Abstract
Aspartic peptidases are proteolytic enzymes present in many organisms like vertebrates, plants, fungi, protozoa and in some retroviruses such as human immunodeficiency virus (HIV). These enzymes are involved in important metabolic processes in microorganisms/virus and play major roles in infectious diseases. Although few studies have been performed in order to identify and characterize aspartic peptidase in trypanosomatids, which include the etiologic agents of leishmaniasis, Chagas’ disease and sleeping sickness, some beneficial properties of aspartic peptidase inhibitors have been described on fundamental biological events of these pathogenic agents. In this context, aspartic peptidase inhibitors (PIs) used in the current chemotherapy against HIV (e.g., amprenavir, indinavir, lopinavir, nelfinavir, ritonavir and saquinavir) were able to inhibit the aspartic peptidase activity produced by different species of Leishmania. Moreover, the treatment of Leishmania promastigotes with HIV PIs induced several perturbations on the parasite homeostasis, including loss of the motility and arrest of proliferation/growth. The HIV PIs also induced an increase in the level of reactive oxygen species and the appearance of irreversible morphological alterations, triggering parasite death pathways such as programed cell death (apoptosis) and uncontrolled autophagy. The blockage of physiological parasite events as well as the induction of death pathways culminated in its incapacity to adhere, survive and escape of phagocytic cells. Collectively, these results support the data showing that parasites treated with HIV PIs have a significant reduction in the ability to cause in vivo infection. Similarly, the treatment of Trypanosoma cruzi cells with pepstatin A showed a significant inhibition on both aspartic peptidase activity and growth as well as promoted several and irreversible morphological changes. These studies indicate that aspartic peptidases can be promising targets in trypanosomatid cells and aspartic proteolytic inhibitors can be benefic chemotherapeutic agents against these human pathogenic microorganisms.
Keywords: Alternative chemotherapy, aspartic peptidases, Chagas’ disease, HAART, HIV, HIV peptidase inhibitors, Leishmania, leishmaniasis, pathogenesis, peptidases, proteolytic inhibitors, Trypanosoma, virulence
1. THE TRYPANOSOMATIDAE FAMILY
The Trypanosomatidae family (order Kinetoplastida) comprises a large group of flagellate parasitic protozoa that causes infections in humans, animals and plants [1-3]. Individuals of this family have a ubiquitous distribution in nature and are easily distinguished from other protozoa by their unique ultrastructure, which exhibits a net of subpellicular microtubules [4], located below the cytoplasmic membrane, firmly attached to the inner surface of the membrane, accounting for the typical morphology, as well as a unique mitochondrion, branched throughout parasite cytoplasm, presenting a typical region of DNA condensation, known as kinetoplast, a giant network of thousands of concatenated circular mitochondrial DNAs (kDNA) [4].
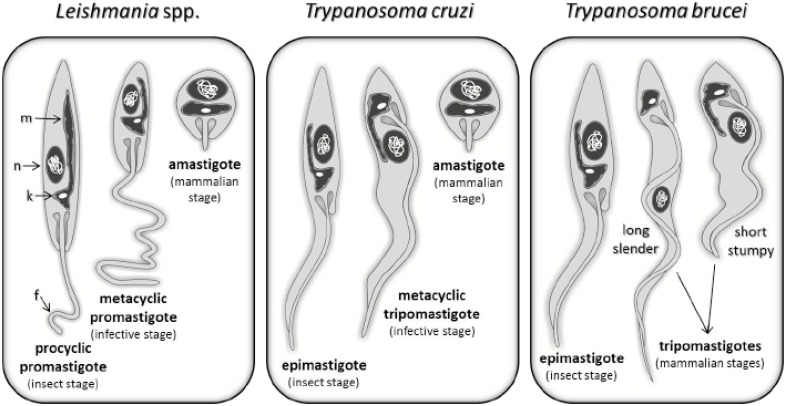
A huge variety of lifestyles and adaptations to parasitism are observed in representatives of this family, as well as in the entire Order, which constitutes a very ancient group of organisms in the phylogenetic tree of eukaryotes. Individuals of this family are divided into two major groups according to their ability to infect vertebrate and/or invertebrate hosts. The heteroxenic group comprises parasites that, during their life cycle, interact with both invertebrate and vertebrate hosts, while parasites of the monoxenic group typically interact solely with invertebrate hosts. Classically, the taxonomy of trypanosomatids is defined by lifestyle, host, clinical manifestation (if applicable), and typical morphotypes. The morphotypes are defined by the position of the kinetoplast relative to the nucleus and the point at which the flagellum emerges from the parasite cell (Fig. 1). The distinct morphological stages are closely correlated to the genera and host-specific stages of trypanosomatids (Fig. 1) [5, 6].
Fig. (1).
Trypanosomatid morphotypes from Leishmania spp, Trypanosoma cruzi and Trypanosoma brucei. Trypanosomatid forms are defined by cell shape, point of flagellum (f) emergence and position of the kinetoplast (k) in relation to the nucleus (n). For epimastigote, promastigote and amastigote forms, the kinetoplast is located in an anterior position relative to the nucleus, while for trypomastigote form the kinetoplast is located in a posterior position. Epimastigote and trypomastigote forms present an undulating membrane connecting to their flagella, once the flagellum is attached to the cell membrane, while other forms have a free flagellum, with the exception of amastigote forms, which presents a very small flagellum. In all morphotypes, only one large mitochondria (m) is observed, which is ramified through the cell body, being the region where the kinetoplast is located.
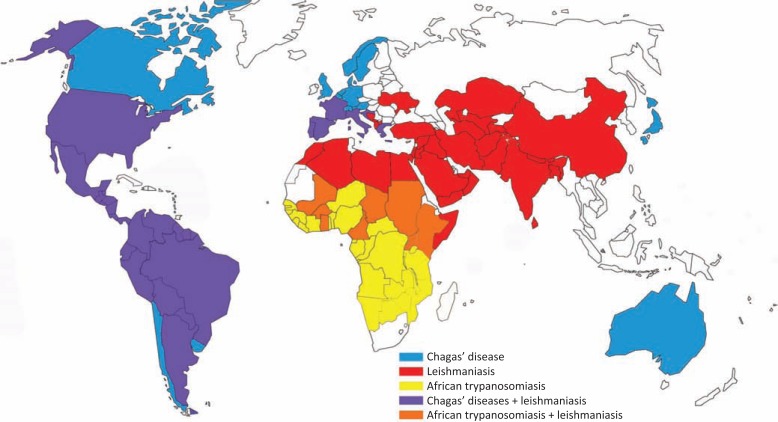
Few members of this family are responsible for major important human diseases, which are collectively referred as the most neglected human diseases by the World Health Organization (WHO) (Table 1). These diseases are Chagas’ disease and sleeping sickness, caused respectively by Trypanosoma cruzi, in South and Central America, and Trypanosoma brucei, in Africa; as well as leishmaniasis, caused by many species from the Leishmania genus, present in tropical and subtropical regions of the world (Fig. 2) [3, 7-12]. Infections in humans occur primarily through blood-sucking insects, such as triatomines, in the case of T. cruzi, tsetse flies for T. brucei and different phlebotomine sand flies species for the Leishmania genus [7-12]. The spread of these diseases all over the world, to many developed, non-endemic countries, is related to the globalization process and the movement of unknowingly infected people (Fig. 2).
Table 1.
Diseases Caused by Trypanosomatids of Human Medical Importance
| Trypanosomatid | Disease | Vector | People at risk (millions) | Prevalence (millions) | Available chemotherapy |
|---|---|---|---|---|---|
| Leishmania spp. | cutaneous, mucocutaneous or visceral leishmaniasis | phlebotomine sand fly | 350 | 12 | amphotericin B, miltefosine, paromomycin, sodium stibogluconate, meglumine antimoniate |
| Trypanosoma cruzi | Chagas’ disease | triatomine | 100 | 10 | nifurtimox, benznidazole |
| Trypanosoma brucei | human sleeping sickness | tsetse fly | 70 | 0,03* | pentamidine, suramine, melarsoprol, eflornithine, nifurtimox |
Although 30,000 cases are reported annually, WHO estimates that about 300,000 infected individuals remain ignored in the field, due to difficulties in diagnosis and remoteness of affected areas.
Fig. (2).
Geographic distribution of cases reported for African trypanosomiasis, Chagas’ disease and leishmaniasis around the world. Data collected from the WHO web site (http://www.who.int/en).
According to WHO, in 2010 an estimated 10 million people were infected by T. cruzi and roughly 100 million were at risk of the disease worldwide, mostly but not restricted to Latin America. It was estimated that more than 10,000 individuals died of Chagas’ disease in 2008. For leishmaniasis, in 2010, 350 million people were considered at risk of contracting the disease, and about 2 million cases occur annually, of which 0.5 million correspond to visceral leishmaniasis (Table 1 and Fig. 2) [13].
Although affecting many people around the world, the major diseases caused by parasites from the Trypanosomatidae family have no efficient treatment or vaccination. The available drugs (Table 1) are expensive, toxic and many parasites have already developed resistance to the chemotherapy, resulting in an urgent need to identify new targets for therapeutic alternatives [7, 8, 11, 14-16]. In this sense, this review will describe the current knowledge on trypanosomatids’ aspartic peptidases and their inhibitors, since there is substantial data indicating that they can be a promising target for chemotherapy.
2. PEPTIDASES
Peptidases, proteinases or proteases are enzymes that catalyze the hydrolysis of peptide bonds or, in other words, proteins able to hydrolyze other proteins or peptides. These enzymes were initially classified into exopeptidases or endopeptidases according to the reaction catalyzed. Exopeptidases are capable of hydrolyzing peptide bonds at the ends of a polypeptide chain, releasing single amino acid, dipeptide or tripeptide residues, while endopeptidases preferentially act on peptide bonds in the inner regions of a polypeptide [17, 18].
The availability of structural and mechanistic information on these enzymes led to improvements on the classification schemes. According to the nature of the catalytic site, peptidases can be classified as aspartic, cysteine, metallo, serine, threonine, glutamic and asparagine type [17-19]. The intensive research on peptidases generates a wide amount of information, requiring a system of classification for the comprehensive study of this diversity. Recently, a new method of classification was introduced and can be easily accessed in the MEROPS database server [19]. In this system, peptidases of the different classes can be further grouped into families on the basis of statistically significant similarities in amino acid sequence. For nomenclature, each family is identified by a letter that represents the catalytic domain, where A is used for aspartic type, C for cysteine type, M for metallo type, S for serine type, T for threonine type, G for glutamic type, N for asparagine type and U for unknown type; followed by a characteristic number. Families that are thought to be homologous, and had arisen from a single evolutionary origin, are grouped together in a clan. It represents one or more families that show evidence on evolutionary relationship by similar tertiary structures, order of catalytic site residues in the polypeptide chain and their common sequence motifs around the catalytic site. For clan representation, two letters are used, being the first related to the family [19-22].
Peptidases, among many other molecules, have been evaluated with respect to its potential as new therapeutic targets. Central roles in physiological processes are carried out by peptidases, which can be found in all domains of life: Eukarya, Bacteria and Archaea, as well as in virus [23]. The importance of peptidases in biological systems is easily recognized, since all proteins of a cell need to be proteolytically processed and/or degraded at some point of cell development. The regulation of protein localization, mobility and activity, as well as modulation of protein-protein interactions, contribution for cellular information processing, generation, transduction and amplification of molecular signals are all coordinated by peptidase activity [23].
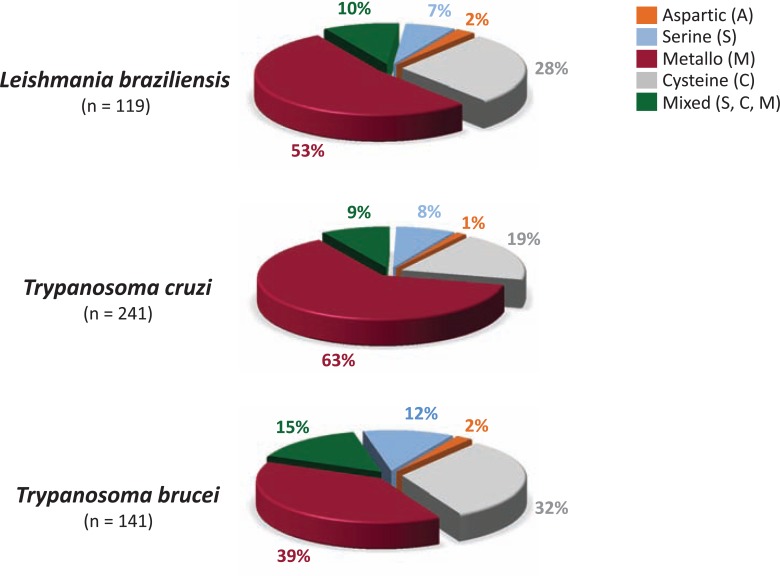
In infectious organisms, peptidases play crucial roles as virulence factors, besides its involvement in basic cellular functions. For instance, peptidases are necessary for colonization, invasion, dissemination and evasion from the host immune system [23, 24]. There are a number of excellent reviews on the functions and exploitation of trypanosomatids’ peptidases as chemotherapeutic targets. Cysteine and metallo-type peptidases are the most abundant and well-studied peptidases in trypanosomatids, followed by serine peptidases. In (Fig. 3), we can see the distribution of the peptidase classes in L. braziliensis, T. cruzi and T. brucei.
Fig. (3).
Distribution of the different peptidase catalytic types in three representatives species belonging to the Trypanosomatidae family: Leishmania braziliensis, Trypanosoma cruzi and Trypanosoma brucei. The data was extracted from MEROPS - The Peptidase Database [19], release 9.6; URL: http://merops.sanger.ac.uk/
3. ASPARTIC PEPTIDASES
The aspartic peptidases are endopeptidases (Fig. 4) present in a wide range of organisms: vertebrates, plants, fungi, protozoa, prokaryotes and retroviruses [25-27]. The aspartic peptidases have attracted intense attention in the scientific community because of their potential for application in the food industry and as a therapeutic target for important human diseases [24, 28]. These include pepsin in peptic ulcer disease, renin in hypertension, plasmepsins in malaria, cathepsin D in metastasis of different types of cancer cells, human immunodeficiency virus (HIV) peptidase in acquired immune deficiency syndrome (AIDS), and b-site amyloid precursor protein cleaving enzyme (BACE) in Alzheimer’s disease [24, 29-34].
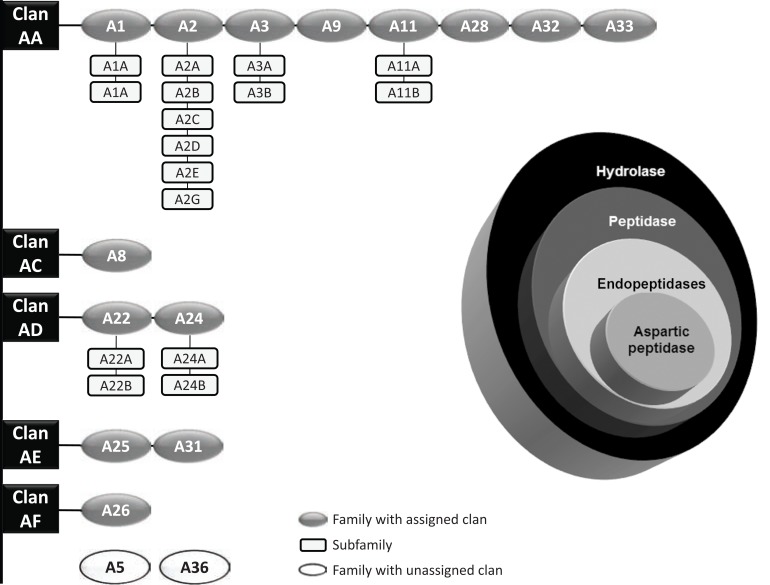
Fig. (4).
Classification of aspartic peptidases according to the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology. Aspartic peptidases (EC 3.4.23.X) are ordered in subgroup 4 (peptidases) of group 3 (hydrolases) (figure on the right side). Overview of the aspartic peptidase clans, families and subfamilies according to the MEROPS Database [19]. The clan (black boxes) contains enzymes that have arisen from a single evolutionary origin of peptidases and represents one or more families (dark grey circles) that show evidence of their evolutionary relationship. The white circles represent two families with unassigned clans. In addition, some families are divided into subfamilies (light grey boxes) since there is evidence of a very ancient divergence within the family.
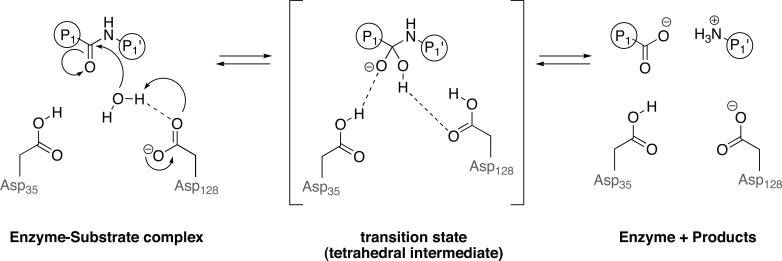
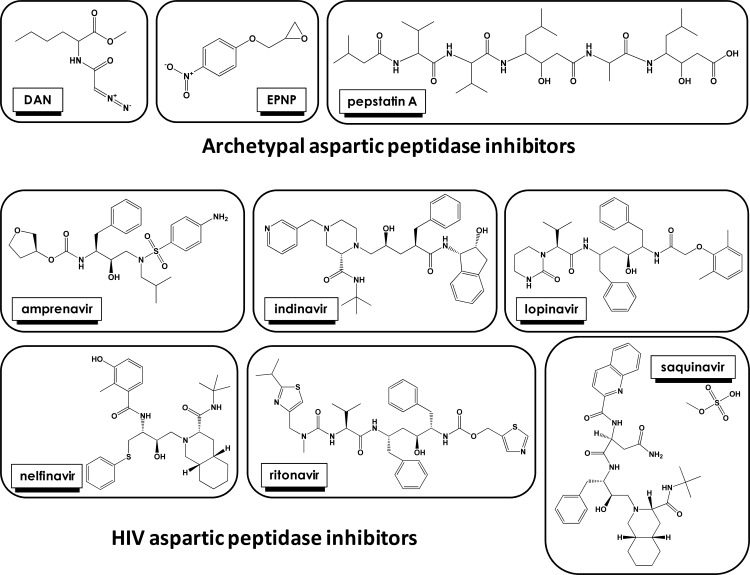
Most of the aspartic peptidases are characterized by two aspartic acid carboxyl groups as key catalytic groups at their active site (Fig. 5) [35-37], three-dimensional structure similarity, low optimal pH value for best hydrolytic activity, and a scission preference between large and hydrophobic amino acids [38-40]. Although most aspartic peptidases fit in these characteristics, considerable differences exist in terms of catalytic properties, cellular localization, biological functions and inhibition by the microbial peptide pepstatin A, which is a prototype inhibitor of aspartic peptidases (Fig. 6) [41].
Fig. (5).
Catalytic mechanism of aspartic peptidase proposed by Nguyen and colleagues 2008 [52]. The water molecule is partly activated by an aspartate and makes a nucleophilic attack at a specific carbonyl carbon in the substrate. The carbonyl oxygen captures a proton from another aspartic acid in the active site, resulting in a tetrahedral intermediate (transition state). Restabilizing from the transition state, the amino moiety from the substrate becomes a better leaving group, and the substrate is cleaved. Dashed lines indicate hydrogen bonds.
Fig. (6).
Aspartic peptidase inhibitors effective against human pathogenic trypanosomatids.
Overall, the aspartic peptidases are synthesized as inactive precursors, which are converted to the active form of the enzyme by acid-triggered, autocatalytic proteolysis and removal of lengths of polypeptides chains that are N-terminal extensions [41, 42]. Most eukaryotic aspartic peptidases are monomeric and consist of a single polypeptide chain that forms two similar domains with the active site cleft located between them; each domain provides an aspartic acid carboxyl group as key catalytic group to form the active site. In contrast, retroviral aspartic peptidases are dimeric, consisting of two identical subunits, each roughly equivalent to one domain of a eukaryotic aspartic peptidase [25-27, 29, 38, 43-45].
In the active enzyme, the two aspartic acid residues are geometrically closer and one aspartate is ionized, whereas the second one is unionized at the optimal pH [46-47]. The most widely accepted mechanism of action of the aspartic peptidases is an acid-base catalysis, which may be called a “push-pull” mechanism involving two active aspartic acid residues in the active site and a water molecule that resides between them. These two aspartic acid residues act as a proton donor and acceptor, to catalyze the hydrolysis of peptide bonds in substrates. The water molecule is partly activated by an aspartate and makes a nucleophilic attack at a specific carbonyl carbon in the substrate. The carbonyl oxygen, in turn, captures a proton from another aspartic acid in the active site, resulting in a noncovalent neutral tetrahedral intermediate. This intermediate is the crucial transition state. Restabilizing from the transition state, the amino moiety from the substrate becomes a better leaving group, and the substrate is cleaved (Fig. 5) [47-52].
The aspartic peptidases are hierarchically classified into five distinct clans (AA, AC, AD, AE and AF), according to the MEROPS database. There are sixteen different peptidase families, of which two have not been assigned to any of the existing clans, and sixteen subfamilies belonging to clans AA (twelve subfamilies) and AD (four subfamilies) [17-22, 53]. An organogram of families and clans of aspartic peptidases, focus of this review article, can be seen in (Fig. 4). In addition, the chemical structure of potent aspartic peptidase inhibitors can be seen in (Fig. 6).
The aspartic peptidases found in the Trypanosomatidae family belong to clans AA (family A28) and AD (families A22A and A22B) (Table 2, Fig. 4). Clan AA contains the classical aspartic peptidases and clan AD comprises aspartic peptidases that hydrolyze peptide bonds within biological membranes. Clan AA is further divided into eight families, including A1 family, whose members are all-beta proteins consisting of two similar beta barrel domains, which both contribute to the formation of the active site [54], and the A2 family that is composed of proteins containing a single beta barrel domain, so dimerization must occur to form an active peptidase [55]. Family A1 contains pepsin-like enzymes such as pepsin, gastricin, rennin, cathepsin D and E, plasmepsins (PMs), and histo-aspartic peptidase (HAP). The family A2, also termed the retropepsin family, includes HIV retropepsin [19, 21, 22, 53, 54]. In clan AD, all members have transmembrane domains that are presumed to be helical, so the protein fold must be different from the all-beta folds found in members of clan AA. Unlike members of clan AC, which also contains membrane-bound proteins, the active site is on the cytoplasmic side of the cell membrane [56]. Presenilin (A22 family), representative of clan AD, forms the catalytic core of the gamma-secretase complex required for intramembrane proteolysis of type I transmembrane proteins such as the amyloid precursor protein [28, 57]. An analysis of the occurrence of aspartic peptidase families in distinct taxonomic groups reveals interesting information, for instance, family A24 (clan AD) are found almost exclusively in Archaea and Bacteria. Family A22 (clan AD), which is found in trypanosomatids, is still undetected in Bacteria, while A28 family (clan AA) is restricted to Eukarya (Table 3).
Table 2.
List of the Aspartic Peptidases from Three Representatives of the Trypanosomatidae Family, Showing their Respective ID, Family and Clan
| Trypanosomatid | Aspartic peptidase classification | ||
|---|---|---|---|
| Clan | Family* | MEROPS ID | |
| Leishmania braziliensis | AA | A28 | MER242455 |
| AD | A22(A) | MER184693 | |
| AD | A22(B) | MER124926 | |
| Trypanosoma cruzi | AA | A28 | MER242463 |
| AD | A22(A) | MER049314 | |
| AD | A22(B) | MER054238 | |
| Trypanosoma brucei | AA | A28 | MER242469 |
| AD | A22(A) | MER049280 | |
| AD | A22(B) | MER048243 | |
Family prototypes: A28 - DNA-damage inducible protein 1 (ddi-1) (Saccharomyces cerevisiae); A22 - presenilin 1 (Homo sapiens). Source: MEROPS - The Peptidase Database (Rawlings et al., 2012), release 9.6; URL: http://merops.sanger.ac.uk/
Table 3.
Distribution of Aspartic Peptidase Families from clans AA and AD Among Distinct Taxonomic Groups
| Aspartic peptidase family | Archaea | Bacteria | Eukarya | Virus | |||||
|---|---|---|---|---|---|---|---|---|---|
| Clan AAA1A2A3A9A11A28A32A33Clan ADA22A24 | Protista Fungi Animalia Plantae | ||||||||
| - | 11 | 240 | 853 | 899 | 888 | - | |||
| - | 1 | - | 28 | 676 | 43 | 233 | |||
| - | - | - | - | - | 4 | 31 | |||
| - | - | - | - | - | - | 10 | |||
| - | - | 2 | 240 | 139 | 1203 | 2 | |||
| - | - | 39 | 71 | 71 | 15 | - | |||
| - | 355 | - | - | - | - | - | |||
| - | - | 1 | - | 17 | 9 | - | |||
| 66 | - | 54 | 53 | 394 | 97 | - | |||
| 158 | 1188 | - | - | 1 | 1 | - | |||
Source: MEROPS - The Peptidase Database (Rawlings et al., 2012), release 9.6; URL: http://merops.sanger.ac.uk/
Although some aspartic peptidases have been identified in members of the Trypanosomatidae family, based on different properties, including molecular-level criteria, such as the reaction catalyzed, the chemical mechanism of catalysis, and the homology relationships revealed by sequence and structure similarity analyses, very little is known about the evolutionary history of aspartic peptidases in this group of parasites. However, the phylogenetic relationship of aspartic peptidase members of the A28 and A22 families recognized by the MEROPS database shows that each family (or subfamily) displays distinct evolutionary histories among distantly related eukaryotic lineages (as one may expect). In addition, trypanosomatid enzymes are consistently separated in two different groups, which indicate that the most recent common ancestor of aspartic peptidases of the A28 and A22 families in Trypanosoma and Leishmania genera are not the same (Fig. 7).
Fig. (7).
Phylogenetic relationship aspartic peptidase members of the A28 and A22 families from trypanosomatids. Peptidase sequences obtained from the MEROPS database release 9.6 [19] were aligned with the program ClustalW version 2.1 [122]; phylogram was constructed with the software MEGA version 5 [123] after 1,000 bootstraps with the neighbor-joining algorithm [124] (A) and (B) denote different subfamilies.
Up to now, the products of the aspartic peptidase genes in trypanosomatids were poorly or indirectly characterized, either by demonstration of degradation of aspartic peptidase substrates in crude extracts followed by inhibition by selective aspartic peptidase inhibitors (Fig. 6), or through the demonstration of the effect of these inhibitors on parasite growth, viability, ultrastructure and infectivity [58-63]. These approaches unequivocally illustrate the presence and relevance of this enzymatic class in the Trypanosomatidae family, and point out to the necessity to further characterize these enzymes. In the sections below, we will describe the data on the literature regarding the inhibition of aspartic peptidases produced by human pathogenic trypanosomatids, especially Leishmania spp. and T. cruzi, which highlight the possibility of an alternative target for chemotherapy.
4. ASPARTIC PROTEOLYTIC INHIBITORS AS PROSPECTIVE CHEMOTHERAPEUTIC AGENTS
There are a number of comprehensive reviews on the applicability of proteolytic inhibitors as chemotherapeutic agents [64-81]. Here, we will shortly illustrate this potentiality, focusing on aspartic peptidase inhibitors. Several proteolytic inhibitors have already been used in the clinic with considerable success to treat hypertension, coagulation disorders, cancer and diabetes. They include angiotensin converting enzyme (ACE) inhibitors for treating high blood pressure, thrombin inhibitors for treating stroke, and an elastase inhibitor for treating systemic inflammatory response syndrome (SIRS) [72]. The example of greater impact and success in treatment of an infectious disease with proteolytic inhibitors is the highly active antiretroviral therapy (HAART) used to treat the acquired immunodeficiency syndrome (AIDS) (Fig. 6). The HAART has led to a marked improvement in the life expectancy of AIDS sufferers by the fall of HIV viremia and by restoring the immune responses with an increase in the number of CD4+ T lymphocytes and with an effective stimulation in the survival and activation of neutrophils, monocytes, endothelial and dendritic cells. All these beneficial properties of HAART culminated in a drastic reduction of opportunistic infections [82-88]. This reduction seems to be based not only on the immune system restoration, but also on the direct inhibition of aspartic peptidases produced by opportunistic pathogens, as demonstrated in some bacteria, fungi and protozoa [62, 63, 89-93].
The difficulty in treating parasitic diseases is partly due to the complexity of biological organisms responsible by these pathologies. Thus, there are several chemotherapeutic approaches being developed [94-95], including the use of proteolytic inhibitors to treat malaria, leishmaniasis and trypanosomiasis. For instance, the Plasmodium parasite, the causative agent of malaria, has proteolytic enzymes that play key roles in hemoglobin hydrolysis and this process appears to involve multiple catalytic classes of peptidases, including cysteine, metallo and aspartic peptidases. Among such enzymes, PMs and, especially, falcipains (cysteine peptidases) are highly promising antimalarial drug targets [96].Two HIV peptidase inhibitors (HIV PIs), saquinavir and ritonavir, have been established as antimalarials in clinical use in combination with chloroquine and mefloquine [97]. Looking for an example in fungi, where the effectiveness of HAART has been more extensively explored, Candida albicans can be selected as a prototypal microorganism [89-93, 98-100]. C. albicans is part of the normal human respiratory, genital and gastrointestinal tracts flora and the major cause of opportunistic fungal infections in immunocompromised people. The secreted aspartic peptidases (Saps) are recognized as the main virulence factor of Candida and they belong to the same superfamily of HIV aspartic peptidase. Thus, studies have been conducted and confirmed the effect of HIV PIs on Sap activity, fungal proliferation, morphogenesis, adhesion to mammalian and experimental candidiasis infection as well as synergistic drug properties with classical antifungals [101]. The main example of beneficial effects of HIV PIs on bacteria was demonstrated against Mycobacterium spp. Studies have demonstrated a decline in the tuberculosis rate coincident with the introduction of HAART [102-104]. Kabbesh and colleagues [105] showed that ritonavir was able to significantly diminish the synthesis of cell wall lipids, suggesting a loss in the function of this fundamental mycobacterial structure. Collectively, these published reports exemplify the wide range of action of HIV PIs against phylogenetic distinct classes of microorganisms.
5. ANTI-TRYPANOSOMATID PROPERTIES OF ASPARTIC PEPTIDASE INHIBITORS
5.1. Leishmania
Peptidases have been extensively studied in Leishmania, and a simple analysis on the number of representative genes from each proteolytic class indicates a clear prevalence of metallopeptidases, followed by cysteine- and serine-type peptidases (Fig. 3). This scenario is reinforced by the extensive reports on Leishmania cysteine peptidases CPA, CPB and CPC, and the metallopeptidase GP63, which accounts for about 1% of the organism’s total protein content [for a comprehensive review, see 67]. There is a general lack of knowledge about aspartic peptidases in Leishmania. Currently, there are only few studies describing aspartic peptidase activities in soluble fractions of crude Leishmania extracts, by means of selective substrates and inhibitors (Fig. 6) to this enzymatic class [58, 60, 62, 63, 106]. L. amazonensis soluble crude extract presents an acidic hydrolytic activity able to degrade the renin synthetic substrate NCbz-Pro-Phe-His-Leu-Leu-Val-Tyr-Ser-b-naphthylamide [58], the HIV-1 aspartic peptidase substrate Arg-Glu(EDANS)-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-Lys(DABCYL)-Arg [62] and the cathepsin D substrate 7-methoxycoumarin-4-acetyl-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys(DNP)-D-Arg-amide [106], these activities were inhibited by pepstatin A and HIV PIs (Fig. 6) [58, 62]. Also, L. mexicana soluble crude extracts were shown to degrade the substrate benzoyl-Arg-Gly-Phe-Phe-Leu-4-methoxy-b-naphthylamide optimally at pH 3.8. Although the substrate degradation was not fully inhibited by pepstatin A (only 45% of inhibition), diazo-acetyl-norleucinemethylester (DAN), another selective aspartic peptidase inhibitor (Fig. 6), virtually abolished the enzymatic activity. Peptidase inhibitors selective for other peptidase classes (E-64, leupeptin and 1,10-phenanthroline) presented only marginal effects on the hydrolysis [60]. Envisaging a possible exploitation of aspartic peptidases as a target for chemotherapy, the authors also demonstrated the anti-proliferative effect of DAN on promastigotes, showing a 50% lethal dose of 22 µM after 72 h of in vitro cultivation [60]. Moreover, DAN induced significant alterations in the shape of promastigotes of L. mexicana, from a long slender form to a spherical one with at least two nuclei per parasite suggesting the blockage of cell division [60]. It was also demonstrated that the aspartic peptidase activity is down regulated during the L. amazonensis promastigote into amastigote differentiation in vitro [58].
Although aspartic peptidases were never purified and fully characterized in Leishmania, several studies have been conducted in order to test the effects of HIV PIs on these parasites. These studies were driven by the drastic reduction in the incidence, morbidity and mortality of AIDS co-infections after the introduction of HIV PIs in the antiretroviral therapy. These inhibitors were able to promote a series of damaging effects on parasite proliferation and ultrastructure, as well as a reduced ability of Leishmania to infect and survive within host cell macrophages [59-63, 106]. In this sense, Savoia and colleagues [59] were the first to describe a dose-dependent effect of HIV PIs on two Leishmania species. The 50% lethal dose (LD50) after incubation of L. major for 24 h with indinavir and saquinavir was shown to be 8.3 µM and 7.0 µM, respectively. The inhibitory effect was more protuberant for this species, which causes cutaneous leishmaniasis, than for L. infantum promastigotes, a causative agent of visceral leishmaniasis. In the latter, the highest concentration tested (50 µM) did not achieve an inhibition of 50% [59]. The HIV PIs present an irreversible effect, since parasites do not resume growth when subcultured into fresh medium [59].
Some years later, Trudel and colleagues [61] reported that ritonavir, saquinavir and nelfinavir presented no inhibitory effect on the growth of L. infantum promastigotes. Almost at the same time, our research group reported the anti-promastigote activity of lopinavir, nelfinavir and amprenavir against L. amazonensis promastigotes, while saquinavir and indinavir presented only negligible inhibition [62]. Following this publication, Valdivieso and colleagues [63] reinforced the data presented by Savoia and colleagues [59], showing the anti-promastigote activity of nelfinavir and saquinavir against a panel of L. infantum isolates [63]. They also showed that Leishmania species associated with cutaneous manifestations present values of IC50 for nelfinavir and saquinavir slightly lower than those of species associated with visceral manifestations, as previously reported by Savoia and colleagues [59, 63]. There seems to be a lack of consensus on the literature about the susceptibility of different Leishmania species to the available HIV PIs. It is yet unclear if these discrepancies are indeed due to the wide genetic variability among Leishmania strains, isolates and species, or if it is due to methodological and reagent differences. This prompted our research group to challenge L. amazonensis, L. braziliensis, L. donovani, L. major and L. infantum with nelfinavir and saquinavir for 72 h in order to compare the results under the same standardized conditions [106]. Saquinavir was capable of statistically inhibiting only L. donovani growth [106], while nelfinavir inhibited in more than 90% all the species tests, except for L. major, which presented an inhibition of 50%. A systematic review on Leishmania inhibition by HIV PIs is a difficult task due to discrepancies in methodological design, data analysis and representation, strains and species assessed, and reagents employed. Nevertheless, in (Table 4), we tried to summarize the available information.
Table 4.
Systematic Review of the Data Available on the Susceptibility of Leishmania Species and Isolates to HIV PIs.
| nelfinavir | saquinavir | Ref. | |||
|---|---|---|---|---|---|
| inhibition | conc. | inhibition | conc. | ||
|
L. amazonensis (MHOM/BR/77/LTB016) |
50% (48h) | 15.12 | Virtually no inhibition1 | [62] | |
|
L. amazonensis (MHOM/BR/77/LTB016) |
95% (72h) | 25 | 5% | 25 | [106] |
|
L. amazonenzis (IFLA/BR/67/PH8) |
50% 3 | 13.36 | 50% 3 | 40 | [63] |
|
L. major (LRC-L137) |
49% (24h) | 6.25 | [59] | ||
|
L. major (MHOM/SU/73/5-ASKH) |
50% 3 | 13.37 | 50% 3 | 46.95 | [63] |
|
L. major (MHOM/IL/1980/FRIEDLIN) |
50% (72h) | 25 | 21.2% (72h) | 25 | [106] |
|
L. infantum (MHM/TN/80/IPT1) |
31% (24h) | 50 | [59] | ||
|
L. infantum (MHOM/MA/67/ITMAP-263) |
Virtually no inhibition2 | Virtually no inhibition2 | [61] | ||
|
L. infantum (MHOM/FR/78/LEM-75) |
50% 3 | 16.46 | 50% 3 | 53.97 | [63] |
|
L. infantum (MCAN/ES/98/LLM-724) |
50% 3 | 17.59 | 50% 3 | 50.87 | [63] |
|
L. infantum (MCAN/VE/98/IBO-78) |
50% 3 | 14.05 | 50% 3 | 55.12 | [63] |
|
L. infantum (MHOM/ES/95/LLM-480) |
50% 3 | 18.21 | 50% 3 | 48.04 | [63] |
|
L. infantum (MHOM/ES/98/LLM-759)4 |
50% 3 | 26.89 | 50% 3 | 64.464 | [63] |
|
L. infantum (MHOM/BR/1974/PP75) |
96.2% (72h) | 25 | 0% (72h) | 25 | [106] |
|
L. infantum (MHOM/BR/2009/ANC)4 |
96.6% (72h) | 25 | 0.4% (72h) | 25 | [106] |
|
L. infantum (MHOM/BR/2009/LCS)4 |
96.3% (72h) | 25 | 0% (72h) | 25 | [106] |
|
L. infantum (MHOM/BR/2009/VCF)4 |
0% (72h) | 25 | 0% (72h) | 25 | [106] |
| L. donovani (MHOM/IN/80/DD8) | 50% 3 | 14.1 | 50% 3 | 51.89 | [63] |
|
L. donovani (MHOM/ET/1967/L82) |
94% (72h) | 25 | 62% (72h) | 25 | [106] |
|
L. mexicana (MHOM/VE/80/NR) |
50% 3 | 9.85 | 50% 3 | 42.08 | [63] |
|
L. mexicana (MHOM/ES/2002/LLM-1162) |
50% 3 | 12. 44 | 50% 3 | 40.67 | [63] |
|
L. mexicana (MHOM/BZ/82/BEL21) |
50% 3 | 10.25 | 50% 3 | 39.54 | [63] |
|
L. brazilensis (MHOM/BR/75/M2903) |
50% 3 | 14. 6 | 50% 3 | 36 | [63] |
|
L. braziliensis (MCAN/BR/1998/619) |
95% (72h) | 25 | 13% (72h) | 25 | [106] |
INHIBITON – Inhibition in relation to control. The number in brackets corresponds to the time when the inhibition was assayed.
Conc. – Concnetration of the inhibitor in µM.
The drugs were screened from 15 to 500 µM and the growth followed from 24 to 96 h. The inhibition observed was only marginal.
The drugs were screened at 12.5 and 25 µM and the growth compared to the control group after 72 h of incubation, no inhibition was observed.
The methodology described in ref [82] was elusive in relation to the time of incubation where the IC50 was calculated.
Strains isolated from HIV/Leishmania co-infected patient.
Although there is some discrepancy on the susceptibility of Leishmania promastigotes to certain HIV PIs, a careful analysis of the data depicted in (Table 4) clearly indicates the anti-proliferative action of these inhibitors against promastigote forms. The next line of evidence on the potentiality of HIV PIs for leishmaniasis chemotherapy was shown by the ability of these inhibitors to impair parasite development in macrophages, which was published almost simultaneously by Trudel and colleagues [61] and our research group [62]. The work of Trudel and colleagues [61] demonstrated that, although under the conditions employed, nelfinavir, ritonavir and saquinavir did not exert an inhibitory action on promastigotes, these inhibitors exerted pronounced effects against the intracellular parasites in two in vitro infection cell systems: phorbolmyristate acetate-differentiated THP-1 macrophages and human primary monocyte-derived macrophages (MDM). Importantly, the efficacy of HIV PIs to reduce the intracellular growth of Leishmania parasites is also observed in MDMs-coinfected with HIV-1 [61]. Also, a field isolate of Leishmania donovani resistant to sodium stibogluconate, one of the drugs most commonly used to treat leishmaniasis, is equally susceptible to the tested PIs compared with a sensitive strain, thus suggesting that resistance to sodium stibogluconate does not result in cross-resistance to HIV PIs [61]. Our research group demonstrated that the HIV PIs can interfere in the early steps of parasite infection in macrophages, since the inhibitors were added exclusively to Leishmania promastigotes and that the interaction process was stopped with only 1 hour. As expected, the treatment of previously infected macrophages with HIV PIs notably reduced the association indexes, in a dose-dependent manner [62]. It is interesting to note that the HIV PIs efficacy is higher for amastigotes inside macrophages than for extracellular amastigotes or promastigotes [61-63]. This could be explained by a combination of factors: a direct anti-amastigote activity together with a modulation of the killing capability of the macrophages and a concentration of the drugs inside the macrophages.
The effectiveness of HIV PIs in treating parasitic infections may be associated to their capacity to modulate or block the cell proteasome or to promote apoptosis [78]. Alternatively, it could act directly on aspartic peptidases produced by protozoa. Our research group was the first to demonstrate that the HIV PIs are capable of inhibiting, in a dose-dependent manner, the degradation of a HIV-1 peptidase substrate at acidic pH by L. amazonensis [62]. This was the first line of evidence that the intracellular target of the HIV PIs in Leishmania could be an aspartic peptidase. It should be pointed out that the HIV PIs were designed to fit viral peptidase and may thus have a lower affinity for Leishmania aspartic peptidase. As a matter of fact, the HIV PIs belongs to family A2, clan AA, while the identified aspartic peptidases in Leishmania genome belongs to family A28 and A22, clans AA and AD, respectively (Table 2, Fig. 4). Although the viral and leishmanial peptidases belong to clan AA, similarity searches using HIV aspartic peptidase sequences against the entire set of annotated proteins encoded in Leishmania genomes reveal no statistically significant hit (data not shown). However, Perteguer and collaborators [108] have recently isolated a full-length cDNA encoding a 49-kDa protein from L. major, which exhibited significant deduced amino acid sequence homology with the annotated Leishmania sp. DNA damage-inducible (Ddi1-like) protein, as well as with the ddi1 protein from Saccharomyces cerevisiae. The protein exhibited an additional fragment at the N-terminal end, homologous to the ubiquitin-like (UBL) domain of this family of proteins described in other organisms, which had not been previously reported for the L. major Ddi1-like protein. In addition, the cloning, expression, and functional characterization of the L. major recombinant Ddi1-like protein demonstrated the proteolytic activity of this protein [108]. Another piece of evidence strongly suggests that Leishmania aspartic peptidases are the intracellular target of the HIV PIs. A Saccharomyces cerevisiae knockout for ddi-1, an orthologous of Leishmania aspartic peptidase (MEROPS ID MER242455, family A28, clan AA, Table 2), was functionally complemented with the Leishmania orthologous, reverting the phenotype to the wild one. This phenotype reversion was also induced in the wild yeast by HIV PIs [105]. In addition, two Leishmania strains isolated from HIV-Leishmania coinfected patients under HAART treatment exhibited lower sensibility to HIV PIs in vitro, as demonstrated by two independent research groups [63, 106]. It is interesting to note that the parasite isolated from a patient under treatment with HIV PIs presented considerably less aspartic peptidase activity than isolates from patients untreated or treated only with reverse transcriptase inhibitors [106]. The cultivation of this isolate in the presence of nelfinavir induced a further reduction in the aspartic peptidase activity, which suggests that these enzymes are the target of the HIV PIs and are down-regulated by the selective pressure induced by the drug [106]. Indeed, it was recently demonstrated that a leishmanial aspartic peptidase can be the intracellular target of the HIV PIs [108]. The cloned Did-1 like protein from L. major that was expressed in baculovirus/insect cells readily hydrolyzed a synthetic substrate for the HIV peptidase [Arg-Glu(EDANS)-Ser-Gln-Asn-Tyr-Pro- Ile-Val-Gln-Lys(DABCYL)-Arg] at acidic pH. This activity was inhibited in 70 and 95% by pepstatin A at 15 mM and DAN at 500 mM, respectively. The HIV PI nelfinavir at 20 µM reduced the activity in 60%, while E-64 and 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF) presented no significant effect on the hydrolytic activity. Two other synthetic substrates specific for cathepsin D, Bz-Arg-Gly-Phe-Phe-Leu-4MbNA, and Bz-Arg-Gly-Phe-Phe-Pro-4MbNA·HCl, were hydrolyzed at lower rates and were inhibited by pepstatin A and DAN, while the inhibition by HIV PIs were not assessed [108]. A 3D model of the Ddi-1 like protein from L. major suggests that it can accommodate bulkier substrates than those accessible to HIV-1 aspartic peptidase [108].
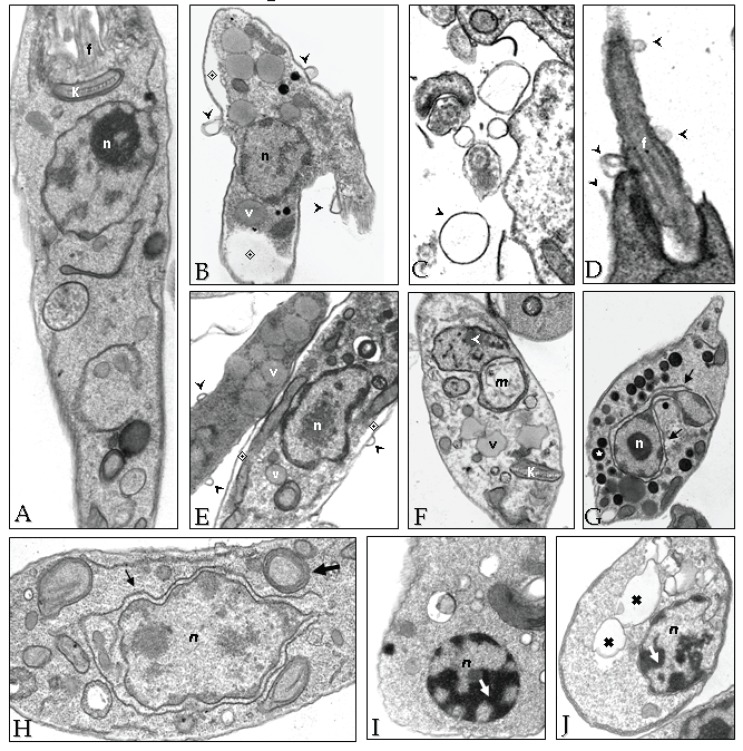
Although it seems reasonable to assume that the HIV PIs target Leishmania aspartic peptidase, the possibility of nonspecific or generally toxic effects of the drugs on parasite cells should not be ruled out. In this context, electron micrographic examination of L. amazonensis cells exposed to nelfinavir or lopinavir revealed some peculiar alterations in vital cellular structures, such as cytoplasmic membrane and internal cellular structures, suggesting irreversible metabolic injuries that culminate in the parasite cell death [62] (Fig. 8). An interesting finding was the increase in the number of vesi- cles, which according to their electron-density, probably corre- sponds to lipid-containing compartments [62] (Fig. 8). Indeed, the HIV PIs are capable of altering the lipid composition in leishmanial cells (unpublished data). A well-known side effect of HIV PIs in humans is the lipodistrophy, which is an abdominal adiposity [109]. Also interesting to note is that some of the ultrastructural alterations observed in L. amazonensis, such as increase in the number of vesi- cles and wrapping of the nucleus by the endoplasmic reticulum, are suggestive of autophagy [62] (Fig. 8). Accordingly, it was later demonstrated that lopinavir is effective in generating oxidative stress in Leishmania, leading to altered physiological parameters such as increase in the sub-G1 DNA content, nuclear DNA frag- mentation and loss of mitochondrial potential, which are all characteristics of apoptosis [110]. Interestingly, HIV PIs also induced a significant increase in the expression of virulence factors (CPB and GP63) by L. amazonensis, when parasites were subjected to HIV PIs. One hypothesis could be that the HIV PIs are inhibiting an aspartic peptidase that should be otherwise degrading some of the GP63 and CPB peptidases. An alternative hypothesis could be that the HIV PIs are exerting stress, or some other non-specific effect (Fig. 8), on the promastigotes that leads to changes in parasite gene expression [62].
Fig. (8).
Ultrastructural changes observed in L. amazonensis after HIV PIs treatment. Parasites (108 cells) from 48-h cultures were inoculated in fresh medium in the absence (A) or in the presence of nelfinavir (B-H) or lopinavir (I-J) at the IC50 concentration, and incubated for 4 h (B-E), 6 h (F-G), 8 h (H) and 24 h (I-J). Subsequently, cells were processed for transmission electron microscopy. An intense flagellar and plasma membrane shedding (black arrowheads) was seen after 4 hours of treatment with both inhibitors (B-D). Some effects were exclusive of nelfinavir, such as cytoplasm shrink (B and E, ◈), increase in the number of intracellular vesicles, resembling acidocalcisomes (G, ★) and lipid inclusions (E and F, v). Both drugs induced nuclear wrapping by the endoplasmic reticulum (G and H, black arrows), mithocondrial swelling (F, white arrowheads) and myelin-like structures (H, larger arrow). In lopinavir treated cells, blocks of condensed chromatin were observed close to the nuclear envelope (I, white arrow), as well as enlarged vesicles (J, ✖). n - nucleus; k - kinetoplast; f - flagellum and m - mithocondrion. The ultrastructural alterations described for nelfinavir (B-H) were also visualized with lopinavir. Reprinted from PLoS One. 2009; 4(3): e4918. doi:10.1371/journal.pone.0004918.
More recently, the effect of the HIV PIs indinavir and ritonavir was also tested in vivo using BALB/c mice infected with L. amazonensis in the footpad followed by oral treatment for 30 days. Antiretroviral-treated mice had a significant reduction in the footpad thickness after the third week of indinavir treatment and after the fifth week of ritonavir treatment. However, there was no reduction in the parasite load [111]. It is yet unclear why the HIV PIs have a poor efficacy in infection experiments with mice, since all data pointed towards a different view, i.e., after the introduction of PIs in the antiretroviral therapy for HIV, the number of coinfected cases reported fell sharply; PIs present an anti-promastigote activity; PIs reduce the infection in macrophages; and an aspartic peptidase seems to be the specific target of the PIs. Nevertheless, these studies strongly suggest that aspartic peptidase(s), together with HIV PIs and/or specific inhibitors to leishmanial aspartic peptidase(s), represent a promising strategy for leishmaniasis chemotherapy improvement.
5.2. Trypanosoma
Different peptidases have been extensively studied in the last decades in T. cruzi, with special emphasis in cysteine peptidases, represented mainly by cruzipain and the 30-kDa cathepsin B, but metallo and serine peptidases were also investigated and play critical functions for the parasite [for comprehensive reviews see 67, 112]. The group of aspartic peptidases, on the contrary, has been only recently analyzed. In this group, two aspartic peptidase activities were identified and isolated from T. cruzi epimastigote forms (Y strain): cruzipsin-I (CZP-I) and cruzipsin-II (CZP-II) [113]. The enzymes were purified by affinity chromatography through the use of the classical aspartic peptidase inhibitor, pepstatin A, coupled to agarose. Interestingly, CZP-I was isolated from cell pellets after freezing-thawing and centrifugation, followed by solubilization with the non-ionic detergent CHAPS, while CZP-II was isolated from the soluble fraction after cells lysis. The molecular mass of both peptidases was estimated to be 120-kDa by HPLC gel filtration, and the proteolytic activity of both enzymes was detected as a doublet of bands (56- and 48-kDa) by gelatin-containing sodium dodecyl sulfate polyacrylamide gel electrophoresis, which suggested that the active T. cruzi hydrolases are dimeric proteins composed of identical subunits of 56-60 kDa associated by bonds, similar to the vertebrate aspartic peptidases [113].
The identification of CZP-I and CZP-II as aspartic peptidases was achieved through distinct methods [113]. At first, substrate specificity studies indicated that the enzymes showed maximal proteolytic activity over the cathepsin D substrate Phe-Ala-Ala-Phe-(4-NO2)-Phe-Val-Leu-O4MP at pH 3.5-4.0, but failed to hydrolyze serine and other peptidase substrates. In addition, the proteolytic activities of the CZP-I and CZP-II fractions were strongly inhibited by pepstatin A and the aspartic active site labeling agent 1,2-epoxy-3-(phenyl-nitrophenoxy) propane (EPNP) (Fig. 6), but not by various other inhibitors of serine, metallo or cysteine peptidases. The authors emphasized that the selective inhibition by EPNP indicates that both T. cruzi proteolytic activities possess the dual aspartates at the active site, the signature configuration of aspartic peptidases belonging to clan AA, family A1, in which pepsin is the family-type peptidase. In this sense, it is worth mentioning that the T. cruzi Genome Project [114] reported only three aspartic peptidases, two of which belong to clan AD, family A22, being presenilin 1 the family-type peptidase, classically inhibited by pepstatin A. All members of this clan have transmembrane domains [19]. The third member of this group found in T. cruzi genome belongs to clan AA, family A28, in which the family-type peptidase is the Ddi-1 from Saccharomyces cerevisiae (Table 2). This protein was previously found to be the ligand for nelfinavir in L. major, affecting growth, proliferation and survival [107, 108]. No peptidase activity has been shown for any member of this family [19]. Interestingly, genes predicting enzymes belonging to the A1 family have not been found yet in the T. cruzi genome. As pointed out by Alvarez and colleagues [112], since no amino acid sequences were reported for CZP-I and CZP-II it is not possible to link these enzymes to any of the genes detected in T. cruzi. Nevertheless, Pinho and colleagues [113] reinforced that there are several sequences that could not be correctly identified in the T. cruzi genome due to difficulties in correlating homologous genes by using the current computer techniques. T. brucei Genome Project [114] also presented the same aspartic peptidase sequences detected for T. cruzi and Leishmania (Table 2), although no study concerning this class of proteolytic enzymes has been performed yet in African trypanosomes. Also, the efficacy of aspartic peptidases inhibitors, including HIV PIs, in T. brucei is a rich unexplored area, which is a point of interest of our research group.
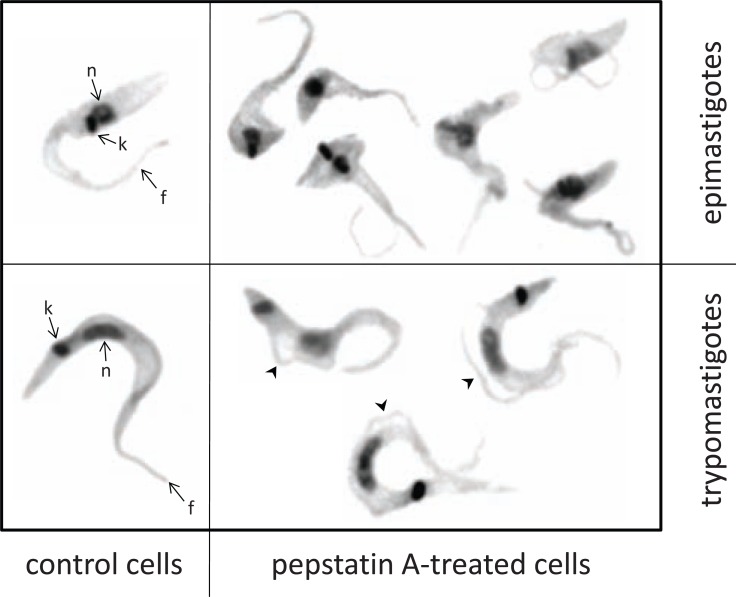
Our group has recently begun to work with the effects of the aspartic peptidase inhibitor pepstatin A against T. cruzi clone Dm28c epimastigote forms [115]. Pepstatin A arrested the parasite proliferation in both dose- and time-dependent manner, resulting in significant morphological alterations, including reduction of the cell size and detachment of parts or the whole flagellum from the cell body (Fig. 9), though cell lysis was not observed. Curiously, the aspartic peptidase inhibitor induced the metacyclogenesis process, which may be connected to the stress promoted in the parasite cells. The epimastigote-to-trypomastigote differentiation was stimulated in a dose-dependent manner, but approximately 45% of the trypomastigotes had their flagellum detached from the cell body (Fig. 9). The treatment of epimastigotes with pepstatin A at the IC50 value (36.2 µM) induced an increase of 54% and 98%, respectively, in the surface expression of gp63- and calpain-related molecules in epimastigotes, but not in the cruzipain level. As previously reported by Santos and colleagues [62], the blockage of a class of peptidase by a proteolytic inhibitor can induce an augment in the expression of distinct classes of peptidase in order to compensate its function.
Fig. (9).
Morphological alterations observed on both epimastigotes and trypomastigotes of Trypanosoma cruzi after 72 h of treatment with 50 µM of pepstatin A. Comparing with untreated epimastigotes, pepstatin A induced the swollen in cell body and detachment of the flagellum. In trypomastigotes the most evident effect was partial and whole flagellum detachment (arrowheads). k, kinetoplast; n, nucleus; f, flagellum.
Growth inhibition in the presence of the aspartic peptidase inhibitor was also observed in T. cruzi strains belonging to distinct phylogenetic lineages: similar levels of inhibition were obtained between clone Dm28c (DTU I) and strains CL Brener (DTU VI) and 4167 (DTU IV), while inhibition of Y strain (DTU II) and 3663 strain (DTU III) was lower in comparison to clone Dm28c [115]. These results are in accordance to the great heterogeneity of natural populations of T. cruzi in biological, biochemical, immunological and molecular features, which must be correlated to distinct clinical manifestations and chemotherapy response [116]. For instance, T. cruzi strains are able to express different amounts of peptidases, including the major cysteine peptidase cruzipain [117, 118].
The possibility of aspartic peptidase activity as the intracellular target of this inhibitor was suggested by the hydrolysis of a cathepsin D fluorogenic substrate (7-methoxy-coumarin-4-acetyl-Gly-Lys-Pro-Ile-Leu-Phe-Phe-Arg-Leu-Lys(DNP)-Arg-amide) by T. cruzi epimastigote extract and the inhibition of its hydrolysis by pepstatin A [115]. These results opened the possibility of exploiting aspartic peptidases as promising targets to treat Chagas’ disease. As a matter of fact, our group started recently to explore the effects of HIV PIs against T. cruzi epimastigotes, and we observed a strong anti-proliferative effect, with IC50 values much lower than pepstatin A, some of them in the nanomolar range (unpublished results). Although there are no studies in the literature about the effect of aspartic peptidase inhibitors on amastigotes or trypomastigotes of T. cruzi, evidences show that compounds may be more effective depending on the developmental stage of the parasite. In Leishmania, for instance, a considerable difference in susceptibility was observed between promastigotes and amastigotes in vitro [61-63]. Also, leishmania-infected mice treated with HIV PIs presented a modest reduction in footpad thickness, and no reduction in parasite load [111]. In this sense, it is interesting to assess the effect of HIV PIs against the clinically relevant forms of the T. cruzi.
CONCLUDING REMARKS
Leishmania spp. and Trypanosoma spp. are responsible for substantial global morbidity, mortality and economic adversity in tropical and subtropical regions, especially affecting the less developed countries. Environmental changes, drug resistance and immunosuppression contribute to the emergence and spread of these diseases. For instance, the HIV pandemic has modified the immunopathogenic, epidemiological and therapeutic aspects of these human parasitic diseases. Corroborating these outcomes, the current therapeutic arsenal against the human pathogenic trypanosomatids T. brucei, T. cruzi and Leishmania spp. is clearly inadequate and underscores the urgent need to develop new effective, safe and cost-effective drugs.
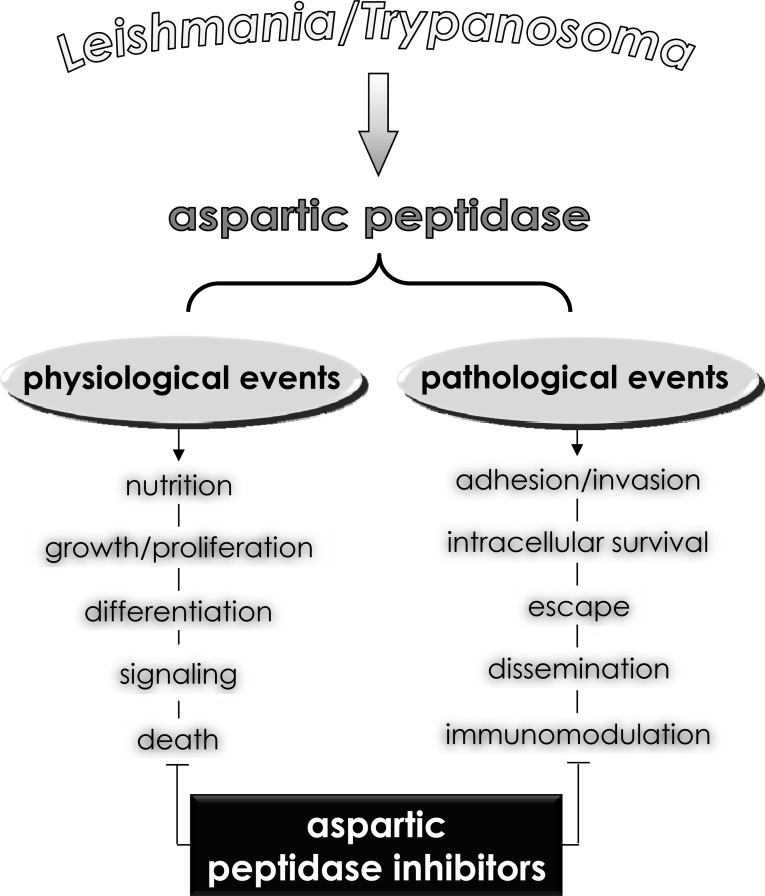
In view of this, a number of new strategies to obstruct trypanosomatid biological processes have emerged; one of them is focused on peptidase inhibition. This particular class of hydrolytic enzymes cleaves peptide bonds in proteinaceous substrates, a reaction extremely important in maintaining the physiology of all living cells (Fig. 10), also peptidases are essential virulence factors for these protozoa during all stages of the infection process (Fig. 10), which make them potential targets for the development of anti-trypanosomatid drugs (Fig. 10). Supporting this view, a sharp decrease in the incidence of visceral leishmaniasis in Europe and Africa was observed following the widespread use of HAART, particularly after the introducing of aspartic peptidase inhibitors to the cocktail, further supporting the notion that HAART helps to prevent visceral leishmaniasis in individuals co-infected with Leishmania and HIV [118-121]. These clinical records led the researchers around the world to focus in the possibility to test aspartic peptidase inhibitors against human pathogenic trypanosomatids.
Fig. (10).
Possible roles played by aspartic-type peptidases produced by human pathogenic trypanosomatids belonging to the Leishmania and Trypanosoma genera. Aspartic peptidases contribute to maintaining basic metabolic processes in trypanosomatid cells, which govern crucial biological events including proliferation, differentiation as well as signaling and death pathways. In addition, aspartic peptidases can also participate in different contexts of the trypanosomatid-host interface, facilitating some pathogenic events such as dissemination, adhesion, escape, nutrition and immunomodulation of the host immune responses. Consequently, aspartic peptidase inhibitors are able to block one or several of these fundamental events, reducing the ability of these trypanosomatids in causing infections.
The attenuation of parasitic infections in HIV-infected individuals might not solely have resulted from improved immunological status, but also as a result of direct inhibition of aspartic peptidases produced by parasites. Taking it in consideration, some researchers showed that T. cruzi and different species belonging to the Leishmania genus are able to produce cell-associated aspartic peptidases sensible to classical aspartic proteolytic inhibitors (e.g., pepstatin A, DAN and EPNP) and HIV PIs (Fig. 6) used in the polichemotherapy administered to the HIV-infected individuals. The inhibition of trypanosomatids’ aspartic peptidases was capable in interfering with fundamental events of these microorganisms. In this context, several studies described the inhibitory effects of HIV PIs on (i) crucial physiological processes including loss of viability/motility, blockage of proliferation/growth, failure to maintain both morphology and cellular homeostasis, and induction of an augmentation in the level of reactive oxygen species that triggers two distinct death pathways, apoptosis and autophagy, (ii) relevant steps of trypanosomatid-host relationships such as inability to either adhere or survive inside of phagocytic cells (Fig. 10). Together, all these beneficial effects culminate in death of the microorganism and/or its inadequate ability to develop an efficient and successful infection in murine model.
Regarding to the future, the purification of aspartic peptidases produced by trypanosomatids, the more accurate knowledge of its biochemical properties and the crystallization of the tertiary structure will contribute to better understanding of the functioning of these proteolytic enzymes as well as allowing the design of more specific inhibitors. It is advisable to focus drug discovery efforts towards new mechanisms of action, in order to be successful at circumventing the problem with existing resistances, and aspartic peptidases can be a real possibility.
ACKNOWLEDGEMENTS
This study was supported by grants from the Brazilian Agencies: Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundação de Amparo à Pesquisa no Estado do Rio de Janeiro (FAPERJ), Conselho de Ensino e Pesquisa para Graduados da Universidade Federal do Rio de Janeiro (CEPG-UFRJ) and Fundação Oswaldo Cruz (FIOCRUZ). André L.S. Santos, Marta H. Branquinha and Claudia M. d’Avila-Levy were supported by CNPq and FAPERJ fellowships.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflicts of interest.
ABBREVIATIONS
- ACE
= Angiotensin-converting enzyme
- AEBSF
= 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride
- AIDS
= Acquired immune deficiency syndrome
- BACE
= β-site amyloid precursor protein cleaving enzyme
- CZP
= Cruzipsin
- DAN
= Diazo-acetyl-norleucinemethylester
- Ddi-1
= DNA-damage inducible protein 1
- EPNP
= 1,2-epoxy-3-(phenyl-nitrophenoxy) propane
- HAP
= Histo-aspartic peptidase
- HAART
= Highly active antiretroviral therapy
- HIV
= Human immunodeficiency virus
- kDNA
= Kinetoplast DNA
- LD50
= 50% lethal dose
- MDM
= Human primary monocyte-derived macrophages
- PIs
= Peptidase inhibitors
- PMs
= Plasmepsins
- SAPs
= Secreted aspartic peptidases
- SIRS
= Systemic inflammatory response syndrome
- WHO
= World Health Organization
REFERENCES
- 1.Wallace FG. The trypanosomatid parasites of insects and arachnids. Exp. Parasitol. 1966;18:124–193. doi: 10.1016/0014-4894(66)90015-4. [DOI] [PubMed] [Google Scholar]
- 2.Mcghee RB, Cosgrove WB. Biology and physiology of the lower Trypanosomatidae. Microbiol. Rev. 1980;44:140–173. doi: 10.1128/mr.44.1.140-173.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vickerman K. The evolutionary expansion of the trypanosomatid ?agellates. Int. J. Parasitol. 1994;24:1317–1331. doi: 10.1016/0020-7519(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 4.De Souza W, Attias M. In: Structures and organelles in pathogenic protists; Microbiology Monographs, Ed. Springer: Brlin. 2010;17:28–30. [Google Scholar]
- 5.Hoare CA, Wallace FG. Developmental stages of trypanosomatid flagellates: a new terminology. Nature. 1966;212:1385–1386. [Google Scholar]
- 6.Svobodová M, Zídková L, Cepicka I, Oborník M, Lukes J, Votýpka J. Sergeia podlipaevi gen.nov sp nov. (Trypanosomatidae Kinetoplastida) a parasite of biting midges (Ceratopogonidae Diptera). Int J Syst Evol Microbiol. 2007;57:423–432. doi: 10.1099/ijs.0.64557-0. [DOI] [PubMed] [Google Scholar]
- 7.Barret MP, Curchmore JS, Stich A, Lazzari JO, Frasch AC, Cazzulo JJ, Krishna S. The trypanosomiases. The Lancet . 2003;362:1469–1480. doi: 10.1016/S0140-6736(03)14694-6. [DOI] [PubMed] [Google Scholar]
- 8.Stuart K, Brun R, Croft S, Fairlamb A, Gürtler RE, McKerrow J, Reed S, Tarleton R. Kinetoplastids: related protozoan pathogens different diseases. J Clin Invest. 2008;118:1301–1310. doi: 10.1172/JCI33945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bates PA. Leishmania sandfly interaction: progress and challenges. Curr. Opin. Microbiol. 2008;11:340–344. doi: 10.1016/j.mib.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bates PA, Rogers ME. New insights into the developmental biology and transmission mechanisms of Leishmania. Curr Mol Med. 2004;4:601–609. doi: 10.2174/1566524043360285. [DOI] [PubMed] [Google Scholar]
- 11.Coura JR, Borges-Pereira J. Chagas disease: 100 years after its discovery. A systemic review. Acta Trop. 2010;115:5–13. doi: 10.1016/j.actatropica.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Welburn SC, Maudlin I. Tsetse-trypanosome interactions: rites of passage. Parasitol. Today. 1999;15:399–403. doi: 10.1016/s0169-4758(99)01512-4. [DOI] [PubMed] [Google Scholar]
- 13.World Health Organization Control of the leishmaniasis World Health Organ. Tech Rep. Ser xii-xiii 1-186 back cover. 2010;949 [PubMed] [Google Scholar]
- 14.Cavali A, Bolognesi L. Neglected tropical diseases: multi-target-directed ligands in the search for novel lead candidates against Trypanosoma and Leishmania. J. Glob Infect Dis. 2010;2:167–176. doi: 10.1021/jm9004835. [DOI] [PubMed] [Google Scholar]
- 15.Chakravarty J, Sundar S. Drug resistance in leishmaniasis. J Glob Infect Dis. 2010;2:167–176. doi: 10.4103/0974-777X.62887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhandari V, Kulshrestha A, Deep DK, Stark O, Prajapati VK, Ramesh V, Sundar S, Schonian G, Dujardin JC, Salotra P. Drug susceptibility in Leishmania isolates following miltefosine treatment in cases of visceral leishmaniasis and post kala-azar dermal leishmaniasis. PLoS. Negl Trop Dis. 2012;6:e1657. doi: 10.1371/journal.pntd.0001657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett AJ. Classification of peptidases. Meth. Enzymol. 1994;244:1–15. doi: 10.1016/0076-6879(94)44003-4. [DOI] [PubMed] [Google Scholar]
- 18.Beynon RJ, Bond JS. Proteolytic Enzymes: A Practical Approach 2nd ed. Oxford University Press: London. 2001 [Google Scholar]
- 19.Rawlings ND, Barret AJ, Baterman A. MEROPS: the database of proteolytic enzymes their substrates and inhibitors. Nucleic Acids Res. 2012;40:D343–350. doi: 10.1093/nar/gkr987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrett AJ, Hopsu-Havu VK. In Proteolysis in Cell Functions IOS Press. Amsterdam. 1997;13:3–8. [Google Scholar]
- 21.Barret AJ, Rawlings ND, O`Brien EA. The MEROPS database as a peptidase information system. J. Struct. Biol. 2001;134:95–102. doi: 10.1006/jsbi.2000.4332. [DOI] [PubMed] [Google Scholar]
- 22.Barret AJ, Tolle DP, Rwalings ND. Managing peptidases in the genomic era. Biol Chem. 2003;384:873–882. doi: 10.1515/BC.2003.098. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Otín C, Bond JS. Proteases: multifunctional enzymes in life and disease. J Biol Chem. 2008;283:30433–30437. doi: 10.1074/jbc.R800035200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vermelho AB, Melo ACN, Branquinha MHS, Santos ALS, d’Ávila-Levy CM, Couri S, Bom EPS. In Enzimas em Biotecnologia-Produção, Aplicações e Mercado Interciência. Rio de Janeiro. 2008;100:273–287. [Google Scholar]
- 25.Hill J, Phylip L. Bacterial aspartic proteinases. FEBS Lett. 1997;409:357–360. doi: 10.1016/s0014-5793(97)00547-4. [DOI] [PubMed] [Google Scholar]
- 26.James M. In Structure and Function of Aspartic Protease: Retroviral and Cellular Enzymes. Plenum Press New York. 1998; 43600:1–481. [Google Scholar]
- 27.Dash C, Kulkarni A, Dunn B, Rao M. Aspartic peptidase inhibitors implications in drug development. Crit. Rev. Biochem. Mol. Biol. 2003;38:89–119. doi: 10.1080/713609213. [DOI] [PubMed] [Google Scholar]
- 28.Horimoto Y, Dee DR, Yada RY. Multifunctional aspartic peptidase prosegments. New Biotechnol. 2009;25:318–324. doi: 10.1016/j.nbt.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 29.Cooper JB. Aspartic proteinases in disease a structural perspective. Curr. Drug Targets. 2002;3:155–173. doi: 10.2174/1389450024605382. [DOI] [PubMed] [Google Scholar]
- 30.Scott BB, McGeehan GM, Harrison RK. Development of inhibitors of the aspartic protease renin for the treatment of hypertension. Curr. Protein Pept. Sci. 2006;7:241–254. doi: 10.2174/138920306777452330. [DOI] [PubMed] [Google Scholar]
- 31.Coombs GH, Goldberg DE, Klemba M, Berry C, Kay J, Mottram JC. Aspartic proteases of Plasmodium falciparum and other parasitic protozoa as drug targets. Trends Parasitol. 2001;17:532–537. doi: 10.1016/s1471-4922(01)02037-2. [DOI] [PubMed] [Google Scholar]
- 32.Benes P, Vetvicka V, Funsek M. Cathepsin D many functions of aspartic protease. Crit. Rev. Oncol. Hematol. 2008;68:12–28. doi: 10.1016/j.critrevonc.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vassar R. Beta-Secretase (BACE) as a drug target for Alzheimer’s disease. Adv. Drug Deliv. Rev. 2002;54:1589–1602. doi: 10.1016/s0169-409x(02)00157-6. [DOI] [PubMed] [Google Scholar]
- 34.Dominguez DI, Hartmann D, DeStrooper B. BACE1 and presenilin two unusual aspartic proteases involved in Alzheimer’s disease. Neurodegener. Dis. 2004;1:168–174. doi: 10.1159/000080982. [DOI] [PubMed] [Google Scholar]
- 35.James MN, Sielecki A, Salituro F, Rich DH, Hofmann T. Conformational flexibility in the active sites of aspartic proteinases revealed by a pepstatin fragment binding to penicillopepsin. Proc. Natl. Acad. Sci. 1982;79:6137–6141. doi: 10.1073/pnas.79.20.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyland LJ, Tomaszek TA, Meek TD. Human immunodeficiency virus-1 protease 2.Use of pH rate studies and solvent kinetic isotope effects to elucidate details of chemical mechanism. Biochemistry. 1991;30:8454–8463. doi: 10.1021/bi00098a024. [DOI] [PubMed] [Google Scholar]
- 37.Tossi A, Bonin I, Antcheva N, Norbedo S, Benedetti F, Miertus S, Nair AC, Maliar T, Dal Bello F, Palù G, Ro-meo D. Aspartic protease inhibitors.An integrated approach for the design and synthesis of diaminodiol based peptidomimetics. Eur. J. Biochem. 2000;267:1715–1722. doi: 10.1046/j.1432-1327.2000.01164.x. [DOI] [PubMed] [Google Scholar]
- 38.Davies DR. The structure and function of the aspartic proteinases. Annu. Rev. Biophys. Chem. 1990;19:189–215. doi: 10.1146/annurev.bb.19.060190.001201. [DOI] [PubMed] [Google Scholar]
- 39.Blundell TL, Johnson MS. Catching a common fold. Protein Sci. 1993;2:877–883. doi: 10.1002/pro.5560020602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coates L, Erskine PT, Mall S, Gill R, Wood SP, Myles DA, Cooper JB. X-ray neutron and NMR studies of the catalytic mechanism of aspartic proteinases. Eur Biophys J. 2006;35:559–566. doi: 10.1007/s00249-006-0065-7. [DOI] [PubMed] [Google Scholar]
- 41.Koelsch G, Mares M, Metcalf P, Fusek M. Multiple functions of pro-parts of aspartic proteinase zymogens. FEBS Lett. 1994;343:6–10. doi: 10.1016/0014-5793(94)80596-2. [DOI] [PubMed] [Google Scholar]
- 42.Foltmann B. Structure and function of proparts in zymogens for aspartic proteinases. Biol Chem Hoppe-Seyler. 1988;369:311–314. [PubMed] [Google Scholar]
- 43.Rao MB, Tanksale AM, Ghatge M, Deshpande V. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 1998;62:597–635. doi: 10.1128/mmbr.62.3.597-635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi K. In: Aspartic peptidases structure function biology and biomedical implications. Plenum Press New York. 1995 [Google Scholar]
- 45.Fruton J. A history of pepsin and related enzymes. Quart. Rev. Biol. 2002;77:127–147. doi: 10.1086/340729. [DOI] [PubMed] [Google Scholar]
- 46.Sielecki R, Fujinanga M, Read RJ, James MNG. Refined structure of porcine pepsinogenat 1. resolution. J. Mol. Biol. 1991;219:671–692. doi: 10.1016/0022-2836(91)90664-r. [DOI] [PubMed] [Google Scholar]
- 47.Blundell T, Cooper J, Sali A, Zhu Z, Dunn BN. In Structure and function of the aspartic proteases. Plenum Press New York. 1991;30600:443. [Google Scholar]
- 48.Holm I, Ollo R, Panthier J, Rougeon F. Evolution of aspartic proteases by gene duplication the mouse renin gene is organized in two homologous clusters of four exons. EMBO J. 1984;3:557–562. doi: 10.1002/j.1460-2075.1984.tb01846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Veerapandian B, Cooper J, Sali A, Blundell T, Rosati R, Dominy B, Damon D, Hoover D. Direct observation by X-ray analysis of the tetrahedral intermediate of aspartic proteinases. Protein Sci. 1992;1:322–328. doi: 10.1002/pro.5560010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Northrop D. Follow the protons: a low-barrier hydrogen bond unifies the mechanisms of the aspartic proteases. Accounts Chem. Res. 2001;34:790–797. doi: 10.1021/ar000184m. [DOI] [PubMed] [Google Scholar]
- 51.Dunn B. Structure and mechanism of the pepsin-like family of aspartic peptidases. Chem. Rev. 2002;102:4431–4458. doi: 10.1021/cr010167q. [DOI] [PubMed] [Google Scholar]
- 52.Nguyen JT, Hamada Y, Kimura T, Kiso Y. Design of potent aspartic protease inhibitors to treat various diseases. Arch. Pharm. Chem. Life Sci. 2008;341:523–535. doi: 10.1002/ardp.200700267. [DOI] [PubMed] [Google Scholar]
- 53.MEROPS - The peptidase database: Realease 9 6. http://merops.sanger.ac.uk/. Accessed June 26. 2012 [Google Scholar]
- 54.Fujinaga M, Chernaia MM, Tarasova NI, Mosimann SC, James MN. Crystal structure of human pepsin and its complex with pepstatin. Protein Sci. 1995;4:960–972. doi: 10.1002/pro.5560040516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jaskólski M, Tomasselli AG, Sawyer TK, Staples DG, Heinrikson RL, Schneider J, Kent SB, Wlodawer A. Structure at 2 5 A resolution of chemically synthesized human immunodeficiency virus type 1 protease complexed with a hydroxyethylene-based inhibitor. Biochemistry. 1991;30:1600–1609. doi: 10.1021/bi00220a023. [DOI] [PubMed] [Google Scholar]
- 56.Steiner H, Kostka M, Romig H Basset, G. Pesold, B. Hardy, J. Capell, A. Meyn, L. Grim ML, Baumeister R, Fechteler K, Haass C. Glycine 384 is required for presenilin-1 function and is conserved in bacterial polytopic aspartic proteases. Nat Cell Biol . 2000;2:848–851. doi: 10.1038/35041097. [DOI] [PubMed] [Google Scholar]
- 57.Laudon H, Winblad B, Näslund J. The Alzheimer’s diseases-associated b-secretase complex: functional domains in the presenilin 1 protein. Physiol Behav. 2007;92:115–120. doi: 10.1016/j.physbeh.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 58.Alves CR, Corte-Real S, Bourguignon SC, Chaves CS, Saraiva EM. Leishmania amazonensis: early proteinase activities during promastigote-amastigote differentiation in vitro. Exp. Parasitol. 2005;109:38–48. doi: 10.1016/j.exppara.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 59.Savoia D, Allice Tovo, P. Antileishmanial activity of HIV protease inhibitors. Int. J. Antimicrob. Agents. 2005;26:92–94. doi: 10.1016/j.ijantimicag.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 60.Valdivieso E, Dagger F, Rascón A. Leishmania mexicana: identification and characterization of an aspartic proteinase activity. Exp. Parasitol. 2007;116:77–82. doi: 10.1016/j.exppara.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 61.Trudel N, Garg R, Messier N, Sundar S, Ouellette M, Tremblay MJ. Intracellular survival of Leishmania species that cause visceral leishmaniasis is significantly reduced by HIV-1 protease inhibitors. J. Infect Dis. 2008;198:1292–1299. doi: 10.1086/592280. [DOI] [PubMed] [Google Scholar]
- 62.Santos LO, Marinho FA, Altoé EF, Vitório BS, Alves CR, Britto C, Motta MC, Branquinha MH, Santos ALS, d'Avila-Levy CM. HIV aspartic peptidase inhibitors interfere with cellular proliferation, ultrastructure and macrophage infection of Leishmania amazonensis. PLoS One. 2009;4:e4918. doi: 10.1371/journal.pone.0004918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valdivieso E, Rangel A, Moreno J, Saugar JM, Cañavate C, Alvar J, Dagger F. Effects of HIV aspartic-proteinase inhibitors on Leishmania sp. Exp. Parasitol. 2010;126:557–563. doi: 10.1016/j.exppara.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 64.Sabotic J, Kos J. Microbial and fungal protease inhibitors: current and potential applications. Appl. Microbiol. Biotechnol. 2012;93:1351–1375. doi: 10.1007/s00253-011-3834-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Santos ALS. Aspartic proteases of human pathogenic fungi are prospective targets for the generation of novel and effective antifungal inhibitors. Curr. Enz. Inhib. 2011;7: 96–118. [Google Scholar]
- 66.Santos ALS. HIV aspartic protease inhibitors as promising compounds against Candida albicans. World J. Biol. Chem. 2010;1:21–30. doi: 10.4331/wjbc.v1.i2.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vermelho AB, Branquinha MH, d’Avila-Levy CM, dos Santos ALS, Paraguai de Souza Dias E, Nogueira de Melo AC. Biological roles of peptidases in trypanosomatids. Open Parasitol. J. 2010;4:5–23. [Google Scholar]
- 68.Santos ALS, d’Avila-Levy CM, Branquinha MH, Cohen JB, Ryseck LP. In Calpain-like proteins in trypanosomatids: effects of calpain inhibitors on the parasites’ physiology and motivations for their possible application as chemotherapeutic agents. Nova Science Publishers: New York. 2011:77–104. [Google Scholar]
- 69.Santos ALS, d’Avila-Levy CM, Branquinha MH. In Anti-trypanosomatid properties of cystatin superfamily: implications on parasite development and virulence. Nova Science Publishers New York . 2011:41–76. [Google Scholar]
- 70.Santos ALS. In Aspartic peptidase inhibitors as potential bioactive pharmacological compounds against human fungal pathogens. 2010: 289–326. [Google Scholar]
- 71.Vermelho AB, Giovanni De Simone S, d'Avila-Levy CM, Santos ALS, Nogueira de Melo AC, Silva-Junior FP, Bom EP, Branquinha MH. Trypanosomatidae peptidases: a target for drugs development. Curr. Enz. Inhib. 2007;3:19–48. [Google Scholar]
- 72.Abbenante G, Fairlie DP. Protease inhibitors in the clinic. Med. Chem. 2005;1:71–104. doi: 10.2174/1573406053402569. [DOI] [PubMed] [Google Scholar]
- 73.Flexner C. HIV-protease inhibitors. N. Engl. J. Med. 1998;338:1281–1292. doi: 10.1056/NEJM199804303381808. [DOI] [PubMed] [Google Scholar]
- 74.McKerrow JH, Engel JC, Caffrey CR. Cysteine protease inhibitors as chemotherapy for parasitic infections. Bioorg Med Chem. 1999;7:639–644. doi: 10.1016/s0968-0896(99)00008-5. [DOI] [PubMed] [Google Scholar]
- 75.Coombs GH, Goldberg DE, Klemba M, Berry C, Kay J, Mottram JC. Aspartic proteases of Plasmodium falciparum and other parasitic protozoa as drug targets. Trends Parasitol. 2001;17:532–537. doi: 10.1016/s1471-4922(01)02037-2. [DOI] [PubMed] [Google Scholar]
- 76.Stewart K, Abad-Zapatero C. Candida proteases and their inhibition: prospects for antifungal therapy. Curr Med Chem. 2001;8: 941–948. doi: 10.2174/0929867013372698. [DOI] [PubMed] [Google Scholar]
- 77.Dash C, Kulkarni A, Dunn B, Rao M. Aspartic peptidase inhibitors: implications in drug development. Crit. Rev. Biochem. Mol. Biol. 2003;38:89–119. doi: 10.1080/713609213. [DOI] [PubMed] [Google Scholar]
- 78.Pozio E, Morales MA. The impact of HIV-protease inhibitors on opportunistic parasites. Trends Parasitol. 2005;21:58–63. doi: 10.1016/j.pt.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 79.Mastrolorenzo A, Rusconi S, Scozzafava A, Barbaro G, Supuran CT. Inhibitors of HIV-1 protease: current state of the art 10 years after their introduction.From antiretroviral drugs to antifngal antibacterial and antitumor agents based on aspartic protease inhibitors. Curr. Med. Chem. 2007;14:2734–2748. doi: 10.2174/092986707782360141. [DOI] [PubMed] [Google Scholar]
- 80.Nguyen JT, Hamada Y, Kimura T, Kiso Y. Design of potent aspartic protease inhibitors to treat various diseases. Arch. Pharm. (Weinheim) 2008;341:523–535. doi: 10.1002/ardp.200700267. [DOI] [PubMed] [Google Scholar]
- 81.McKerrow JH, Rosenthal PJ, Swenerton R, Doyle P. Development of protease inhibitors for protozoan infections. Curr. Opin. Infect. Dis. 2008;21:668–672. doi: 10.1097/QCO.0b013e328315cca9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Powderly WG, Landay A, Lederman MM. Recovery of the immune system with antiretroviral therapy. The end of opportunism?. JAMA. 1998;280:72–77. doi: 10.1001/jama.280.1.72. [DOI] [PubMed] [Google Scholar]
- 83.Mastroianni CM, Lichtner M, Mengoni F, D'Agostino C, Forcina G, d'Ettorre G, Santopadre P, Vullo V. Improvement in neutrophil and monocyte function during highly active antiretroviral treatment of HIV-1 infected patients. AIDS. 1999;13:883–890. doi: 10.1097/00002030-199905280-00003. [DOI] [PubMed] [Google Scholar]
- 84.Mezzaroma I, Carlesimo M, Pinter E, Alario C, Sacco G, Muratori DS, Bernardi ML, Paganelli R, Aiuti F. Long-term evaluation of T-cell subsets and T-cell function after HAART in advanced stage. HIV-1 disease AIDS. 1999;13:1187–1193. doi: 10.1097/00002030-199907090-00006. [DOI] [PubMed] [Google Scholar]
- 85.Malhotra U, Berrei MM, Huang Y, Maree J, Brown DJ, Ap S, Musey L, Schacker T, Corey L, McElrath MJ. Effect of combination antiretroviral therapy on T-cell immunity in acute human immunodeficiency virus type 1 infection. J Infect Dis. 2000;181:121–131. doi: 10.1086/315202. [DOI] [PubMed] [Google Scholar]
- 86.Alvar J, Aparício P, Aseffa A, Den Bôer M, Cañavate C, Dedet JP, Gradoni L, TerHorst R, López-Vélez R, Moreno J. The relationship between leishmaniasis and AIDS the second 10 years. J. Clin. Microbiol. Rev. 2008;21:334–359. doi: 10.1128/CMR.00061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nissapatorn V, Sawangjaroen N. Parasitic infections in HIV infected individuals: Diagnostic & therapeutic challenges. Indian J. Med. Res. 2011;134:878–897. doi: 10.4103/0971-5916.92633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Palella FJJr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection.HIV Outpatient Study Investigation. N Engl J Med. 1998;338:853–886. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 89.Casolari C, Rossi T, Baggio G, Coppi A, Zandomeneghi G, Ruberto AI, Farina C, Fabio G, Zanca A, Castelli M. Interaction between saquinavir and antimycotic drugs on C.albicans and C. neoformans strains. Pharmacol. Res. 2004;50:605–610. doi: 10.1016/j.phrs.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 90.Falkensammer B, Pilz G, Bektic J, Imwidthaya P, Jöhrer K, Speth Absent reduction by HIV protease inhibitors of Candida albicans adhesion to endothelial cells. Mycoses. . 2007; 50:172–177. doi: 10.1111/j.1439-0507.2006.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cenci E, Francisci D, Belfiori B, Pierucci S, Baldelli F, Bistoni F, Vecchiarelli A. Tipranavir exhibits different effects on opportunistic pathogenic fungi. J. Infect. 2008;56:58–64. doi: 10.1016/j.jinf.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 92.Tsang CS, Hong I. HIV protease inhibitors differentially inhibit adhesion of Candida albicans to acrylic surfac-es. Mycoses. 2010;53:488–494. doi: 10.1111/j.1439-0507.2009.01743.x. [DOI] [PubMed] [Google Scholar]
- 93.Braga-Silva LA, Mogami SS, Valle RS, Silva-Neto ID, Santos ALS. Multiple effects of amprenavir against Candida albicans. FEMS Yeast Res. 2010;10:221–224. doi: 10.1111/j.1567-1364.2009.00595.x. [DOI] [PubMed] [Google Scholar]
- 94.Moreno M, Gonzalez VM. Advances on Aptamers Targeting Plasmodium and Trypanosomatids. Curr. Med. Chem. 2011;18:5003–5010. doi: 10.2174/092986711797535218. [DOI] [PubMed] [Google Scholar]
- 95.Peyrottes S, Caldarelli S, Wein S, Périgaud C, Pellet A, Vial H. Choline Analogues in Malaria Chemotherapy. Curr. Pharm. Desgn. 2012;18:3454–3466. doi: 10.2174/138161212801327338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Teixeira C, Gomes JRB, Gomes P. Falcipains Plasmodium falciparum Cysteine proteases as key drug targets against malaria. Curr. Med. Chem. 2011;18:1555–1572. doi: 10.2174/092986711795328328. [DOI] [PubMed] [Google Scholar]
- 97.Alam A, Goyal M, Iqbal MS, Pal C, Dey S, Bindu S, Maity P, Bandyopadhyay U. Novel antimalarial drug targets: hope for new antimalarial drugs. Expert. Rev. Clin. Pharmacol. 2009;2:469–489. doi: 10.1586/ecp.09.28. [DOI] [PubMed] [Google Scholar]
- 98.Bektic J, Lell CP, Fuchs A, Stoiber H, Speth C, Lass-Flörl C, Borg-von Zepelin M, Dierich MP, Würzner R. HIV protease inhibitors attenuate adherence of Candida albicans to epithelial cells in vitro. FEMS Immunol. Med. Microbiol. 2001;31:65–71. doi: 10.1111/j.1574-695X.2001.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 99.Borg-von Zepelin M, Meyer I, Thomssen R, Würzner R, Sanglard D, Telenti A, Monod M. HIV-Protease inhibitors reduce cell adherence of Candida albicans strains by inhibition of yeast secreted aspartic proteases. J. Invest. Dermatol. 1999;113:747–751. doi: 10.1046/j.1523-1747.1999.00747.x. [DOI] [PubMed] [Google Scholar]
- 100.Monod M, Borg-von Zepelin M, Telenti A, Sanglard D. The inhibition of Candida-albicans-secreted aspartic proteases by three different HIV protease inhibitors. Dermatology. 1999;198:412–414. [PubMed] [Google Scholar]
- 101.Braga-Silva LA, Santos ALS. Aspartic protease inhibitors as potential anti-Candida albicans drugs: impacts on fungal biology, virulence and pathogenesis. Curr. Med. Chem. 2011;18:2401–2419. doi: 10.2174/092986711795843182. [DOI] [PubMed] [Google Scholar]
- 102.Girardi E, Antonucci G, Vanacore P, Libanore M, Errante I, Matteelli A, Ippolito G. Impact of combination antiretroviral therapy on the risk of tuberculosis among persons with HIV infection. Gruppo Italiano di Studio Tubercolosi e AIDS (GISTA). AIDS. 2000;14:1985–1991. doi: 10.1097/00002030-200009080-00015. [DOI] [PubMed] [Google Scholar]
- 103.Kirk O, Gatell JM, Mocroft A, Pedersen C, Proenca R, Brettle RP, Barton SE, Sudre P, Phillips AN. Infections with Mycobacterium tuberculosis and Mycobacterium avium among HIV-infected patients after the introduction of highly active antiretroviral therapy.EuroSIDA Study Group JD. Am. J. Respir. Crit. Care Med 2000 162 865-872. Erratum in: Am. J. Respir. Crit. Care Med. 2003;167:1719. doi: 10.1164/ajrccm.162.3.9908018. [DOI] [PubMed] [Google Scholar]
- 104.Shen Y, Shen L, Sehgal P, Zhou D, Simon M, Miller M, Enimi EA, Henckler B, Chalifoux L, Sehgal N, Gastron M, Letvin NL, Chen ZW. Antiretroviral agents restore Mycobacterium-specific T cell immune responses and facilitate controlling a fatal tuberculosis-like disease in Macaques coinfected with simianimmunodeficiency virus and Mycobacterium bovis BCG. J Virol. 2001;75:8690–8696. doi: 10.1128/JVI.75.18.8690-8696.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kabbesh M, Koomer A, Molteni A, Amajoyi R, Quinn T, Herndon B. The effect of treatment with a protease inhibitor on mycobacterial infection. Diagn. Microbiol. Infect. Dis. 2005;51:251–258. doi: 10.1016/j.diagmicrobio.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 106.Santos LO, Vitório BS, Branquinha MH, Pedrozo C, Santos ALS, d’Avila-Levy CM. Nelfinavir, an HIV aspartyl peptidase inhibitor, is effective in inhibiting the multiplication and aspartyl peptidase activity of several Leishmania species, including strains obtained from HIV-positive patients. J. Antmicrob. Chemother. 2013;68:348–353. doi: 10.1093/jac/dks410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.White RE, Powell DJ, Berry C. HIV proteinase inhibitors target the Ddi1-like protein of Leishmania parasites. FASEB J. 2011;25:1729–1736. doi: 10.1096/fj.10-178947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Perteguer MJ, Gómez-Puertas P, Cañavate C, Dagger F, Gárate T, Valdivieso E. Ddi1-like protein from Leishmania major is an active aspartyl proteinase. Cell Stress Chaperones. 2012 doi: 10.1007/s12192-012-0368-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Falutz J. Management of fat accumulation in patients with HIV infection. Curr. HIV/AIDS Rep. 2011;8:200–208. doi: 10.1007/s11904-011-0087-3. [DOI] [PubMed] [Google Scholar]
- 110.Kumar P, Lodge R, Trudel N, Ouellet M, Ouellette M, Tremblay MJ. Nelfinavir an HIV-1 protease inhibitor induces oxidative stress-mediated caspase-independent apoptosis in Leishmania amastigotes. PLoS. Negl. Trop. Dis. 2010;4:e642. doi: 10.1371/journal.pntd.0000642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Demarchi IG, Silveira TG, Ferreira IC, Lonardoni MV. Effect of HIV protease inhibitors on New World Leishmania. Parasitol Int. 2012;61:538–544. doi: 10.1016/j.parint.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 112.Alvarez VE, Niemirowicz GT, Cazzulo JJ. The peptidases of Trypanosoma cruzi digestive enzymes virulence factors and mediators of autophagy and programmed cell death. Biochim. Biophys. Acta. 2012;1824:195–206. doi: 10.1016/j.bbapap.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 113.Pinho RT, Beltramini LM, Alves CR, De-Simone SG. Trypanosoma cruzi: isolation and characterization of aspartic proteases. Exp. Parasitol. 2009;122:128–133. doi: 10.1016/j.exppara.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 114.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, Westenberger SJ, Caler E, Cerqueira GC, Branche C, Haas B, Anupama A, Arner E, Aslund L, Attipoe P, Bontempi E, Bringaud F, Burton P, Cadag E, Campbell DA, Carrington M, Crabtree J, Darban H, da Silveira JF, de Jong P, Edwards K, Englund PT, Fazelina G, Feldblyum T, Ferella M, Frasch AC, Gull K, Horn D, Hou L, Huang Y, Kindlund E, Klingbeil M, Kluge S, Koo H, Lacerda D, Levin MJ, Lorenzi H, Louie T, Machado CR, McCulloch R, McKenna A, Mizuno Y, Mottram JC, Nelson S, Ochaya S, Osoegawa K, Pai G, Parsons M, Pentony M, Pettersson U, Pop M, Ramirez JL, Rinta J, Robertson L, Salzberg SL, Sanchez DO, Seyler A, Sharma R, Shetty J, Simpson AJ, Sisk E, Tammi MT, Tarleton R, Teixeira S, Van Aken S, Vogt C, Ward PN, Wickstead B, Wortman J, White O, Fraser CM, Stuart KD, Andersson B. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas’ disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 115.Sangenito LS, Gonçalves KC, Abi-Chacra EA, Sodré CL, d'Avila-Levy CM, Branquinha MH, Santos ALS. Multiple effects of pepstatin A on Trypanosoma cruzi epimastigote forms. Parasitol. Res. 2012;110:2533–2540. doi: 10.1007/s00436-011-2796-3. [DOI] [PubMed] [Google Scholar]
- 116.Andrade ZA. Immunopathology of Chagas disease. Mem. Inst. Oswaldo Cruz. 1999;1:71–80. doi: 10.1590/s0074-02761999000700007. [DOI] [PubMed] [Google Scholar]
- 117.Fampa P, Santos ALS, Ramirez MI. Trypanosoma cruzi ubiquity expression of surface cruzipain molecules in TCI and TCII field isolates. Parasitol. Res. 2010;107:443–447. doi: 10.1007/s00436-010-1888-9. [DOI] [PubMed] [Google Scholar]
- 118.Kikuchi SA, Sodré CL, Kalume DE, Elias CG, Santos ALS, deNazaréSoeiro M, Meuser M, Chapeaurouge A, Perales J, Fernandes O. Proteomic analysis of two Trypanosoma cruzi zymodeme 3 strains. Exp. Parasitol. 2010;126:540–551. doi: 10.1016/j.exppara.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 119.Tumbarello M, Tacconelli E, Bertagnolio S, Cauda R. Highly active antiretroviral therapy decreases the incidence of visceral leishmaniasis in HIV-infected individuals. AIDS. 2000;14:2948–2950. doi: 10.1097/00002030-200012220-00021. [DOI] [PubMed] [Google Scholar]
- 120.Rosenthal E, Tempesta S, del Giudice P, Marty P, Desjeux P, Pradier C, Le Fichoux Y, Jill-Patrice C. Declining incidence of visceral leishmaniasis in HIV-infected individuals in the era of highly active antiretroviral therapy. AIDS. 2001;15:1184–1185. doi: 10.1097/00002030-200106150-00017. [DOI] [PubMed] [Google Scholar]
- 121.ter Horst R, Collin SM, Ritmeijer K, Bogale A, Davidson RN. Concordant HIV infection and visceral leishmaniasis in Ethiopia: the influence of antiretroviral treatment and other factors on outcome. Clin. Infect. Dis. 2008;46:1702–1709. doi: 10.1086/587899. [DOI] [PubMed] [Google Scholar]
- 122.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2. . Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 123.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]