Abstract
Rationale
Vascular smooth muscle cell (VSMC) differentiation from neural crest cells (NCCs) is critical for cardiovascular development, but the mechanisms remain largely unknown.
Objective
TGF-β function in VSMC differentiation from NCCs is controversial. We therefore determined the role and the mechanism of a TGF-β downstream signaling intermediate Smad2 in NCC differentiation to VSMCs.
Methods and Results
By using Cre/loxP system, we generated NCC tissue-specific Smad2 knockout mouse model and found that Smad2 deletion resulted in defective NCC differentiation to VSMCs in aortic arch arteries during embryonic development and caused vessel wall abnormality in adult carotid arteries where the VSMCs are derived from NCCs. The abnormalities included missing one layer of VSMCs in the media of the arteries with distorted and thinner elastic lamina, leading to a thinner vessel wall as compared to the wild type vessel. Mechanistically, Smad2 interacted with MRTFB to regulate VSMC marker gene expression. Smad2 was required for TGF-β-induced MRTFB nuclear translocation whereas MRTFB enhanced Smad2 binding to VSMC marker promoter. Moreover, we found that Smad2, but not Smad3, was a progenitor-specific transcription factor mediating TGF-β-induced VSMC differentiation from NCCs. Smad2 appeared to be also involved in determining the physiological differences between NCC- and mesoderm-derived VSMCs.
Conclusions
Smad2 is an important factor in regulating progenitor-specific VSMC development and physiological differences between NCC- and mesoderm-derived VSMCs.
Keywords: Smad2, MRTFB, smooth muscle differentiation, neural crest
INTRODUCTION
Vascular Smooth muscle cells (VSMCs) play a pivotal role in angiogenesis and vasculogenesis during embryonic development. Abnormal VSMC differentiation or modulation leads to the development of several prominent cardiovascular diseases including congenital heart diseases, atherosclerosis, hypertension, and restenosis after angioplasty.1-3 During embryonic development, VSMCs originate from at least eight progenitors including neural crest cells (NCC), secondary heart field, somites, mesoangioblasts, proepicardium, splanchnic mesoderm, mesothelium, and various stem cells.4 Interestingly, VSMCs from different progenitors display distinct functional properties. For instance, neural crest-derived VSMCs and mesoderm-derived VSMCs show dramatically different responses to the stimulation of morphogenetic factors such as transforming growth factor-β (TGF-β).4 In fact, these two VSMC subtypes exhibit opposite responses to TGF-β regarding cell growth, extracellular protein expression, and gene promoter activation.5 Moreover, mesoderm-derived VSMCs were unable to rescue the outflow tract defects observed in neural crest-ablated chicken embryos.6 Studies from several laboratories demonstrate that embryonically distinct subpopulations of VSMCs are not functionally equivalent and that VSMCs within different vascular beds utilize distinct cis-elements and control regions to regulate VSMC marker gene activation.7-11 These studies suggest that VSMC differentiation is controlled by different intracellular mechanisms among distinct VSMC subtypes. The molecular mechanisms governing the progenitor-specific regulation of VSMC differentiation, however, remain largely unknown.
The NCC is a multipotent cell population that develops in the dorsal neural tube and then migrates throughout the embryo and differentiates into numerous tissue types. In response to poorly understood developmental cues, a subpopulation of NCCs called cardiac NCCs migrate ventrally and populate aortic arch arteries and cardiac outflow tract. The NCC derivatives then differentiate into VSMCs forming the media layers of the aortic arch, ascending aorta, and carotid arteries, etc. Notch signaling and myocardin-related transcription factor B (MRTFB) have been shown to be important for VSMC differentiation of NCC.12, 13 Although TGF-β induces SMC differentiation from NCC in vitro, the in vivo findings are controversial.14-19 Wurdak et al reported that knockout of TGF-β type II receptor (TβRII) in NCCs abolishes the development of VSMC at embryonic day 10.5.14 However, a subsequent study using the same strategy to mutate TβRII in NCCs failed to identify VSMC defects, which is likely due to an in vivo compensatory mechanism or the use of a different TβRII-floxed mouse line, as discussed by the authors.20 Endoglin, a TGF-β accessory protein, appears to be also required for VSMC differentiation from NCCs, but Endoglin function is independent of TGF-β/Smad signaling.18 Knockout of a type I receptor of TGF-β family Alk2 leads to outflow track and VSMC defects.17 However, ALk2 is a receptor of bone morphogenetic proteins (BMP) that signal through Smad1/3/5. Therefore, the role of TGF-β/Smad2 signaling in NCC differentiation and the progenitor-specific regulator for VSMC differentiation from NCC has not been established.
In the present study, we identified Smad2, one of the TGF-β downstream signaling molecules, as a critical regulator of VSMC differentiation from NCCs. NCC-specific knockout of Smad2 leads to a defective VSMC differentiation in aortic arch arteries and reduced layers of VSMCs and distortion of elastic lamina in the media of adult mouse carotid arteries. Smad2 interacts with MRTFB to activate VSMC marker promoter activity. Mechanistically, Smad2 mediates MRTFB nuclear translocation whereas MRTFB enhances Smad2 binding to VSMC marker promoter. Although both Smad2 and Smad3 are important intermediates for TGF-β signaling, Smad2 plays a more important role than Smad3 in VSMC differentiation from NCCs while Smad3 is more important than Smad2 in VSMC differentiation from mesenchymal progenitors. Moreover, Smad2 appears to be also involved in determining the physiological differences between NCC- and mesoderm-derived VSMCs.
METHODS
Cell culture and reagents
Neural crest Monc-1 cells and mesenchymal C3H10T1/2 (10T1/2) cells were cultured as previously described.21, 22 Smad3 expression plasmids were previously described.23, 24 Smad2 expression plasmid was a generous gift from Dr. Ying Zhang.25 MRTFB expression plasmid was provided by Dr. Joseph Miano. Smooth muscle α-actin (α-SMA) and SM22α promoter-luciferase constructs were previously described.21, 26
Generation of Smad2 NCC-specific knockout mice
All animal experiments were approved by the Institutional Animal Care and Use Committee at the University of Georgia. Smad2-floxed (Smad2fl/fl) and Wnt1-Cre mice were previously described.27, 28 To generate Smad2 deletion in NCCs, Wnt1-Cre male mice were cross-breed with Smad2fl/fl mice to produce Wnt1-Cre;Smad2fl/+ mice. The male and female Wnt1-Cre;Smad2fl/+ mice were then cross-breed to produce Wnt1-Cre;Smad2fl/fl mice. Wnt1-Cre;Smad2+/+ littermates serve as controls. For in vivo fate mapping of NCCs, Wnt1-Cre mice heterozygous for Smad2-floxed allele (Wnt1-Cre;Smad2fl/+) were mated with Smad2fl/fl mice carrying a ROSA26 Cre reporter (R26R) allele, which expresses β-galactosidase (β-Gal) upon Cre-mediated recombination. Genotyping and X-gal staining for NCC fate mapping were performed as described.27, 29-31
Histomorphometric analysis and immunohistochemistry (IHC) staining
11.5 days of mouse embryos or carotid arteries from adult mice were fixed with 4% paraformaldehyde and paraffin-embedded. Embryos or vessels were cut by serial sectioning (5 μm). The sections were stained with hematoxylin and eosin for structural observation or Elastica van Gieson for elastin. For IHC, sections were rehydrated, blocked with 5% goat serum and permeabilized with 0.01% Triton X-100 in PBS, and incubated with rabbit anti-Smad2 or α-SMA antibody overnight at 4 followed by incubation with HRP-conjugated secondary antibody. α-SMA staining was visualized by the Vectastain method using NovaRed as a substrate. Smad2 staining was visualized by using Vectastain ABC-AP kit by following the manufacturer’s protocol (Vector Laboratories). The vessel sections were counterstained with hematoxylin.
Preparation of shRNA adenoviral vector
Adenoviral short hairpin RNA (shRNA) target sequences were ATG GAG CTG GTG GAG AAG AA for MRTFB and TGG TGT TCA ATC GCA TAC TAT for Smad2. Double-stranded DNAs coding MRTFB shRNAs were cloned into pRNAT-H1.1/Adeno shuttle vector (Genscript). Adenovirus expressing Flag-tagged MRTFB was constructed by cloning human MRTFB cDNA into the Xho I site of pShuttele-IRES-hrGFP-1 (Agilent) and was confirmed by sequencing. Adenovirus was packaged in 293 cells (Agilent) and purified by CsCl2 gradient ultracentrifugation as previously described.32, 33 Viral particle titer was determined by plaque assay. Adenovirus expressing Smad3 shRNA was previously described.34 For adenoviral transduction, Monc-1 cells were transduced with 100 moi of adenovirus expressing control, MRTFB, or shRNA for 24 to 48 hours.
Reverse Transcription-PCR (RT-PCR) and quantitative PCR (qPCR)
Total RNA was extracted using TRIzol reagent (Invitrogen) following the manufacturer’s instruction. cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad). RT-PCR was performed using Bio-Rad C1000 thermal cycler. qPCR was performed in MX3000P qPCR machine using SYBR Green qPCR Mastermix (Agilent). The primer sequences were as follows: Smad2: 5′-CCG GCT GAA CTG TCT CCT AC -3′ (forward) and 5′-GCA GAA CCT CTC CGA GTT TG -3′ (reverse); Smad3: 5′-CTG GGC CTA CTG TCC AAT GT -3′ (forward) and 5′-GCA GCA AAT TCC TGG TTG TT-3′ (reverse). The primers used for VSMC markers were described previously.22, 35
Western blotting
Monc-1 cells were lysed in RIPA lysis buffer. Carotid arteries were homogenized using a tissue homogenizer in RIPA buffer containing protease inhibitor mix (Sigma). Samples were separated on SDS-polyacrylamide gels, and electrotransferred onto PVDF membranes (Bio-Rad). The membranes were incubated at 4°C for 16 h with antibodies against α-SMA (Millipore), SM22α (Abcam), Smad2 (Cell Signaling), or α-tubulin (Sigma) in blocking buffer containing 5% milk followed by incubation with HRP-conjugated secondary antibody (Sigma).
Transient transfection and luciferase assay
Monc-1 and 10T1/2 cells were plated at 2×105 cells/well in 12-well plates and incubated at 37°C in a 5% CO2 incubator until 80% confluence. Cells were then transiently transfected (in triplicate) with LipofectAMINE LTX Plus according to the manufacturer’s recommendation (Invitrogen, Carlsbad, CA). Luciferase assay was performed as described previously.21, 36
Co-immunoprecipitation assay (Co-IP) and immunoblotting analysis
Monc-1 cells were transduced with adenovirus expressing Flag-tagged MRTFB followed by vehicle or TGF-β treatment for 2 h. Cells were then lysed with ice-cold lysis buffer containing protease inhibitor mix. The lysates were incubated with IgG or Flag antibody for one hour and then protein-A/G agarose at 4°C for 12 hours. The immunoprecipitates were pelleted, washed and subjected to immunoblotting using Flag or Smad2 antibody as described previously.34, 37
Immunofluorescent staining
Adenovirus expressing Flag-tagged MRTFB was transduced with adenovirus expressing GFP or Smad2 shRNA into Monc-1 cells for two days followed by vehicle or 5 ng/ml of TGF-β treatment for 2 hours. The cells were then fixed and incubated with rabbit anti-Flag antibody (Sigma), followed by incubation with TRITC-conjugated secondary goat anti-rabbit IgG as described previously.36, 38 MRTFB nuclear translocation was observed with fluorescent microscopy (Nikon). DAPI stains nuclei.
Chromatin immunoprecipitation assay (ChIP)
ChIP assays were performed as described previously.34 Monc-1 cells were transduced with adenovirus expressing GFP, MRTFB, or MRTFB shRNA followed by TGF-β treatment for 2 hours. Chromatin complexes were immunoprecipitated with 3 μg of Smad2 antibody or IgG (negative control). Semi-quantitative PCR and qPCR were performed to amplify the SM22α promoter region containing functional Smad binding element (SBE) using the following primer set: 5′- TCT GCC CCA GCC CAG ACA CC -3′ (forward) and 5′- CCC ACA GCC CTT CTG CTC CC -3′ (reverse).
Statistical analysis
All values are expressed as mean ± SEM. Data were analyzed using ANOVA with pairwise comparisons between groups. P values < 0.05 were considered statistically significant.
RESULTS
NCC-specific deletion of Smad2 blocked VSMC differentiation in aortic arch arteries but did not affect NCC migration
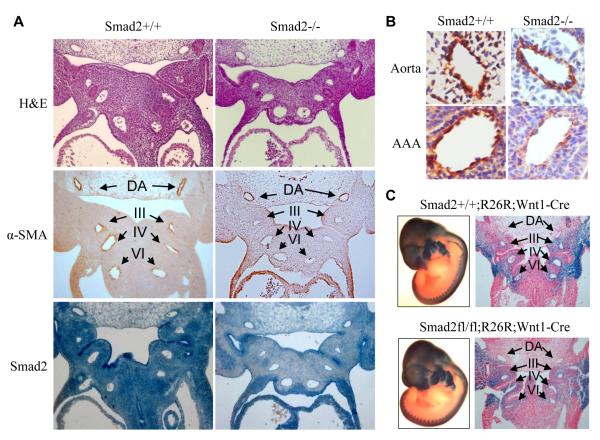
To address the role of Smad2 in VSMC differentiation from NCCs in vivo, we used the cre/loxP system to conditionally delete Smad2 gene in NCCs. Crossing of Wnt1-Cre mouse with Smad2-floxed mouse generated Smad2 knockout in NCCs and NCC-derived tissues including VSMCs (Online Figure I). At embryonic day 11.5 (E11.5), Smad2 mutant embryos did not display obvious malformations. However, Smad2 NCC-specific deletion blocked VSMC differentiation in aortic arch arteries III, IV, and VI (Figure 1A). In the wild type mouse embryos (Figure 1A, left panels), expression of VSMC marker α-actin (α-SMA) was detected in both dorsal aorta and aortic arch arteries (Figure 1A, left middle panel). Smad2 deletion diminished α-SMA expression in aortic arch arteries, but not in dorsal aorta (Figure 1A, right middle panel). It is known that aortic arch VSMCs are derived from NCCs while dorsal aortic VSMCs are derived from mesoderm. Higher magnification of dorsal aorta and the 4th aortic arch artery images showed that the numbers of cells expressing α-SMA were significantly reduced in Smad2-deleted 4th aortic arch artery (Figure 1B), suggesting a defective VSMC differentiation. This differentiation impairment was not due to a defect in NCC migration because Smad2-deficient NCCs migrated correctly to the cardiac outflow tract, as evaluated by in vivo fate mapping using the ROSA26 Cre reporter mouse line (Figure 1C). These data demonstrate that Smad2 is important for VSMC differentiation from NCCs in vivo.
Figure 1. Smad2 NCC-specific deletion blocked VSMC differentiation in aortic arch arteries.
(A) Smad2 deletion in NCCs blocked VSMC marker gene expression in aortic arch arteries. Sagittal sections of wild type (Smad2+/+, left panels) or Smad2 NCC-specific deleted E11.5 mouse embryos (Smad2−/−, right panels) were stained with H&E or immunostained with α-SMA or Smad2 antibody as indicated. α-SMA staining in dorsal aorta (DA) and aortic arch arteries III, IV and VI are indicated by arrows, respectively. Smad2 deletion in NCCs blocked α-SMA expression in aortic arch arteries but not in dorsal aorta (40x). (B) Higher magnification of α-SMA expression in wild type or Smad2-NCC deleted mouse aorta and the IV aortic arch arteries (AAA). α-SMA was significantly blocked in the AAA with Smad2 deletion in NCC but not in the aorta (400x). (C) Smad2 deletion in NCCs did not alter NCC migration. E11.5 wild type (upper panels) and Smad2 NCC-specifically deleted (lower panels) mouse embryos carrying R26R reporter were stained with X-gal as described in Methods (left panels). The embryos were then paraffin-embedded and sectioned to observe LacZ staining in dorsal aorta (DA) and aortic arch arteries III, IV and VI as indicated (right panel). Smad2 deletion did not affect NCC migration to aortic arch arteries.
Smad2 deficiency in NCCs caused reduced VSMC layers, distorted and thinner elastic lamina, resulting in a thinner vessel wall in adult mouse carotid arteries
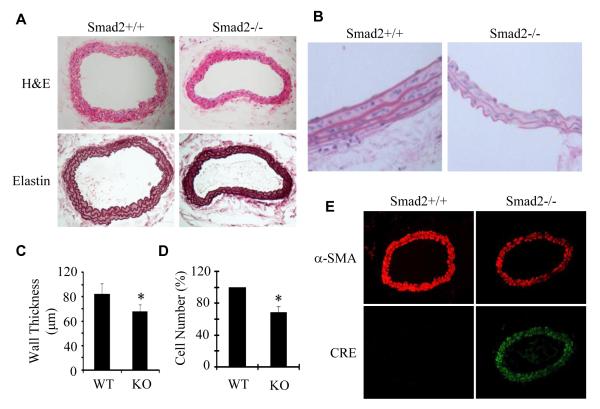
Although NCC-specific deletion of Smad2 impaired VSMC differentiation in aortic arch arteries, most Smad2 mutant mice were born normally and can survive until adulthood. We then examined whether Smad2 deletion causes defects in adult blood vessels in which VSMCs are derived from NCCs. We found that the vessel walls of Smad2-deficient carotid arteries were thinner than those in the wild type mice (Figure 2A-2D). A detailed examination revealed that one layer of VSMCs in the media was disappeared in Smad2-deficient arteries, which led to a significant reduction of VSMC numbers in the tunica media of Smad2-deleted vessels (Figure 2B and 2D). Moreover, Smad2-deficient vessels exhibited abnormality in elastic lamina including reduced layer and content, and distorted structure compared to wild type vessels (Figure 2B). These data demonstrate that defective VSMC differentiation due to Smad2 deletion significantly alters the normal vessel wall structure. To determine whether or not the cells in Smad2-deleted artery are SMCs, we stained the expression of α-SMA in the artery sections and found that Smad2−/− cells in the arteries displayed a reduced level of α-SMA (Fig 2E), suggesting that they retained SMC phenotype. To determine if these SMCs were derived from NCCs or other origins, we detected the Cre protein expression because the Cre was driven by Wnt1 promoter and thus only expressed in NCCs or NCC-derived cells in our system, we found that Smad2−/− SMCs expressed Cre, indicating that SMCs in the Smad2−/−artery were derived from NCCs.
Figure 2. Smad2 NCC-specific knockout caused reduced layers and numbers of VSMCs in the media, distorted and thinner elastic lamina, resulting in a decreased vessel wall thickness of carotid arteries.
(A) Cross sections of carotid arteries from wild type littermates (Smad2+/+ or WT) and Smad2 NCC-specifically deleted mice (Smad2−/− or KO) were stained with hematoxylin and eosin (H&E) or Elastica van Gieson (Elastin). (B) Higher magnification of H&E-stained vessel wall showed less number of VSMCs, thinner wall, and distorted and thinner elastic lamina in Smad2 KO carotid arteries as compared to the normal littermates. (C) Vessel wall thickness was measured. Mean thickness of the vessel walls was calculated with 12 sections covering different areas of common carotid arteries from 4 mice for each group. *P<0.05 compared to the WT arteries. (D) Quantification of VSMC numbers in the carotid arteries. The percentage reduction in KO artery was shown. *P<0.05 compared to WT arteries (n=12). (E) α-SMA immunostaining showed that Smad2−/− caused a reduction of VSMC marker gene expression. Cre immunostaining indicated that VSMCs in Smad2−/− arteries were derived from NCCs.
Smad2 and MRTFB cooperatively regulate VSMC marker gene expression in NCCs
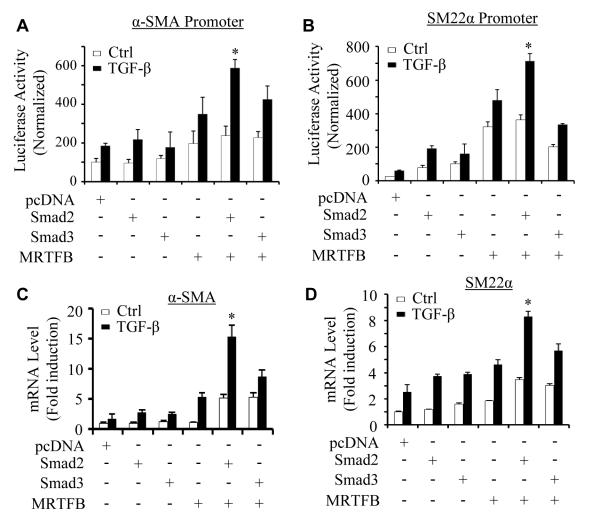
Previous studies have shown that MRTFB is specifically important for VSMC differentiation from NCCs.12, 13 To determine the mechanisms underlying Smad2 function in VSMC differentiation from NCCs, we explored if Smad2 and MRTFB coordinately regulate NCC differentiation to VSMC. MRTFB is induced in Monc-1 cells 2 h after TGF-β treatment (Online Figure II). We did not explore the function of myocardin or MRTFA in NCC differentiation because MRTFA and myocardin were not induced in Monc-1 cells until 48 h of TGF-β induction (Online Figure II), a time after the activation of SMC marker genes including SMMHC.23 By co-expressing MRTFB with Smad2 or Smad3 in Monc-1 cells, we found that MRTFB and Smad2, but not Smad3, cooperatively up-regulated TGF-β-induced α-SMA and SM22α promoter activities as compared to the effect of individual proteins (Figure 3A-3B). MRTFB and Smad2 also synergistically enhanced the endogenous mRNA expression of both α-SMA and SM22α genes (Figure 3C-3D). MRTFB appears to also increase VSMC marker promoter activity in the absence of TGF-β (Figure 3A-3B), probably because the transfected promoters were located in the cytoplasm where a certain level of MRTFB was present when the cells were in the basal status.
Figure 3. Smad2 and MRTFB cooperatively regulated VSMC marker gene expression in NCCs.
(A-B) Smad2 and MRTFB synergistically enhanced TGF-β-induced VSMC marker gene promoter activity. pcDNA, Smad2, Smad3, or MRTFB cDNA was co-transfected individually or in combination with α-SMA (A) or SM22α promoter (B) as indicated followed by vehicle (Ctrl) or TGF-β treatment (5 ng/ml) for 16 hours. Luciferase assays were performed. Luciferase activity was normalized to renilla activity. *P<0.01 compared to Smad2 or MRTFB alone-transfected group with TGF-β treatment in A and B, respectively. (C-D) Smad2 and MRTFB synergistically enhanced TGF-β-induced mRNA expression of VSMC markers. Monc-1 cells were transfected with pcDNA, Smad2, Smad3, or MRTFB cDNA individually or in combination followed by vehicle (Ctrl) or TGF-β treatment (5 ng/ml) for 8 hours. α-SMA (C) and SM22α (D) mRNA expression was detected by qPCR and normalized to GAPDH expression. *P<0.01 compared to Smad2 or MRTFB alone-transfected group treated with TGF-β in both C and D, respectively.
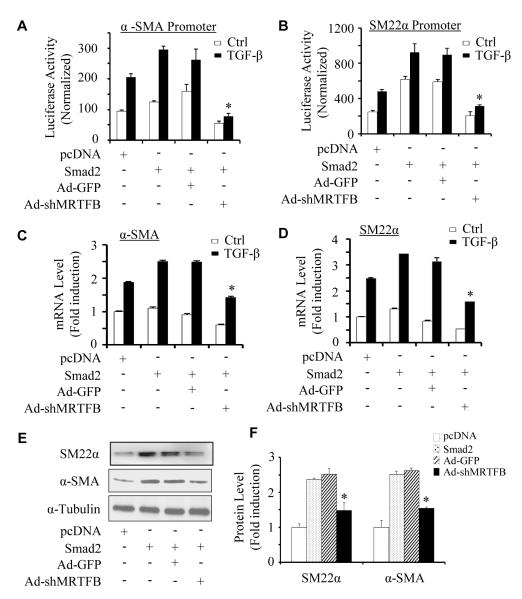
To determine if MRTFB is required for Smad2 function in VSMC differentiation, we used adenovirus-expressing shRNA to knock down MRTFB expression in Monc-1 cells. These cells were then co-transfected with Smad2 and SMC marker gene promoters. We found that knockdown of MRTFB completely blocked Smad2-mediated increase in α-SMA and SM22α promoter activities in both vehicle and TGF-β-treated cells (Figure 4A-4B). Importantly, knockdown of MRTFB also significantly blocked Smad2-induced mRNA (Figure 4C-4D) as well as protein expression (Figure 4E-4F) of both α-SMA and SM22α genes. These data demonstrate that MRTFB is essential for Smad2 function in VSMC differentiation from NCCs.
Figure 4. MRTFB was essential for Smad2-mediated VSMC marker expression in NCCs.
(A-B) MRTFB was required for Smad2-mediated VSMC marker gene transcription. Monc-1 cells were transduced with adenovirus expressing GFP (Ad-GFP) or MRTFB shRNA (Ad-shMRTFB) and co-transfected with α-SMA (A) or SM22α (B) promoter construct and pcDNA or Smad2 cDNA as indicated followed by vehicle (Ctrl) or TGF-β treatment (5 ng/ml) for 16 hours. Luciferase assays were performed. Luciferase activity was normalized to renilla activity. *P<0.01 compared to Ad-GFP groups with TGF-β. (C-E) MRTFB was required for Smad2-mediated VSMC marker mRNA and protein expression. Monc-1 cells were treated as in A and B but without promoter transfection. α-SMA (C) and SM22α (D) mRNA expression was detected by qPCR and normalized to GAPDH expression. Ttheir protein expression was detected by western blot (E). *P<0.01 compared to Ad-GFP group treated with TGF-β for both α-SMA (C) and SM22α (D), respectively. (F) Quantitative analysis of protein expression in E by normalized to α-tubulin. *P<0.01 compared to Ad-GFP group for both α-SMA and SM22α, respectively.
To determine if Smad2/Smad3 is required for MRTFB function in VSMC differentiation from NCCs, we used adenovirus-expressing shRNA to knock down Smad2 or Smad3 in Monc-1 cells. The cells were then cotransfected with MRTFB and VSMC marker gene promoters. We found that knockdown of Smad3 weakly while knockdown of Smad2 strongly inhibited MRTFB-induced α-SMA and SM22α promoter activities in both vehicle and TGF-β-treated cells (Figure 5A-5B). However, knockdown of Smad2, but not Smad3, significantly blocked MRTFB-induced mRNA (Figure 5C-5D) and protein expression (Figure 5E-5F) of both α-SMA and SM22α genes. Similar to the results in Figure 3A and 3B, MRTFB induced the activity of exogenously-introduced VSMC marker gene promoters in the absence of TGF-β because transfected promoters were located in the cytoplasm where MRTFB was present in the basal status (see Figure 6 below for MRTFB cellular location). These data demonstrate that Smad proteins are essential for the full function of MRTFB in VSMC differentiation from NCCs. Smad2 appears to be more important than Smad3 in mediating MRTFB function in NCCs.
Figure 5. Smad2 was essential for MRTFB-mediated VSMC marker expression in NCCs.
(A-B) Smad2 was required for MRTFB-mediated VSMC marker gene transcription. Monc-1 cells were transduced with adenovirus expressing GFP (Ad-GFP), Smad2 (Ad-shS2), or Smad3 shRNA (Ad-shS3) and co-transfected with α-SMA (A) or SM22α (B) promoter construct and pcDNA or MRTFB cDNA as indicated followed by vehicle (Ctrl) or TGF-β treatment (5 ng/ml) for 16 hours. Luciferase assays were performed. Luciferase activity was normalized to renilla activity. *P<0.01, #P=0.043, &P=0.048 compared to Ad-GFP groups with TGF-β for both A and B, respectively. (C-D) Smad2 was required for MRTFB-mediated VSMC marker mRNA expression. Monc-1 cells were treated as in A and B but without promoter transfection. α-SMA (C) and SM22α (D) mRNA expression was detected by qPCR and normalized to GAPDH expression. *P<0.05, @P>0.05 compared to Ad-GFP group treated with TGF-β for both C and D. (E) Smad2 was required for MRTFB-mediated VSMC marker protein expression. Monc-1 cells were transduced with Ad-GFP or Ad-shS2 as indicated and treated with TGF-β. α-SMA and SM22α protein expression was detected by western blot. (F) Quantitative analysis of the protein expression in E by normalized to α-tubulin. *P<0.05 compared to Ad-GFP group for both α-SMA and SM22α, respectively.
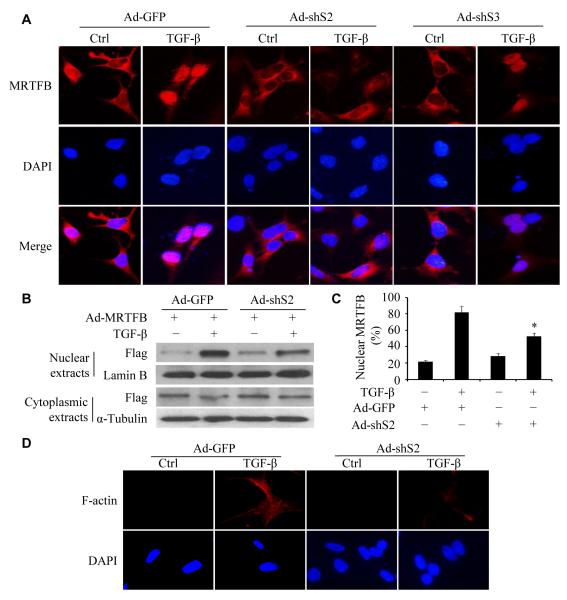
Figure 6. Smad2 was essential for MRTFB nuclear translocation.
(A) Knockdown of Smad2 blocked MRTFB nuclear translocation. Monc-1 cells were transduced with adenovirus expressing Flag-tagged MRTFB and GFP (Ad-GFP), Smad2 (Ad-shS2) or Smad3 shRNA (Ad-shS3) as indicated for 2 days followed by vehicle (Ctrl) or TGF-β induction for 2 hours. MRTFB expression and its cellular location were detected by immunostaining with anti-Flag antibody. DAPI stains nuclei. TGF-β induced MRTFB nuclear translocation, which was blocked by Ad-ShS2, but not Ad-shS3. (B) MRTFB cellular location detected by western blot. Monc-1 cells were treated the same as in A, and cytoplasmic or nuclear MRTFB was detected with Flag antibody. (C) Nuclear MRTFB in B was quantified and shown as percentage of the total MRTFB. Prior to the calculation, nuclear MRTFB was normalized to Lamin B, and cytoplasmic MRTFB was normalized to α-Tubulin. *P<0.05 compared to Ad-GFP-transduced cells treated with TGF-β. (D) Knockdown of Smad2 blocked F-actin formation. Monc-1 cells were transduced with Ad-GFP or Ad-ShS2 followed by vehicle (Ctrl) or TGF-β treatment. F-actin was stained with Phalloidin.
Smad2 is essential for MRTFB nuclear translocation in NCCs
Consistent with previous report,39 MRTFB is predominantly localized in the cytoplasm of Monc-1 cells (Figure 6A). However, MRTFB nuclear translocation is essential for the activation of VSMC endogenous gene transcription. We found that TGF-β induced MRTFB nuclear translocation. To determine if Smad2 and Smad3 affect MRTFB nuclear translocation, we knocked down Smad2 and Smad3 in Monc-1 cells using Smad2 and Smad3 shRNA, respectively. As shown in Figure 6A, deficiency of Smad2, but not Smad3, blocked TGF-β-induced MRTFB nuclear translocation. To quantify the nuclear translocation of MRTFB, we separated cytoplasmic and nuclear proteins of vehicle and TGF-β-treated Monc-1 cells and examined the MRTFB expression in each portion. We found that only 20% of the MRTFB was located in the nuclei of vehicle-treated Monc-1 cells (Fig 6B-6C). TGF-β induction increased the nuclear portion to 80% of the total MRTFB (Fig 6B-6C). Smad2 knockdown by shRNA, however, significantly reduced the nuclear MRTFB level down to 50% of the total in TGF-β-treated cells (Fig 6B-6C). Previous studies have shown that MRTF nuclear translocation was mediated by cytoskeleton reorganization.40 We therefore sought to determine if Smad2 mediates MRTFB nuclear translocation through facilitating F-actin formation. As shown in Figure 6D, knockdown of Smad2 blocked TGF-β-induced F-actin formation, suggesting that Smad2 indeed mediated MRTFB nuclear translocation by regulating F-actin formation. Together, these data demonstrate that Smad2 regulates MRTFB function by mediating its nuclear translocation.
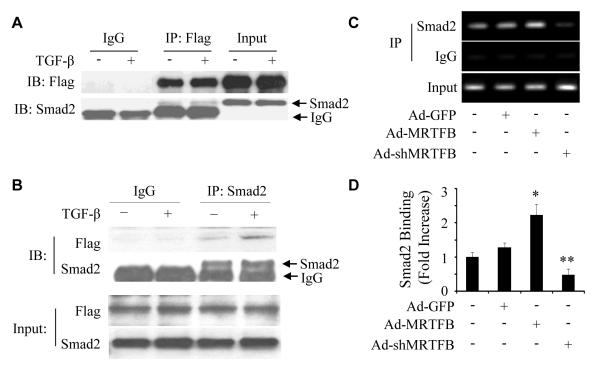
MRTFB physically interacts with Smad2 and facilitates Smad2 binding to VSMC marker gene promoter
In addition to being a TGF-β signaling molecule, Smad2 is also a transcription factor regulating TGF-β target genes. To further determine the mechanism by which Smad2 regulates VSMC differentiation from NCCs, we examined if Smad2 physically interacts with MRTFB. Since high quality MRTFB antibody is not available, we used adenoviral vector to express Flag-tagged MRTFB in Monc-1 cells and performed co-immunoprecipitation of MRTFB with endogenous Smad2 proteins. As shown in Fig 7A, endogenous Smad2 formed a complex with Flag-tagged MRTFB, indicating a physical interaction between Smad2 and MRTFB. TGF-β induction appeared to enhance Smad2-MRTFB interaction. To confirm the MRTFB-Smad2 interaction, we reversed the order of Co-IP and immunoblotting by using Smad2 antibody co-IP with MRTFB. We found that MRTFB was in the complex immunoprecipitated by Smad2 antibody as shown in the blotting with Flag antibody (Fig 7B). TGF-β treatment clearly enhanced their interaction (Fig 7B). These data demonstrate that Smad2 physically interacts with MRTFB in Monc-1 cells.
Figure 7. MRTFB physically interacted with Smad2 and enhanced Smad2 binding to VSMC marker promoter.
(A-B) Co-IP of Smad2 with MRTFB. Monc-1 cells were transduced with adenovirus expressing Flag-tagged MRTFB for 2 days followed by vehicle (-) or TGF-β treatment (+) for 2 h. Cell lysates were immunoprecipitated with Flag (A) or Smad2 (B) antibody and blotted with Flag or Smad2 antibody as indicated. Input was 5% of the cell lysates. (C) MRTFB enhanced Smad2 binding to SM22α promoter. Monc-1 cells were transduced with adenovirus expressing GFP (Ad-GFP), MRTFB (Ad-MRTFB) or MRTFB shRNA (AdshMRTFB) followed by TGF-β treatment (5 ng/ml) for 2 hours. CHIP was performed as described in Methods. Smad2 binding enrichment was detected by RT-PCR using primers amplifying the functional SBE element in SM22α promoter. (D) Smad2 binding to SM22α promoter was quantified by qPCR, and the binding fold increase was shown. *P<0.001, **P<0.001 compared to Ad-GFP group.
Smad2 regulates VSMC marker gene transcription by binding to VSMC marker promoters. To determine how MRTFB regulated Smad2 activity, we tested if MRTFB alters Smad2 interaction with SM22α promoter in Monc-1 cells. We found that in TGF-β-treated Monc-1 cells, MRTFB overexpression significantly increased Smad2 binding to SM22α promoter while shRNA knockdown of MRTFB completely blocked Smad2 binding to the promoter (Figure 7C-7D). These data demonstrate that MRTFB serves as a Smad2 coactivator to mediate and enhance Smad2 binding to VSMC marker promoters, leading to activation of VSMC marker gene transcription.
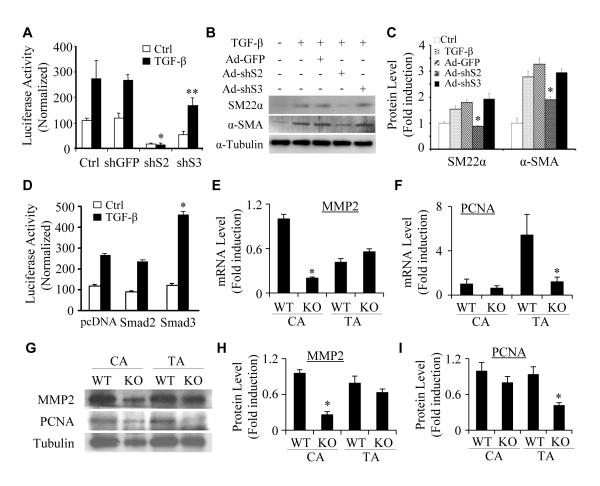
Smad2 is more important than Smad3 in mediating VSMC differentiation from NCCs while Smad3 is important in the differentiation from mesenchymal progenitors
TGF-β signaling is transduced predominantly by both Smad2 and Smad3. Previous studies from our and other laboratories have shown that Smad3 plays an important role in TGF-β-induced VSMC differentiation from mesenchymal progenitor cells.21, 41, 42 Our present study demonstrates that Smad2 is essential for VSMC differentiation from NCCs. These results led us to hypothesize that Smad2 and Smad3 are involved in progenitor-specific regulation of VSMC differentiation. To test this hypothesis, we knocked down the expression of endogenous Smad2 and Smad3 individually in Monc-1 cells (Online Figure III), and then transfected the cells with SM22α promoter.43 Luciferase assay showed that knockdown of Smad3 slightly inhibits both the basal and TGF-β-induced SM22α promoter activity, but did not alter TGF-β-induced fold induction of the promoter activity. However, knockdown of Smad2 completely abolishes the promoter function (Figure 8A). Moreover, knockdown of Smad2 blocked TGF-β-induced protein expression of endogenous α-SMA and SM22α while knockdown of Smad3 had no effect (Figure 8B-8C). These data demonstrate that Smad2, but not Smad3, plays a major role in TGF-β-induced VSMC differentiation from NCCs. Interestingly, when we overexpressed Smad2 and Smad3 individually in mesenchymal progenitor C3H10T1/2 cells, Smad3, but not Smad2, enhanced TGF-β-induced SM22α promoter activity (Figure 8D). These data, together with the previously published results suggest that Smad proteins serve as progenitor-specific regulators in VSMC fate determination or VSMC differentiation from at least some of the progenitors, i.e. Smad2 regulates VSMC differentiation from NCCs, while Smad3 controls VSMC differentiation from mesenchymal progenitors.
Figure 8. Progenitor-specific roles of Smad2 and Smad3 in regulating VSMC marker gene transcription and protein expression.
(A) Smad2 was more important than Smad3 in regulating VSMC marker gene promoter activity in NCCs. Monc-1 cells were transduced with adenovirus expressing GFP (shGFP), Smad2 (shS2), or Smad3 (shS3) shRNA for 2 days and transfected with SM22α promoter followed by vehicle (Ctrl) or TGF-β (5 ng/ml) treatment. Luciferase assays were performed. Luciferase activity was normalized to renilla activity. *P<0.001, **P<0.05 compared to shGFP/TGF-treated groups. (B) Smad2, but not Smad3, was essential for VSMC mark protein expression in NCCs. Monc-1 cells were treated similarly as described in A without promoter transfection. α-SMA and SM22α protein expression was detected by western blot. (C) Quantification of protein expression shown in B by normalized to α-tubulin. *P<0.01 compared to Ad-GFP group for α-SMA and SM22α, respectively. (D) Smad3 was more important than Smad2 in VSMC differentiation from mesenchymal progenitors. 10T1/2 cells were co-transfected with SM22α promoter and pcDNA, Smad2, or Smad3 cDNA followed by vehicle (Ctrl) or TGF-β treatment. Luciferase assays were performed. Luciferase activity was normalized to renilla activity. *P<0.01 compared to pcDNA/TGF-β-treated group. (E and F) Smad2 was deleted in SMCs by crossing Smad2floxed mice with SM22α-Cre mice. Carotid (CA) or thoracic artery (TA) media layers from wild type littermates (WT) or SMC-specific Smad2 knockout mice (KO) were homogenized, and total RNA was extracted. qPCR were performed to detect MMP2 (E) and PCNA expression (F) and fold inductions were shown. *P<0.01 compared to the corresponding WT SMCs. (G) MMP2 and PCNA protein expression in WT and Smad2 KO SMCs was examined by western blot. (H-I) MMP2 (H) and PCNA expression (I) was normalized to α-tubulin. *P<0.01 compared to the corresponding WT SMCs.
Since VSMCs from different progenitors display distinct functional properties, and our results suggest that Smad2 is important for NCC, but not mesenchymal progenitor, differentiation to VSMC, we sought to determine if Smad2 is involved in the physiological differences between NCC- and mesoderm-derived VSMCs. To this end, we crossed SM22α-Cre mice with Smad2fl/fl mice to knock out Smad2 in VSMCs derived from NCC as well as mesoderm. To test if Smad2 affects the properties of these VSMCs, we isolated the media layers of carotid arteries (SMCs originate from NCC) and thoracic aortas (SMCs originate from mesoderm) that contains mainly SMCs, prepared the total RNA and proteins, and detected the expression of MMP2 and proliferating cell nuclear antigen (PCNA), a well-known cell proliferation marker. It appeared that the expression levels of these two proteins were different in NCC-derived and mesoderm-derived VSMCs (Figure 8E-8I). Importantly, Smad2 was essential for MMP2 mRNA and protein expression in NCC-derived VSMCs, but not in mesoderm-derived VSMCs (Figure 8E, 8G and 8H). However, Smad2 appeared not to affect PCNA expression in NCC-derived VSMCs, but was required for PCNA expression in mesoderm-derived VSMCs (Figure 8F, 8G and 8I), which is consistent with the critical role of Smad2 in VSMC differentiation of NCCs, but not mesenchymal progenitor. Collectively, our data suggest that Smad2 is involved in defining the physiological difference between NCC- and mesoderm-derived VSMCs.
DISCUSSION
By using tissue-specific knockout mouse model, we demonstrate for the first time that Smad2 is critical for VSMC differentiation from NCCs in vivo. The results are very specific because Smad2 deletion in NCCs only blocks VSMC marker gene expression in aortic arch arteries, but not in the dorsal aorta. Although previous studies have tried to link Smad2 function with NCC differentiation to VSMCs or endothelial cells,22, 44, 45 the results are limited to the detection of Smad2 expression or use of in vitro systems. Our results represent the first direct evidence showing the critical role of Smad2 in VSMC differentiation from NCCs in vivo. Interestingly, Smad proteins appear to play a role in the progenitor-specific regulation of VSMC differentiation. Although both Smad2 and Smad3 mediate TGF-β function, Smad3 is important for the differentiation of mesenchymal-originated progenitors,21, 41, 42 while Smad2 plays a major role in VSMC differentiation from NCCs. Smad3 knockout mice are viable and show no abnormality in cardiovascular or VSMC development,46, 47 suggesting that loss of Smad3 function in these mice may be compensated by Smad2. However, Smad3 is unable to compensate for the loss of Smad2 function because Smad2 knockout mice die around gastrulation before the cardiovascular system starts to develop.48 Our study further demonstrates that Smad3 cannot make up for the loss of Smad2 function in NCC differentiation to VSMCs in vivo because deletion of Smad2 in NCCs resulted in VSMC defects.
Both TGF-β signaling and MRTFB play critical roles in regulating VSMC differentiation from NCCs.12-14 It is unclear, however, how these two independent pathways contribute to the same biological process. Our study has provided a novel mechanism underlying the functions of TGF-β signaling and MRTFB in VSMC differentiation from NCCs. It appears that MRTFB and TGF-β downstream signaling protein Smad2 cooperatively regulate VSMC marker gene transcription. In fact, Smad2 and MRTFB are interdependent in regulating VSMC marker gene expression. Knockdown of Smad2 impairs MRTFB regulation of VSMC gene expression whereas knockdown of MRTFB blocks Smad2 activity in this process. More importantly, we have identified a novel function of Smad2 in promoting MRTFB nuclear translocation. Smad2 likely binds to MRTFB upon TGF-β induction in order to facilitate MRTFB nuclear translocation. Indeed, Smad2 physically interacts with MRTFB, and this interaction is enhanced by TGF-β. Importantly, MRTFB-Smad2 interaction not only results in transporting MRTFB into nuclei, but also facilitates Smad2 binding to VSMC marker promoter, which is essential for TGF-β induction of VSMC differentiation.
Previous studies have shown that a muscle-specific actin-binding protein named striated muscle activator of Rho signaling (STARS) activates serum response factor (SRF) by inducing the nuclear translocation of MRTFA and MRTFB, and the STARS-dependent nuclear import of MRTFs requires RhoA and actin polymerization.39 Smad2 appears to mediate MRTFB nuclear translocation through regulating actin polymerization. Since TGF-β activates RhoA; and RhoA crosstalks with Smad signaling in Monc-1 cells,36 it is likely that RhoA, STARS, and Smad2 work together to coordinate MRTFB nuclear translocation. Interestingly, TGF-β increases MRTFA nuclear localization but has no effect on MRTFB nuclear location in 10T1/2 cells.49 The present study shows that TGF-β induces MRTFB nuclear translocation in Monc-1 cells, which is likely due to a progenitor-specific effect. 10T1/2 cells are derived from mesoderm and considered as mesenchymal progenitors, in which Smad2 is not important for their differentiation to VSMCs. However, in NCCs, Smad2 plays a major role in the differentiation, and mediates MRTFB nuclear translocation. These results further support the novel concept that Smad proteins may be important mediators for progenitor-specific regulation of VSMC differentiation.
Smad2 knockout in NCCS not only causes defective VSMC differentiation during embryonic development, but also leads to abnormal structure in the wall of the adult vasculature where VSMCs are derived from NCCs. These abnormalities, i.e., less numbers of VSMCs, distorted and thinner elastic lamina, and thinner vessel walls are likely due to the defective VSMC differentiation, leading to reduced numbers of VSMCs in the tunica media. VSMCs have been shown to produce elastin, the major component of the elastic fibers.50 It is unknown, however, how Smad2 deletion causes abnormal elastin expression in VSMCs and distorts the structure of the elastic lamina in the tunica media of the carotid artery, which requires an extensive future study.
Although it is well recognized that SMCs derived from different origins display distinct physiological properties, the molecular mechanisms controlling these differences or regulatory factors involved in defining these variances remain largely unknown. Smad2 appears to be one of these factors because Smad2 not only play distinct roles in regulating SMC gene expression in different progenitors, but also discretely regulates the properties of NCC- and mesoderm-derived VSMCs including the production of extracellular matrix protein regulator MMP2 and proliferation-related gene PCNA. Although the physiological properties are not extensively studied, our results provide initial evidence that Smad2 is involved in defining the different properties of VSMCs derived from NCC and mesoderm.
In summary, we have found that Smad2 is essential for VSMC differentiation from NCCs in vivo and contributes to progenitor-specific regulation of VSMC differentiation as well as the physiological difference of SMCs derived from NCCs and mesoderm. Smad2 interaction with MRTFB appears to be a novel and important mechanism underlying NCC differentiation to VSMCs. Smad2 facilitates MRTFB nuclear translocation while MRTFB enhancing Smad2 binding to VSMC marker promoter, leading to activation of VSMC gene transcription.
Supplementary Material
Novelty and Significance.
What Is Known?
TGF-β signaling plays an important role in vascular smooth muscle cell (VSMC) differentiation. TGF-β receptors and its accessory protein, Endoglin, are implicated in VSMC differentiation from neural crest cells (NCC).
During embryonic development, VSMCs originate from at least eight different progenitors. VSMCs from different progenitors display distinct functional properties, and utilize distinct cis-elements and control regions to regulate their marker gene activation.
Myocardin-related transcription factor B (MRTFB) regulates VSMC differentiation from NCC. MRTFB is normally located in cytoplasm and translocates into the nucleus to exert its function as a coactivator.
What New Information Does This Article Contribute?
TGF-β downstream signaling protein Smad2 is essential for VSMC differentiation from NCC in vivo. Deletion of Smad2 in NCC results in VSMC defects in aortic arch arteries and abnormal carotid arterial structure.
Smad2 regulates VSMC marker gene transcription by interacting with MRTFB. Smad2 mediates MRTFB nuclear translocation upon TGF-β induction. MRTF-B enhances Smad2 binding to VSMC marker promoter.
Smad proteins play roles in progenitor-specific regulation of VSMC differentiation. Smad2 is more important than Smad3 in VSMC differentiation from NCC while Smad3 is more important than Smad2 in VSMC differentiation from mesenchymal progenitors.
Previous studies have shown that both TGF-β signaling and MRTFB play essential roles in VSMC differentiation from NCC. Here, we used tissue-specific Smad2 knockout mouse model as well as molecular and cellular analyses to identify novel mechanisms by which TGF-β regulates NCC differentiation to VSMC. Our results show that TGF-β downstream signaling intermediate Smad2 is essential for VSMC differentiation from NCC. We found that Smad2 deletion in NCC results in defective VSMC differentiation in aortic arch arteries during embryonic development and blood vessel wall abnormality in adult arteries where VSMCs are derived from NCCs. The abnormalities include a missing layer of VSMCs and distorted elastic fibers, resulting in a thinner vessel wall compared to the wild type vessel. It appears that Smad2 interacts with MRTFB to regulate VSMC marker gene expression. Smad2 mediates TGF-β-induced MRTFB nuclear translocation while MRTFB enhancing Smad2 binding to VSMC marker promoter. Importantly, our studies indicate that Smad2 is a progenitor-specific regulator for VSMC differentiation from NCCs while Smad3 is more important than Smad2 in mediating VSMC differentiation from mesenchymal progenitors. These findings suggest that TGF-β signaling is involved in the determination of VSMC diversity.
Acknowledgments
SOURCES OF FUNDING This study was supported by grant from National Institutes of Health (HL093429 and HL107526 to S.Y.C).
Nonstandard Abbreviations and Acronyms
- MRTFB
myocardin-related transcription factor B
- TGF-β
transforming growth factor-β
- NCC
neural crest cells
- TβRII
TGF-β type II receptor
- VSMC
vascular smooth muscle cell
- α-SMA
smooth muscle α-actin
- IHC
immunohistochemistry
- shRNA
small hairpin RNA
- ChIP
chromatin immunoprecipitation assay
- Co-IP
co-immunoprecipitation assay
Footnotes
DISCLOSURES None.
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Guo DC, Pannu H, Tran-Fadulu V, Papke CL, Yu RK, Avidan N, Bourgeois S, Estrera AL, Safi HJ, Sparks E, Amor D, Ades L, McConnell V, Willoughby CE, Abuelo D, Willing M, Lewis RA, Kim DH, Scherer S, Tung PP, Ahn C, Buja LM, Raman CS, Shete SS, Milewicz DM. Mutations in smooth muscle alpha-actin (acta2) lead to thoracic aortic aneurysms and dissections. Nat Genet. 2007;39:1488–1493. doi: 10.1038/ng.2007.6. [DOI] [PubMed] [Google Scholar]
- 2.Milewicz DM, Guo DC, Tran-Fadulu V, Lafont AL, Papke CL, Inamoto S, Kwartler CS, Pannu H. Genetic basis of thoracic aortic aneurysms and dissections: Focus on smooth muscle cell contractile dysfunction. Annu Rev Genomics Hum Genet. 2008;9:283–302. doi: 10.1146/annurev.genom.8.080706.092303. [DOI] [PubMed] [Google Scholar]
- 3.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 4.Majesky MW. Developmental basis of vascular smooth muscle diversity. Arterioscler Thromb Vasc Biol. 2007;27:1248–1258. doi: 10.1161/ATVBAHA.107.141069. [DOI] [PubMed] [Google Scholar]
- 5.Topouzis S, Majesky MW. Smooth muscle lineage diversity in the chick embryo. Dev Biol. 1996;178:430–445. [PubMed] [Google Scholar]
- 6.Rosenquist TH, Beall AC. Elastogenic cells in the developing cardiovascular system. Smooth muscle, nonmuscle, and cardiac neural crest. Ann N Y Acad Sci. 1990;588:106–119. doi: 10.1111/j.1749-6632.1990.tb13201.x. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Ip HS, Lu MM, Clendenin C, Parmacek MS. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol Cell Biol. 1997;17:2266–2278. doi: 10.1128/mcb.17.4.2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, Miano JM, Mercer B, Olson EN. Expression of the sm22alpha promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J Cell Biol. 1996;132:849–859. doi: 10.1083/jcb.132.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lilly B, Olson EN, Beckerle MC. Identification of a carg box-dependent enhancer within the cysteine-rich protein 1 gene that directs expression in arterial but not venous or visceral smooth muscle cells. Dev Biol. 2001;240:531–547. doi: 10.1006/dbio.2001.0507. [DOI] [PubMed] [Google Scholar]
- 10.Manabe I, Owens GK. Carg elements control smooth muscle subtype-specific expression of smooth muscle myosin in vivo. J Clin Invest. 2001;107:823–834. doi: 10.1172/JCI11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manabe I, Owens GK. The smooth muscle myosin heavy chain gene exhibits smooth muscle subtype-selective modular regulation in vivo. J Biol Chem. 2001;276:39076–39087. doi: 10.1074/jbc.M105402200. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Zhu X, Chen M, Cheng L, Zhou D, Lu MM, Du K, Epstein JA, Parmacek MS. Myocardin-related transcription factor b is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc Natl Acad Sci U S A. 2005;102:8916–8921. doi: 10.1073/pnas.0503741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh J, Richardson JA, Olson EN. Requirement of myocardin-related transcription factor-b for remodeling of branchial arch arteries and smooth muscle differentiation. Proc Natl Acad Sci U S A. 2005;102:15122–15127. doi: 10.1073/pnas.0507346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wurdak H, Ittner LM, Lang KS, Leveen P, Suter U, Fischer JA, Karlsson S, Born W, Sommer L. Inactivation of tgfbeta signaling in neural crest stem cells leads to multiple defects reminiscent of digeorge syndrome. Genes Dev. 2005;19:530–535. doi: 10.1101/gad.317405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah NM, Anderson DJ. Integration of multiple instructive cues by neural crest stem cells reveals cell-intrinsic biases in relative growth factor responsiveness. Proc Natl Acad Sci U S A. 1997;94:11369–11374. doi: 10.1073/pnas.94.21.11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.High FA, Zhang M, Proweller A, Tu L, Parmacek MS, Pear WS, Epstein JA. An essential role for notch in neural crest during cardiovascular development and smooth muscle differentiation. J Clin Invest. 2007;117:353–363. doi: 10.1172/JCI30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaartinen V, Dudas M, Nagy A, Sridurongrit S, Lu MM, Epstein JA. Cardiac outflow tract defects in mice lacking alk2 in neural crest cells. Development. 2004;131:3481–3490. doi: 10.1242/dev.01214. [DOI] [PubMed] [Google Scholar]
- 18.Mancini ML, Verdi JM, Conley BA, Nicola T, Spicer DB, Oxburgh LH, Vary CP. Endoglin is required for myogenic differentiation potential of neural crest stem cells. Dev Biol. 2007;308:520–533. doi: 10.1016/j.ydbio.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah NM, Groves AK, Anderson DJ. Alternative neural crest cell fates are instructively promoted by tgfbeta superfamily members. Cell. 1996;85:331–343. doi: 10.1016/s0092-8674(00)81112-5. [DOI] [PubMed] [Google Scholar]
- 20.Choudhary B, Ito Y, Makita T, Sasaki T, Chai Y, Sucov HM. Cardiovascular malformations with normal smooth muscle differentiation in neural crest-specific type ii tgfbeta receptor (tgfbr2) mutant mice. Dev Biol. 2006;289:420–429. doi: 10.1016/j.ydbio.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, Kulik M, Lechleider RJ. Smad proteins regulate transcriptional induction of the sm22alpha gene by tgf-beta. Nucleic Acids Res. 2003;31:1302–1310. doi: 10.1093/nar/gkg224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S, Lechleider RJ. Transforming growth factor-beta-induced differentiation of smooth muscle from a neural crest stem cell line. Circ Res. 2004;94:1195–1202. doi: 10.1161/01.RES.0000126897.41658.81. [DOI] [PubMed] [Google Scholar]
- 23.de Caestecker MP, Parks WT, Frank CJ, Castagnino P, Bottaro DP, Roberts AB, Lechleider RJ. Smad2 transduces common signals from receptor serine-threonine and tyrosine kinases. Genes Dev. 1998;12:1587–1592. doi: 10.1101/gad.12.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Caestecker MP, Yahata T, Wang D, Parks WT, Huang S, Hill CS, Shioda T, Roberts AB, Lechleider RJ. The smad4 activation domain (sad) is a proline-rich, p300-dependent transcriptional activation domain. J Biol Chem. 2000;275:2115–2122. doi: 10.1074/jbc.275.3.2115. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Feng XH, Derynck R. Smad3 and smad4 cooperate with c-jun/c-fos to mediate tgf-beta-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida T, Sinha S, Dandre F, Wamhoff BR, Hoofnagle MH, Kremer BE, Wang DZ, Olson EN, Owens GK. Myocardin is a key regulator of carg-dependent transcription of multiple smooth muscle marker genes. Circ Res. 2003;92:856–864. doi: 10.1161/01.RES.0000068405.49081.09. [DOI] [PubMed] [Google Scholar]
- 27.Ju W, Ogawa A, Heyer J, Nierhof D, Yu L, Kucherlapati R, Shafritz DA, Bottinger EP. Deletion of smad2 in mouse liver reveals novel functions in hepatocyte growth and differentiation. Mol Cell Biol. 2006;26:654–667. doi: 10.1128/MCB.26.2.654-667.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 29.Ito Y, Yeo JY, Chytil A, Han J, Bringas P, Jr., Nakajima A, Shuler CF, Moses HL, Chai Y. Conditional inactivation of tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development. 2003;130:5269–5280. doi: 10.1242/dev.00708. [DOI] [PubMed] [Google Scholar]
- 30.Chai Y, Jiang X, Ito Y, Bringas P, Jr., Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 31.Oka K, Oka S, Sasaki T, Ito Y, Bringas P, Jr., Nonaka K, Chai Y. The role of tgf-beta signaling in regulating chondrogenesis and osteogenesis during mandibular development. Dev Biol. 2007;303:391–404. doi: 10.1016/j.ydbio.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi N, Xie WB, Chen SY. Cell division cycle 7 is a novel regulator of transforming growth factor-beta-induced smooth muscle cell differentiation. J Biol Chem. 2012;287:6860–6867. doi: 10.1074/jbc.M111.306209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang JN, Shi N, Xie WB, Guo X, Chen SY. Response gene to complement 32 promotes vascular lesion formation through stimulation of smooth muscle cell proliferation and migration. Arterioscler Thromb Vasc Biol. 2011;31:e19–26. doi: 10.1161/ATVBAHA.111.230706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie WB, Li Z, Miano JM, Long X, Chen SY. Smad3-mediated myocardin silencing: A novel mechanism governing the initiation of smooth muscle differentiation. J Biol Chem. 2011;286:15050–15057. doi: 10.1074/jbc.M110.202747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li F, Luo Z, Huang W, Lu Q, Wilcox CS, Jose PA, Chen S. Response gene to complement 32, a novel regulator for transforming growth factor-beta-induced smooth muscle differentiation of neural crest cells. J Biol Chem. 2007;282:10133–10137. doi: 10.1074/jbc.C600225200. [DOI] [PubMed] [Google Scholar]
- 36.Chen S, Crawford M, Day RM, Briones VR, Leader JE, Jose PA, Lechleider RJ. Rhoa modulates smad signaling during transforming growth factor-beta-induced smooth muscle differentiation. J Biol Chem. 2006;281:1765–1770. doi: 10.1074/jbc.M507771200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Xie WB, Escano CS, Asico LD, Xie Q, Jose PA, Chen SY. Response gene to complement 32 is essential for fibroblast activation in renal fibrosis. J Biol Chem. 2011;286:41323–41330. doi: 10.1074/jbc.M111.259184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo X, Jose PA, Chen SY. Response gene to complement 32 interacts with smad3 to promote epithelial-mesenchymal transition of human renal tubular cells. Am J Physiol Cell Physiol. 2011;300:C1415–1421. doi: 10.1152/ajpcell.00204.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuwahara K, Barrientos T, Pipes GC, Li S, Olson EN. Muscle-specific signaling mechanism that links actin dynamics to serum response factor. Mol Cell Biol. 2005;25:3173–3181. doi: 10.1128/MCB.25.8.3173-3181.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control srf activity by regulation of its coactivator mal. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 41.Qiu P, Ritchie RP, Fu Z, Cao D, Cumming J, Miano JM, Wang DZ, Li HJ, Li L. Myocardin enhances smad3-mediated transforming growth factor-beta1 signaling in a carg box-independent manner: Smad-binding element is an important cis element for sm22alpha transcription in vivo. Circ Res. 2005;97:983–991. doi: 10.1161/01.RES.0000190604.90049.71. [DOI] [PubMed] [Google Scholar]
- 42.Qiu P, Feng XH, Li L. Interaction of smad3 and srf-associated complex mediates tgf-beta1 signals to regulate sm22 transcription during myofibroblast differentiation. J Mol Cell Cardiol. 2003;35:1407–1420. doi: 10.1016/j.yjmcc.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Moessler H, Mericskay M, Li Z, Nagl S, Paulin D, Small JV. The sm 22 promoter directs tissue-specific expression in arterial but not in venous or visceral smooth muscle cells in transgenic mice. Development. 1996;122:2415–2425. doi: 10.1242/dev.122.8.2415. [DOI] [PubMed] [Google Scholar]
- 44.Molin DG, Poelmann RE, DeRuiter MC, Azhar M, Doetschman T, Gittenberger-de Groot AC. Transforming growth factor beta-smad2 signaling regulates aortic arch innervation and development. Circ Res. 2004;95:1109–1117. doi: 10.1161/01.RES.0000150047.16909.ab. [DOI] [PubMed] [Google Scholar]
- 45.Huang WY, Xie W, Guo X, Li F, Jose PA, Chen SY. Smad2 and pea3 cooperatively regulate transcription of response gene to complement 32 in tgf-beta-induced smooth muscle cell differentiation of neural crest cells. Am J Physiol Cell Physiol. 2011;301:C499–506. doi: 10.1152/ajpcell.00480.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Datto MB, Frederick JP, Pan L, Borton AJ, Zhuang Y, Wang XF. Targeted disruption of smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol Cell Biol. 1999;19:2495–2504. doi: 10.1128/mcb.19.4.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashcroft GS, Yang X, Glick AB, Weinstein M, Letterio JL, Mizel DE, Anzano M, Greenwell-Wild T, Wahl SM, Deng C, Roberts AB. Mice lacking smad3 show accelerated wound healing and an impaired local inflammatory response. Nat Cell Biol. 1999;1:260–266. doi: 10.1038/12971. [DOI] [PubMed] [Google Scholar]
- 48.Heyer J, Escalante-Alcalde D, Lia M, Boettinger E, Edelmann W, Stewart CL, Kucherlapati R. Postgastrulation smad2-deficient embryos show defects in embryo turning and anterior morphogenesis. Proc Natl Acad Sci U S A. 1999;96:12595–12600. doi: 10.1073/pnas.96.22.12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hinson JS, Medlin MD, Lockman K, Taylor JM, Mack CP. Smooth muscle cell-specific transcription is regulated by nuclear localization of the myocardin-related transcription factors. Am J Physiol Heart Circ Physiol. 2007;292:H1170–1180. doi: 10.1152/ajpheart.00864.2006. [DOI] [PubMed] [Google Scholar]
- 50.Narayanan AS, Sandberg LB, Ross R, Layman DL. The smooth muscle cell. Iii. Elastin synthesis in arterial smooth muscle cell culture. J Cell Biol. 1976;68:411–419. doi: 10.1083/jcb.68.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.