Abstract
PCSK9 has exploded onto center stage of plasma cholesterol metabolism, raising hopes for a new strategy to treat hypercholesterolemia. PCSK9 in a plasma protein that triggers increased degradation of the LDL receptor. Gain-of-function mutations in PCSK9 reduce LDL receptor levels in the liver, resulting in high levels of LDL cholesterol in the plasma and increased susceptibility to coronary heart disease. Loss-of-function mutations lead to higher levels of the LDL receptor, lower LDL cholesterol levels, and protection from coronary heart disease. Two papers in this issue of the Journal of Lipid Research exemplify the rapid pace of progress in understanding PCSK9 molecular interactions and physiology. Dr. Shilpa Pandit and coworkers from Merck Research Laboratories describe the functional basis for the hypercholesterolemia associated with gain-of-function missense mutations in PCSK9. Dr. Jay Horton’s group at UT Southwestern describe the kinetics and metabolism of PCSK9 and the impact of PCSK9 on LDL receptors in the liver and adrenal gland.
During the past few years, the proprotein convertase subtilisin kexin 9 (PCSK9) field has been red hot, fueled by the realization that PCSK9 is a key player in plasma cholesterol metabolism and by a hope, shared by scientists in academia and industry alike, that PCSK9 is a target for treating hypercholesterolemia. PCSK9 regulates the levels of the LDL receptor (1–3), which is a plasma membrane glycoprotein that removes cholesterol-rich LDL particles from the plasma (4, 5). Gain-of-function mutations in PCSK9 reduce LDL receptor levels in the liver, resulting in high levels of LDL cholesterol in the plasma and increased susceptibility to coronary heart disease (6). Loss-of-function mutations lead to higher levels of the LDL receptor, lower LDL cholesterol levels, and protection from coronary heart disease (7–11). The loss of PCSK9 appears to have no adverse consequences (11). Thus, interest in PCSK9 as a cholesterol-lowering target has been high, and an army of investigators is now working to elucidate PCSK9 molecular interactions and physiology. In this issue of the Journal of Lipid Research (JLR), two leading research groups describe their recent efforts. Dr. Shilpa Pandit et al. (12) from Merck Research Laboratories describe the functional basis for the hypercholesterolemia associated with gain-of-function missense mutations in PCSK9. Grefhorst et al. (13) at UT Southwestern describe the kinetics and metabolism of recombinant PCSK9 and the impact of PCSK9 on LDL receptors in the liver and adrenal gland.
PCSK9 was initially identified as a new member of the proprotein convertase family and suggested to have a role in liver regeneration and the differentiation of cortical neurons (14). Its relevance to cholesterol metabolism was suggested by a pair of unrelated observations. The first was the discovery, by Abifadel et al. (6), that heterozygosity for specific PCSK9 missense mutations cause autosomal dominant hypercholesterolemia. For most enzymes, the loss of a single allele is inconsequential, so the authors immediately raised the possibility that PCSK9 missense mutations might increase plasma cholesterol levels via a gain-of-function mechanism (6). The second observation was that PCSK9 mRNA levels are responsive to cellular cholesterol levels through the transcription factor SREBP-2 (15, 16).
The human genetic and gene-expression discoveries sparked interest in defining PCSK9 function. Mouse experiments quickly established a link between PCSK9 and levels of LDL receptors in the liver. Adenoviral overexpression of PCSK9 in mice resulted in higher LDL cholesterol levels and lower levels of LDL receptors— without changing LDL receptor mRNA levels (1, 3, 17). Similarly, PCSK9 transgenic mice exhibited increased LDL cholesterol levels, and LDL receptors in the liver were virtually eliminated (18). Parabiosis studies established that the PCSK9 from a transgenic mouse reduced LDL receptor levels in the liver of a paired nontransgenic mouse (18). Thus, circulating PCSK9 lowers LDL receptor levels. Conversely, knocking out PCSK9 increased hepatic LDL receptors and reduced plasma cholesterol levels (19).
The loss-of-function findings in mice are complemented by human data. Nearly 1 in 50 African-Americans has one of two nonsense mutations in PCSK9, which lowers LDL cholesterol levels by ∼30% (9, 10, 20). A common missense mutation in Caucasians lowers LDL levels by ∼15% (7–9, 20, 21). The lower cholesterol levels are accompanied by a strikingly reduced frequency of coronary heart disease (9, 22). Homozygous loss of PCSK9 results in LDL cholesterol levels of ∼15 mg/dl but no other detectable abnormalities (11, 23).
So, what is PCSK9 and why does it lower hepatic LDL receptor levels? PCSK9 is a 72-kd protease, expressed highly in liver, with three recognizable domains—an N-terminal prodomain, a catalytic domain, and a carboxyl-terminal domain of unknown function. Crystallographic studies uncovered similarities between the carboxyl-terminal domain and resistin (24), an inflammatory cytokine associated with insulin resistance; but the significance of this homology is unknown. Following its synthesis, PCSK9 undergoes an autocatalytic cleavage reaction that clips off the prodomain, but the prodomain remains attached to the catalytic and resistin-like domains (3, 14). The autocatalytic processing step is required for the secretion of PCSK9 (14), likely because the prodomain serves as a chaperone and facilitates folding. The continued attachment of the prodomain partially blocks the substrate binding pocket of PCSK9 (24–26).
The observation that PCSK9 reduces hepatic LDL receptors naturally suggested the hypothesis that the LDL receptor is cleaved and degraded by PCSK9. This hypothesis was elegantly disproved by the laboratory of Dr. Jay Horton (27). They developed a clever system for expressing a catalytically inactive form of PCSK9 and then showed that the catalytically dead protein retained the ability to lower LDL receptor levels (27).
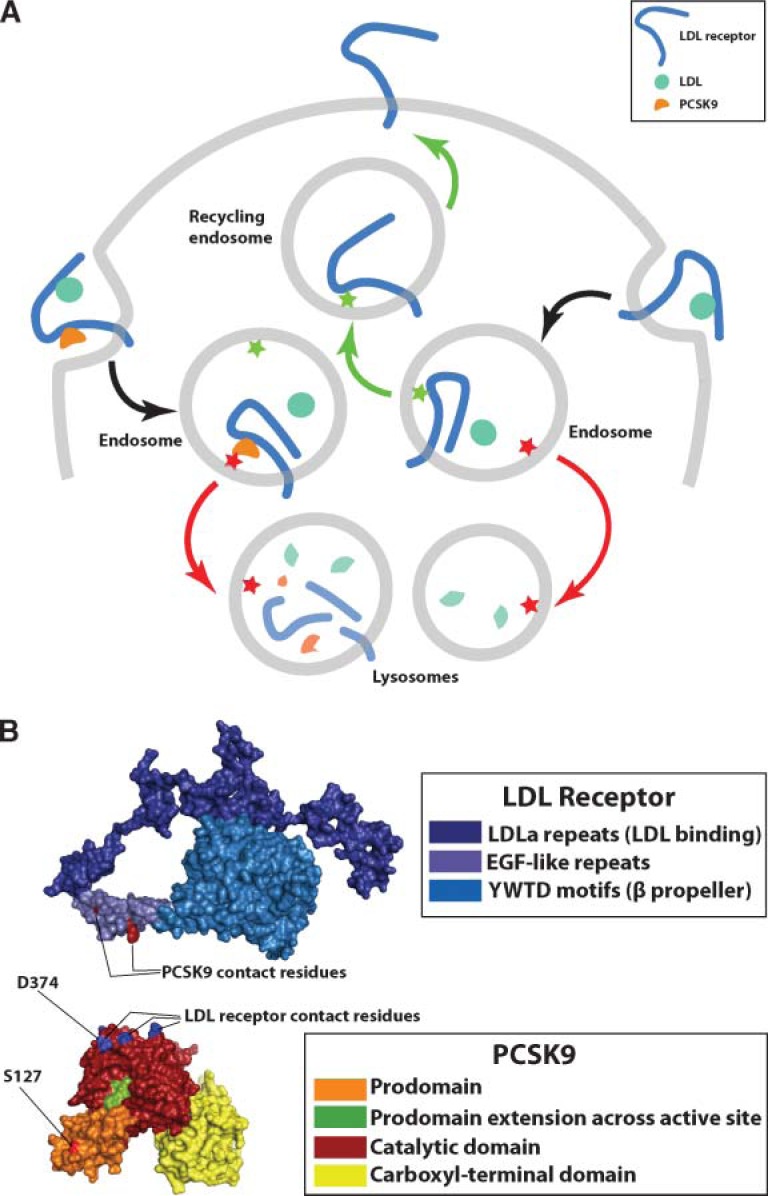
How PCSK9 lowers LDL receptor levels was utterly mysterious for several years, but recent studies are beginning to uncloak the mechanisms (see Fig. 1A). PCSK9 binds to the LDL receptor on the surface of cells, but the removal of LDL receptors requires internalization of the PCSK9–LDL receptor complex (18). After reaching the acidic environment of the endosome, the affinity of PCSK9 for the LDL receptor increases by nearly 150-fold (28, 29). PCSK9 binds to the epidermal growth factor (EGF)-like repeat A of the LDL receptor (28), a region known to be crucial for recycling of the LDL receptor from endosomes to the cell surface (30–32). A crystal structure of PCSK9 bound to the EGF-like repeat A revealed that the binding site on PCSK9 for the LDL receptor is distant from its catalytic site (26). Interestingly, the binding of PCSK9 to the LDL receptor results in the redistribution of LDL receptors from the cell surface to lysosomes (17, 18). Thus, PCSK9 appears to change the itinerary of the LDL receptor, diverting internalized LDL receptors to degradation in lysosomes and preventing them from recycling to the cell surface (17, 18, 27–29). The molecular mechanisms remain unclear, but it seems possible that the binding of PCSK9 produces conformational changes in the LDL receptor that render it incapable of being sorted to recycling endosomes. Alternatively, the PCSK9–LDL receptor complex could be actively recognized and directed toward lysosomes (Fig. 1A).
Fig. 1.
A: Model of PCSK9-mediated sorting of LDL receptors to lysosomes. The EGFa domain of the LDL receptor is required for proper sorting of the LDL receptor back to the cell surface (30–32). The EGFa domain may contain a sorting signal that interacts with an endosomal protein (green star), directing the LDL receptor back to the cell surface on recycling endosomes (green arrows). Binding of PCSK9 might interfere with that signal, preventing the LDL receptor from returning to the cell surface. Alternatively, PCSK9 could contain a distinct sorting signal (red star) that results in the sorting of the PCSK9–LDL receptor complex (red arrows) to lysosomes. The gain-of-function mutation involving S127 of PCSK9 may enhance the sorting of the PCSK9–LDL receptor complex to lysosomes (12). B: The structure of the LDL receptor and PCSK9 at endosomal pH. The LDL receptor is folded back upon itself at low pH (31); however, the face of the EGFa domain that binds PCSK9 (31) is exposed. The LDL receptor–binding site on PCSK9 is at the apex of a roughly triangular structure formed by the tripartite domain structure of PCSK9 (26). The D374 residue that is altered in gain-of-function PCSK mutants is located within the apical LDL receptor–binding site, whereas the S127 residue is quite distant from the binding interface. S127 mutations do not affect binding of PCSK9 to the LDL receptor (12). Gain-of-function mutations affecting residue 127 may reduce LDL receptors by enhancing the sorting of LDL receptors to lysosomes, rather than by affecting the strength of PCSK9–LDL receptor interactions (12).
The advances in the structural and cellular biology of PCSK9 have focused attention on the mechanisms by which PCSK9 gain-of-function mutations cause autosomal dominant hypercholesterolemia. In the current issue of the JLR, Dr. Shilpa Pandit et al. (12) examine mechanisms by which missense mutations at residues 374 and 127 (e.g., D374Y and S127R) reduce LDL receptors. In the case of residue 374, the results seem straightforward. Any of seven different amino acids (aside from aspartate) at residue 374 increased the affinity of PCSK9 for the LDL receptor, and this difference correlated with PCSK9’s potency in reducing LDL receptors. The PCSK9 crystal structure (26) has placed these biochemical findings in context (Fig. 1B). D374 is buried in the interface between PCSK9 and the LDL receptor, and changing that residue alters LDL receptor binding by altering hydrogen bonding and other interactions or simply by reducing the energetic cost of burying a charged residue. It is intriguing that the wild-type amino acid at position 374, aspartate, exhibited the lowest affinity for the LDL receptor (12), suggesting that evolution has selected against higher affinity interactions.
The S127R mutation is more enigmatic. Located in the prodomain, residue 127 is more than 40Å from the LDL receptor–binding site (Fig. 1B) (24–26), and biochemical studies revealed that this mutation has little or no impact on PCSK9’s affinity for the LDL receptor (12). Nevertheless, the S127R mutant (like the D374Y mutant) potently reduced LDL receptor numbers (12, 21). The S127R mutation did inhibit the efficiency of the autocatalytic processing step, but it is not at all clear that this finding explains its ability to reduce LDL receptors. Rather, it seems possible that the S127R mutant could modulate interactions with the cellular machinery that sorts LDL receptors to lysosomes. In any case, it is clear that the mechanisms underlying the S127R and D374Y mutations are distinct. Interestingly, the impact of the two amino acid substitutions on LDL receptors is additive; a mutant PCSK9 protein carrying both substitutions is 70-fold more potent than wild-type PCSK9 in reducing LDL receptors (12).
Understanding PCSK9 physiology in mammals has been boosted by a trio of practical advances—sensitive monoclonal-based assays for measuring plasma PCSK9 levels (18, 33), expression systems to generate large amounts of PCSK9 protein, and a clever approach for expressing catalytically inactive PCSK9 (27). These tools have made it possible to consider key questions regarding the function of PCSK9 in vivo, including: Are the plasma levels of PCSK9 in humans sufficient to account for the regulation of LDL receptors? What is the half-life of PCSK9 in the plasma, and is its turnover affected by LDL receptors? Is a gain-of-function PCSK9 mutant more potent in lowering LDL receptors in vivo, and is the turnover of a gain-of-function mutant more rapid? Can a catalytically dead PCSK9 reduce LDL receptors in vivo?
The paper by Dr. Horton’s group (13) in this issue of the JLR addresses all of these questions. By infusing recombinant human PCSK9 into mice, they established that the levels of PCSK9 found in human plasma are indeed sufficient to reduce hepatic LDL receptors. A gain-of-function mutant, PCSK9 (D374Y), was even more potent. Also, a catalytically dead PCSK9 functioned perfectly well in reducing LDL receptors, dashing any remaining hope that an inhibitor of PCSK9’s catalytic activity would prevent PCSK9 in the plasma from lowering LDL receptors. The clearance of PCSK9 from the plasma was retarded in LDL receptor–knockout mice, showing that the LDL receptor is a key factor in controlling PCSK9 levels in the plasma. The clearance of the PCSK9 (D374Y) mutant was more rapid, consistent with its higher affinity for the LDL receptor.
Dr. Horton’s group found that infusions of PCSK9 into mice, even at high levels, had little effect on LDL receptors in the adrenal gland—an organ with high levels of LDL receptors (13). This intriguing observation suggested that the cellular machinery for PCSK9-dependent removal of LDL receptors differs in the liver and extrahepatic tissues.
Dr. Horton’s group also provided an intriguing speculation regarding the regulation of PCSK9 expression. Cholesterol depletion in the liver, via SREBP-2, simultaneously upregulates the expression of the LDL receptor and upregulates PCSK9—a molecule that in turn lowers LDL receptors. What is the “physiologic rationale” for this peculiar regulation? SREBP-2 activation is accompanied by increased lipid synthesis and VLDL secretion. Dr. Horton’s group proposed that short-term downregulation of LDL receptors in the liver, via PCSK9, might channel newly secreted hepatic lipoproteins away from the liver, allowing time for these lipoproteins to unload their cargo in peripheral tissues.
Much of the excitement surrounding PCSK9 stems from its attractiveness as a cholesterol-lowering target. There is little doubt that inhibitors of PCSK9 function would lower plasma cholesterol levels, and there is no reason to suspect that the loss of PCSK9 would be harmful. Also, inhibition of PCSK9 should potentiate the effects of statins. Statins actually upregulate PCSK9 (19, 34, 35), which puts the brakes on their principal mode of action, which is to increase LDL receptors in the liver.
Several approaches for inhibiting PCSK9 function are theoretically feasible. Because autocatalytic cleavage is required for the maturation of PCSK9, a small-molecule inhibitor of autocatalysis might be useful (3), provided that it was specific for PCSK9 processing and did not lead to a toxic accumulation of misfolded PCSK9. Small molecules that block the PCSK9–LDL receptor interactions would likely be efficacious, although designing inhibitors of protein–protein interactions is a tall order. Antisense approaches pioneered by Isis Pharmaceuticals (Carlsbad, CA) are well suited for liver targets (36, 37), and studies in mice suggest that this approach is efficacious for PCSK9(38). Finally, there is considerable interest in developing antibody therapeutics to inhibit PCSK9–LDL receptor interactions. Pharma and biotech are hard at work! Over the next few years, the pages of the JLR will likely contain many preclinical and clinical studies on inhibitors of PCSK9 function.
With the search for drugs underway, academic and industry scientists will have their hands full investigating enigmas in PCSK9 biology. Why is PCSK9 more effective in lowering LDL receptors in the liver than in the adrenal gland? What are the molecular mechanisms for the redistribution of LDL receptors to lysosomes? How does the S127R mutation reduce LDL receptor numbers? With luck, answering these questions could lead to the discovery of new molecules controlling LDL receptors (and new targets). Do garden-variety hyperlipidemias, the metabolic syndrome, diabetes mellitus, endocrine disorders, and commonly used medications perturb PCSK9 metabolism? If PCSK9 is truly dispensable, why has it been conserved in evolution? Why are PCSK9 nonsense mutations common in African-Americans? Presumably, evolution selected for these mutations, but no one knows why. Many laboratories will be taking their best shot at these issues and their efforts are likely to fill the pages of the JLR over the next few years. Stay tuned.
Acknowledgments
We thank Stuart Bunting, Daniel Kirchhofer, and James Ernst for helpful discussions.
Footnotes
Supported by NIH grants U01 HL66621 and R01 HL087228 (to SGY).
REFERENCES
- 1.Park SW, Moon YA, Horton JD. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J. Biol. Chem. 2004;279:50630–50638. doi: 10.1074/jbc.M410077200. [DOI] [PubMed] [Google Scholar]
- 2.Maxwell KN, Breslow JL. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc. Natl. Acad. Sci. USA. 2004;101:7100–7105. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjannet S, Rhainds D, Essalmani R, Mayne J, Wickham L, Jin W, Asselin MC, Hamelin J, Varret M, Allard D, et al. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 2004;279:48865–48875. doi: 10.1074/jbc.M409699200. [DOI] [PubMed] [Google Scholar]
- 4.Brown MS, Goldstein JL. Lipoprotein receptors in the liver. Control signals for plasma cholesterol traffic. J. Clin. Invest. 1983;72:743–747. doi: 10.1172/JCI111044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein JL, Brown MS. The low-density lipoprotein pathway and its relation to atherosclerosis. Annu. Rev. Biochem. 1977;46:897–930. doi: 10.1146/annurev.bi.46.070177.004341. [DOI] [PubMed] [Google Scholar]
- 6.Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, et al. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003;34:154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 7.Berge KE, Ose L, Leren TP. Missense mutations in the PCSK9 gene are associated with hypocholesterolemia and possibly increased response to statin therapy. Arterioscler. Thromb. Vasc. Biol. 2006;26:1094–1100. doi: 10.1161/01.ATV.0000204337.81286.1c. [DOI] [PubMed] [Google Scholar]
- 8.Kotowski IK, Pertsemlidis A, Luke A, Cooper RS, Vega GL, Cohen JC, Hobbs HH. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am. J. Hum. Genet. 2006;78:410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 10.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 11.Zhao Z, Tuakli-Wosornu Y, Lagace TA, Kinch L, Grishin NV, Horton JD, Cohen JC, Hobbs HH. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am. J. Hum. Genet. 2006;79:514–523. doi: 10.1086/507488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pandit S, Wisniewski D, Santoro JC, Ha S, Ramakrishnan V, Cubbon RM, Cummings RT, Wright SD, Sparrow CP, Sitlani A, Fisher TS. Functional Analysis of Sites within PCSK9 Responsible for Hypercholesterolemia. J. Lipid Res. 2008;49:1333–1343. doi: 10.1194/jlr.M800049-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Grefhorst A, McNutt MC, Lagace TA, Horton JD. Plasma PCSK9 Preferentially Reduces Liver LDL Receptors in Mice. J. Lipid Res. 2008;49:1303–1311. doi: 10.1194/jlr.M800027-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seidah NG, Benjannet S, Wickham L, Marcinkiewicz J, Jasmin SB, Stifani S, Basak A, Prat A, Chretien M. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA. 2003;100:928–933. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxwell KN, Soccio RE, Duncan EM, Sehayek E, Breslow JL. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J. Lipid Res. 2003;44:2109–2119. doi: 10.1194/jlr.M300203-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Maxwell KN, Fisher EA, Breslow JL. Overexpression of PCSK9 accelerates the degradation of the LDLR in a post-endoplasmic reticulum compartment. Proc. Natl. Acad. Sci. USA. 2005;102:2069–2074. doi: 10.1073/pnas.0409736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagace TA, Curtis DE, Garuti R, McNutt MC, Park SW, Prather HB, Anderson NN, Ho YK, Hammer RE, Horton JD. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Invest. 2006;116:2995–3005. doi: 10.1172/JCI29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rashid S, Curtis DE, Garuti R, Anderson NN, Bashmakov Y, Ho YK, Hammer RE, Moon YA, Horton JD. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc. Natl. Acad. Sci. USA. 2005;102:5374–5379. doi: 10.1073/pnas.0501652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hallman DM, Srinivasan SR, Chen W, Boerwinkle E, Berenson GS. Relation of PCSK9 mutations to serum low-density lipoprotein cholesterol in childhood and adulthood (from The Bogalusa Heart Study) Am. J. Cardiol. 2007;100:69–72. doi: 10.1016/j.amjcard.2007.02.057. [DOI] [PubMed] [Google Scholar]
- 21.Cameron J, Holla OL, Ranheim T, Kulseth MA, Berge KE, Leren TP. Effect of mutations in the PCSK9 gene on the cell surface LDL receptors. Hum. Mol. Genet. 2006;15:1551–1558. doi: 10.1093/hmg/ddl077. [DOI] [PubMed] [Google Scholar]
- 22.Brown MS, Goldstein JL. Biomedicine. Lowering LDL–not only how low, but how long? Science. 2006;311:1721–1723. doi: 10.1126/science.1125884. [DOI] [PubMed] [Google Scholar]
- 23.Hooper AJ, Marais AD, Tanyanyiwa DM, Burnett JR. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis. 2007;193:445–448. doi: 10.1016/j.atherosclerosis.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 24.Hampton EN, Knuth MW, Li J, Harris JL, Lesley SA, Spraggon G. The self-inhibited structure of full-length PCSK9 at 1.9 A reveals structural homology with resistin within the C-terminal domain. Proc. Natl. Acad. Sci. USA. 2007;104:14604–14609. doi: 10.1073/pnas.0703402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piper DE, Jackson S, Liu Q, Romanow WG, Shetterly S, Thibault ST, Shan B, Walker NP. The crystal structure of PCSK9: a regulator of plasma LDL-cholesterol. Structure. 2007;15:545–552. doi: 10.1016/j.str.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Kwon HJ, Lagace TA, McNutt MC, Horton JD, Deisenhofer J. Molecular basis for LDL receptor recognition by PCSK9. Proc. Natl. Acad. Sci. USA. 2008;105:1820–1825. doi: 10.1073/pnas.0712064105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McNutt MC, Lagace TA, Horton JD. Catalytic activity is not required for secreted PCSK9 to reduce low density lipoprotein receptors in HepG2 cells. J. Biol. Chem. 2007;282:20799–20803. doi: 10.1074/jbc.C700095200. [DOI] [PubMed] [Google Scholar]
- 28.Zhang DW, Lagace TA, Garuti R, Zhao Z, McDonald M, Horton JD, Cohen JC, Hobbs HH. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat A of low density lipoprotein receptor decreases receptor recycling and increases degradation. J. Biol. Chem. 2007;282:18602–18612. doi: 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- 29.Fisher TS, Lo Surdo P, Pandit S, Mattu M, Santoro JC, Wisniewski D, Cummings RT, Calzetta A, Cubbon RM, Fischer PA, et al. Effects of pH and low density lipoprotein (LDL) on PCSK9-dependent LDL receptor regulation. J. Biol. Chem. 2007;282:20502–20512. doi: 10.1074/jbc.M701634200. [DOI] [PubMed] [Google Scholar]
- 30.Davis CG, Goldstein JL, Sudhof TC, Anderson RG, Russell DW, Brown MS. Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region. Nature. 1987;326:760–765. doi: 10.1038/326760a0. [DOI] [PubMed] [Google Scholar]
- 31.Rudenko G, Henry L, Henderson K, Ichtchenko K, Brown MS, Goldstein JL, Deisenhofer J. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 32.van der Westhuyzen DR, Stein ML, Henderson HE, Marais AD, Fourie AM, Coetzee GA. Deletion of two growth-factor repeats from the low-density-lipoprotein receptor accelerates its degradation. Biochem. J. 1991;277:677–682. doi: 10.1042/bj2770677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alborn WE, Cao G, Careskey HE, Qian YW, Subramaniam DR, Davies J, Conner EM, Konrad RJ. Serum proprotein convertase subtilisin kexin type 9 is correlated directly with serum LDL cholesterol. Clin. Chem. 2007;53:1814–1819. doi: 10.1373/clinchem.2007.091280. [DOI] [PubMed] [Google Scholar]
- 34.Careskey HE, Davis RA, Alborn WE, Troutt JS, Cao G, Konrad RJ. Atorvastatin increases human serum levels of proprotein convertase subtilisin/kexin type 9. J. Lipid Res. 2008;49:394–398. doi: 10.1194/jlr.M700437-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Dubuc G, Chamberland A, Wassef H, Davignon J, Seidah NG, Bernier L, Prat A. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler. Thromb. Vasc. Biol. 2004;24:1454–1459. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- 36.Crooke RM, Graham MJ, Lemonidis KM, Whipple CP, Koo S, Perera RJ. An apolipoprotein B antisense oligo-nucleotide lowers LDL cholesterol in hyperlipidemic mice without causing hepatic steatosis. J. Lipid Res. 2005;46:872–884. doi: 10.1194/jlr.M400492-JLR200. [DOI] [PubMed] [Google Scholar]
- 37.Kastelein JJ, Wedel MK, Baker BF, Su J, Bradley JD, Yu RZ, Chuang E, Graham MJ, Crooke RM. Potent reduction of apolipoprotein B and low-density lipoprotein cholesterol by short-term administration of an antisense inhibitor of apolipoprotein B. Circulation. 2006;114:1729–1735. doi: 10.1161/CIRCULATIONAHA.105.606442. [DOI] [PubMed] [Google Scholar]
- 38.Graham MJ, Lemonidis KM, Whipple CP, Subramaniam A, Monia BP, Crooke ST, Crooke RM. Antisense inhibition of proprotein convertase subtilisin/kexin type 9 reduces serum LDL in hyperlipidemic mice. J. Lipid Res. 2007;48:763–767. doi: 10.1194/jlr.C600025-JLR200. [DOI] [PubMed] [Google Scholar]