To the Editor: Mycobacterium yongonense is a recently described species (1) that belongs to the M. avium complex (MAC) and is associated with pulmonary infection. The strain on which the description of species was based was isolated in South Korea from the sputum of a patient with unspecified pulmonary disease. We describe 2 M. yongonense strains isolated from patients in Italy.
Patient 1 was a 74-year-old woman who had experienced fatigue, diarrhea, and weight loss. Her medical history included liver cirrhosis resulting from hepatitis C virus infection and surgery for colon cancer; the patient also reported tuberculosis in childhood. Chest radiograph revealed a cavitary lesion, a finding confirmed by computed tomography scan (Figure). Cultures in liquid and solid media grew a nonchromogenic mycobacterium from sputum and stool samples; results were negative for urine samples.
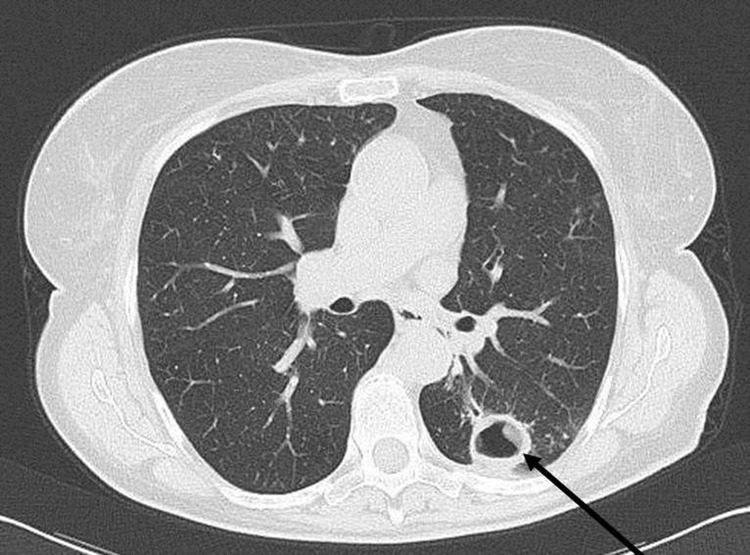
Figure.
Computed tomography scan showing a cavity (arrow) in the left lung of a 74-year-old woman (patient 1) in Italy. Laboratory testing suggests that the woman was infected with Mycobacterium yongonense.
The patient was treated with clarithromycin, rifabutin, and ethambutol and showed some improvement. A bronchoscopic investigation was performed, and microscopic examination of bronchoalveolar lavage samples revealed the presence of acid-fast bacilli that subsequently were grown in culture. The patient began improving markedly starting with the second month of treatment, which will be continued for a total of 18 months.
Patient 2 was a 74-year-old woman, living in a community of nuns, who reported cough and dyspnea. Her medical history included renal failure and surgery for breast cancer. A bronchoalveolar lavage was performed; samples yielded in culture Pseudomonas aeruginosa and a nonchromogenic mycobacterium. The patient was treated with cefepime, to which P. aeruginosa was susceptible in vitro, and rapidly improved. The isolation of the nontuberculous mycobacterium was considered irrelevant, and no specific treatment was undertaken.
To determine the specific mycobacteria species isolated from these patients, we conducted a commercial line-probe assay (GenoType Mycobacterium CM; Hain Lifesciences, Nehren, Germany). Both strains were identified as M. intracellulare. However, the known cross-reaction of M. intracellulare probe with most MAC species (2) led us to determine the complete sequence of the 16S rRNA gene. Both strains showed 100% similarity with M. yongonense and M. marseillense (3) strains.
To confirm this unusual finding, we investigated other genetic regions. We detected 100% identity with M. yongonense in the internal transcribed spacer 1 region and in a 1,384-bp region of the hsp65 gene and found 2 mismatches in a 420-bp fragment of the sodA gene (99.5% similarity). In contrast, M. marseillense showed 6 mismatches (98.6% similarity) in the internal transcribed spacer 1 region and 24 (98.3% similarity) in hsp65; no sodA sequence is available in GenBank for this species. Partial sequencing of other genetic targets not available in GenBank for M. yongonense enabled us to confirm the close relatedness of the strains to M. intracellulare (100% similarity in dnaK gene; 99.3% identity in gyrB and gyrC genes).
The finding of the same novel Mycobacterium species in these 2 unrelated patients reflects variability in the significance of nontuberculous mycobacteria isolated from clinical specimens. M. yongonense was probably a contaminant in the second case, but in the first, its involvement as causative agent of disease seems incontrovertible. The specific criteria of the American Thoracic Society (4) were fulfilled: radiographic imaging clearly documented the presence of a cavitary pulmonary lesion, no other pathogen possibly responsible of disease was detected by bronchoscopic investigation, and the same mycobacterium was isolated repeatedly from sputum (its presence in stool probably results from swallowed sputum) and bronchoalveolar lavage samples. Confirmation is further provided by the response to the specific therapy, according to international guidelines (4,5), for MAC pulmonary disease (MICs were 2, 1, and 8 µg/mL for clarithromycin, rifabutin, and ethambutol, respectively).
The initial description of M. yongonense noted that it has a distinct rpoB sequence (1), identical to that of a distantly related scotochromogenic species, M. parascrofulaceum. In a more recent article (6), the same authors investigated 2 more strains of M. yongonense with similar characteristics and suggested that the recent acquisition of the rpoB gene resulted from a lateral gene transfer event from M. parascrofulaceum. The rpoB genes of the strains we investigated, however, were substantially different from that of M. scrofulaceum and were instead related to that of M. intracellulare (99.4% similarity) and, less closely, to that of other species belonging to the MAC, including M. marseillense (97.4%). Discrepancy in the rpoB sequence means some uncertainty remains that our strains are M. yongonense, but the 100% identity in major phylogenetically relevant regions strongly supports this hypothesis and suggests the possibility of a variant of the species preceding the acquisition of the rpoB gene from M. parascrofulaceum. Less evidence exists for identifying the strains as M. marseillense because of the clear divergence in the genes investigated, other than 16S rRNA.
The complete epidemiology of M. youngonense is unknown, in part because few strains have been identified. However, as in the cases we describe, use of suboptimal identification methods may mean that some isolates have been misidentified as other mycobacteria species.
Acknowledgments
GenBank accession numbers for the M. yongonense strains identified in this study (FI-13004 and FI-13005) are KF224989–KF224999.
Footnotes
Suggested citation for this article: Tortoli E, Mariottini A, Pierotti P, Simonetti TM, Rossolini GM. Mycobacterium yongonense in pulmonary disease, Italy [letter]. Emerg Infect Dis [Internet]. 2013 Nov [date cited]. http://dx.doi.org/10.3201/eid1911.130911
References
- 1.Kim BJ, Math RK, Jeon CO, Yu HK, Park YG, Kook YH, et al. Mycobacterium yongonense sp. nov., a slow-growing non-chromogenic species closely related to Mycobacterium intracellulare. Int J Syst Evol Microbiol. 2013;63:192–9. 10.1099/ijs.0.037465-0 [DOI] [PubMed] [Google Scholar]
- 2.Tortoli E, Pecorari M, Fabio G, Messinò M, Fabio A. Commercial DNA probes for mycobacteria incorrectly identify a number of less frequently encountered species. J Clin Microbiol. 2010;48:307–10. 10.1128/JCM.01536-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben Salah I, Cayrou C, Raoult D, Drancourt M. Mycobacterium marseillense sp. nov., Mycobacterium timonense sp. nov. and Mycobacterium bouchedurhonense sp. nov., novel species in the Mycobacterium avium complex. Int J Syst Evol Microbiol. 2009;59:2803–8. 10.1099/ijs.0.010637-0 [DOI] [PubMed] [Google Scholar]
- 4.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, et al. An official ATS/IDSA statement: diagnosis, treatment and prevention of nontuberculous mycobacterial disease. Am J Respir Crit Care Med. 2007;175:367–416. 10.1164/rccm.200604-571ST [DOI] [PubMed] [Google Scholar]
- 5.Clinical Laboratory and Standards Institute. Susceptibility testing of mycobacteria, nocardiae and other aerobic actinomycetes; approved standard. 2nd edition, M24–A2. Wayne (PA); The Institute; 2011. [PubMed] [Google Scholar]
- 6.Kim BJ, Hong SH, Kook YH, Kim BJ. Molecular evidence of lateral gene transfer in rpoB gene of Mycobacterium yongonense strains via multilocus sequence analysis. PLoS ONE. 2013;8:e51846. 10.1371/journal.pone.0051846 [DOI] [PMC free article] [PubMed] [Google Scholar]