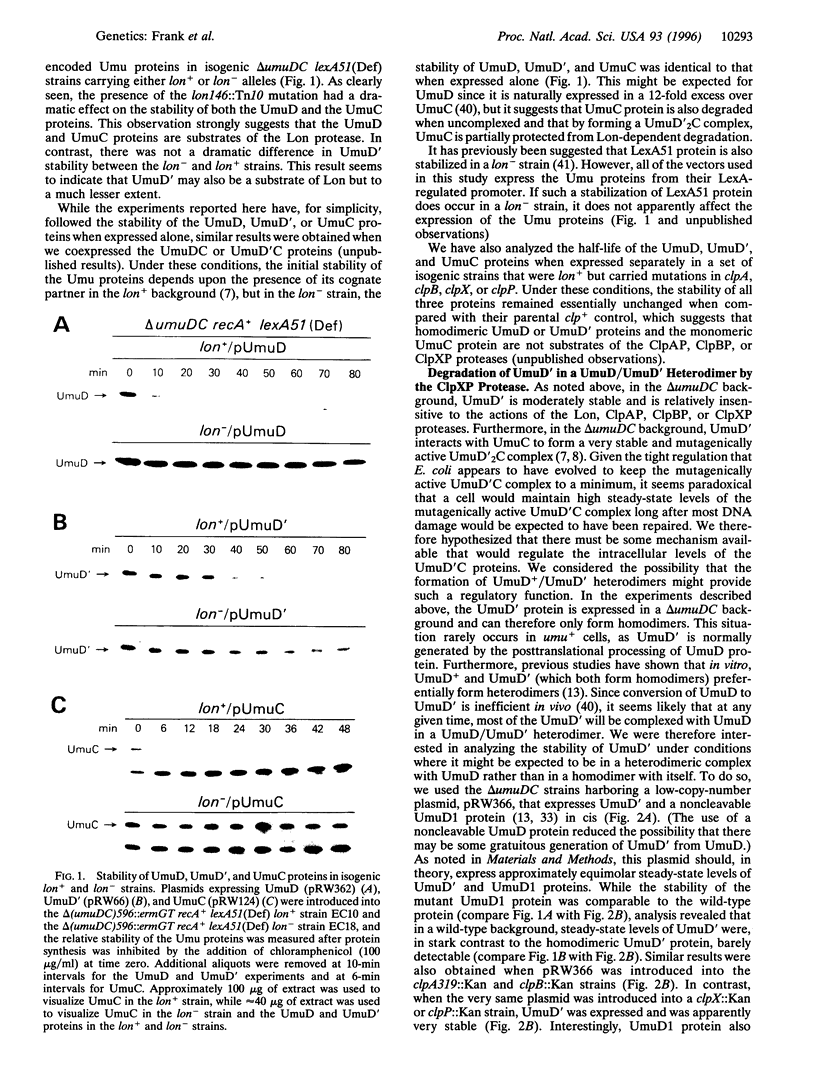
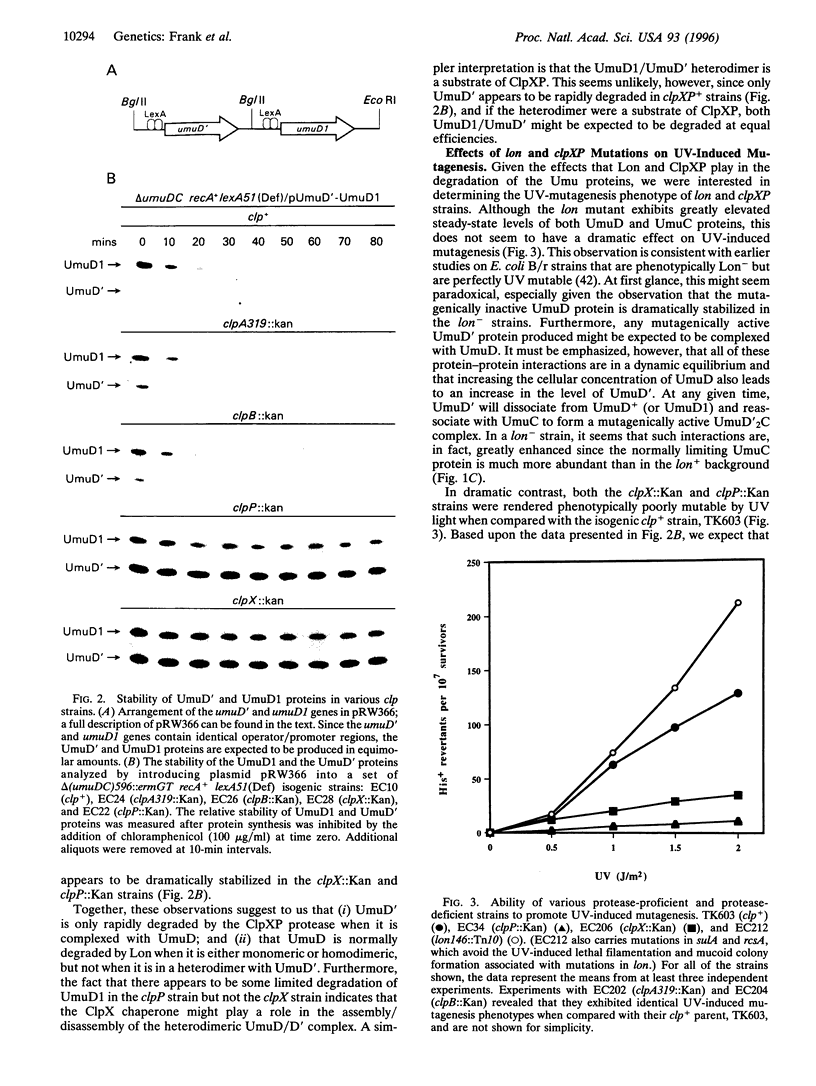
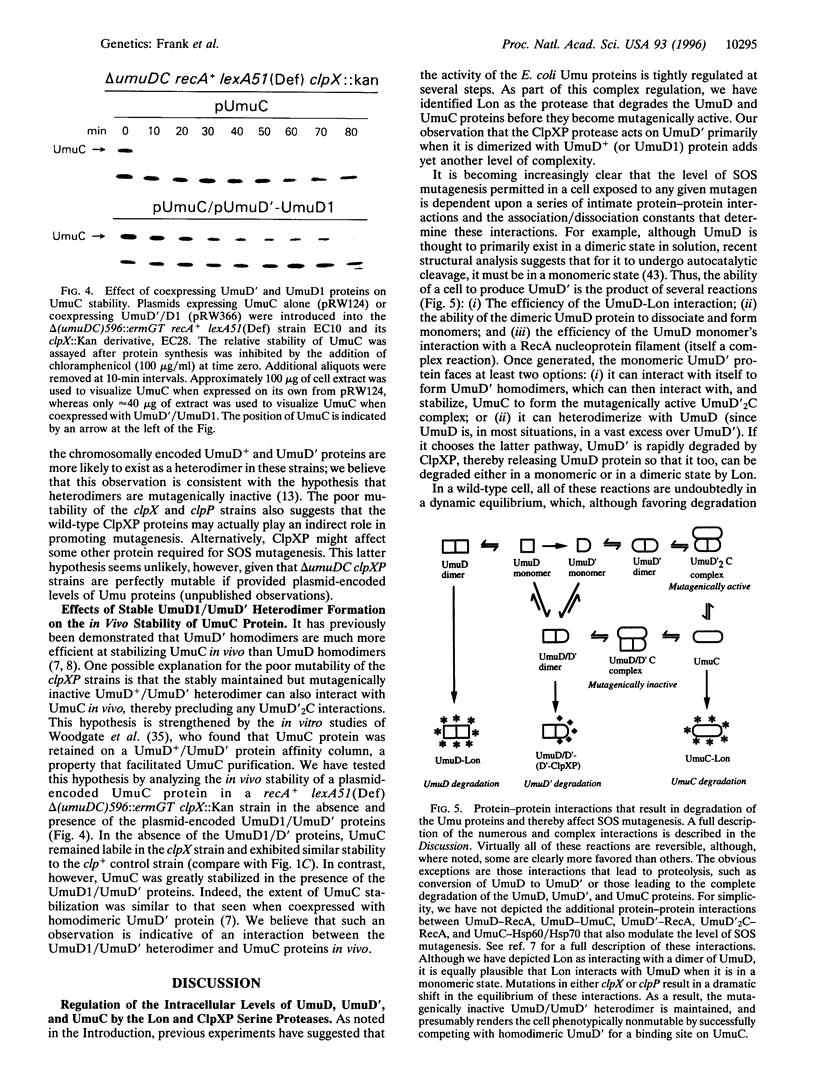
Abstract
DNA damage-inducible mutagenesis in Escherichia coli is largely dependent upon the activity of the UmuD (UmuD') and UmuC proteins. The intracellular level of these proteins is tightly regulated at both the transcriptional and the posttranslational levels. Such regulation presumably allows cells to deal with DNA damage via error-free repair pathways before being committed to error-prone pathways. We have recently discovered that as part of this elaborate regulation, both the UmuD and the UmuC proteins are rapidly degraded in vivo. We report here that the enzyme responsible for their degradation is the ATP-dependent serine protease, Lon. In contrast, UmuD' (the posttranslational product and mutagenically active form of UmuD) is degraded at a much reduced rate by Lon, but is instead rapidly degraded by another ATP-dependent protease, ClpXP. Interestingly, UmuD' is rapidly degraded by ClpXP only when it is in a heterodimeric complex with UmuD. Formation of UmuD/UmuD' heterodimers in preference to UmuD' homodimers therefore targets UmuD' protein for proteolysis. Such a mechanism allows cells to reduce the intracellular levels of the mutagenically active Umu proteins and thereby return to a resting state once error-prone DNA repair has occurred. The apparent half-life of the heterodimeric UmuD/D' complex is greatly increased in the clpX::Kan and clpP::Kan strains and these strains are correspondingly rendered virtually UV non-mutable. We believe that these phenotypes are consistent with the suggestion that while the UmuD/D' heterodimer is mutagenically inactive, it still retains the ability to interact with UmuC, and thereby precludes the formation of the mutagenically active UmuD'2C complex.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Battista J. R., Ohta T., Nohmi T., Sun W., Walker G. C. Dominant negative umuD mutations decreasing RecA-mediated cleavage suggest roles for intact UmuD in modulation of SOS mutagenesis. Proc Natl Acad Sci U S A. 1990 Sep;87(18):7190–7194. doi: 10.1073/pnas.87.18.7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck I., Woodgate R., McEntee K., Goodman M. F. Purification of a soluble UmuD'C complex from Escherichia coli. Cooperative binding of UmuD'C to single-stranded DNA. J Biol Chem. 1996 May 3;271(18):10767–10774. doi: 10.1074/jbc.271.18.10767. [DOI] [PubMed] [Google Scholar]
- Burckhardt S. E., Woodgate R., Scheuermann R. H., Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly C. E., Walker G. C. Coexpression of UmuD' with UmuC suppresses the UV mutagenesis deficiency of groE mutants. J Bacteriol. 1992 May;174(10):3133–3139. doi: 10.1128/jb.174.10.3133-3139.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly C. E., Walker G. C. groE mutants of Escherichia coli are defective in umuDC-dependent UV mutagenesis. J Bacteriol. 1989 Nov;171(11):6117–6125. doi: 10.1128/jb.171.11.6117-6125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H., Goodman M. F. Mutation induced by DNA damage: a many protein affair. Mutat Res. 1990 Sep-Nov;236(2-3):301–311. doi: 10.1016/0921-8777(90)90013-u. [DOI] [PubMed] [Google Scholar]
- Ennis D. G., Levine A. S., Koch W. H., Woodgate R. Analysis of recA mutants with altered SOS functions. Mutat Res. 1995 Jan;336(1):39–48. doi: 10.1016/0921-8777(94)00045-8. [DOI] [PubMed] [Google Scholar]
- Frank E. G., Gonzalez M., Ennis D. G., Levine A. S., Woodgate R. In vivo stability of the Umu mutagenesis proteins: a major role for RecA. J Bacteriol. 1996 Jun;178(12):3550–3556. doi: 10.1128/jb.178.12.3550-3556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuskens V., Mhammedi-Alaoui A., Desmet L., Toussaint A. Virulence in bacteriophage Mu: a case of trans-dominant proteolysis by the Escherichia coli Clp serine protease. EMBO J. 1992 Dec;11(13):5121–5127. doi: 10.1002/j.1460-2075.1992.tb05619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Clark W. P., Maurizi M. R. The ATP-dependent Clp protease of Escherichia coli. Sequence of clpA and identification of a Clp-specific substrate. J Biol Chem. 1990 May 15;265(14):7886–7893. [PubMed] [Google Scholar]
- Gottesman S., Clark W. P., de Crecy-Lagard V., Maurizi M. R. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993 Oct 25;268(30):22618–22626. [PubMed] [Google Scholar]
- Gottesman S., Maurizi M. R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992 Dec;56(4):592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Minimizing proteolysis in Escherichia coli: genetic solutions. Methods Enzymol. 1990;185:119–129. doi: 10.1016/0076-6879(90)85013-e. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Stout V. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol Microbiol. 1991 Jul;5(7):1599–1606. doi: 10.1111/j.1365-2958.1991.tb01906.x. [DOI] [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987 Sep 10;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Kato T., Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977 Nov 14;156(2):121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- Kitagawa Y., Akaboshi E., Shinagawa H., Horii T., Ogawa H., Kato T. Structural analysis of the umu operon required for inducible mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4336–4340. doi: 10.1073/pnas.82.13.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch W. H., Ennis D. G., Levine A. S., Woodgate R. Escherichia coli umuDC mutants: DNA sequence alterations and UmuD cleavage. Mol Gen Genet. 1992 Jun;233(3):443–448. doi: 10.1007/BF00265442. [DOI] [PubMed] [Google Scholar]
- Laachouch J. E., Desmet L., Geuskens V., Grimaud R., Toussaint A. Bacteriophage Mu repressor as a target for the Escherichia coli ATP-dependent Clp Protease. EMBO J. 1996 Jan 15;15(2):437–444. [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. W., Christensen R. UV mutagenesis in radiation-sensitive strains of yeast. Genetics. 1976 Feb;82(2):207–232. doi: 10.1093/genetics/82.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnherr H., Yarmolinsky M. B. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc Natl Acad Sci U S A. 1995 Apr 11;92(8):3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh L., Walker G. C. Cold sensitivity induced by overproduction of UmuDC in Escherichia coli. J Bacteriol. 1985 Apr;162(1):155–161. doi: 10.1128/jb.162.1.155-161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizi M. R. Proteases and protein degradation in Escherichia coli. Experientia. 1992 Feb 15;48(2):178–201. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- Mhammedi-Alaoui A., Pato M., Gama M. J., Toussaint A. A new component of bacteriophage Mu replicative transposition machinery: the Escherichia coli ClpX protein. Mol Microbiol. 1994 Mar;11(6):1109–1116. doi: 10.1111/j.1365-2958.1994.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Murli S., Walker G. C. SOS mutagenesis. Curr Opin Genet Dev. 1993 Oct;3(5):719–725. doi: 10.1016/s0959-437x(05)80089-9. [DOI] [PubMed] [Google Scholar]
- Nohmi T., Battista J. R., Dodson L. A., Walker G. C. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peat T. S., Frank E. G., McDonald J. P., Levine A. S., Woodgate R., Hendrickson W. A. Structure of the UmuD' protein and its regulation in response to DNA damage. Nature. 1996 Apr 25;380(6576):727–730. doi: 10.1038/380727a0. [DOI] [PubMed] [Google Scholar]
- Rajagopalan M., Lu C., Woodgate R., O'Donnell M., Goodman M. F., Echols H. Activity of the purified mutagenesis proteins UmuC, UmuD', and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassanfar M., Roberts J. W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990 Mar 5;212(1):79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- Schweder T., Lee K. H., Lomovskaya O., Matin A. Regulation of Escherichia coli starvation sigma factor (sigma s) by ClpXP protease. J Bacteriol. 1996 Jan;178(2):470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick S. G., Bridges B. A. Survival, mutation and capacity to repair single-strand DNA breaks after gamma irradiation in different Exr - strains of Escherichia coli. Mol Gen Genet. 1972;119(2):93–102. doi: 10.1007/BF00269129. [DOI] [PubMed] [Google Scholar]
- Shinagawa H., Iwasaki H., Kato T., Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires C., Squires C. L. The Clp proteins: proteolysis regulators or molecular chaperones? J Bacteriol. 1992 Feb;174(4):1081–1085. doi: 10.1128/jb.174.4.1081-1085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout V., Torres-Cabassa A., Maurizi M. R., Gutnick D., Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol. 1991 Mar;173(5):1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. W., Maurizi M. R. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J Biol Chem. 1994 Jul 8;269(27):18201–18208. [PubMed] [Google Scholar]
- Torres-Cabassa A. S., Gottesman S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J Bacteriol. 1987 Mar;169(3):981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. SOS-regulated proteins in translesion DNA synthesis and mutagenesis. Trends Biochem Sci. 1995 Oct;20(10):416–420. doi: 10.1016/s0968-0004(00)89091-x. [DOI] [PubMed] [Google Scholar]
- Wawrzynow A., Wojtkowiak D., Marszalek J., Banecki B., Jonsen M., Graves B., Georgopoulos C., Zylicz M. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 1995 May 1;14(9):1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S., Gottesman S., Skowyra D., Hoskins J., McKenney K., Maurizi M. R. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtkowiak D., Georgopoulos C., Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993 Oct 25;268(30):22609–22617. [PubMed] [Google Scholar]
- Woodgate R., Ennis D. G. Levels of chromosomally encoded Umu proteins and requirements for in vivo UmuD cleavage. Mol Gen Genet. 1991 Sep;229(1):10–16. doi: 10.1007/BF00264207. [DOI] [PubMed] [Google Scholar]
- Woodgate R., Rajagopalan M., Lu C., Echols H. UmuC mutagenesis protein of Escherichia coli: purification and interaction with UmuD and UmuD'. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7301–7305. doi: 10.1073/pnas.86.19.7301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgate R., Sedgwick S. G. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol Microbiol. 1992 Aug;6(16):2213–2218. doi: 10.1111/j.1365-2958.1992.tb01397.x. [DOI] [PubMed] [Google Scholar]
- Woodgate R., Singh M., Kulaeva O. I., Frank E. G., Levine A. S., Koch W. H. Isolation and characterization of novel plasmid-encoded umuC mutants. J Bacteriol. 1994 Aug;176(16):5011–5021. doi: 10.1128/jb.176.16.5011-5021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]






