Abstract
Despite recent attempts at sub-categorization, including gene expression profiling into prognostically different groups of “germinal center B-cell type” and “activated B-cell type”, diffuse large B-cell lymphoma (DLBCL) remains a biologically heterogenous tumor with no clear prognostic biomarkers to guide therapy.
Whole genome, high resolution array comparative genomic hybridization (aCGH) was performed on 4 cases of chemoresistant DLBCL and 4 cases of chemo-responsive DLBCL to identify genetic differences which may correlate with response to R-CHOP therapy.
Array CGH analysis identified 7 DNA copy number alteration (CNA) regions exclusive to the chemoresistant group, consisting of amplifications at 1p36.13, 1q42.3, 3p21.31, 7q11.23, and 16p13.3, and loss at 9p21.3, and 14p21.31. Copy number loss of the tumor suppressor genes CDKN2A (p16, p14) and CDKN2B (p15) at 9p21.3 was validated by fluorescence in situ hybridization and immunohistochemistry as independent techniques. In the chemo-sensitive group, 12 CNAs were detected consisting of segment gains on 1p36.11, 1p36.22, 2q11.2, 8q24.3, 12p13.33, and 22q13.2 and segment loss on 6p21.32. RUNX3, a tumor suppressor gene located on 1p36.11 and MTHFR, which encodes for the enzyme methylenetetrahydrofolate reductase, located on 1p36.22 are the only known genes in this group associated with lymphoma.
Whole genome aCGH analysis has detected copy number alterations exclusive to either chemoresistant or chemo-responsive DLBCL that may represent consistent clonal changes predictive for prognosis and outcome of chemotherapy.
Keywords: aCGH, DLBCL, copy number alterations, chemo-sensitive, chemoresponsive
Introduction
Of all non-Hodgkin lymphomas, diffuse large B-cell lymphoma (DLBCL) is the most common, accounting for ~30-40% cases [1,2]. It represents a clinically, morphologically, and genetically heterogenous group of tumors, of which only a subset falls into the more specific categories outlined by the recently revised WHO Classification of Tumors of Hematopoietic and Lymphoid Tissues [1]. The largest proportion of the DLBCL, namely 20-30%, still continues to be defined only by their nuclear size and falls into the indistinct category of DLBCL, not otherwise specified [1,3].
Gene expression profiling performed on untreated, de novo DLBCL recognized two main, prognostically different groups: the “germinal center B-cell like (GCB-like) type”, expressing a gene signature resembling normal germinal center B cells and associated with better event-free and overall survival, and the “activated B-cell like (ABC-like) type” with a gene signature reminiscent of activated peripheral blood B-cells and worse prognosis [4-6]. Despite the overall better survival for patients with the GCB-like type, responsiveness to chemotherapy in this group remains inconsistent, with a significant number of patients succumbing to the disease early in the disease course [7]. Likewise, long-term remissions of greater than five years after treatment have been observed in patients with the ABC-like type DLBCL. Overall, although most patients with a diagnosis of DLBCL respond initially to chemotherapy, only ~40% achieve durable remission [7].
In light of these problems, classification based on predicted response to chemotherapy treatment could be beneficial. Currently, the International Prognostic Index (IPI) is the prognostic score most commonly used to predict outcome in DLBCL [8]. It incorporates five clinical factors and separates patients into four prognostic groups, with disease free survival ranging from 73 to 26%. However, even within a given clinical prognostic group, the rate of disease progression and response to therapy is heterogeneous. This variable clinical course within the different subgroups has prompted research direction towards identifying underlying mechanisms for either resistance or sensitivity to chemotherapy-induced cell death of DLBCL. For instance, studies have shown that the cell death-inducing effect of chemotherapy is dependent on proper activation of the apoptosis cascade [7,9-11]. Alteration in expression levels of key apoptotic proteins is believed to cause inhibition of the apoptosis cascade and consequently may influence sensitivity to drug-induced cytotoxicity [10]. While previous studies have concentrated on selective genetic profiling of mostly apoptosis genes, our study utilized high-resolution array comparative genomic in situ hybridization for global detection of copy number alterations (CNA) exclusive to either chemoresistant or chemo-responsive DLBCL. This discovery set identifies potential clinically-relevant genetic determinants of chemotherapy responsiveness.
Material and Methods
Patients and Tissue Collection
The study protocol was reviewed and approved by the Institutional Review Board of Washington University in St. Louis, Missouri. For this retrospective cohort study, DLBCL specimens collected from 2004-2007 were identified. Specimens were frozen cell pellets stored after flow cytometry was performed on the diagnostic tumor biopsy. Specimens were included in this analysis if they met the following criteria: specimen was collected at diagnosis, prior to any anti-lymphoma therapy; patient was treated with R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone); and greater than 1 year of clinical follow-up was available. DLBCL specimens that met the inclusion criteria were divided into two groups based on the patients’ outcomes. “Chemoresistant” specimens were from patients who failed to achieve a complete response to R-CHOP or who achieved an initial response but relapsed within 1 year of treatment completion, and “chemo-responsive” specimens were from patients who maintained a complete response to R-CHOP for ≥1 year. For each specimen, the percentage of lymphoma cells was calculated from the percentage of CD19+ or CD20+ B-cells reported on flow cytometry analysis, and the subtype (activated B-cell vs. germinal center B-cell) was determined from the CD10, BCL-6, and MUM-1 expression pattern performed by routine immunohistochemistry on the corresponding biopsy specimen at diagnosis for prognostic purposes [12].
DNA Extraction and Array Comparative Genomic Hybridization
Total DNA was extracted with the Gentra Puregene DNA Isolation Kit (Qiagen Inc, Valencia, CA). Isolated DNA of samples was shipped to Roche NimbleGen (Reykjavik, Iceland) for Full Service Workflow including sample labeling, hybridization and data analysis according to Roche NimbleGen protocols. Labeled samples were hybridized to the ultra-high resolution (~5-10 kb resolution) CGH arrays with 2.1 million probes and 1,169 bp median probe spacing (Human CGH 2.1M Whole-Genome Tiling v2.0D) for comprehensive analysis of DNA copy number variation. Normal pooled genomic DNA provided by Roche NimbelGen was used as the reference genome.
Data Analysis of Array Comparative Genomic Hybridization
Normalization and calculation of log2-ratios of the probe signal intensities (Cy3/Cy5) were identified using the segMNT algorithm implemented in NimbleScan software. Raw and processed data including segment start and end positions were provided by Roche NimbleGen to the authors for further analysis. Changes in segment copy number were determined by filtered log2-ratios > 0.2 (copy number gain) or < −0.2 (copy number loss), relative to reference DNA. The cases were screened for DNA copy number variants present in 3 or 4 of the chemoresistant DLBCL samples but not in the chemo-responsive DLBCL samples and vice versa, without considering content overlapping segments. Identification of copy number variation genes that overlap published copy number variant regions was by comparison to the Database of Genomic Variants (http://projects.tcag.ca/variation/). The physical positions of genes overlapping detected copy number variation segments were based on the Human hg18 Assembly (NCBI Build 36.1). Published studies used for comparison were limited to only those studies with at least 10 biological samples utilizing Affymetrix, Agilent or Illumina CGH microarrays with 500K or greater resolution.
Gene Network Analysis and Pathway Overrepresentation Analysis of Chemoresistant CNA Genes
Gene network analysis of CNA genes was performed using the web application GRANITE (Shaik 2010 submitted) Entrez GeneIds for unfiltered chemoresistant (n=37) CNA genes were submitted to GRANITE using the default settings; the output was downloaded and separately visualized in Cytoscape (www.cytoscape.org). DAVID was applied to find biological ontologies and pathways enriched in CNAs. Entrez GeneIDs for unfiltered chemoresistant (n=37) and chemoresponsive (n=87) CNA genes were submitted and analyzed using the default settings on the DAVID website. Results were filtered for Bonferroni-corrected p-value < 0.05 and redundant biological themes with identical list hit gene members were removed leaving the Pathway / Ontology Term with the lowest p-value.
Fluorescence in Situ Hybridization for CDKN2A
FISH was performed on all eight paraffin embedded tissue sections utilizing the dual color Vysis CDKN2A/CEP9 probe kit (Vysis Inc., Downers Grove, IL). According to the manufacturer, the CDKN2A probe is labeled with SpectrumOrange and spans approximately 222 kb containing a number of genetic loci including D9S1749, DS1747, p16 (INK4B), p14 (ARF), D9S1748, p15 (INK4B), and D9S1752. Assay and scoring were strictly performed according to manufacturer’s protocol. In a normal sample, the expected pattern for a nucleus hybridized with this kit is a two orange/two green signal pattern. If a deletion at the 190kb region covered by the CDKN2A probe occurs on one chromosome 9 homolog and both centromeres from chromosome 9 are retained, the one orange/two green signal pattern is expected. A homozygous deletion at the 190 kb region covered by the CDKN2A probe would lead to green only signal pattern.
Histology and Immunohistochemistry
The tissues of the four chemoresistant DLBCL cases were routinely fixed in 10% buffered formalin, embedded in paraffin, and serially sectioned into 4-μm-thick sections for routine hematoxylin and eosin staining and immunohistochemistry. Immunohistochemistry for p16 (CINtec® Histology V-Kit, MTM Laboratories, Inc. Westborough, MA) was performed utilizing an autostainer (Benchmark XT System, Ventana Medical Systems, Tucson, AZ), as per the manufacturer’s instructions. Positive and negative controls were run simultaneously.
Results
Patients
Thirty-five DLBCL specimens were identified, and 13 met the inclusion criteria. A discovery set of 4 chemoresistant and 4 chemo-responsive samples were selected for further analysis based on specimen quality and clearly divergent clinical outcomes (Table 1).
Table 1.
Patient and specimen characteristics
| Patient ID |
Age (yrs) |
Gender | IPI | Stage | Specimen site |
% NHL in specimen |
DLBCL subtype | Outcome |
|---|---|---|---|---|---|---|---|---|
| 3325 | 36 | F | 2 | IV | spleen | 10 | ABC | CR, OS ≥ 53 mo. |
| 3329 | 54 | F | 0 | I | spleen | 15 | GCB | CR, OS ≥ 37 mo. |
| 3330 | 79 | M | 1 | I | lymph node | 64 | GCB | CR, OS ≥ 37 mo. |
| 3333 | 58 | F | 1 | III | orbital mass | 49 | ABC | CR, OS ≥ 33 mo. |
| 3326 | 64 | M | 4 | IV | lymph node | 76 | ABC | refractory disease, OS 5 mo. |
| 3328 | 68 | M | 4 | IV | pleural fluid | 68 | GCB | refractory disease, OS 1 mo. |
| 3331 | 65 | F | 3 | IV | lymph node | 30 | ABC | relapse at 1 mo., OS 9 mo. |
| 3332 | 44 | F | 3 | IV | lymph node | 56 | GCB | relapse at 6 mo., OS ≥ 34 mo. |
F, female; M, male; IPI, International prognostic index; ABC, activated B-cell subtype; GCB, germinal center B-cell subtype; CR, complete remission; OS, overall survival
Unfiltered and Filtered Differences in DNA Copy Number Alterations between Chemoresistant and Chemo-Responsive Diffuse Large B-Cell Lymphomas
Without removing the genes that overlap published copy number variant regions, array CGH analysis identified 7 DNA CNA regions exclusive to the chemoresistant group, consisting of amplifications at 1p36.13, 1q42.3, 3p21.31, 7q11.23, and 16p13.3, and loss at 9p21.3, and 14p21.31 (Table 2). The only gene amplified in all 4 samples was CROCC, which encodes for the ciliary rootlet coiled-coil protein, located on 1p36.13. CROCC does not have a known association with lymphoma. Copy number gain of ABCA3 on 16p13.3, which encodes for ATP-binding cassette (ABC) transporter 3, was observed in 3 of 4 chemoresistant DLBCL.
Table 2.
Imbalances detected by array CGH in the chemoresistant and chemo-responsive diffuse large B-cell lymphomas.
| Chromosome band |
Position (Mb) | Size (Mb) |
Gain/Loss | Genes | |
|---|---|---|---|---|---|
|
Chemo-
resistant |
1p36.13 | 17.08-17.14 | 0.06 | G | CROCC |
| 1q42.3 | 232.91-233.26 | 0.35 | G | ||
| 3p21.31 | 50.13-50.66 | 0.53 | G |
C3orf18 ,C3orf45, CISH, CYP561D2,
GNAI2, GNAT1, HEMK1, HYAL1, IFRD2, MAPKAPK3, NAT6, RASSF1, RBM5 ,SEMA3F, SEMK1, SLC38A3, TMEM115, TUSC2, TUSC4, ZMYND10 |
|
| 7q11.23 | 76.41-76.48 | 0.07 | G | FDPSL2B, PMS2L2 | |
| 9p21.3 | 21.97-22.04 | 0.07 | L | CDKN2A, CDKN2B | |
| 14p21.31 | 19.27-19.47 | 0.20 | L |
OR11K2P, OR4H12P, OR4K2, OR4K3, OR4K4P,
OR4K5, OR4K6P, OR4M1, OR4N2, OR4Q3, ORN1P |
|
| 16p13.3 | 2.31-2.33 | 0.02 | G | ABCA3 | |
|
Chemo-
sensitive |
1p36.11 | 25.06-25.30 | 0.24 | G | RUNX3 |
| 1p36.22 | 10.60-10.73 | 0.13 | G | CASZ1, PEX14 | |
| 1p36.22 | 11.42-12.20 | 0.78 | G |
AGTRAP,C1orf167,C1orf187,CLCN6,FBXO44,
FBX02,FBXO6,KIAA2013,MFN2,NPPA,NPPB,PLOD1, PTCHD2,TNFRSF1B,MAD2L2,MTHFR,TNFRSF8 |
|
| 2q11.2 | 96.53-96.59 | 0.06 | G | ARID5A | |
| 6p21.31 | 32.56-32.71 | 0.14 | L | HLA-DRB1, HLA-DRB5, HLA-DRB6 | |
| 8q24.3 | 145.70-145.75 | 0.05 | G |
C8orf82, GPT, LRRC14, LRRC24, MFSD3,
PPP1R16A, RECQL4 |
|
| 8q24.3 | 142.40-142.51 | 0.11 | G | GPR20, PTP4A3 | |
| 8q24.3 | 144.16-144.20 | 0.04 | G | C8orf31, LY6E | |
| 8q24.3 | 144.96-145.75 | 0.79 | G |
ADCK5,BOP1,C8orf30A,C8orf82,CPSF1,CYC1,CYHR1,DGAT1,EXOSC4,FBXL6,
FOXH1,GPAA1,GPR172A,GPT,GRINA,HSF1,KIAA1875,KIFC2,LRRC14,LRRC24, MAF1,MFSD3,NFKBIL2,NRBP2,OPLAH,PARP10,PLEC1,PPP1R16A,PUF60, RECQL4,SCRIB,SCRT1,SCXB,SHARPIN,SLC39A4,SPATC1,VPS28 |
|
| 8q24.3 | 145.76-146.26 | 0.50 | G |
C8orf33,C8orf77,COMMD5,RPL8,TMED10P,
ZNF7,ZNF16,ZNF34,ZNF250,ZNF251,ZNF517 |
|
| 12q13.2 | 1.78-1.82 | 0.04 | G | CACNA2D4 | |
| 22q13.2 | 41.23-41.29 | 0.06 | G | RRP7A,SERHL,SERHL2 |
Bold genes, genes that remained after filtering out the genes that overlapped published copy number variant regions using the Database of Genomic Variants (http://projects.tcag.ca/variation/. Except for CROCC which was found in all 4 chemoresistant cases, all other genes were detected in 3 of the chemoresistant or chemo-responsive cases.
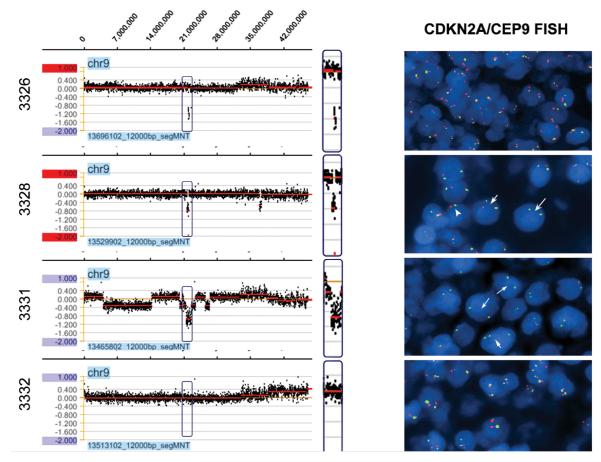
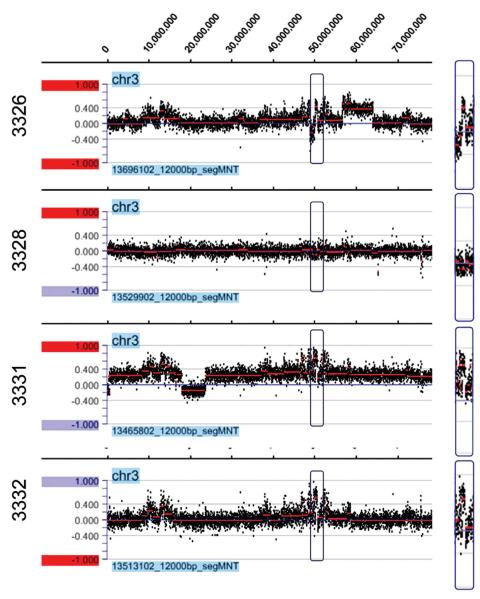
Copy number gain of MAPKAPK3 located on 3p21.31 and copy number loss of CDKN2A and CDKN2B located on 9p21.3 remained the only lymphoma related gene alterations in the chemoresistant group after removal of genes that overlapped published copy number variant regions. Figure 1A demonstrates a deletion region in chromosome 9 associated with CDKN2A and CDKN2B in three of the four cases and Figure 2 demonstrates an amplification region in chromosome 3 involving MAPKAPK3.
Figure 1.
a. Detection of CDKN2A and CDKN2B deletions at 9p21.3, 21.97-22.04 in three of four chemoresistant cases (3326, 3328, and 3331) using the NimbleGen Human CGH 2.1M Whole-Genome Tiling v2.0D Array. Cases 3326 and 3331 appear to have a homozygous deletion. Figure 1b. Representative CDKN2A/CEP9 FISH images of all chemoresistant DLBCL cases. Case 3326 reveals normal CDKN2A/CEP9 signals, likely due to a microdeletion (~130 kb) which was not detected by the probe. Case 3328 reveals hemizygous (short arrow) and, to a lesser extent, homozygous (long arrow) deletions, as well as monosomy 9 (arrowhead). Case 3331 is significant for a homozygous deletion in diploid (long arrows) and polysomy 9 (short arrow) tumor cells. Case 3332 reveals a normal signal pattern. All 4 cases in the chemo-responsive group showed a normal two orange/two green signal pattern (data not shown).
Figure 2.
Detection of MAPKAPK3 gains located on 3p21.31 in three of four chemoresistant cases (3326, 3331, and 3332) using the NimbleGen Human CGH 2.1M Whole-Genome Tiling v2.0D Array.
In the chemo-sensitive group, 12 unfiltered CNAs were detected consisting of segment gains on 1p36.11, 1p36.22, 2q11.2, 8q24.3, 12p13.33, and 22q13.2 and segment loss on 6p21.32 (Table 2). The majority of the genes that remained after genes were filtered out for overlapping published copy number variant regions have no known association with lymphoma, except for RUNX3, a tumor suppressor gene located on 1p36.11 and MTHFR, which encodes for the enzyme methylenetetrahydrofolate reductase, located on 1p36.22.
Gene Network Analysis and Pathway Overrepresentation Analysis of chemoresistant CNA Genes
Supplementary Figure 1 represents the network analysis of the chemoresistant copy number alteration genes showing the relationship between these genes. The interactive network map also includes “discovered” network genes (grey nodes) that are not part of the discovery set, but are known to interact with these genes. These types of discovered interactions, those with “discovered” network genes that do not show copy number variation but interact with CNV genes may be important.
Supplementary Table 1 represents the pathway overrepresentation of chemoresistant CNA genes. The most significant group was the tumor suppressor group including the NAT6, CDKN2A/B, TUSC4, RASSF1, and TUSC2 genes. The NAT6, TUSC4, RASSF1 and TUSC2 genes belong to the 3p21.3 tumor-suppressor gene cluster with deletions of this region leading to development of cancer. The transcript RASSF1A is one of the most frequently methylated genes so far described in human cancer.
Fluorescence in Situ Hybridization for CDKN2A
FISH hybridization signals were interpretable in all eight cases. All 4 patients in the chemo-responsive group showed a normal two orange/two green signal pattern supporting the normal aCGH pattern at this gene locus (data not shown). Abnormal hybridization signals were found in 2 of the 4 chemoresistant diffuse large B-cell lymphoma cases (Figure 1B). Case 3331 revealed a homozygous deletion from both diploid and polysomy 9 tumor cells, which resulted in two or three signals of green, only. Case 3328 revealed predominantly a hemizygous deletion signal pattern as well as monosomy 9 and, in a minor subset, a homozygous deletion pattern, with cells displaying single orange and two green signals (hemizygous), one orange and one green signal (monosomy 9), or two green signals only (homozygous). Case 3326, which showed a homozygous deletion by aCGH, revealed a normal two orange/two green signal pattern. However, the deletion in this case is a microdeletion of ~130 kb and likely was too small for detection by the larger FISH probe (~220 kb) used in this study.
Histology and Immunohistochemistry for p16
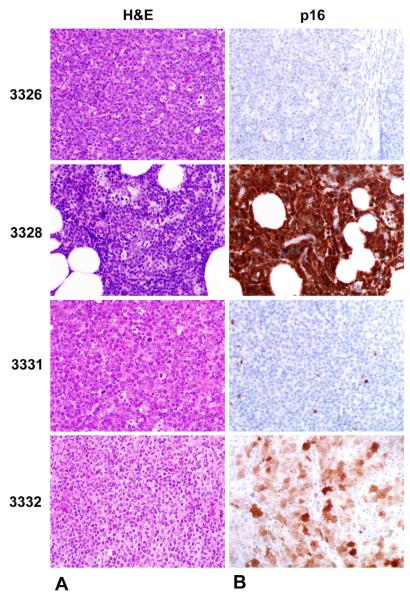
H&E stained sections of the four chemoresistant DLBCL revealed sheets of large neoplastic lymphoid cells (Figure 3A). Neoplastic cells of cases 3326 and 3331 (both homozygously deleted for p16) did not stain for p16 antibody, whereas 3328 (hemizygously deleted for p16) and 3332 (no p16 deletion) stained positively for this antibody in a nuclear and cytoplasmic pattern (Figure 3B).
Figure 3.
a. Representative H&E stained sections of thechemoresistant DLBCL cases revealing sheets of large neoplastic lymphoid cells (500X magnification). Figure 3b. Immunophenotypic expression patterns of p16 in all four chemoresistant DLBCL cases. Whereas cases 3326 and 3332 have the expected loss of p16 expression due to homozygous deletions, case 3328 shows strong nuclear and cytoplasmic p16 staining, likely due to incomplete CDKN2A silencing. Case 3332 reveals nuclear and cytoplasmic staining in tumor cells, an expected finding for cases that display no CDKN2A deletion (500X magnification). Supplementary Figure 2A shows the positive p16 control of an endocervical carcinoma showing strong staining for p16 (100X magnification). 2B reveals the negative control of case 3326 at 200X magnification. No staining cells are seen.
Discussion
In this study, array CGH successfully identified copy number losses and gains exclusive to either chemoresistant or chemo-responsive DLBCL. A striking finding in our analysis is the deletion of CDKN2A and CDKN2B at 9p21.3 in three of four chemoresistant cases. To our knowledge, our study is the first to suggest an association between deletion of CDKN2A and CDKN2B in DLBCL and response to standard R-CHOP immunochemotherapy.
CDKN2A/ p16INK4/p14ARF and CDKN2B/ p15INK4B are tumor suppressor genes that regulate cell cycle progression via the retinoblastoma (Rb) and the p53 apoptosis pathways, and deletions at 9p21 have been reported in many lymphoid malignancies, including acute lymphoblastic leukemia and other non-Hodgkin lymphomas [13,14]. Furthermore, deletions of p16 and p15 have been shown to be associated with inferior survival in follicular lymphoma and mantle cell lymphoma [14-16]. In two studies focused on diffuse large B-cell lymphoma, deletion of 9p21 was reported in 35% of 66 cases and in 16.4% of 64 cases, although both analyses may have missed cases with small deletions due to the use of a relatively low resolution BAC array CGH [17,18]. In neither study was deletion 9p21 associated with overall survival. However, Tagawa et al. did find an association of deletion 9p21 with lower survival in DLBCL in a subset of patients with the ABC-type [19]. Another group found consistent loss of 9p21 in cases of primary cutaneous large B-cell lymphoma, leg type, a particularly aggressive cutaneous B-cell lymphoma [20].
Defective p53/INK/ARF signaling due to deletions involving the 9p21 locus is also thought to represent one of the intrinsic pathway mechanisms of apoptosis resistance in diffuse large B-cell lymphoma. The p16INK4a/p14ARF locus is an important mediator of oncogene-dependant p53 induced apoptosis and disruption of this pathway was noted in ~16-28% of DLBCL [21-23].
In our study, the CDKN2A/B copy numbers detected by aCGH were validated by dual-color FISH analysis. All cases in the chemosensitive group showed a normal FISH signal pattern confirming that there was no copy number loss involving that gene in that group. Two of the three chemo-resistant CDKN2A/B deletions detected by aCGH revealed p16 deletions by FISH analysis. The third case displayed a microdeletion of less than 190 kb in size that was not detected by standard FISH technique. Although FISH is considered reliable and cost-effective, it may overlook microdeletions smaller than the probe size. S. Savola et al. performed 44K aCGH on 37 Ewing sarcoma cases in order to define sizes of 9p21.3 deletions and correlate these with standard FISH analysis using a commercial dual color CDKN2A probe (the same probe was used in our study). 14 of 37 tumor cases had CDKN2A deletions by aCGH of which 4 tumors had microdeletions of up to 185 kb. FISH analysis was only performed on the case with the 58 kb microdeletion and read as a false negative [24].
The presence of strong immunophenotypic p16 expression in the case with the predominantly hemizygous deletion pattern by FISH could be explained by p16 expression from the other allele. The retained allel could have no mutation or a mutation leading to over-expression of a non-functional p16 protein.
The detection of copy number gain of MAPKAPK3 at 3p21.31 in three of four chemoresistant samples may also be of clinical relevance. MAP kinase phosphorylation sites are embedded in the central portion of the BCL-6 proto-oncogene, and they are believed to play a critical role in degradation of BCL-6 [25]. BCL-6 is expressed in GC B-cells. Its expression characterizes the GCB-like type of DLBCL, while the ABC-like type is recognized by increased NFκB activation [26]. Perez-Rosado et al. have proposed that BCL-6 silencing through increased degradation leads to transcriptional up-regulation of multiple NFκB target genes, including MAPKAPK3, causing NFκB activation [25]. They argue that BCL-6 functions as a molecular switch controlling NFκB activation status in normal and malignant B-cells. ABC-like DLBCL are characterized by constitutive activation of the NFκB pathway resulting in high expression levels of many apoptosis inhibiting genes, consequently contributing to apoptosis resistance in these lymphomas [9,25-27]. Two of the three DLBCL with the amplification of MAPKAPK3 revealed BCL-6 negativity by immunohistochemistry (data not shown) and these two DLBCL corresponded to the ABC-like type.
Although copy number gain of the ABCA3 gene at 16p13.3 overlapped with known published copy number variant regions in the Database of Genomic Variants (http://projects.tcag.ca/variation/), several studies have implicated overexpression of this ATP-binding cassette transporter as a possible cause of drug resistance in numerous malignancies, including hematolymphoid malignancies by promoting efflux of drugs out of the tumor cell. There are an increasing number of reports addressing genetic polymorphisms, such as copy number polymorphism (CNP) or single nucleotide polymorphisms (SNP) in drug resistance. Steinbach et al. report in their microarray study that ABCA3 was expressed at three times higher levels in acute myeloid leukemia (AML) blasts of pediatric patients who did not achieve remission after induction therapy [28].
Methylenetetrahydrofolate reductase gene (MTHFR) polymorphisms have been implicated as another example of genetic polymorphisms involved in predisposition to malignancy. In our study, three of the four patients in the chemo-responsive group displayed copy number variations in the MTHFR gene located on 1p36.22. Matsuo et al. outlined in their study from 2004 that individuals in the healthy control group expressed a significantly higher percentage of MTHFR 677T and/or 1298C alleles compared to DLBCL samples [29]. Likewise, these alleles have also been implicated in altering susceptibility to adult acute lymphoblastic leukemia [30]. It would be of interest to genotype all of our samples for these different polymorphisms to establish a possible link between MTHFR polymorphisms and clinical outcome in patients with DLBCL.
Array CGH was not performed on purified tumor samples, so that background (germline) material may have diluted our ability to detect other genetic changes that are specific to the tumor. Deletions in particular may be difficult to detect under these circumstances, which may explain the fact that the vast majority of copy number changes found were amplifications. However, tumor percentage in the chemoresistant samples was at least 30%, with most samples containing more than 55% tumor cells, and the homozygous deletion of CDKN2A and CDKN2B at 9p21 was also detected in the sample with only 30% tumor cells.
In summary, our study represents a discovery set of several interesting genes that may represent consistent genetic determinants of chemotherapy responsiveness. We picked one promising gene, namely CDKN2A, and validated aCGH findings by utilizing fluorescence in situ hybridization and immunohistochemistry as independent techniques. The combination of these more cost-effective and less time consuming techniques to detect p16 deletions may offer guidance in prognosis and outcome of chemotherapy. We recognize that the small case series utilized in this aCGH study raises concern of low detection power and validation studies with a larger sample size are needed to confirm the predictive value of the genes identified in our discovery set.
Supplementary Material
Acknowledgements
This publication is supported by the Alvin J. Siteman Cancer Center Bioinformatics Core (NIH/NCI Grant # P30 CA91842) and the Center for Biomedical Informatics (NCRR Grant # UL1 RR024992), and the Barnes Jewish Foundation K Steinback Cancer Research Fund. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH. Furthermore, we would like to thank Diane H. Robirds, CLSp (CG) from the Barnes Jewish Hospital Clinical FISH Laboratory for the preparation and interpretation of the FISH slides, Kevin Selle, MT, HTL, MBA from the Barnes Jewish Hospital Clinical Histology Laboratory for the preparation of p16 immunohistochemical slides; and Nancy L. Bartlett, MD, for her support in study design and funding.
Funding Grant numbers and sources of support: NIH/NCI Grant # P30 CA91842, NCRR Grant # UL1 RR024992, Barnes Jewish Foundation K Steinback Cancer Research Fund
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Stein H, Warnke RA, Chan WC, Jaffe ES, Chan JKC, Gatter KC, Campo E. Diffuse large B-cell lymphoma, not otherwise specified. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. Tumours of hematopoietic and lymphoid tissues. IARC Press; Lyon, France: 2008. pp. 233–7. [Google Scholar]
- [2].De Paepe P, De Wolf-Peeters Diffuse large B-cell lymphomas: a heterogenous group of non-Hodgkin lymphomas comprising several distinct clinicopathological entities. Leukemia. 2007;21:37–43. doi: 10.1038/sj.leu.2404449. [DOI] [PubMed] [Google Scholar]
- [3].Gurbaxani S, Anastasi J, Hyek E. Diffuse large B-cell lymphoma - more than a diffuse collection of large B cells. Arch Pathol Lab Med. 2009;133:1121–34. doi: 10.5858/133.7.1121. [DOI] [PubMed] [Google Scholar]
- [4].Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tishirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- [5].Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltrane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, Lopez-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM. Lymphoma/Leukemia Molecular Profiling Project. The use of molecular profiling to predict survival after chemotherapy for diffuse large B-cell lymphoma. N Engl J Med. 2002;346:1937–47. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- [6].Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeck M, Angelo M, Reich M, Pinkus GS, Ray TS, Koval MA, Last KW, Norton A, Lister TA, Mesirov J, Neuberg DS, Lander ES, Aster JC, Golup TR. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- [7].Muris JJF, Ylstra B, Cillessen SAGM, Ossenkoppele GJ, Kluin-Nelemans JC, Eijk PP, Nota B, Tijssen M, de Boer WPH, van de Wiel M, van den Ijssel PRLA, Jansen P, de Bruin PC, van Krieken JHJM, Meijer GA, Meijer CJLM, Oudejans JJ. Profiling of apoptosis genes allows for clinical stratification of primary nodal diffuse large B-cell lymphoma. Br J Hematol. 2006;136:38–47. doi: 10.1111/j.1365-2141.2006.06375.x. [DOI] [PubMed] [Google Scholar]
- [8].The International Non-Hodgkin’s Lymphoma Prognostic Factors Project. A predictive model for aggressive non-Hodgkin’s lymphoma. N Engl J Med. 1993;329:987–94. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- [9].Muris JJF, Meijer CJLM, Ossenkoppele GJ, Vos W, Oudejans JJ. Apoptosis resistance and response to chemotherapy in primary nodal diffuse large B-cell lymphoma. Hematol Oncol. 2006;24:97–104. doi: 10.1002/hon.774. [DOI] [PubMed] [Google Scholar]
- [10].Georgakis GV, Li Y, Humphreys R, Andreeff M, O’Brian S, Younes M, Carbone A, Albert V, Younes A. Activity of selective fully human agonistic antibodies to the TRAIL death receptors TRAIL-R1 and TRAIL-R2 in primary and cultured lymphoma cells: induction of apoptosis and enhancement of doxorubicin- and bortezomib-induced cell death. Br J Hematol. 2005;130:501–10. doi: 10.1111/j.1365-2141.2005.05656.x. [DOI] [PubMed] [Google Scholar]
- [11].Gascoyne RD, Adomat SA, Krajewski S, Krajewska M, Horsman DE, Tolcher AW, O’Reilly SE, Hoskins P, Coldman AJ, Reed JC, Connors JM. Prognostic significance of BCL-2 protein expression and BCL-2 gene rearrangement in diffuse aggressive non-Hodgkin’s lymphoma. Blood. 1997;90:244–51. [PubMed] [Google Scholar]
- [12].Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- [13].Quesnel B, Preudhomme C, Philippe N, Vanrumbeke M, Dervite I, Lai JL, Bauters F, Wattel E, Fenaux P. p16 gene homozygous deletions in acute lymphoblastic leukemia. Blood. 1995;85:657–63. [PubMed] [Google Scholar]
- [14].Elenitoba-Johnson KSJ, Gascoyne RD, Lim MS, Chhanabai M, Jajje ES, Raffeld M. Homozygous deletions at chromosome 9p21 involving the p16 and p15 are associated with the histologic progression in follicle center lymphoma. Blood. 1998;91:4677–85. [PubMed] [Google Scholar]
- [15].Schwaenen C, Viardot A, Berger H, Barth TFE, Bentink S, Döhner H, Enz M, Feller AC, Hansmann M-L, Hummel M, Kestler HA, Klapper W, Kreuz M, Lenze D, Loeffler M, Möller P, Müller-Hermelink H-K, Ott G, Rosolowski M, Rosenwald A, Ruf S, Siebert R, Spang R, Stein H, Truemper L, Lichter P, Bentz M, Wessendorf S, for the Molecular Mechanisms in Malignant Lymphomas Network Project of the Deutsche Krebshilfe Microarray-based genomic profiling reveals novel genomic aberrations in follicular lymphoma which associate with patient survival and gene expression status. Genes, Chromosomes & Cancer. 2009;48:39–54. doi: 10.1002/gcc.20617. [DOI] [PubMed] [Google Scholar]
- [16].Jardin F, Picquenot J-M, Parmentier F, Ruminy P, Cornic M, Penther D, Bertrand P, Lanic H, Cassuto O, Humbrecht C, Lemasle E, Wautier A, Bastard C, Tilly H. Detection of gene copy number aberrations in mantle cell lymphoma by a single quantitative multiplex PCR assay: clinicopathological relevance and prognosis value. Br J Haematol. 2009;146:607–618. doi: 10.1111/j.1365-2141.2009.07791.x. [DOI] [PubMed] [Google Scholar]
- [17].Tagawa H, Tsuzuki S, Suzuki R, Karnan S, Ota A, Kameoka Y, Sugoro M, Matsuo K, Yamaguchi M, Okamoto M, Nakamura S, Seto M. Genome-wide array-based comparative genomic hybridization of diffuse large B-cell lymphoma: Comparison between CD5-positive and CD5-negative cases. Cancer Res. 2004;64:5948–5955. doi: 10.1158/0008-5472.CAN-03-4056. [DOI] [PubMed] [Google Scholar]
- [18].Chen W, Houldsworth J, Olshen AB, Nanjangud G, Chaganti S, Venkatraman ES, Halaas J, Teruya-Feldstein J, Zelenetz A, Chaganti RSK. Array comparative genomic hybridization reveals genomic copy number changes associated with outcome in diffuse large B-cell lymphomas. Blood. 2006;107:2477–2485. doi: 10.1182/blood-2005-07-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tagawa H, Suguro M, Tsuzuki S, Matsuo K, Karnan S, Ohshima K, Okamoto M, Morishima Y, Nakamura S, Seto M. Comparison of genome profiles for identification of distinct subgroups of diffuse large B-cell lymphoma. Blood. 2005;106:1770–1777. doi: 10.1182/blood-2005-02-0542. [DOI] [PubMed] [Google Scholar]
- [20].Belaud-Rotureau M-A, Marietta V, Vergier B, Mainhaguiet G, Turmo M, Idrissi Y, Ferrer J, Beylot-Barry M, Dubus P, Merlio J-P. Inactivation of p16 INK4a /CDKN2A gene may be a diagnostic feature of large B cell lymphoma leg type among cutaneous B cell lymphomas. Virchows Arch. 2008;452:607–620. doi: 10.1007/s00428-008-0593-x. [DOI] [PubMed] [Google Scholar]
- [21].Gombart AF, Morosetti R, Miller CW, Said JW, Koeffler HP. Deletions of the cyclin-dependent kinase inhibitor genes p16INK4A and p15INK4B in non-Hodgkin’s lymphomas. Blood. 1995;4:1534–39. [PubMed] [Google Scholar]
- [22].Pinyol M, Cobo F, Bea S, Jares S, Nayach I, Fernandez PL, Montserrat E, Cardesa A, Campo E. p16INK4A gene inactivation by deletions, mutations, and hypermethylation is associated with transformed and aggressive variants of non-Hodgkin’s lymphomas. Blood. 1998;8:2977–84. [PubMed] [Google Scholar]
- [23].Lowe SW, Sherr CJ. Tumor suppression by Inka-Arf: progress and puzzles. Curr Opin Genet Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- [24].Savola S, Nardi F, Scotland K, Picci P, Knuutila S. Microdeletions in 9p21.3 induce false negative results in CDKN2A FISH analysis of Ewing sarcoma. Cytogene Genome Res. 119:21–26. doi: 10.1159/000109614. [DOI] [PubMed] [Google Scholar]
- [25].Perez-Rosado A, Artiga MJ, Vargiu P, Sanchez-Aguilera, Alvarez-Barrientos A, Piris MA. BCL-6 represses NFκB activity in diffuse large B-cell lymphomas. J Pathol. 2008;214:498–507. doi: 10.1002/path.2279. [DOI] [PubMed] [Google Scholar]
- [26].Lam LT, wright G, Davis RE, Lenz G, Farinha P, Dang L, Chan JW, Rosenwald A, Gascoyne RD, Staudt LM. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-κB pathways in subtypes of diffuse large B-cell lymphoma. Blood. 2008;111(7):3701–13. doi: 10.1182/blood-2007-09-111948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes & Development. 1998;12:1953–61. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Steinbach D, Gillet JP, Sauerbrey A, Gruhn B, Dawczynski K, Bertholet V, de Longueville F, Zintl F, Remacle J, Efferth T. ABCA3 as a possible cause of drug resistance in childhood acute myeloid leukemia. Clin Cancer Res. 2006;12:4357–63. doi: 10.1158/1078-0432.CCR-05-2587. [DOI] [PubMed] [Google Scholar]
- [29].Matsuo K, Hamajima N, Suzuki R, Ogura M, Kagami Y, Taji H, Yasue Tetsuo, Mueller NE, Nakamura S, Seto M, Morishima Y, Tajima K. Methylenetetrahydrofolate reductase gene (MTHFR) polymorphisms and reduced risk of malignant lymphoma. Am J Hematol. 77:351–57. doi: 10.1002/ajh.20215. [DOI] [PubMed] [Google Scholar]
- [30].Gemmati D, Ongaro A, Scapoli DL, Della Porta M, Tognazzo S, Serino ML, Di Bona E, Rodeghiero F, Gilli G, Reverberi R, Caruso A, Pasello M, Pellati A, De Mattei M. Common gene polymorphisms in the metabolic folate and methylation pathway and the risk of acute lymphoblastic leukemia and non-Hodgkin’s lymphoma in adults. Cancer Epidemiol Biomarkers Prev. 2004;13(5):787–94. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.