binding site, is the primary target of S-nitrosylation (Fig. 1, line 13, in green) (46). Durham et al. (47) recently showed that RyR1 S-nitrosylation underlies the abnormal temperature sensitivity found with RyR1 mutations associated with malignant hyperthermia. Redox-sensitive cysteines have been identified by Aracena-Parks et al. (48) under conditions in which the channel was primarily in a open conformation (Fig. 1A, line 13) and by Voss et al. (49) under conditions in which the channel was in a primarily closed conformation. Although the differences may be due to differences in labeling reagents, it is also possible that different cysteines are reactive when the channel is in the open versus closed conformation.
Where are the redox-sensitive cysteines in the three-dimensional structure? Several can be identified in the docking model (Fig. 2C). In addition, Cys3635 is in the CaM-binding site and is therefore located in subdomain 3. Other hyper-reactive cysteines are probably located in subdomains 5 and 6 (Fig. 1B) because the region from aa 1727 to 2503 localized to these subdomains (39).
Disease-causing Mutations—Mutations in RyR1 underlie malignant hyperthermia, central core disease, and some cases of multiminicore disease (see recent review by Durham et al. (17)). Although there is some tendency for the mutations to cluster in three regions of RyR1, it now appears that disease-associated mutations can occur throughout the primary sequence (Fig. 1A, line 14). Mutations that produce malignant hyperthermia generally cause the channel to be more sensitive to activators, whereas central core disease mutations either make the channel leaky or block efflux through the channel. Predictions for the location of some of the N-terminal mutations can be made from the docking model (Fig. 2B).
Correlating Structure with Function
Channel opening is associated with significant structural transitions in the CY region, involving clamp subregions 5–7 and 9 (Fig. 2A) (10, 11). Given the three-dimensional locations of the FKBP12- and CaM-binding sites and other important functional sites, the clamp subregions are likely to be involved in allosteric modulation of the channel activity. These findings suggest a direct connection between the conformation of the clamp and channel opening. Consistent with this, many mutations associated with malignant hyperthermia and central core disease in humans, mutations that allow the channel to open more readily, are located in clamp subregions 5, 7, and 9 (Fig. 2) (4).
Near-atomic Resolution Structure of RyR1: Perspectives and Challenges
Although RyR1 was among the first non-icosahedral structures solved by single-particle cryo-EM at 30-Å resolution (50, 51), the RyR assembly remains the most challenging target for structural analysis due to its complex dynamic nature. Recent RyR1 reconstructions at ∼10 Å by two groups have provided important insights into its pore structure, gating, and control by multiple intracellular molecules (2, 3).
Several factors limit obtaining higher resolution structures of RyR1, including variability arising from cryo-specimen preparation (vitrification) of detergent-solubilized membrane proteins, removal of the protein from its natural lipid environment, incomplete saturation with modulators, and mixtures of different conformational states. Approaches are needed to trap the channel population in a single functional state and either to remove all modulators or to saturate all binding sites with modulators. New computational methods are needed to sort heterogeneous molecule populations. To approach near-atomic resolution, the number of particles has to be increased. Thus, automated data collection and the use of charge couple device cameras are needed. Approaches that allow some of these limitations to be bypassed are 1) fitting crystal structures of individual components or their subdomains into the cryo-EM density map and 2) comparative modeling and fitting the models into the cryo-EM map. Despite the difficulties, continued breakthroughs in the resolution of RyR structure are highly likely to occur in the foreseeable future.
Footnotes
This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
The abbreviations used are: RyR, ryanodine receptor; cryo-EM, cryoelectron microscopy; CY, cytoplasmic; TM, transmembrane; aa, amino acids; GFP, green fluorescent protein; IP3, inositol trisphosphate; CaM, calmodulin; FKBP, FK506-binding protein.
REFERENCES
- 1.Serysheva II, Chiu W, Ludtke SJ. Methods Cell Biol. 2007;79:407–435. doi: 10.1016/S0091-679X(06)79016-1. [DOI] [PubMed] [Google Scholar]
- 2.Ludtke SJ, Serysheva II, Hamilton SL, Chiu W. Structure (Camb) 2005;13:1203–1211. doi: 10.1016/j.str.2005.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samso M, Wagenknecht T, Allen PD. Nat Struct Mol Biol. 2005;12:539–544. doi: 10.1038/nsmb938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serysheva II, Ludtke SJ, Baker ML, Cong Y, Topf M, Eramian D, Sali A, Hamilton SL, Chiu W. Proc Natl Acad Sci U S A. 2008;105:9610–9615. doi: 10.1073/pnas.0803189105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Block BA, Franzini-Armstrong C. J Cell Biol. 1988;107:1099–1112. doi: 10.1083/jcb.107.3.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du GG, Sandhu B, Khanna VK, Guo XH, MacLennan DH. Proc Natl Acad Sci U S A. 2002;99:16725–16730. doi: 10.1073/pnas.012688999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. Sciences (N. Y) 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y, Lee A, Chen J, Cadene M, Chait BT, MacKinnon R. Nature. 2002;417:515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 9.Kuo A, Gulbis JM, Antcliff JF, Rahman T, Lowe ED, Zimmer J, Cuthbertson J, Ashcroft FM, Ezaki T, Doyle DA. Sciences (N. Y) 2003;300:1922–1926. doi: 10.1126/science.1085028. [DOI] [PubMed] [Google Scholar]
- 10.Orlova EV, Serysheva II, van Heel M, Hamilton SL, Chiu W. Nat Struct Biol. 1996;3:547–552. doi: 10.1038/nsb0696-547. [DOI] [PubMed] [Google Scholar]
- 11.Serysheva II, Schatz M, van Heel M, Chiu W, Hamilton SL. Biophys J. 1999;77:1936–1944. doi: 10.1016/S0006-3495(99)77035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma MR, Jeyakumar LH, Fleischer S, Wagenknecht T. J Biol Chem. 2000;275:9485–9491. doi: 10.1074/jbc.275.13.9485. [DOI] [PubMed] [Google Scholar]
- 13.Marks AR, Marx SO, Reiken S. Trends Cardiovasc Med. 2002;12:166–170. doi: 10.1016/s1050-1738(02)00156-1. [DOI] [PubMed] [Google Scholar]
- 14.Samso M, Wagenknecht T. J Biol Chem. 2002;277:1349–1353. doi: 10.1074/jbc.M109196200. [DOI] [PubMed] [Google Scholar]
- 15.Stewart R, Zissimopoulos S, Lai FA. Biochem J. 2003;376:795–799. doi: 10.1042/BJ20030597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galvan DL, Mignery GA. J Biol Chem. 2002;277:48248–48260. doi: 10.1074/jbc.M209990200. [DOI] [PubMed] [Google Scholar]
- 17.Durham WJ, Wehrens XHT, Sood S, Hamilton SL. In: Calcium Signaling and Disease. Carafoli E, Brini M, editors. Springer-Verlag New York Inc; New York: 2007. pp. 273–321. [Google Scholar]
- 18.Dirksen RT. Front Biosci. 2002;7:d659–d670. doi: 10.2741/A802. [DOI] [PubMed] [Google Scholar]
- 19.Sheridan DC, Takekura H, Franzini-Armstrong C, Beam KG, Allen PD, Perez CF. Proc Natl Acad Sci U S A. 2006;103:19760–19765. doi: 10.1073/pnas.0609473103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paolini C, Fessenden JD, Pessah IN, Franzini-Armstrong C. Proc Natl Acad Sci U S A. 2004;101:12748–12752. doi: 10.1073/pnas.0404836101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen SR, MacLennan DH. J Biol Chem. 1994;269:22698–22704. [PubMed] [Google Scholar]
- 22.Jiang D, Xiao B, Yang D, Wang R, Choi P, Zhang L, Cheng H, Chen SR. Proc Natl Acad Sci U S A. 2004;101:13062–13067. doi: 10.1073/pnas.0402388101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treves S, Chiozzi P, Zorzato F. Biochem J. 1993;291:757–763. doi: 10.1042/bj2910757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiong H, Feng X, Gao L, Xu L, Pasek DA, Seok JH, Meissner G. Biochemistry. 1998;37:4804–4814. doi: 10.1021/bi971198b. [DOI] [PubMed] [Google Scholar]
- 25.Li P, Chen SR. J Gen Physiol. 2001;118:33–44. doi: 10.1085/jgp.118.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fessenden JD, Feng W, Pessah IN, Allen PD. J Biol Chem. 2004;279:53028–53035. doi: 10.1074/jbc.M411136200. [DOI] [PubMed] [Google Scholar]
- 27.Xiong L, Zhang JZ, He R, Hamilton SL. Biophys J. 2006;90:173–182. doi: 10.1529/biophysj.105.066092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tripathy A, Xu L, Mann G, Meissner G. Biophys J. 1995;69:106–119. doi: 10.1016/S0006-3495(95)79880-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meissner G, Pasek DA, Yamaguchi N, Ramachandran S, Dokholyan NV, Tripathy A. Proteins. 2008 doi: 10.1002/prot.22148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamaguchi N, Xu L, Pasek DA, Evans KE, Chen SR, Meissner G. Biochemistry. 2005;44:15074–15081. doi: 10.1021/bi051251t. [DOI] [PubMed] [Google Scholar]
- 31.Moore CP, Rodney G, Zhang JZ, Santacruz-Toloza L, Strasburg G, Hamilton SL. Biochemistry. 1999;38:8532–8537. doi: 10.1021/bi9907431. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi N, Xu L, Pasek DA, Evans KE, Meissner G. J Biol Chem. 2003;278:23480–23486. doi: 10.1074/jbc.M301125200. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, Zhang JZ, Danila CI, Hamilton SL. J Biol Chem. 2003;278:8348–8355. doi: 10.1074/jbc.M209565200. [DOI] [PubMed] [Google Scholar]
- 34.Marks AR. Physiol Rev. 1996;76:631–649. doi: 10.1152/physrev.1996.76.3.631. [DOI] [PubMed] [Google Scholar]
- 35.Gaburjakova M, Gaburjakova J, Reiken S, Huang F, Marx SO, Rosemblit N, Marks AR. J Biol Chem. 2001;276:16931–16935. doi: 10.1074/jbc.M100856200. [DOI] [PubMed] [Google Scholar]
- 36.Zissimopoulos S, Lai FA. Cell Biochem Biophys. 2005;43:203–219. doi: 10.1385/CBB:43:2:203. [DOI] [PubMed] [Google Scholar]
- 37.Samso M, Shen X, Allen PD. J Mol Biol. 2006;356:917–927. doi: 10.1016/j.jmb.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 38.Feng W, Tu J, Yang T, Vernon PS, Allen PD, Worley PF, Pessah IN. J Biol Chem. 2002;277:44722–44730. doi: 10.1074/jbc.M207675200. [DOI] [PubMed] [Google Scholar]
- 39.Liu Z, Wang R, Zhang J, Chen SR, Wagenknecht T. J Biol Chem. 2005;280:37941–37947. doi: 10.1074/jbc.M505714200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiken S, Lacampagne A, Zhou H, Kherani A, Lehnart SE, Ward C, Huang F, Gaburjakova M, Gaburjakova J, Rosemblit N, Warren MS, He KL, Yi GH, Wang J, Burkhoff D, Vassort G, Marks AR. J Cell Biol. 2003;160:919–928. doi: 10.1083/jcb.200211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benkusky NA, Weber CS, Scherman JA, Farrell EF, Hacker TA, John MC, Powers PA, Valdivia HH. Circ Res. 2007;101:819–829. doi: 10.1161/CIRCRESAHA.107.153007. [DOI] [PubMed] [Google Scholar]
- 42.Carter S, Colyer J, Sitsapesan R. Circ Res. 2006;98:1506–1513. doi: 10.1161/01.RES.0000227506.43292.df. [DOI] [PubMed] [Google Scholar]
- 43.MacDonnell SM, Garcia-Rivas G, Scherman JA, Kubo H, Chen X, Valdivia H, Houser SR. Circ Res. 2008;102:e65–e72. doi: 10.1161/CIRCRESAHA.108.174722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng X, Xiao B, Cai S, Huang X, Li F, Bolstad J, Trujillo R, Airey J, Chen SR, Wagenknecht T, Liu Z. J Biol Chem. 2007;282:25929–25939. doi: 10.1074/jbc.M704474200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones PP, Meng X, Xiao B, Cai S, Bolstad J, Wagenknecht T, Liu Z, Chen SR. Biochem J. 2008;410:261–270. doi: 10.1042/BJ20071257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun J, Xin C, Eu JP, Stamler JS, Meissner G. Proc Natl Acad Sci U S A. 2001;98:11158–11162. doi: 10.1073/pnas.201289098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durham WJ, Aracena-Parks P, Long C, Rossi AE, Goonasekera SA, Boncompagni S, Galvan DL, Gilman CP, Baker MR, Shirokova N, Protasi F, Dirksen R, Hamilton SL. Cell. 2008;133:53–65. doi: 10.1016/j.cell.2008.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aracena-Parks P, Goonasekera SA, Gilman CP, Dirksen RT, Hidalgo C, Hamilton SL. J Biol Chem. 2006;281:40354–40368. doi: 10.1074/jbc.M600876200. [DOI] [PubMed] [Google Scholar]
- 49.Voss AA, Lango J, Ernst-Russell M, Morin D, Pessah IN. J Biol Chem. 2004;279:34514–34520. doi: 10.1074/jbc.M404290200. [DOI] [PubMed] [Google Scholar]
- 50.Radermacher M, Rao V, Grassucci R, Frank J, Timerman AP, Fleischer S, Wagenknecht T. J Cell Biol. 1994;127:411–423. doi: 10.1083/jcb.127.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serysheva II, Orlova EV, Chiu W, Sherman MB, Hamilton SL, van Heel M. Nat Struct Biol. 1995;2:18–24. doi: 10.1038/nsb0195-18. [DOI] [PubMed] [Google Scholar]