The biochemistry of amyloid proteins has been a fascinating and important area of research because of its contribution to our understanding of protein folding dynamics and assembly and of the pathogenetic mechanisms of human disease. One such disease is AD,2 the most common neurodegenerative disorder of aging. In AD, Aβ (Fig. 1A), which is expressed normally and ubiquitously throughout life as a 40–42-residue peptide, forms fibrils that deposit in the brain as “amyloid plaques.” This pathologic deposition process led researchers to investigate fibril formation as a target for therapeutic intervention. In doing so, an increasing number of fibril precursors and non-fibrillar Aβ assemblies have been identified, the majority of which are neurotoxic. These findings have altered prevailing fibril-centered views of the pathobiology of amyloid diseases (1) and intensified efforts to understand the early folding and assembly dynamics of Aβ. In the discussion that follows, we seek to introduce the reader to the complex world of Aβ assembly and biological activity, a goal we hope will provide a conceptual framework upon which further knowledge or experimentation may be built.
FIGURE 1.
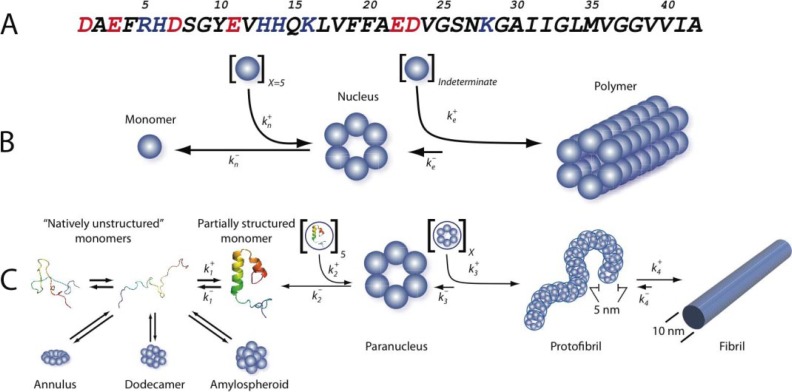
Aβ assembly. A, the sequence of Aβ42 is shown in one-letter amino acid code. The side chain charge at neutral pH is color-coded (red, negative; blue, positive). B, nucleation-dependent polymerization, reflecting the unfavorable self-association (rate constant ) of X natively folded monomers (in this case, six total) to form a fibril nucleus and the favorable addition ( ) of a large indeterminate number of monomers to the nucleus (nascent fibril) during fibril elongation. C, Aβself-assembly. Aβbelongs to the class of “natively disordered” proteins, existing in the monomer state as an equilibrium mixture of many conformers. On-pathway assembly requires the formation of a partially folded monomer that self-associates to form a nucleus for fibril elongation, a paranucleus (in this case, containing six monomers). Nucleation of monomer folding is a process distinct from fibril nucleation (50). Fibril nucleation is unfavorable kinetically ( ), which explains the lag phase of fibrillogenesis experiments, a period during which no fibril formation is apparent. Paranuclei self-associate readily ( ) to form protofibrils, which are relatively narrow (∼5 nm), short (<150 nm), flexible structures. These protofibrils comprise a significant but finite number (X) of paranuclei. Maturation of protofibrils through a process that is kinetically favorable ( ) yields classical amyloid-type fibrils (∼10-nm diameter, indeterminate (but often >1 μm) length). Other assembly pathways produce annular pore-like structures, globular dodecameric (and higher order) structures, and amylospheroids. Annuli and amylospheroids appear to be off-pathway assemblies.
Aβ Fibril Structure
The determination of the structure of fibrils has been an unusually difficult problem because Aβ belongs to a class of proteins that are “natively unfolded” (2) and preferentially form amyloid fibrils rather than protein crystals. This has precluded x-ray diffraction studies of full-length Aβ and made solution NMR studies problematic (3). Nevertheless, site-directed spin labeling and solid-state NMR studies have been informative. The former studies have revealed that Aβ fibrils comprise β-strands organized in a parallel, in-register fashion. The latter studies showed that in Aβ40 fibrils, residues 12–24 and 30–40 form parallel β-sheets and that these two β-strand segments are connected by a turn involving residues 25–29 (4). Hydrogen/deuterium exchange coupled with solution-state NMR revealed a similar, but distinct, segmental arrangement of β-strands within Aβ42 fibrils. Here, residues 18–26 and 31–42 form the β-strands. In both models, salt bridges between Asp23 and Lys28 stabilize the turn region connecting the two β-strands (2, 5). Similar findings have been obtained using other methods (5, 6).
Differences among the studies likely result from the examination of different peptides (Aβ40 versus Aβ42), the absence or presence of Met35(O), or the conditions under which fibrils were formed. All these factors have been shown to affect significantly peptide assembly and biological activity (6, 7). Although no crystal structures have been determined with full-length Aβ, exciting work has been done on microcrystals formed by C-terminal peptides. These microcrystals yield diffraction patterns consistent with an in-register cross-β-organization of two interdigitated β-sheets. This “steric zipper” structure has been found in at least 13 other amyloid protein microcrystals (8). Whether steric zippers exists in Aβ fibrils is unclear.
Pathways of Peptide Assembly
How do monomers form fibrils? This question is fundamental to understanding fibrillogenesis and for identifying assembly steps that could be therapeutic targets. Influential early investigations promulgated the idea that Aβ assembly was a specific example of the general class of nucleation-dependent polymerization reactions (Fig. 1B). These reactions comprise a slow nucleation step, producing a “lag phase” during assembly monitoring, followed by a rapid fibril elongation step. Operating within this paradigm, nucleation (kn) and elongation (ke) rate constants for Aβ fibril formation were determined (9). However, continuing elucidation of this ostensibly classical polymerization process revealed unexpected complexity in the numbers and types (“on-pathway” or “off-pathway” for fibril formation) of assembly paths and the structures resulting therefrom (Fig. 1C and supplemental Table S1).
Protofibrils, Paranuclei, and Monomer Folds
Fig. 1C illustrates one pathway of fibril assembly. The penultimate fibril intermediate, the protofibril, was first identified more than a decade ago (10). Protofibrils were described as beaded chains, each bead of which was ∼5 nm in diameter. The length of these structures generally was <150 nm. Kinetics and solution-phase AFM experiments showed that protofibrils matured into fibrils (10). To understand how protofibrils formed, methods were developed to determine quantitatively the oligomer size distribution in nascent Aβ preparations (11). In Aβ42 assembly, these experiments suggested that a pentamer or hexamer, the “paranucleus,” was the basic unit of the protofibril and that the beaded chains comprising protofibrils formed by the self-association of paranuclei.
To understand the oligomerization process in atomic detail, computer simulations have been done (12). These studies yielded oligomer frequency distributions similar to those determined experimentally, but in addition provided high resolution conformational information. Aβ40 oligomers were more compact than Aβ42 oligomers due to increased conformational freedom of the Aβ42 N termini. This suggested that intermolecular interactions among Aβ42 N termini might facilitate the C-terminal interactions obligatory for fibril formation. The work also revealed the formation of a turn in Aβ42 at Gly37-Gly38 that was not observed in Aβ40 and that thus could be critical in paranucleus formation.
The importance of the C terminus of Aβ in controlling Aβ assembly has also been revealed in experiments involving amino acid substitutions (11). Systematic alterations in residue 41 side chain hydrophobicity showed that Gly or Ala largely eliminated paranucleus formation, whereas amino acids with hydropathic characteristics similar to Ile had no effect. Elimination of the Ala42 side chain blocked paranucleus self-association, whereas insertion of larger apolar side chains facilitated the process. Similar studies examined Met35 polarity, an important question with respect to redox chemistry in AD (5, 11). In these experiments, oxidation of Met35 to Met35(O) or Met35 sulfone had no effect on Aβ40 oligomerization, whereas Aβ42 paranucleus formation was abolished. Interestingly, the modified Aβ42 peptides oligomerized identically to Aβ40.
The relative importance of the C terminus in controlling Aβ assembly was also apparent in studies of Aβ40 and Aβ42 peptides containing substitutions linked to familial forms of AD or CAA. These substitutions (Glu22 → Gln, Glu22 → Gly, Glu22 → Lys, and Asp23 → Asn) produced oligomers of higher order when substituted in Aβ40 but had little effect on Aβ42 oligomerization. Removal of N-terminal residues Asp1–Gly9 in Aβ42 had no effect on its oligomer size distribution, whereas truncation of either the N-terminal two or four residues of Aβ40 produced higher-order oligomers. This observation was consistent with the aforementioned simulation data that suggested that collapse of the N terminus of Aβ40 on the oligomer surface might shield underlying hydrophobic regions of the oligomers that otherwise might interact to form higher-order assemblies (12). In fact, this process was observed in studies of the folding and assembly of urea-denatured Aβ (13). Aβ40 formed an unstable but largely collapsed monomeric species, whereas Aβ42 existed in a trimeric or tetrameric state (13).
The solvent inaccessibility of the Ala21–Ala30 region of Aβ likely results from the formation of a turn-like structure that nucleates monomer folding (14). This decapeptide region initially was identified due to its resistance to proteolysis, a resistance that remained in the isolated decapeptide itself and that allowed NMR and computational determinations of its structure and dynamics (14). Most recently, thermodynamics studies showed that the turn is destabilized by amino acid substitutions that cause AD and CAA (15). Destabilization correlates with accelerated Aβ oligomerization and higher-order assembly and thus provides a mechanistic explanation for these familial forms of AD and CAA.
Other Assembly Pathways
The idea that an Aβ hexamer building block exists is intriguing because at least four other structures, ADDLs, Aβ*56, “globulomers,” and “Aβ oligomers,” comprise multiples of this basic unit (Fig. 1C and supplemental Table S1). ADDLs are dodecamers produced in vitro from Aβ42 using special solvent conditions and appear in AFM studies as globular structures with heights of 5–6 nm (16). Aβ*56 was identified in SDS extracts from brains of Tg2576 transgenic mice (17). The “56” refers to the molecular weight of the oligomer, which is consistent with that of a dodecamer. The morphology of Aβ*56 is a prolate ellipsoid. A third type of dodecamer is the globulomer (so-called because it is a globular oligomer), which is formed by Aβ42 in the presence of SDS (18). Protease digestion, antibody binding, and mass spectrometry studies of globulomers suggest a structural model in which the hydrophobic C terminus (residues 24–42) forms a stable core and the more hydrophilic N terminus is on the surface. Although globulomers have substantial β-sheet content, presumably at the C terminus, they do not form fibrils and thus may be considered an off-pathway assembly (18). A larger species, the Aβ oligomer, also has been produced in vitro (19). Its molecular weight (∼90,000) suggests that its assembly order is ∼15–20, consistent with that of an octadecamer. In addition to assemblies with globular morphology, annular pore-like structures with diameters of 8–12 nm and pore sizes of 2–2.5 nm also have been described (10, 20).
The largest globular assemblies are amylospheroids and β-amyloid balls. Amylospheroids are off-pathway spheroidal structures with diameters of 10–15 nm that are formed by Aβ40 or Aβ42 (21). β-Amyloid balls are very large (20–200 μm) spheroidal structures formed only by Aβ40 at high concentration (300–600 μm) (22). Although such concentrations are non-physiological with respect to the average concentration of soluble Aβ in vivo, β-amyloid balls may be an interesting model of amyloid plaques or of the inclusion bodies formed in Parkinson and Huntington diseases and in the transmissible spongiform encephalopathies.
Assembly Complexity and Provenance
The complexity of Aβ assembly complicates the determination of precursor-product relationships. For example, are the different dodecameric assemblies discussed above really different, or are they all the same entity described in different ways by different investigators? Do the different larger spheroidal assemblies form from the same hexamer building blocks that produce dodecamers and thus belong on the same pathway? We do not know, but the answers to these questions are important because they have implications for the development of therapeutic agents targeting critical steps in the assembly pathways. For example, recent work has shown that compounds exist that can efficiently inhibit fibril formation or oligomerization, but not both (23). The distinction is critical if one assembly is benign and the other toxic.
Aβ Assembly and Disease
Thus far, we have discussed basic aspects of the physical biochemistry of Aβ assembly. However, the most fundamental biological question is, “what is the relationship between Aβ assemblies and AD?” Strong linkage exists between amyloid formation per se and disease (for a comprehensive review, see Ref. 24), and this linkage formed, in part, the foundation for the “amyloid cascade hypothesis,” which posited that amyloid fibril formation was the key pathogenetic process in AD (25). As discussed above, elucidation of the mechanisms of fibril formation unexpectedly revealed a broad range of fibrillar and non-fibrillar structures (supplemental Table S1). Aβ oligomers appear to be particularly important because they are potent neurotoxins and are isolable from AD patients, and their concentrations correlate positively with neuropathology in vivo. These facts have produced a fundamental paradigm shift resulting in a revised amyloid cascade hypothesis (1, 20, 26), one that posits the primacy of oligomeric forms of Aβ in AD causation.
A substantial experimental corpus exists demonstrating that “Aβ” is neurotoxic (27). However, it was not until approximately a decade ago, with the discovery and characterization of protofibrils and ADDLs, that a more structurally precise definition of Aβ could be made, one that in turn enabled more precise structure-neurotoxicity correlations to be established (16, 28). Each new assembly subsequently discovered also was toxic. An important goal of current research is to better define the mechanisms of this toxicity, a variety of which we now discuss.
Membrane Effects
Aβ is an amphipathic peptide (Fig. 1A). The side chains of 16 of the first 28 residues are polar; 12 are charged at neutral pH. The remaining 12 (Aβ40) or 14 (Aβ42) side chains are apolar. Structures such as these can form micelles (29) or interact with membranes directly. Recent work has shown that Aβ40 inserts into membranes of hippocampal neurons from AD brains (30). Membrane insertion can perturb plasma membrane structure and function. For example, conformational analysis of the C-terminal domain of Aβ (residues 29–40/2) has shown it to have properties similar to those of fusion peptides of viral proteins. Insertion of these fragments in a tilted manner in the membrane is thought to disrupt the parallel symmetry of the fatty acyl chains, altering the curvature of the membrane surface and destabilizing the membrane. Consistent with this prediction, Aβ (22–42) induces membrane fusion and permeabilizes lipid vesicles that mimic neuronal membranes (31).
Aβ oligomers have also been shown to increase the conductance of lipid bilayers and living cell membranes by lowering the “dielectric barrier,” possibly by increasing the membrane dielectric constant, introducing localized structural defects, or thinning the membrane (thereby facilitating charge translocation across the bilayer) (32). These effects may be related to oligomer-induced release of membrane components, including cholesterol, phospholipids, and monosialogangliosides, which in turn may lead to tau hyperphosphorylation and neurodegeneration (30, 33).
Structured membrane reorganization may also occur. Aβ40 oligomers form cation-sensitive ion channels in neuronal plasma membranes and liposomes (30, 34). These channels may comprise four to six subunits, each of which is an Aβ oligomer of order four to six, and thus the channels comprise a total of 16–36 Aβ monomers. The channels are quasi-stable, suggesting that channel formation is a dynamic process (31). For example, Arispe et al. (31) have shown that Aβ40 channel activity in planar lipid bilayers results in spontaneous transitions to higher conductances. AFM images of Aβ-treated reconstituted bilayers have revealed disk-like structures with pore-like concavities of 8–12-nm outside diameter and 1–2-nm inside diameter. However, pore formation remains a contentious issue. Some believe that Aβ oligomer-mediated interference with the surface packing of lipid headgroups effectively thins the membrane, reduces effective membrane conductance, and may produce the appearance of pores. Time-lapse AFM experiments have revealed that Aβ aggregates ∼500 nm in size form along the edges of bilayer defects, a result that could be misinterpreted as pore formation (35). Consistent with this interpretation are recent results suggesting that oligomers alter membrane conductivity without forming discrete pores (32).
We note that two general classes of Aβ/membrane interaction may occur: 1) nonreceptor-mediated structural interactions of the type discussed above; and 2) specific receptor-mediated interactions. These latter interactions may involve fibrillar and oligomeric forms of Aβ that act either as agonists or antagonists. Many membrane Aβ receptors have been identified (30), but the important question that remains unanswered is whether these interactions are physiologically relevant or serendipitous.
Metals, Aggregation, and Radicals
Evidence exists that metals are involved in the pathogenesis of AD. However, this is a contentious issue that remains unresolved. We present here a number of prominent mechanistic hypotheses.
In vitro results indicate that physiological concentrations of Zn2+ and Cu2+ can accelerate Aβ aggregation and increase Aβ toxicity (36, 37). Aβ has a strong positive reduction potential and displays high-affinity binding for Cu2+, Zn2+, and Fe3+ ions (34). Solution-state NMR and EPR have suggested that the three His residues in Aβ, His6, His13, and His14, coordinate Cu2+. This metalloenzyme-like complex has been proposed to catalyze Fenton chemistry (Equation 1),
| (Eq. 1) |
which yields toxic hydroxyl (OH•) and peroxide (OOH•) radicals. Fe2+ is also thought to participate in this chemistry. In addition to its postulated catalytic role in Fenton chemistry, it has been suggested that Aβ-linked inhibition of catalase increases H2O2 production (Equation 2) (38).
| (Eq. 2) |
A second center for redox chemistry is Met35 (39). The generation of reactive oxygen species by Aβ requires reduction of Cu2+ or Fe3+, a reaction that may proceed through the oxidation of Met35 to its corresponding sulfide radical cation. Cu+ or Fe2+ produced in this way may react with molecular oxygen and biological reducing agents (e.g.; cholesterol, vitamin C, or catecholamine) to yield H2O2 and the starting Cu+ cation (40). The H2O2 thus produced can further oxidize Met35 to its sulfoxide form and also react with superoxide anion ( ) in a Haber-Weiss reaction to produce OH• (Equation 3).
| (Eq. 3) |
Interestingly, the Met35(O) and Met35 sulfone forms of Aβ do not assemble as does the wild-type peptide (11, 41). Hou et al. (41) have reported that oxidation of Met35 to Met35(O) significantly reduces the rate of amyloid formation and alters fibril morphology. Bitan and Teplow (11) reported similar findings and found that Met35(O) Aβ42 does not form paranuclei, but rather oligomerizes similarly to Aβ40. These in vitro observations are consistent with the strong negative correlation that exists between oxidative damage and Aβ deposition in AD (11, 39).
Murakami et al. (42) have proposed that Tyr10 is also involved in redox chemistry. They suggested that H2O2 produced by Aβ-metal complexes oxidizes Tyr10 to produce the tyrosyl radical, which then attacks the thioether of Met35 and yields an S-oxidized radical cation. A turn at Gly38–Val39 brings the C-terminal carboxylate anion proximate to the radical, stabilizing it and simultaneously creating a hydrophobic subdomain facilitating peptide oligomerization, fibril formation, and longer lasting oxidative stress.
Mitochondrial Effects
Mitochondrial dysfunction has been linked directly to the aging process (43), a process that is the largest single risk factor for AD. Exacerbation of age-related dysfunction by toxic Aβ assemblies may explain the linkage of both age and Aβ to AD. Increasing evidence suggests that Aβ-induced mitochondrial dysfunction does in fact occur. The interaction of full-length Aβ or truncated forms with mitochondria causes potent inhibition of electron transport chain enzyme complexes and reductions in the activities of tricarboxylic acid cycle enzymes, leading to inhibition of ATP production, mitochondrial swelling, cytochrome c release, caspase activation, transition pore opening, increased mitochondrial reactive oxygen species production, and decreased mitochondrial membrane potential and respiration rates (43, 44). Complexation of Aβ with Aβ-binding alcohol dehydrogenase, a mitochondrial matrix enzyme, or with endoplasmic reticulum-associated Aβ-binding protein also produces this type of damage (45).
Apoptosis
A common final pathway of Aβ-induced neuronal dysfunction is apoptosis. This pathway is particularly likely to occur following mitochondrial compromise. Aβ40 and Aβ42 oligomers also have been shown recently to activate sphingomyelinases, which results in apoptotic cell death through a redox-sensitive cytosolic phospholipase A2/arachidonic acid-dependent pathway (46). In rat hippocampal neuron cultures, activation of ERK1/2 (extracellular signal-regulated kinase-1/2) by Aβ oligomers results in caspase-3 activation, tau cleavage, dysregulation of cell structure, and finally apoptosis (47). Transforming growth factor-β1 has been found to exacerbate Aβ-induced toxicity through Smad7 and β-catenin interactions and nuclear localization. Aβ40 also can activate the NF-κB apoptosis pathway by selectively inducing the nuclear translocation of the NF-κB p65 and p50 subunits. For this reason, p65 and p50 have been suggested as AD therapeutic targets. The connection of apoptosis with Aβ assemblies is supported by the observation that up-regulation in neurons of peroxisome proliferator-activated receptor-γ, which increases expression of the anti-apoptotic protein Bcl-2, protects these cells against Aβ-induced toxicity (48).
An Explication
The impetus for studies of Aβ structure, dynamics, and bio-activity has been the causal link of Aβ to AD. The result of these studies has been an extraordinary expansion of knowledge. The rapidly increasing number of clinical trials of mechanistically novel AD therapies suggests that this knowledge has been of value (49). However, a consensus does not exist regarding either the biophysical or biological behavior of Aβ. For academics, rigorous experiments done in well controlled systems provide reliable, although not necessarily clinically relevant, information. However, for AD patients, their families, and the treating clinicians, relevance is paramount. For their sake, it is hoped that the information presented here will stimulate current and especially new researchers to conceive of novel experimental approaches seeking to answer three fundamental questions. 1) Is Aβ, in fact, the proximate etiologic agent of AD; 2) if so, what is the structure of the proximate neurotoxic Aβ assembly; and 3) if not, what is?
Footnotes
This work was supported, in whole or in part, by National Institutes of Heath Grants NS038328 and AG027818. This work was also supported by the Jim Easton Consortium for Alzheimer’s Drug Discovery and Biomarkers at UCLA and State of California Alzheimer’s Disease Research Fund Grant 07-65806. This is the seventh article of eleven in the Thematic Minireview Series on the Molecular Basis of Alzheimer Disease. This minireview will be reprinted in the 2009 Minireview Compendium, which will be available in January, 2010.
The abbreviations used are: AD, Alzheimer disease; Aβ, amyloid β-protein; Met35(O), Met35 sulfoxide; AFM, atomic force microscopy; CAA, cerebral amyloid angiopathy; ADDLs, Aβ-derived diffusible ligands.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and additional references.
Supplemental material: http://www.jbc.org/content/suppl/2008/10/10/R800036200.DC1.html
REFERENCES
- 1.Kirkitadze MD, Bitan G, Teplow DB. J Neurosci Res. 2002;69:567–577. doi: 10.1002/jnr.10328. [DOI] [PubMed] [Google Scholar]
- 2.Nelson R, Eisenberg D. Curr Opin Struct Biol. 2006;16:260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Teplow DB. Methods Enzymol. 2006;413:20–33. doi: 10.1016/S0076-6879(06)13002-5. [DOI] [PubMed] [Google Scholar]
- 4.Tycko R. Methods Enzymol. 2006;413:103–122. doi: 10.1016/S0076-6879(06)13006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finder VH, Glockshuber R. Neurodegener Dis. 2007;4:13–27. doi: 10.1159/000100355. [DOI] [PubMed] [Google Scholar]
- 6.Fändrich M. CMLS Cell Mol Life Sci. 2007;64:2066–2078. doi: 10.1007/s00018-007-7110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kodali R, Wetzel R. Curr Opin Struct Biol. 2007;17:48–57. doi: 10.1016/j.sbi.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, Madsen A, Riekel C, Eisenberg D. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 9.Teplow DB. Amyloid. 1998;5:121–142. doi: 10.3109/13506129808995290. [DOI] [PubMed] [Google Scholar]
- 10.Caughey B, Lansbury PT. Annu Rev Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 11.Bitan G, Teplow DB. Acc Chem Res. 2004;37:357–364. doi: 10.1021/ar000214l. [DOI] [PubMed] [Google Scholar]
- 12.Urbanc B, Cruz L, Yun S, Buldyrev SV, Bitan G, Teplow DB, Stanley HE. Proc Natl Acad Sci U S A. 2004;101:17345–17350. doi: 10.1073/pnas.0408153101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y-R, Glabe CG. J Biol Chem. 2006;281:24414–24422. doi: 10.1074/jbc.M602363200. [DOI] [PubMed] [Google Scholar]
- 14.Teplow DB, Lazo ND, Bitan G, Bernstein S, Wyttenbach T, Bowers MT, Baumketner A, Shea J-E, Urbanc B, Cruz L, Borreguero J, Stanley HE. Acc Chem Res. 2006;39:635–645. doi: 10.1021/ar050063s. [DOI] [PubMed] [Google Scholar]
- 15.Grant MA, Lazo ND, Lomakin A, Condron MM, Arai H, Yamin G, Rigby AC, Teplow DB. Proc Natl Acad Sci U S A. 2007;104:16522–16527. doi: 10.1073/pnas.0705197104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL. Proc Natl Acad Sci U S A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lesné S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 18.Gellermann GP, Byrnes H, Striebinger A, Ullrich K, Mueller R, Hillen H, Barghorn S. Neurobiol Dis. 2008;30:212–220. doi: 10.1016/j.nbd.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 19.Deshpande A, Mina E, Glabe C, Busciglio J. J Neurosci. 2006;26:6011–6018. doi: 10.1523/JNEUROSCI.1189-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haass C, Selkoe DJ. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 21.Hoshi M, Sato M, Matsumoto S, Noguchi A, Yasutake K, Yoshida N, Sato K. Proc Natl Acad Sci U S A. 2003;100:6370–6375. doi: 10.1073/pnas.1237107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westlind-Danielsson A, Arnerup G. Biochemistry. 2001;40:14736–14743. doi: 10.1021/bi010375c. [DOI] [PubMed] [Google Scholar]
- 23.Necula M, Kayed R, Milton S, Glabe CG. J Biol Chem. 2007;282:10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- 24.Sipe JC, editor. Amyloid Proteins: The Beta Sheet Conformation and Disease. Wiley-VCH; Weinheim, Germany: 2005. [Google Scholar]
- 25.Hardy J. Ann Med. 1996;28:255–258. doi: 10.3109/07853899609033127. [DOI] [PubMed] [Google Scholar]
- 26.Hardy J, Selkoe DJ. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 27.Yankner BA, Lu T. J Biol Chem. 2009;284:4755–4759. doi: 10.1074/jbc.R800018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh DM, Hartley DM, Kusumoto Y, Fezoui Y, Condron MM, Lomakin A, Benedek GB, Selkoe DJ, Teplow DB. J Biol Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 29.Lomakin A, Chung DS, Benedek GB, Kirschner DA, Teplow DB. Proc Natl Acad Sci U S A. 1996;93:1125–1129. doi: 10.1073/pnas.93.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verdier Y, Zarndi M, Penke B. J Pept Sci. 2004;10:229–248. doi: 10.1002/psc.573. [DOI] [PubMed] [Google Scholar]
- 31.Arispe N, Diaz JC, Simakova O. Biochim. Biophys. Acta. 2007;1768:1952–1965. doi: 10.1016/j.bbamem.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 32.Sokolov Y, Kozak JA, Kayed R, Chanturiya A, Glabe C, Hall JE. J Gen Physiol. 2006;128:637–647. doi: 10.1085/jgp.200609533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tashima Y, Oe R, Lee S, Sugihara G, Chambers EJ, Takahashi M, Yamada T. J Biol Chem. 2004;279:17587–17595. doi: 10.1074/jbc.M308622200. [DOI] [PubMed] [Google Scholar]
- 34.Kawahara M, Arispe N, Kuroda Y, Rojas E. Biophys J. 1997;73:67–75. doi: 10.1016/S0006-3495(97)78048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Green JD, Kreplak L, Goldsbury C, Blatter XL, Stolz M, Cooper GS, Seelig A, Kistler J, Aebi U. J Mol Biol. 2004;342:877–887. doi: 10.1016/j.jmb.2004.07.052. [DOI] [PubMed] [Google Scholar]
- 36.Jun S, Saxena S. Angew Chem Int Ed Engl. 2007;46:5251–5263. doi: 10.1002/anie.200700318. [DOI] [PubMed] [Google Scholar]
- 37.Bush AI. Trends Neurosci. 2003;26:207–214. doi: 10.1016/S0166-2236(03)00067-5. [DOI] [PubMed] [Google Scholar]
- 38.Behl C, Davis JB, Lesley R, Schubert D. Cell. 1994;77:817–827. doi: 10.1016/0092-8674(94)90131-7. [DOI] [PubMed] [Google Scholar]
- 39.Butterfield DA. Curr Med Chem. 2003;10:2651–2659. doi: 10.2174/0929867033456422. [DOI] [PubMed] [Google Scholar]
- 40.Crouch PJ, Harding S-ME, White AR, Camakaris J, Bush AI, Masters CL. Int J Biochem Cell Biol. 2008;40:181–198. doi: 10.1016/j.biocel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Hou L, Kang I, Marchant RE, Zagorski MG. J Biol Chem. 2002;277:40173–40176. doi: 10.1074/jbc.C200338200. [DOI] [PubMed] [Google Scholar]
- 42.Murakami K, Irie K, Ohigashi H, Hara H, Nagao M, Shimizu T, Shirasawa T. J Am Chem Soc. 2005;127:15168–15174. doi: 10.1021/ja054041c. [DOI] [PubMed] [Google Scholar]
- 43.Crouch PJ, Cimdins K, Duce JA, Bush AI, Trounce IA. Rejuvenation Res. 2007;10:349–357. doi: 10.1089/rej.2007.0592. [DOI] [PubMed] [Google Scholar]
- 44.Mancuso C, Scapagini G, Curr D, Stella AMG, Marco CD, Butterfield DA, Calabrese V. Front Biosci. 2007;12:1107–1123. doi: 10.2741/2130. [DOI] [PubMed] [Google Scholar]
- 45.Chen JX, Yan SD. J Alzheimer’s Dis. 2007;12:177–184. doi: 10.3233/jad-2007-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malaplate-Armand C, Florent-Bchard S, Youssef I, Koziel V, Sponne I, Kriem B, Leininger-Muller B, Olivier J-L, Oster T, Pillot T. Neurobiol Dis. 2006;23:178–189. doi: 10.1016/j.nbd.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 47.Chong YH, Shin YJ, Lee EO, Kayed R, Glabe CG, Tenner AJ. J Biol Chem. 2006;281:20315–20325. doi: 10.1074/jbc.M601016200. [DOI] [PubMed] [Google Scholar]
- 48.Fuenzalida K, Quintanilla R, Ramos P, Piderit D, Fuentealba RA, Martinez G, Inestrosa NC, Bronfman M. J Biol Chem. 2007;282:37006–37015. doi: 10.1074/jbc.M700447200. [DOI] [PubMed] [Google Scholar]
- 49.Yamin G, Ono K, Inayathullah M, Teplow DB. Curr Pharm Des. 2008;14:3231–3246. doi: 10.2174/138161208786404137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lazo ND, Grant MA, Condron MC, Rigby AC, Teplow DB. Protein Sci. 2005;14:1581–1596. doi: 10.1110/ps.041292205. [DOI] [PMC free article] [PubMed] [Google Scholar]