Abstract
Mitochondrial morphology and intracellular organization are tightly controlled by the processes of mitochondrial fission–fusion. Moreover, mitochondrial movement and redistribution provide a local ATP supply at cellular sites of particular demands. Here we analysed mitochondrial dynamics in isolated primary human pancreatic cells. Using real time confocal microscopy and mitochondria-specific fluorescent probes tetramethylrhodamine methyl ester and MitoTracker Green we documented complex and novel patterns of spatial and temporal organization of mitochondria, mitochondrial morphology and motility. The most commonly observed types of mitochondrial dynamics were (i) fast fission and fusion; (ii) small oscillating movements of the mitochondrial network; (iii) larger movements, including filament extension, retraction, fast (0.1–0.3 μm/sec.) and frequent oscillating (back and forth) branching in the mitochondrial network; (iv) as well as combinations of these actions and (v) long-distance intracellular translocation of single spherical mitochondria or separated mitochondrial filaments with velocity up to 0.5 μm/sec. Moreover, we show here for the first time, a formation of unusual mitochondrial shapes like rings, loops, and astonishingly even knots created from one or more mitochondrial filaments. These data demonstrate the presence of extensive heterogeneity in mitochondrial morphology and dynamics in living cells under primary culture conditions. In summary, this study reports new patterns of morphological changes and dynamic motion of mitochondria in human pancreatic cells, suggesting an important role of integrations of mitochondria with other intracellular structures and systems.
Keywords: mitochondrial dynamics, fusion and fission, mitochondrial morphology, mitochondrial motility, pancreatic cells
Introduction
Mitochondria have been recognized in recent years not only as main cellular source of energy in the form of ATP for normal cell function and viability, but also as integral part of multiple cell signalling and a major controller in a variety of important cellular functions [1–4]. These organelles synthesize different metabolites, control cellular redox potential and participate in different ionic regulations, in particular in calcium homeostasis [5] and play a central role in apoptotic cell death [4, 6]. Our understanding of mitochondrial intracellular distribution, morphology and dynamics has been enhanced by advances in fluorescent confocal microscopy that permits real time imaging of these organelles in living cells [7–10]. This technique has demonstrated that mitochondrial morphology can vary from small spheres or short rod-like shape to long tubules; from interconnected networks to separated distinct electrically non-connected mitochondria [8, 11, 12]. In the last few years, confocal imaging studies revealed that in many organisms ranging from yeast to human beings, mitochondria are highly dynamic, motile and undergo frequent fission and fusion processes [7]. Recently developed methods include also fluorescence resonance energy transfer and recording the image data in three spatial dimensions over time (i.e. 4D-imaging), as well as 4Pi- and STED-microscopy [9, 13]. Having higher spatial resolutions (∼100 nm), these techniques possess rather limited time resolution necessary for the studies of fast changes in mitochondrial network and morphology [13].
It is known that specific mitochondrial distribution and organization in the cell can be achieved by the movement of these organelles along the cytoskeleton filaments (e.g. actin or microfilaments) and attachment to the cytoskeleton using specific motor and connector proteins [10, 14, 15]. Importantly, defects in mitochondrial dynamics may contribute to a variety of human diseases, in particular neurological diseases [16–19]. Mitochondrial movement is metabolically regulated and can be rather rapid in some cells like neurons and serve to direct mitochondria to cellular regions of locally high ATP demand to provide energy (and Ca2+ buffering capacity), or to transport mitochondria destined for elimination [14, 20]. Alternatively, mitochondria can be rather fixed in their positions in cells such as adult cardiomyocytes or skeletal muscles, pointing to the cell type specificity of mitochondrial dynamics. However, all aspects of mitochondrial dynamics mentioned above were not studied in pancreatic cells. Among metabolic diseases, mitochondrial diabetes is the consequence of dysfunction of pancreatic cells caused by mutations in mitochondrial DNA [21, 22]. The impact of such mutations on β-cells function reflects the importance of mitochondria in the control of insulin secretion. The mitochondria of these cells serve as fuel sensors, generating factors that couple nutrient metabolism to the exocytosis of insulin-containing vesicles. The latter process requires an increase in cytosolic Ca2+, which, in turn, depends on ATP synthesized by the mitochondria [22].
Most mitochondrial imaging studies were performed on tumour derived cells (e.g. HeLa cells), which may have abnormal bioenergetic properties with possible consequences also for mitochondrial dynamics. In our study, using fast imaging technique and specific fluorescent probes we analysed dynamics of mitochondria in intact cells isolated from human pancreas, including transient changes in mitochondrial morphology, organization and position. Our data demonstrated a highly dynamic mitochondrial behaviour and complex patterns of morphological changes and intracellular motions of mitochondria comparable with mitochondrial dynamics reported in neurons [14, 15], showing, however, absolutely new variations in mitochondrial shapes and dynamics in pancreatic cells. This provides further competence of mitochondria to modify their metabolism by flexible remodelling of the mitochondrial network for multiple, heterogeneous and possibly region-specific cellular functional requirements and for the integration of mitochondria with other intracellular organelles and systems (endoplasmic reticulum, Golgi, nucleus, cytoskeleton, etc.) [23–28].
Materials and methods
Materials
All chemicals, including the mitochondria specific fluorescent dye tetramethylrhodamine methyl ester (TMRM), were obtained from Sigma (St. Louis, MO, USA) except MitoTracker® Green (Molecular Probes, Eugene, OR, USA).
Islet isolation
Human pancreata were retrieved from heart-beating cadaveric donors at the time of multi-organ harvest for transplantation. Research use of tissue was according to Austrian Law. Following vascular flushing with University of Wisconsin solution (UW), the pancreas was removed and used for the islet isolation. Human islets were isolated using the method of Ricordi et al.[29] with slight modifications as described elsewhere [30]. Briefly, a cold solution of 2 mg/ml liberase enzyme blend (Roche Molecular, Indianapolis, IN, USA) in Hank’s balanced salt solution (HBSS) (PAN Biotech GmbH, Aidenbach, Germany) supplemented with 0.2 mg/ml DNase I (Boehringer-Mannheim, Montreal, Quebec, Canada) was infused into the main pancreatic duct of head and tail using a pressure- and temperature-control perfusion apparatus. The distended pancreas was placed in a sterilized stainless steel digestion chamber (Oberhammer, Innsbruck, Austria), through which HBSS was recirculated at 37°C, for 20 min. Extent of tissue digestion was assessed by staining aliquots with dithizone (Sigma). Islets were visualized under an inverted light microscope (IX-70, Olympus, Vienna, Austria). The digestion process was terminated by cooling the circuit to 5°C to 10°C when 50% of islets were free of surrounding acinar tissue. Then islets were collected, centrifuged (400 ×g) and washed three times in HBSS. After that islets were purified on a continuous Euro-Ficoll density gradient (Biocoll Biochrome, Berlin, Germany) using a COBE 2991 Cell Processor (COBE BCT, Denver, CO, USA). Immediately after isolation, islets were cultured (under adherent conditions) on eight-chambered Lab-Tek Coverglasses (Nalge Nunc International, Rochester, NY, USA) in 400 μl of RPMI-1640 medium (PAA, Linz, Austria) supplemented with 10% FCS (Life Science, Milan, Italy), 2 g/l (11.1 mM) glucose, penicillin (100 U/ml), streptomycin (100 μg/ml; PAA) and L-glutamine (20 μg/ml; Gibco/Invitrogen, Carlsbad CA, USA) at 37°C and 5% CO2. After 72 hrs cultivation TMRM (0.1 μM) was added directly to the chambers.
Confocal microscopy
To investigate mitochondrial distribution, morphology and movement, the mitochondria-specific fluorescent dye TMRM was used. TMRM is a lipophilic, cell permeable, cationic, nontoxic, fluorescent dye that specifically stains live mitochondria [31]. TMRM is accumulated in mitochondria depending on the inner membrane potential. Cultured cells were loaded with TMRM (0.1 μM) by incubating them for 30 min. at room temperature (21–22°C) in serum-free culture medium with 2 g/l glucose. In separate series of experiments cells were loaded also with another established mitochondrial specific probe MitoTracker® Green (0.2 μM, Molecular Probes). Incubation and confocal imaging analysis of cells was performed in Lab-Tek eight-chambered coverglasses (Nalge Nunc International, Naperville, IL, USA). Mitochondrial imaging in live cells was acquired with a microlens-enhanced Nipkow disk-based confocal system UltraVIEW RS (Perkin Elmer, Wellesley MA, USA) mounted on an Olympus IX-70 inverse microscope (Olympus, Nagano, Japan) with a 40× water immersion objective (LUM-PlanFI/IR, NA 0.8). In time series, mitochondrial images were collected every second during 5–10 min. (300–600 images were collected in each experimental set). Minimum laser power was used to minimize photobleaching. Nevertheless, the fluorescence intensity of TMRM was slightly decreased over long observation time, showing yet no oscillations of the signal (Fig. 1). The MitoTracker® Green fluorescence signal was monitored with the 488 nm line of a laser for excitation and TMRM fluorescence was analysed using 546 nm for excitation. Image acquisition and analysis were done using the UltraVIEW RS software. All images were corrected by subtraction of background fluorescence.
Fig 1.
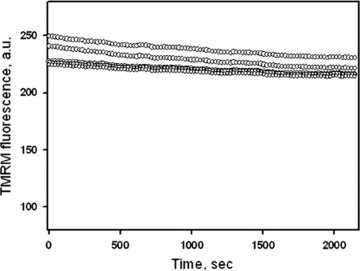
TMRM fluorescence in human pancreatic cells over time shows a slight decline and no oscillations of the signal intensity. Note that time of this test (∼2000 sec.) exceeds markedly a typical observation time used in our study. Data of five different cells are presented.
Results
Intracellular organization of pancreatic mitochondria
In our study mitochondrial imaging in pancreatic cells was performed by the mitochondria-specific dye TMRM, which is widely used for the analysis of static and dynamic mitochondrial arrangement in various cell types [8, 11, 30–34]. Mitochondrial imaging was also performed using additional mitochondria-specific probe MitoTracker Green. In experiments combining MitoTracker Green and TMRM fluorescence co-localization of these mitochondria-specific probes was demonstrated (data not shown). Also, at the concentrations used in this study, both TMRM and MitoTracker® Green had no effect on mitochondrial dynamics and cell viability over long time (up to 2 days) incubation of human pancreatic cells. They also did not change functional parameters of mitochondria as measured by high-resolution respirometry (data not shown). Moreover, no oscillations (flickering) of the TMRM signal were observed (Fig. 1), which are early signs of permeability transition [35]. All these data confirm that fully functional mitochondria were studied during entire time of observation.
Figure 2 demonstrates the typical arrangement of mitochondria in living human primary pancreatic cells visualized by TMRM, showing an intensely branched network of thread-shaped organelles, and sometimes also very long network resembling wire-based structures (Fig. 2, right panel). Notably, mitochondrial filaments may differ by a factor of 100 in length (ranging from less than 0.4 to greater than 40 μm), but at the same time they have similar diameter (about 0.2 μm). Subsets of separated short tubular mitochondria or even individual spherical organelles were also observed (Fig. 2), reflecting thus heterogeneity in mitochondrial morphology. In some cells a dense mitochondrial network uniformly covered the whole cell (arrows in Fig. 2). Other cells showed a less dense network with more elongated mitochondrial threads surrounding nuclei (N).
Fig 2.
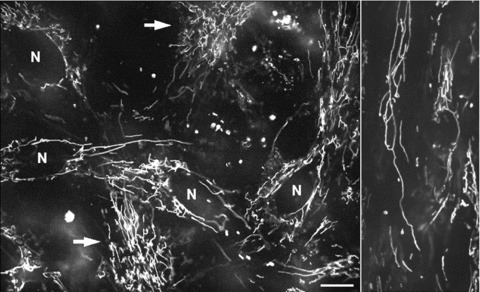
Typical mitochondrial network in human pancreatic cells visualized by mitochondrial fluorescent probe TMRM. In some cells the mitochondrial network covers the whole cell (left panel, arrows), whereas other cells show elongated mitochondrial threads surrounding nuclei (N). Note that mitochondrial morphology vary from small, sphere like mitochondria to very long up to 30 μm long mitochondrial filaments (right panel). Scale bar, 10 μm.
Mitochondrial fission/fusion
Time series imaging of pancreatic mitochondria by collecting 300–600 images revealed that these organelles continually divide and fuse to form a dynamic interconnecting network. Figure 3 demonstrates that the formation of short tubular mitochondria can be a result of fragmentation (fission) of long mitochondrial threads (cf. Fig. 3A and dashed box in Fig. 3B), where mitochondrial network collapses into shorter separated filaments. Figure 3C shows that this process can be fast and one mitochondrial thread may be divided into three fragments during about 100 sec. (see also supplementary video file: Kuz 1 Fission). Another example of the fission/fusion dynamics is shown in Fig. 4. Again, the branched mitochondrial network was divided into three fragments (Fig. 4A), whereas Fig. 4B shows the opposite process of fusion of two network branches (or even two separated networks, see also supplementary video file: Kuz 2 Fusion).
Fig 3.
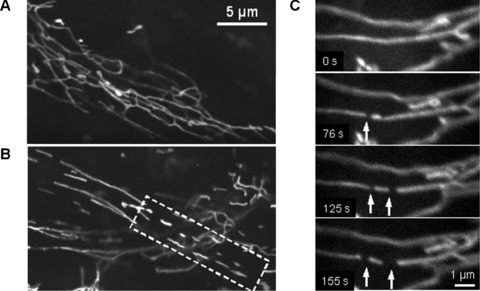
Fragmentation of mitochondrial network filaments in human pancreatic cells. (A) Continuous mitochondrial network. (B) Partially fragmented mitochondrial network with shorter separated filaments (dashed box). (C) Dynamics of mitochondrial fission. Fast division of long filament into three fragments (arrows) occurred during about 100 sec. (A) and (B): Scale bar, 5 μm. (C): Scale bar, 1 μm. The video file for Fig. 2C is available under ‘Supporting Information’.
Fig 4.
Remodelling of mitochondrial network in human pancreatic cells. Dynamics of network fragmentation (A) and fusion of two network branches (B) is shown (arrows). Note that similar time (∼100 sec.) is required for both fission and fusion processes (cf. Fig. 2C). Scale bar, 1 μm.
Mitochondrial motions
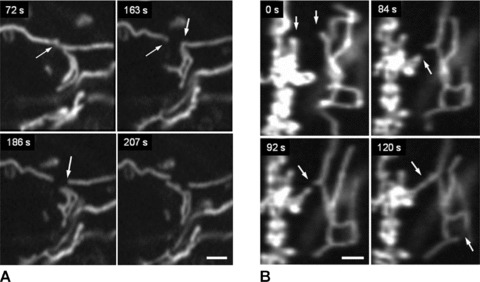
In addition to permanent mitochondrial fission/fusion, highly dynamic multidirectional movement of TMRM stained mitochondria was observed in human pancreatic cells. While some mitochondria were rather fixed in their position, others were in fast and independent movement (frequently changing their direction) or changed their state between movement and immobility. Figure 5 demonstrates an example of displacement of one small ring-shaped mitochondrion (arrow) with a velocity of ∼0.2 μm/sec. and, in the same time, another similar size and shape mitochon drion (asterisk) which is fixed over the entire observation time. Figure 6 shows the combination of small amplitude fluctuations in mitochondrial network, long-distance (∼20 μm) occasional translocation of single mitochondrion (circles, dotted arrows) at a velocity comparable to the motility of mitochondria in human fibroblasts [36], together with fast ‘back and forth’ fluctuating branching (arrows) in mitochondrial network (see supplementary video file: Kuz 3 Branching). Moreover, we observed also new unusual types of mitochondrial shape and dynamics. In human pancreatic cells mitochondria may rapidly form structures such as separated rings (Fig. 5), rings or loops at the end (Figs 7 and 8) or in the middle of mitochondrial filament (Fig. 5). The internal loops can be created by the fusion of two parallel threads located close to each other, and a division of such a structure from both sides may lead to formation of small mitochondrial rings. The dynamic of the formation of mitochondrial rings at the end of a filament is shown in Fig. 7 (as well as their opening, see dashed circle). It can be seen that formation of such ring structures takes again about 100 sec. and can be in combination with mitochondrial branch creation/extension (Fig. 8).
Fig 5.
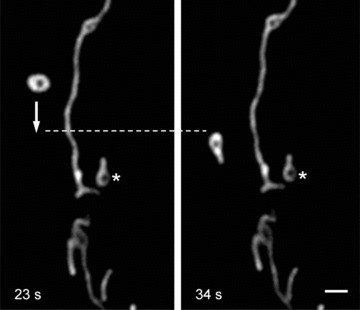
Mitochondrial dynamic motion in human pancreatic cells. Rapid intracellular displacement (move and stop) of individual mitochondrion (arrow) with velocity of ∼0.25 μm/sec. can be seen. Note that another similar size and shape mitochondrion (asterisk) is fixed in its position. Scale bar, 1 μm. The video file for this figure is available under ‘Supporting Information’.
Fig 6.
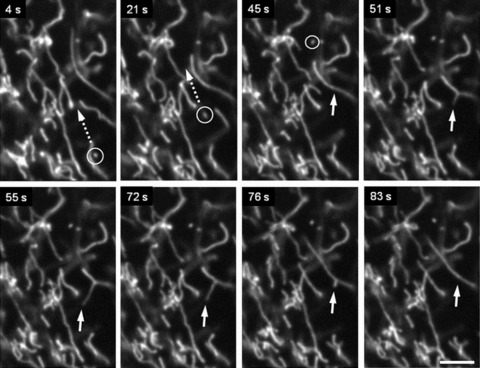
Complex pattern of mitochondrial motility in human pancreatic cells. Combination of (i) small amplitude fluctuations, (ii) long-distance (∼20 μm) occasional translocation of single mitochondrion (circles, dotted arrows) and (iii) fast ‘back and forth’ branching in mitochondrial network (arrows) is shown. Scale bar, 5 μm. The video file for this figure is available under ‘Supporting Information’.
Fig 7.
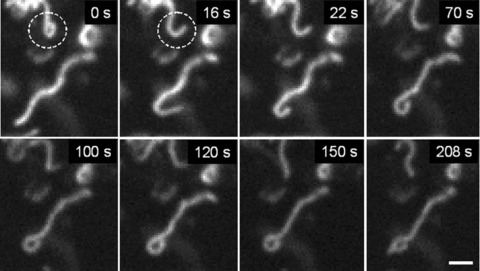
Dynamic ring formation at the end of mitochondrial filament. Note that this process can be reversible (dashed circle). Scale bar, 1 μm.
Fig 8.
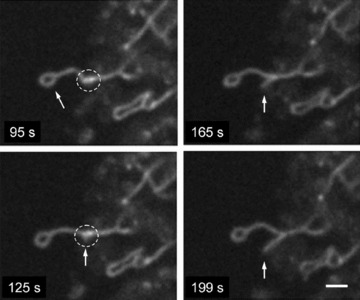
Combination of ring at the end of mitochondrial filament with fast branch formation. Note that more intense signal is observed in the sites of mitochondrial branch formation (dashed circle). Scale bar, 1 μm.
Formation of mitochondrial knots
Besides the very interesting phenomenon of high amplitude forward and backward branching of mitochondrial network found in our study (Fig. 6), the most exiting observation was an occasional formation of highly dynamic mitochondrial ‘knots’ created from one (Fig. 9) (for more detailed information see supplementary video-movie file: Kuz 4 Knot) or even more (Fig. 10) mitochondrial threads. This type of mitochondrial behaviour in pancreatic cells includes very complicated multidirectional movements of opposite ends of individual mitochondria for ‘knot’ formation (arrows in Fig. 9 indicate the direction of the filament movement) and represents to our knowledge the most complex type of mitochondrial dynamic motion in living cells.
Fig 9.
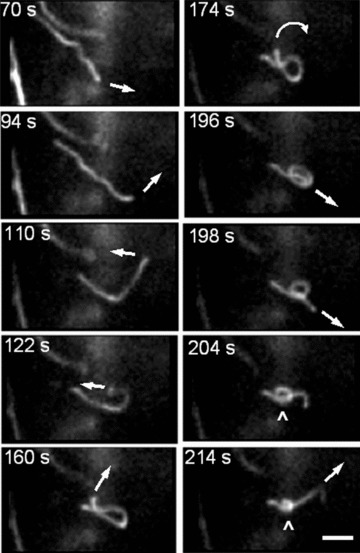
New level of the complexity of mitochondrial dynamics in human pancreatic cells. Dynamic formation of mitochondrial ‘knot’ from single mitochondrial filament. Complex multidirectional movement of mitochondrial thread is indicated by arrows. Scale bar, 1 μm. The video file for this figure is available under ‘Supporting Information’.
Fig 10.
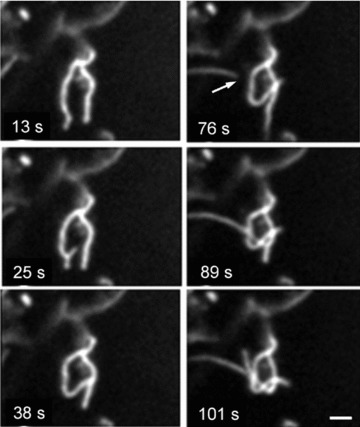
Formation of mitochondrial dynamic ‘knot’ from separated mitochondrial threads (arrow). Scale bar, 1 μm. The video file for this figure is available under ‘Supporting Information’.
Discussion
Recent studies have implicated an important role of mitochondrial dynamics in living cells and ability of mitochondrial network for rapid remodelling, suggesting important roles of multiple interactions of mitochondria with other cellular structures, and demonstrating also tissue-specific patterns of their intracellular organization [20, 23–28, 37]. Mitochondrial morphology is different in various cell types, and may vary from small spheres, short rod-like shape, spaghetti-like shape and long branched tubules. In some cell types mitochondria may be single evenly distributed within the entire cell [8], whereas other cells may display tightly clustered organelles [12, 33] or complex and dynamic networks of interconnected mitochondria with very long filaments (Fig. 1, right panel). This is consistent with the idea of several levels of mitochondrial networks [20], that may serve as an efficient system to deliver energy between different regions of the cell and such electrical connections across a mitochondrial reticulum may be crucial for mitochondrial synchronization and cellular physiology [11, 12]. In addition, a continuous mitochondrial network may provide a mechanism for Ca2+ tunnelling. Our imaging study of the morphology and distribution of the mitochondria in primary human pancreatic cells under standard cell culture conditions showed the presence of both branched tubular network and short separated mitochondria (see Fig. 2, 3B and 5) which may constitute 2–5% of the total mitochondrial population. In most of the cells, long perinuclear threads of mitochondria or interconnected network surrounding nucleus (N) can be clearly seen (Fig. 2, left panel), which is rather usual also for some other cell types [7, 8, 33, 38]. It has been suggested that such an organization may help to generate energy (ATP) in vicinity of the nucleus [39], providing basis for the integrated phosphotransfer network and metabolic channelling between mitochondria and nuclei [24], playing thus an important role in the mechanisms for nuclear import and for regulating a variety of other nuclear functions.
Fission/fusion
Previous studies have demonstrated also that in many cell types mitochondria may be very dynamic, rapidly changing their morphology by fission–fusion [7], depending on metabolic state [20, 40–42]. Fusion and fission continuously change mitochondrial shape under physiological (e.g. cellular division) and pathophysiological conditions [18, 40, 41], participating also in the control of apoptosis [43]. The precise balance between these two processes might therefore play a key role in mitochondrial and cellular function. In our study, a great variety of mitochondrial shapes (Fig. 2) and network remodelling (Fig. 3) thought to be a result of fast and continuous fission and fusion (Figs 3C and 4). For example Fig. 4A shows network fragmentation and Fig. 4B opposite process of fusion of two separated networks, demonstrating also that similar time may be required for both events. These results suggest that complex intracellular organization of human pancreas mitochondria is dynamically regulated by continuous fusion and fission. Notably, the time scale of mitochondrial dynamics in human pancreatic cells (Figs 3C and 4) is comparable with recently reported data on mitochondrial fission/fusion in INS1 and COS7 cells, describing ‘kiss and run’ mitochondrial behaviour with brief fusion (during about 100 sec.) followed by fission [44, 45].
Interestingly, a more intense TMRM signal can be observed in sites of mitochondrial branch formation (e.g. Fig. 8, dashed circle). This is consistent with the phenomenon of heterogeneous distribution of membrane potential along single mitochondrial filament described in the recent review of Benard and Rossignol [20] and may reflect region-specific increase in membrane potential (probably necessary for fusion or branch formation). It is thus possible that remodelling of mitochondrial network may, at least in part, be modulated by membrane potential, in line with its role also for mitochondrial movement [46].
Mitochondrial motility
Mitochondrial movement within the cell thought to supply more ATP at sites of high ATP demands [14]. In addition, mitochondria may be recruited to provide Ca2+-buffering capacity for local calcium regulations [14]. Many diseases, in particular neurological diseases, are associated with mutations in proteins that control mitochondrial dynamics [16–19, 40, 41]. The most common types of mitochondrial dynamic motion observed in our study were: (i) small oscillatory movements in the mitochondrial network, (ii) long-distance intracellular translocation of separated mitochondria (Figs 5 and 6) and (iii) larger movements in network including filament extension, retraction, fast branching (Fig. 8) as well as combinations of these actions (Fig. 6), demonstrating thus a heterogeneity of mitochondrial dynamics. For example it can be repeatedly seen that, whereas one mitochondrion rapidly move and stop (arrow), other similar size/shape organelle (*) is fixed during entire period of observation (Fig. 5). Similar to morphological alterations (fusion), intracellular movement of mitochondria can be significantly inhibited by mitochondrial membrane depolarization or under conditions of inhibited respiration and ATP synthesis [46].
Rapid branching of mitochondrial network, unusual shapes of mitochondria
Interestingly, branches can be rapidly and often formed from the main mitochondrial trunk (arrows in Figs. 6 and 8), or by fast extensive filament elongation, extending its length up to 10 folds. These processes occur without visible changes in thickness of tubular mitochondria (filaments were not observed becoming thinner upon their extensive elongation). It seems also that processes of branch formation/elongation observed in pancreas mitochondria are usually faster than fission/fusion (e.g. it may take only ∼10 sec., see Fig. 6). This fast formation, elongation and/or ‘backward and forward’ oscillations of mitochondrial branches are thought to provide a mechanism for searching potentially important connections of mitochondria with each other or with other cellular structures like ER and elements of cytoskeleton, which could be crucial for organization of the optimal network [20, 23–27, 37]. Similarly, unusual shapes of mitochondria like rings and loops at the end of mitochondrial filament frequently seen in pancreatic cells (Figs. 7 and 8) and in some other studies [20] could serve to envelop small vesicles and structures (e.g. fragmented Golgi), and this wrapping can be reversible (cf. Fig. 7, dashed circle).
Knots formation
Anterograde and retrograde mitochondrial movements along microtubules and actin filaments are known for various cell types (neurons, budding yeast, etc.) [14, 15, 40, 41, 47]. In human pancreatic cells however we occasionally observed more complex mitochondrial movements. In these cells, for the first time, we were able to detect astonishing formations of knots created from one or more mitochondrial threads as a result of complex movements (forward/backward) of opposite ends of individual filaments (Figs. 9 and 10). This first evidence for the very complicated dynamic behaviour of mitochondria indicates that our knowledge regarding general mechanisms of mitochondrial intracellular motility in mammalian cells is far from its full understanding and that the patterns of mitochondrial dynamics in mammalian cells may be much more complex as it has been considered. More attempts, including simultaneous imaging of other intracellular structures like cytoskeleton, Golgi, etc., will be required in the future to identify and characterize complex mitochondrial interactions. It is also interesting and unclear if some elements of the complex mitochondrial behaviour are a feature of only human pancreatic cells or represent a rather general phenomenon for cells with similar mitochondrial morphology and arrangement.
In summary, we report here new patterns and new level of complexity of highly dynamic morphological changes and intracellular movements of mitochondria in living human pancreatic cells. Our data demonstrate also heterogeneity of mitochondria under normal growth conditions, which at least in part may be caused by their complex dynamics.
Acknowledgments
This work was supported in part by the research grant from the Austrian Cancer Society/Tyrol. We also gratefully acknowledge support provided by the Allianz Elementar Versicherungs-AG.
References
- 1.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–60. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 3.Newmeyer DD, Ferguson-Miller S. Mitochondria: Releasing power for life and unleashing the machineries of death. Cell. 2003;112:873. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 4.Kroemer G, Reed JC. Mitochondrial control of cell death. Nat Med. 2000;6:513–9. doi: 10.1038/74994. [DOI] [PubMed] [Google Scholar]
- 5.Toescu EC, Verkhratsky A. Ca2+ and mitochondria as substrates for deficits in synaptic plasticity in normal brain ageing. J Cell Mol Med. 2004;8:181–90. doi: 10.1111/j.1582-4934.2004.tb00273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reeve JLV, Szegezdi E, Logue SE, et al. Distinct mechanisms of cardiomyocyte apoptosis induced by doxorubicin and hypoxia converge on mitochondria and are inhibited by Bcl-X-L. J Cell Mol Med. 2007;11:509–20. doi: 10.1111/j.1582-4934.2007.00042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- 8.Collins TJ, Berridge MJ, Lipp P, et al. Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 2002;21:1616–27. doi: 10.1093/emboj/21.7.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egner A, Jakobs S, Hell SW. Fast 100-nm resolution three-dimensional microscope reveals structural plasticity of mitochon dria in live yeast. Proc Natl Acad Sci USA. 2002;99:3370–5. doi: 10.1073/pnas.052545099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fehrenbacher KL, Yang HC, Gay AC, et al. Live cell imaging of mitochondrial movement along actin cables in budding yeast. Curr Biol. 2004;14:1996–2004. doi: 10.1016/j.cub.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 11.De Giorgi F, Lartigue L, Ichas F. Electrical coupling and plasticity of the mitochondrial network. Cell Calcium. 2000;28:365–70. doi: 10.1054/ceca.2000.0177. [DOI] [PubMed] [Google Scholar]
- 12.Skulachev VP. Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends Biochem Sci. 2001;26:23–9. doi: 10.1016/s0968-0004(00)01735-7. [DOI] [PubMed] [Google Scholar]
- 13.Plecitá-Hlavatá L, Lessard M, Santorová J, et al. Mitochondrial oxidative phosphorylation and energetic status are reflected by morphology of mitochondrial network in INS-1E and HEP-G2 cells viewed by 4Pi microscopy. Biochim Biophys Acta. 2008;1777:834–46. doi: 10.1016/j.bbabio.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Hollenbeck PJ, Saxton WM. The axonal transport of mitochondria. J Cell Sci. 2005;118:5411–9. doi: 10.1242/jcs.02745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J Cell Biol. 1995;131:1315–26. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delettre C, Lenaers G, Griffoin JM, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–10. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 17.Olichon A, Baricault L, Gas N, et al. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J Biol Chem. 2003;278:7743–6. doi: 10.1074/jbc.C200677200. [DOI] [PubMed] [Google Scholar]
- 18.Olichon A, Guillou E, Delettre C, et al. Mitochondrial dynamics and disease, OPA1. Biochim Biophys Acta. 2006;1763:500–9. doi: 10.1016/j.bbamcr.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Zuchner S, Mersiyanova IV, Muglia M, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–51. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 20.Benard G, Rossignol R. Ultrastructure of the mitochondrion and its bearing on function and bioenergetics. Antioxid Redox Signal. 2008;10:1313–42. doi: 10.1089/ars.2007.2000. [DOI] [PubMed] [Google Scholar]
- 21.Maassen JA, ‘t Hart LM, Van EE, et al. Mitochondrial diabetes: molecular mechanisms and clinical presentation. Diabetes. 2004;53:S103–9. doi: 10.2337/diabetes.53.2007.s103. [DOI] [PubMed] [Google Scholar]
- 22.Maechler P, Wollheim CB. Mitochondrial function in normal and diabetic beta-cells. Nature. 2001;414:807–12. doi: 10.1038/414807a. [DOI] [PubMed] [Google Scholar]
- 23.Anesti V, Scorrano L. The relationship between mitochondrial shape and function and the cytoskeleton. Biochim Biophys Acta. 2006;1757:692–9. doi: 10.1016/j.bbabio.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Dzeja PP, Bortolon R, Perez-Terzic C, et al. Energetic communication between mitochondria and nucleus directed by catalyzed phosphotransfer. Proc Natl Acad Sci USA. 2002;99:10156–61. doi: 10.1073/pnas.152259999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaasik A, Veksler V, Boehm E, et al. Energetic crosstalk between organelles: architectural integration of energy production and utilization. Circ Res. 2001;89:153–9. doi: 10.1161/hh1401.093440. [DOI] [PubMed] [Google Scholar]
- 26.Milner DJ, Mavroidis M, Weisleder N, et al. Desmin cytoskeleton linked to muscle mitochondrial distribution and respiratory function. J Cell Biol. 2000;150:1283–98. doi: 10.1083/jcb.150.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizzuto R, Pinton P, Carrington W, et al. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–6. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 28.Seppet EK, Kaambre T, Sikk P, et al. Functional complexes of mitochondria with Ca,MgATPases of myofibrils and sarcoplasmic reticulum in muscle cells. Biochim Biophys Acta. 2001;1504:379–95. doi: 10.1016/s0005-2728(00)00269-3. [DOI] [PubMed] [Google Scholar]
- 29.Ricordi C, Lacy PE, Finke EH, et al. Automated-method for isolation of human pancreatic-islets. Diabetes. 1988;37:413–20. doi: 10.2337/diab.37.4.413. [DOI] [PubMed] [Google Scholar]
- 30.Hermann M, Pirkebner D, Draxl A, et al. “Real-time” assessment of human islet preparations with confocal live cell imaging. Transplant Proc. 2005;37:3409–11. doi: 10.1016/j.transproceed.2005.09.076. [DOI] [PubMed] [Google Scholar]
- 31.Hermann M, Margreiter R, Hengster P. Molecular and cellular key players in human islet transplantation. J Cell Mol Med. 2007;11:398–415. doi: 10.1111/j.1582-4934.2007.00055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuznetsov AV, Usson Y, Leverve X, et al. Subcellular heterogeneity of mitochondrial function and dysfunction: evidence obtained by confocal imaging. Mol Cell Biochem. 2004;256–257:359–65. doi: 10.1023/b:mcbi.0000009881.01943.68. [DOI] [PubMed] [Google Scholar]
- 33.Kuznetsov AV, Troppmair J, Sucher R, et al. Mitochondrial subpopulations and heterogeneity revealed by confocal imaging: possible physiological role? Biochim Biophys Acta. 2006;1757:686–91. doi: 10.1016/j.bbabio.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Scaduto RC, Grotyohann LW. Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys J. 1999;76:469–77. doi: 10.1016/S0006-3495(99)77214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buckman JF, Reynolds IJ. Spontaneous changes in mitochondrial membrane potential in cultured neurons. J Neurosci. 2001;21:5054–65. doi: 10.1523/JNEUROSCI.21-14-05054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koopman WJ, Distelmaier F, Hink MA, et al. Inherited complex I deficiency is associated with faster protein diffusion in the matrix of moving mitochondria. Am J Physiol Cell Physiol. 2008;294:C1124–32. doi: 10.1152/ajpcell.00079.2008. [DOI] [PubMed] [Google Scholar]
- 37.Saks VA, Kaambre T, Sikk P, et al. Intracellular energetic units in red muscle cells. Biochem J. 2001;356:643–57. doi: 10.1042/0264-6021:3560643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bereiter-Hahn J. Behavior of mitochondria in the living cell. Int Rev Cytol. 1990;122:1–63. doi: 10.1016/s0074-7696(08)61205-x. [DOI] [PubMed] [Google Scholar]
- 39.Bruce JI, Giovannucci DR, Blinder G, et al. Modulation of [Ca2+]i signaling dynamics and metabolism by perinuclear mitochondria in mouse parotid acinar cells. J Biol Chem. 2004;279:12909–17. doi: 10.1074/jbc.M309070200. [DOI] [PubMed] [Google Scholar]
- 40.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–52. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 41.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–9. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 42.Rossignol R, Gilkerson R, Aggeler R, et al. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–93. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- 43.Perfettini JL, Roumier T, Kroemer G. Mitochondrial fusion and fission in the control of apoptosis. Trends Cell Biol. 2005;15:179–83. doi: 10.1016/j.tcb.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008;1777:1092–7. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Twig G, Elorza A, Molina AJ, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–46. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller KE, Sheetz MP. Axonal mitochondrial transport and potential are correlated. J Cell Sci. 2004;117:2791–804. doi: 10.1242/jcs.01130. [DOI] [PubMed] [Google Scholar]
- 47.Yaffe MP. Dynamic mitochondria. Nat Cell Biol. 1999;1:E149–50. doi: 10.1038/14101. [DOI] [PubMed] [Google Scholar]


