Abstract
Incubation experiments using filtered waters from Lake Kasumigaura were conducted to examine bacterial contribution to a dissolved organic carbon (DOC) pool. Bacterial abundance, bacterial production, concentrations of DOC, total dissolved amino acids (TDAA), and total dissolved neutral sugars (TDNS) were monitored during the experiments. Bacterial production during the first few days was very high (20 to 35 μg C liter−1 day−1), accounting for 40 to 70% of primary production. The total bacterial production accounted for 34 to 55% of the DOC loss during the experiment, indicating high bacterial activities in Lake Kasumigaura. The DOC degradation was only 12 to 15%, whereas the degradation of TDAA and TDNS ranged from 30 to 50%, suggesting the preferential usage of TDAA and TDNS. The contribution of bacterially derived carbon to a DOC pool in Lake Kasumigaura was estimated using d-amino acids as bacterial biomarkers and accounted for 30 to 50% of the lake DOC. These values were much higher than those estimated for the open ocean (20 to 30%). The ratio of bacterially derived carbon to bulk carbon increased slightly with time, suggesting that the bacterially derived carbon is more resistant to microbial degradation than bulk carbon. This is the first study to estimate the bacterial contribution to a DOC pool in freshwater environments. These results indicate that bacteria play even more important roles in carbon cycles in freshwater environments than in open oceans and also suggests that recent increases in recalcitrant DOC in various lakes could be attributed to bacterially derived carbon. The potential differences in bacterial contributions to dissolved organic matter (DOM) between freshwater and marine environments are discussed.
INTRODUCTION
Heterotrophic bacteria play critical roles in carbon and nitrogen cycles in aquatic environments. They are the integral components of a microbial loop in which they assimilate dissolved organic matter (DOM) and contribute to secondary production. They are consumed by ciliates and protozoa, which eventually are grazed by organisms at higher trophic levels. Their production has been reported to account for about 10 to 30% of primary production in open oceans (1, 2) and even higher in coastal and freshwater environments (2, 3). Heterotrophic bacteria are also considered to be major contributors to the DOM pool in the ocean (4). Contributions of bacterially derived carbon to particulate organic carbon (POC) and dissolved organic carbon (DOC) pools are in the range of 20 to 40% in open oceans (5, 6). Thus, their contribution to aquatic carbon cycles is significant and should be included in carbon and energy flow models.
5-Bromo-2′-deoxyuridine (BrdU) has been known to be used as a thymidine (Thy) analogue in immunochemical technical fields (7). Since BrdU is not produced by any organism, BrdU taken up by bacteria instead of Thy can be used and considered bacterial activity (8). Hamasaki et al. (9) and Nelson and Carlson (10) further improved the method so as to distinguish activities of individual bacterial groups and to increase the sensitivity. However, BrdU labeled with a radioisotope is still required to convert immunochemical fluorescence into concentrations. Thus, the number of bacterial production measurements, especially in freshwater environments, has remained severely limited.
Heterotrophic bacteria have several biomolecules that are unique to bacteria. d-Amino acids (4) and muramic acids (11) in peptidoglycan, specific bacterial membrane proteins, such as porin P (12), and specific lipid components of membranes, lipopolysaccharides (13), have been found in marine DOM. These studies demonstrate the ubiquitous nature of bacterially derived DOM in aquatic environments. Recent studies used d-amino acids to estimate the bacterial contribution to organic carbon and nitrogen pools (5, 6). These studies found that bacterially derived organic matter accounted for 20 to 40% of dissolved and particulate organic carbon and ∼50% of dissolved and particulate organic nitrogen. Their results indicate that bacterially derived organic matter could be a major source of organic matter in marine environments. However, similar studies have not been conducted in freshwater environments.
In this study, we carried out incubation experiments to estimate bacterial production and changes in the DOC concentration, chemical composition of total dissolved amino acids (TDAA) and neutral sugars (TDNS), and d-amino acids. From these results, the total bacterial contribution to the carbon cycle of Lake Kasumigaura was evaluated.
MATERIALS AND METHODS
Sample collection and experimental setup.
Water samples were collected in pre-acid-washed 20-liter carboys at the lake center sampling station in Lake Kasumigaura on 25 August 2009 (experiment I [Exp I]) and 14 April 2010 (experiment II [Exp II]). Lake Kasumigaura, on the eastern Kanto Plain in Japan, has a surface area of 171 km2 and a watershed of about 2,200 km2. Its mean and maximum depths are 4 m and 7.4 m, respectively. About 1 million people live in the Lake Kasumigaura watershed, and about 50% of the land area is used for agriculture, such as paddy fields and upland fields (14).
After collection, samples were immediately brought back to the National Institute for Environmental Studies (NIES) Lake Kasumigaura Water Research Station. The sample water was filtered through a 0.65-μm cartridge filter (TCS-G065-S1FS; Advantec) using an extrusion pump (Nez-500 Valve-T; Meito Kakoki Co., Ltd.) directly into a 10-liter pre-acid-washed polycarbonate bottle (Nalgene) for the incubation experiment. Right after filtration, day 0 samples for bacterial abundance (BA) and bacterial production (BP) were collected, and day 0 samples for dissolved organic carbon (DOC), d and l enantiomers of total dissolved amino acids (TDAA), and total dissolved neutral sugars (TDNS) were collected after being filtered through GF/F filters. The filtered water was further filtered by the GF/F filter to remove aggregates, because the aggregation of DOM sometimes occurs after the water samples from Lake Kasumigaura are filtered. The incubation bottle was placed on a rotator at 70 rpm in dark at a constant temperature (20°C). Each incubation experiment was run in duplicate.
On days 1, 2, 3, 5, 9, 14, 28, and 60 for Exp I and days 0.125 (3 h), 1, 3, 5, 9, and 36 for Exp II, subsamples for BA, BP, DOC, TDAA, and TDNS were collected. The subsample for BA was collected directly from the sampling bottle, and 20% paraformaldehyde (final concentration, 2%) was added. It was kept at 4°C until measurement. The subsample for BP was collected in a 60-ml Nalgene PC bottle and incubated at room temperature for 1 h after the addition of BrdU (final concentration, 1 μmol liter−1). After 1 h, thymidine (Thy) (final concentration, 1 mmol liter−1) was added for termination of BrdU uptake, and the sample was stored in a freezer (−30°C) until analysis. The subsamples for DOC, TDAA, and TDNS were collected after being filtered through precombusted (450°C for 4 h) GF/F filters and stored in a freezer (−30°C) until analysis.
Sample analyses.
Bacterial abundance (BA) was determined by epifluorescence microscopy using the 4′-6-diamidino-2-phenylidole (DAPI) method (15). Bacterial samples were stained with DAPI and then filtered through 0.2-μm-pore-size black Millipore polycarbonate filters mounted on prewetted 0.45-μm-pore-size MF Millipore filters with a low vacuum pressure (<25 kPa). After 5 min, the samples were filtered with gentle vacuum (<25 kPa). At least 300 bacteria per sample were counted. For the bacterial production (BP) measurements, BrdU incorporation and following bacterial production were calculated using the method of Hamasaki (16). BP was measured only in Exp I.
DOC was measured using a Shimadzu TOC-V analyzer, using the method of Benner and Strom (17). d and l enantiomers of total dissolved amino acids (TDAA) were determined using reverse-phase high-performance liquid chromatography (HPLC) and precolumn derivatization with o-phthaldialdehyde (OPA) and N-isobutyryl l-cysteine (18). Ascorbic acid (0.12 μmol liter−1) was added to all samples before hydrolysis. Racemization of enantiomers during hydrolysis was corrected as described by Kaiser and Benner (18). Total dissolved neutral sugars (TDNS) were determined using a Dionex 500 high-performance anion exchange chromatography (HPAEC) system with pulsed amperometric detection (PAD), following the procedure described by Shinohara et al. (19).
Calculations.
The percentage of individual d-amino acids (d-AA), such as d-Ala, was calculated as follows: % d-Ala = ([d-Ala]/[d-Ala + l-Ala]) × 100, where [d-Ala] is the concentration of d-Ala, and [d-Ala + l-Ala] is the sum of the d- and l-Ala concentrations.
The degradation index (DI) of organic matter using mol% amino acid composition was calculated by the following formula proposed by Dauwe et al. (20): DI = ∑i[(vari − AVG vari)/STD vari] × fac.coeff.i, where vari is the mol% of the individual amino acid, AVG vari and STD vari are the mean mol% and standard deviation for each amino acid, respectively, and fac.coeff.i is the factor coefficient from Table 1 in the work of Dauwe et al. (20). Kaiser and Benner (21) also presented their own DI parameters. The DI values from both studies were presented.
Table 1.
Initial conditions of sample waters used in incubation experiments
The total amount of BP during the experiment was calculated using the equation BPtotal = ∑i[AvgBPi,i + 1 × (Dayi + 1 − Dayi)], where BPtotal is the total amount of BP during the 60-day experiment, AvgBPi,i + 1 is the average BP between a time point (i) and the next time point (i + 1), and “Dayi + 1 − Dayi” is a length of time between two time points.
The contribution of bacterially derived DOM in Lake Kasumigaura was calculated using the following formula, described by Kaiser and Benner (21): bacterially derived carbon (%) = (BiomarkerDOM/BiomarkerBacterial DOM) × 100, where BiomarkerDOM and BiomarkerBacterial DOM are the carbon-normalized yields of specific d-amino acids in sample DOM and freshly produced bacterial DOM, respectively. BiomarkerBacterial DOM values were obtained from data by Kaiser and Benner (5) and Kawasaki et al. (6).
RESULTS
Initial conditions of sample waters collected from Lake Kasumigaura.
Initial conditions of DOM for two incubation experiments are shown in Table 1. Bacterial abundances (BA) were 19.4 × 105 and 17.0 × 105 ml−1 in Exp I and II, respectively. Concentrations of DOC, TDAA, and TDNS were 316 μmol liter−1, 5.71 μmol liter−1, and 3.71 μmol liter−1 for Exp I and 245 μmol liter−1, 3.77 μmol liter−1, and 3.38 μmol liter−1 for Exp II, respectively. The carbon-normalized yields of TDAA (TDAA %DOC) and TDNS (TDNS %DOC) were relatively high, ranging between 5.6 and 7.8%. The degradation index (DI) was −0.44 and 0.23 in Exp I and II, respectively, by Dauwe's calculation and 3.78 and 4.47 in Exp I and II, respectively, by Kaiser and Benner's calculation. Most of the initial values except DI and TDNS %DOC were higher in Exp I than in Exp II.
Changes in BA and BP during incubation experiments.
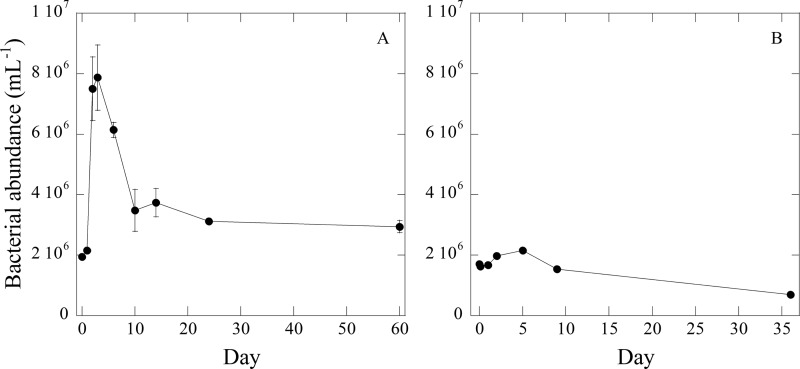
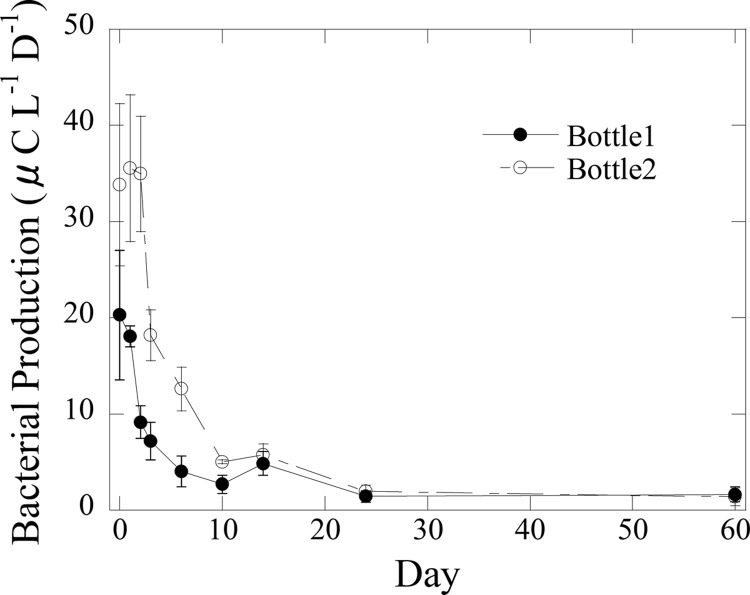
BA increased during the first 3 days in both experiments (Fig. 1). The average BA in Exp I rapidly increased from 1.9 × 106 ml−1 to 7.9 × 106 ml−1 on day 3, while the average BA in Exp II increased a little, from 1.7 × 106 ml−1 to 2.2 × 106 ml−1 on day 3. After day 3, BA decreased slowly between day 3 and day 9 and became constant at ∼3 × 106 ml−1 for Exp I and ∼1 × 106 ml−1 for Exp II after day 10. Bacterial production (BP) was measured only in Exp I (Fig. 2). BP in the bottle 1 was ∼ 35 μg C liter−1 day−1 during the first 2 days, while BP in bottle 2 was ∼20 μg C liter−1 day−1. After day 2, BP sharply decreased in both bottles, becoming less than 5 μg C liter−1 day−1 after day 9. The total bacterial production (BPtotal) in bottle 1 and bottle 2 was 0.31 mg C liter−1 and 0.17 mg C liter−1, respectively. These values accounted for 51% and 34% of the total decreased DOC in bottles 1 and 2, respectively.
Fig 1.
Bacterial abundance during incubation experiment I (A) or II (B).
Fig 2.
Bacterial production during incubation experiment in Exp I.
Changes in DOC and TDAA and TDNS concentrations during incubation experiments.
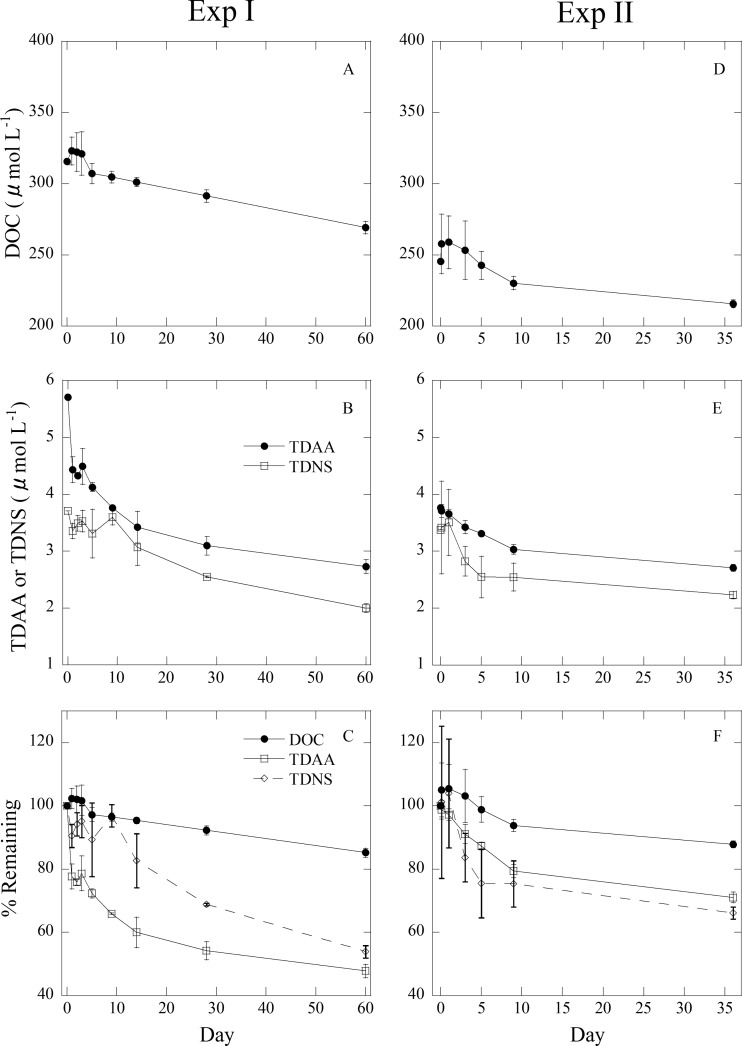
Concentrations of DOC, TDAA, and TDNS slowly decreased during incubation experiments. In Exp I, the concentrations of DOC, TDAA, and TDNS decreased from 316 to 269 μmol liter−1, 5.71 to 2.73 μmol liter−1, and 3.71 to 2.00 μmol liter−1, respectively, during the 60-day incubation (Fig. 3). In Exp II, the concentrations of DOC, TDAA, and TDNS decreased from 245 to 216 μmol liter−1, 3.77 to 2.67 μmol liter−1, and 3.38 to 2.23 μmol liter−1, respectively, during the 36-day incubation experiment (Fig. 3). The remaining DOC, TDAA, and TDNS proportions were 85.2, 47.8, and 53.9%, respectively, in Exp I and 87.9, 70.3, and 66.1%, respectively, in Exp II (Fig. 3).
Fig 3.
Concentrations of DOC (Exp I [A] or II [D]) and TDAA and TDNS in DOC (Exp I [B] or II [E]) and the percentages of remaining DOC, TDAA, and TDNS (Exp I [C] or II [F]) during incubation experiments.
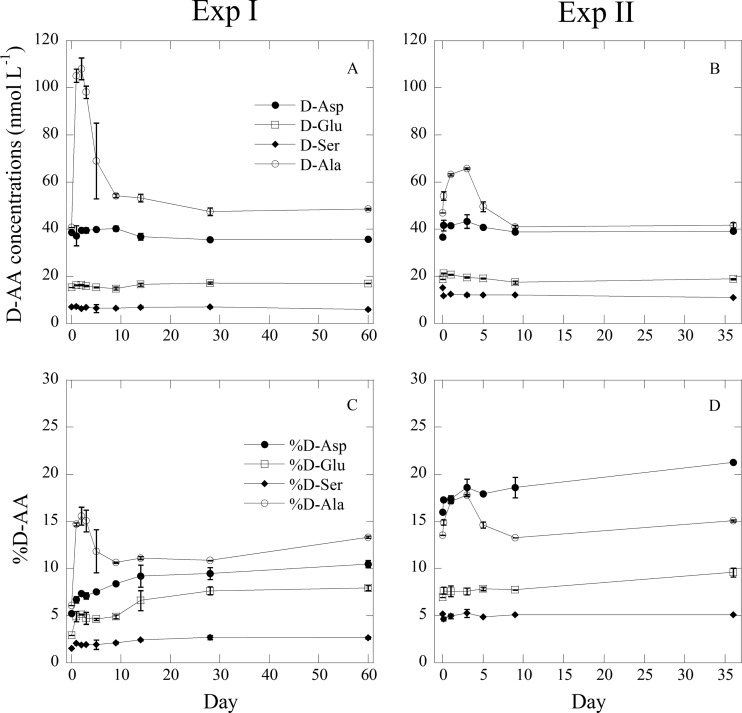
The concentrations of d and l enantiomers of aspartic acid (Asp), glutamic acid (Glu), serine (Ser), and alanine (Ala) were measured. The data on concentrations of d-amino acids (d-AA) showed that only d-Ala concentrations increased during the exponential growth of bacteria (Fig. 4A and B). The d-Ala concentrations in Exp I and II increased from 40.8 to 107.1 nmol liter−1 and from 47.0 to 66.2 nmol liter−1, respectively. After the exponential growth of bacteria, d-Ala concentrations declined to the initial values. The other d-AA concentrations remained constant or decreased slightly during the experiment. d-Asp, d-Glu, and d-Ser percentages increased slightly with time, while the d-Ala percentage increased rapidly during exponential growth. After the rapid decrease, it gradually increased again (Fig. 4C and D).
Fig 4.
Concentrations of d-amino acids of d-aspartic acid (d-Asp), d-glutamic acid (d-Glu), d-serine (d-Ser), and d-alanine (d-Ala) (Exp I [A] or II [C]) and a percentage of each respective d-amino acid in relation to total amino acids (d + l) for d-Asp, d-Glu, d-Ser, and d-Ala (Exp I [B] or II [D]) during incubation experiments.
The mole percentages of the individual TDAA and TDNS are shown in Table S1 in the supplemental material. For TDAA, Gly was the dominant amino acid, and followed by Ala. The concentrations of all amino acids gradually decreased with time during the incubation experiments. The mole percentage of amino acids showed that there were amino acid groups of increasing, decreasing, and almost unchanged mole percentages through the experiments. The amino acids which showed a notable increase in mole percentage during the two incubation experiments were Gly and the nonprotein amino acid beta-alanine (β-Ala). The amino acids with decreasing mole percentages included leucine (Leu), tyrosine (Tyr), and phenylalanine (Phe) in both experiments. The other amino acids showed inconsistent increases or decreases between the two experiments. For the initial conditions for TDNS, rhamnose (Rha) and glucose were dominant in Exp I, and glucose and mannose (Man) were dominant in Exp II. The chemical compositions of TNDS were quite different between the two incubation experiments. The concentrations of all TDNS decreased with time. However, the decreasing rates were different among individual neutral sugars, resulting in the change in mole percentages of neutral sugars. The mole percentages of glucose and Man decreased during the two incubation experiments, while those of arabinose (Ara) and xylose (Xyl) increased steadily. The mole percentages of the other neutral sugars showed inconsistent changes during the incubation experiments.
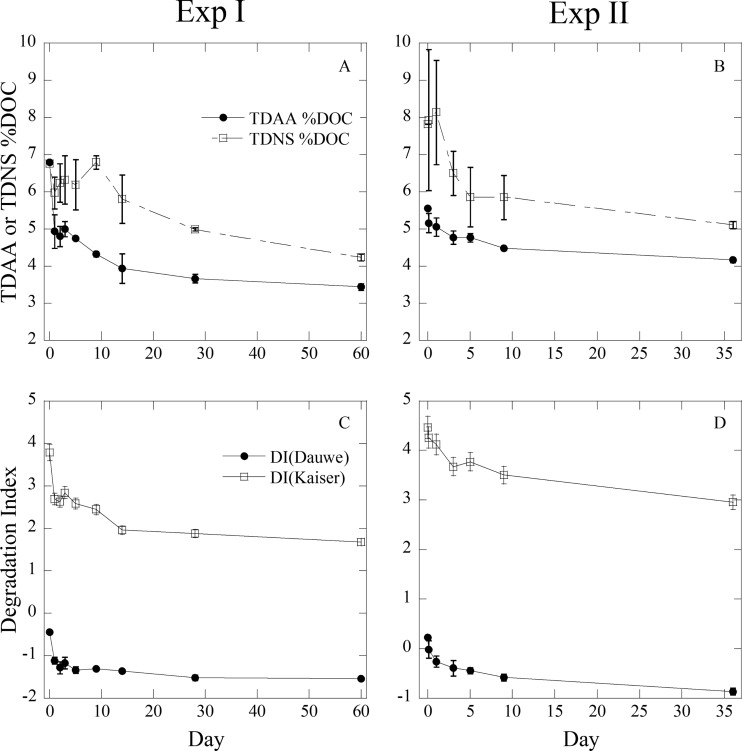
The carbon-normalized yields of TDAA and TDNS (TDAA %DOC and TDNS %DOC) also decreased with time. The TDAA %DOC in Exp I and II decreased from 6.8 to 3.4% and from 5.6 to 4.2%, respectively (Fig. 5A). The TDNS %DOC in Exp I and II decreased from 6.8 to 4.2% and 7.8 to 5.1%, respectively (Fig. 5B). The DI in Exp I and II decreased from −0.44 to −1.54 and from 0.23 to −0.86, respectively, in Dauwe's calculation and from 3.78 to 1.67 and from 4.47 to 2.96, respectively, in Kaiser and Benner's calculation (Fig. 5C and D). Both calculations showed the similar trend, but the DI determined by the method of Kaiser and Benner (21) showed wider changes and captured a small change compared to the DI determined by the method of Dauwe et al. (20), possibly suggesting the more sensitive formula for observing diagenetic alternations.
Fig 5.
Carbon-normalized yields of TDAA and TDNS (TDAA %DOC or TDNS %DOC) (Exp I [A] or II [C]) and degradation index using a formula by Dauwe et al. (20) [DI(Dauwe)] and Kaiser and Benner (21) [DI(Kaiser)] during incubation experiments (Exp I [B] or II [D]).
Bacterial contribution to DOM at Lake Kasumigaura.
To estimate the bacterial contribution to DOM, it is necessary to obtain the carbon-normalized yields of d-amino acids from the freshly produced bacterial DOM (BiomarkerBacterial DOM). Since the BiomarkerBacterial DOM in Lake Kasumigaura was not estimated in this study, the carbon-normalized yields of d-Asp, Glu, and Ala from the study by Kaiser and Benner (5), as well as unpublished data of an estuary measured on the South Carolina coast, were used (Table 2). The average BiomarkerBacterial DOM values of d-Asp, d-Glu, and d-Ala were 25.2, 16.3, and 29.7 nmol mg C−1, respectively (Table 2). d-Ser is another measurable d-amino acid, but the value was excluded from the calculation because the d-Ser concentrations were much lower than the others. The carbon-normalized yields of d-Asp, d-Glu, and d-Ala during the two incubation experiments were also calculated (Table 3). Using these values, the contributions of bacterially derived carbon in Lake Kasumigaura was estimated. The average bacterial contributions to initial Lake Kasumigaura DOM in Exp I and II were 33.8 and 47.4%, respectively. The bacterial contribution using d-Ala values increased rapidly due to the increase in the d-Ala concentration during bacterial growth, but the values dropped quickly after the growth. The average bacterial contributions to final Lake Kasumigaura DOM in Exp I and II were 42.3 and 53.0%, respectively.
Table 2.
Carbon-normalized yields of d-amino acids in freshly produced bacterially derived DOC
| Site | Carbon-normalized yield of d-amino acid (nmol mg C−1) in freshly produced bacterial DOC |
Reference | ||
|---|---|---|---|---|
| d-Asp | d-Glu | d-Ala | ||
| Estuary | 27.9 | 15.9 | 13.9 | Unpublished data |
| Atlantic coast | 21.1 | 17.6 | 27.0 | Kaiser and Benner (5) |
| North Pacific I | 24.9 | 8.9 | 38.0 | Kaiser and Benner (5) |
| North Pacific II | 26.9 | 23.0 | 40.1 | Kaiser and Benner (5) |
| Avg | 23.3 | 14.2 | 25.6 | |
Table 3.
Carbon-normalized yields of d-amino acids in DOC during incubation experiments in this study and percentages of bacterial contribution to DOC
| Expt and day | Carbon-normalized yield of d-amino acid (nmol mg C−1) in this expt |
% bacterial contribution |
|||||
|---|---|---|---|---|---|---|---|
| d-Asp | d-Glu | d-Ala | d-Asp | d-Glu | d-Ala | Avg | |
| Exp I | |||||||
| 0 | 10.2 | 4.0 | 10.8 | 40.6 | 24.7 | 36.2 | 33.8 |
| 1 | 9.6 | 4.2 | 27.2 | 38.2 | 25.6 | 91.3 | 51.7 |
| 2 | 10.2 | 4.2 | 28.0 | 40.6 | 25.9 | 94.0 | 53.5 |
| 3 | 10.3 | 4.1 | 25.5 | 40.7 | 25.4 | 85.8 | 50.7 |
| 6 | 10.8 | 4.2 | 18.7 | 43.0 | 25.4 | 62.9 | 43.8 |
| 10 | 11.0 | 4.1 | 14.8 | 43.7 | 24.9 | 49.9 | 39.5 |
| 14 | 10.2 | 4.6 | 14.7 | 40.4 | 28.1 | 49.6 | 39.4 |
| 24 | 10.2 | 4.9 | 13.6 | 40.4 | 30.2 | 45.7 | 38.8 |
| 60 | 11.1 | 5.3 | 15.1 | 43.9 | 32.3 | 50.6 | 42.3 |
| Exp II | |||||||
| 0 | 12.4 | 6.4 | 15.9 | 49.4 | 39.2 | 53.6 | 47.4 |
| 0.125 | 13.5 | 6.9 | 17.6 | 53.5 | 42.2 | 59.1 | 51.6 |
| 1 | 13.4 | 6.7 | 20.4 | 53.1 | 40.9 | 68.5 | 54.2 |
| 2 | 14.3 | 6.5 | 21.7 | 56.6 | 39.6 | 73.0 | 56.4 |
| 5 | 14.0 | 6.6 | 17.1 | 55.7 | 40.2 | 57.3 | 51.1 |
| 9 | 14.1 | 6.3 | 14.9 | 55.9 | 38.6 | 50.0 | 48.2 |
| 36 | 15.2 | 7.3 | 16.1 | 60.2 | 44.7 | 54.2 | 53.0 |
DISCUSSION
Chemical and biological properties of DOM in Lake Kasumigaura.
Dissolved organic matter (DOM) in Lake Kasumigaura was relatively rich in amino acids and neutral sugars, indicating that it contained freshly derived bioavailable components. Imai and Matsushige (22) reported that the origin of recalcitrant DOM, which was comprised of >80% DOM in Lake Kasumigaura, came from riverine input (∼40%), pore water (∼25%), sewage treatment plant effluent (STPE) (∼20%), and water column production (∼15%) at the center of the lake, although the contribution of the DOM sources varied depending on sampling sites. For example, the contribution of the STPE was drastically increased at the site near the point of effluent discharge. Planktonic organisms, sediment pore water, riverine input, and human activities contribute to the DOM reservoir in most freshwater environments.
Changes in bacterial abundance and production during incubation experiments.
Bacterial production (BP) at Lake Kasumigaura (20 to 35 μg C liter−1 day−1) was found to be relatively high compared to that in other aquatic environments, such as open oceans (∼5 μg C liter−1 day−1) and coastal oceans (∼10 μg C liter−1 day−1), and similar to that in estuary regions (∼50 μg C liter−1 day−1) (see reference 23 and references therein). This result suggests that bacterial activities appear to be high in Lake Kasumigaura.
Bacterial activities were compared with primary production (PP). A few observations of PP from Lake Kasumigaura were made in the 1980s (24, 25), but a recent study using a fast-repetition-rate fluorometry (FRRF) method showed the PP at a lake center sampling point from the GEMS/Water Trend Monitoring Project at Lake Kasumigaura in 2012 and was used in this study. It ranged between 39.9 and 208.5 g C m−2 year−1, with the average of 126 g C m−2 year−1 (K. Komatsu, unpublished data). BP estimated in this study ranged between 48.1 and 80.3 mg C m−2 year−1, accounting for 38 to 64% of PP. T. Tsuchiya, K. Kamatsu, N. Kawasaki, T. Toda, and A. Imai (unpublished data) measured in situ BP in Lake Kasumigaura in 2011, and their values ranged between 75.5 and 133.3 g C m−2 year−1, with the average of 105.5 gC m−2 year−1, slightly higher than the values estimated in this study. Our BP values could be underestimated because some of bacteria were removed during the filtration and many grazers were removed by filtration before the experiments. Obernosterer et al. (26) conducted a bacterial production study and found that BP was reduced significantly when bacteria were separated from their grazers. Therefore, BP measurements in this study showed reasonable values, and our results showed that bacteria play critical roles in carbon and nitrogen cycles in Lake Kasumigaura.
The total amount of BP during the incubation experiment accounted for 51% and 34% of the total decreased DOC in bottles 1 and 2, respectively. Bacterial growth efficiency (BGE) is derived from the ratio of BP to the sum of BP and bacterial respiration (BR). Supposing that BGE in Lake Kasumigaura were assumed to range from 20 to 30% (3), the BR values would become 3 to 4 times more than the BP values, resulting in total bacterial activity (BP plus BR) ranging from 180% to 360% of the total decreased DOC. This result indicates that bacteria utilized about at least twice to 3.5 times more carbon than the decreased DOC, and thus DOM must be recycled in order to maintain the high bacterial activity. As del Giorgio and Cole (3) pointed out, it is very difficult to measure BGE; these values should contain various factors of errors and uncertainties. However, these values indicate that bacteria could efficiently recycle a small part of available DOM.
Bacterial abundance and growth were significantly different between the two experiments, and bacterial production was also very different between the two bottles. The growth difference could be due to the different amounts of available DOM, grazing activities, or viral lysis. In Exp I, bacterial abundance rapidly increased, but bacterial abundance increased a little in Exp II. Physical conditions such as rainfall may cause the temporal changes in the chemical composition of DOM. However, the total of 15 mm rain was observed about 36 h before the sampling in Exp I, while the total of 35 mm rain was observed in the previous 2 days in Exp II, suggesting that the physical conditions may not be the major issue in the growth difference. Although not examined in this study, grazing and viral lysis activities could be high in both experiments, especially high enough to suppress bacterial growth in Exp II. Grazing pressure could be different between the two experiments, although the grazers were not examined in this study. As for viral infection, since the infection rate of viruses for phytoplankton and bacteria is likely to be constant (27), their contribution to the differences in bacterial abundance and growth could be minor.
Degradation of DOM by bacteria.
The concentration of DOM decreased by about 12 to 15% during the 36- or 60-day incubation experiment. Imai and Matsushige (22) reported that the average degradation rate of DOC during 100-day incubation experiments conducted monthly for Lake Kasumigaura samples between 1997 and 2001 was about 11%. Our results were slightly higher than theirs. However, since their data showed that the degraded DOM fractions during spring and summer were relatively higher than those in the other seasons, our values were considered very comparable to theirs. Søndergaard and Middelboe (28) compiled studies of the labile fraction of DOC and found that the average labile fraction of DOC in 27 lakes studied was about 14%. Our results were also similar to their average. Weiss and Simon (29) also described degradation experiments with lake DOM and found that the degradation rate of DOM varied between 8 and 57%. However, the high rate was observed only during the phytoplankton bloom. The average DOM degradation rate excluding this exceptionally high rate for their study was ∼15%.
The observed bioavailable fraction of DOC was relatively low despite the high concentrations and yields of TDAA and TDNS. Even at the end of the experiments, TDAA %DOC and TDNS %DOC ranged from 3.4% to 5.2%. These values were higher than those found in the surface and deep oceans (21). Typical ranges of TDAA %DOC and TDNS %DOC in the ocean were 1 to 2% and 2 to 4%, respectively (30). This might be due to a short incubation period, different microbial community, or difference in properties of DOM in Lake Kasumigaura compared to oceanic environments. According to Imai, the DOM degradation percentages were similar for an even longer incubation period (data not shown), suggesting that the incubation period may not be a major issue.
Bacterial composition between freshwater and marine environments is known to be quite different. While the betaproteobacteria are dominant in freshwater environments (31, 32), the alphaproteobacteria are a main bacterial group in marine environments (33). According to Watanabe et al. (34), 84% of bacteria at Lake Kasumigaura were composed of betaproteobacteria. Hobbie (35) reported that most bacterial activities are similar between freshwater and marine bacteria, although some physiological differences can be found, such as requiring sodium and adapting to high pressure. However, Watanabe et al. (34) further reported that most of the betaproteobacteria assimilated only a few types of amino acids or carbohydrates and preferred to assimilate organic acids. This result indicates that freshwater bacteria may not consume amino acids and carbohydrates as intensively as marine bacteria, resulting in the higher fractions of TDAA and TDNS in freshwater recalcitrant DOM. This may also suggest that the chemical compositions of recalcitrant DOM could be different between freshwater and marine environments, although no studies have been conducted to address this yet. Baldock et al. (36) reported that decomposition-induced changes in DOM molecular composition were very different in freshwater and marine systems, based on 13C nuclear magnetic resonance (NMR) analyses of major classes of compounds. Therefore, the chemical compositions of recalcitrant DOM could be different between oceans and lakes, such as Lake Kasumigaura, resulting in relatively low biodegradation despite high yields of TDAA and TDNS.
Most DOM in the ocean is of phytoplankton origin, but various DOC sources contributed to DOM in Lake Kasumigaura. The major difference in DOC sources between the open and freshwater environments may be terrestrial humic substances. The humic substances are derived from decaying plant debris and are major components of organic matter in soil and freshwater environments. Imai et al. (37) reported that about 32% of DOC consisted of humic substances in Lake Kasumigaura. In the open ocean, the fraction of humic substances in DOC appears to be much lower (∼10%) (38). According to Opsahl and Benner (39), terrigenous DOM only accounts for 0.7 to 2.4% of total DOM in the ocean. Bacterial processes could also contribute to the production of humic substances (40, 41), but their contribution could also be minor. The source difference could also result in a different chemical composition of recalcitrant DOM at Lake Kasumigaura compared to that in oceans.
d-Amino acid concentrations and estimation of bacterially derived DOM at Lake Kasumigaura.
Four d-amino acids, d-Asp, d-Glu, d-Ser, and d-Ala, were measured, and only d-Ala concentrations increased during bacterial growth. Kawasaki and Benner (42) conducted similar incubation experiments and also found an increase in d-Ala during exponential growth of bacteria. They suggested the possible release of d-Ala during cell growth and division. The increased d-Ala was quickly removed after exponential growth, suggesting that it was taken up by bacteria promptly. Therefore, the increased d-Ala during exponential growth is labile and rapidly utilized.
The high bacterial contribution to Lake Kasumigaura DOM could be due to differences in DOM sources, different microbial composition, or both. The riverine input, pore water, and sewage treatment plant effluent are the main DOM sources of Lake Kasumigaura. DOM from these sources should have been extensively altered by bacteria before coming into the lake, resulting in a high bacterial contribution to Lake Kasumigaura DOM. As stated earlier, the dominant betaproteobacteria prefer to assimilate organic acids (34) and could leave more amino acids in recalcitrant DOM, indicating that the carbon-normalized yields of d-amino acids in the bacterial DOM (BiomarkerBacterial DOM) could be different between freshwater and marine bacteria. If so, it is necessary to use different BiomarkerBacterial DOM values in freshwater environments. This needs to be further addressed. Another possibility, suggested by Kaiser and Benner (5), is that biomarker approaches could result in underestimation of bacterial contribution because structures of biomarkers could be altered by diagenesis and no longer recognized. Watanabe et al. (43) reported that the average DOM radiocarbon age of Lake Kasumigaura ranged between 200 and 700 years old, much younger than oceanic DOM (4,000 to 6,000 years old) (44). If this is true, the bacterial contribution to marine DOM could be higher than those estimated and could be matched to the values estimated in this study. These age differences could also contribute to higher bacterial contribution at Lake Kasumigaura than for oceanic DOM.
The contribution of bacterial DOM increased with time during the incubation experiments. Bacterial contribution using d-Ala increased or even exceeded 100% during cell growth due to the specific release of d-Ala during cell growth (42), but the values quickly decreased after growth. The final bacterial contribution was slightly increased from that at the beginning, indicating that bacterially derived carbon is less reactive than bulk carbon, resulting in the accumulation of bacterially derived carbon. The degradation index, as well as the increase in the mole percentage of glycine (Gly), also supported that the DOM was becoming more diagenetically altered and more recalcitrant during the incubation experiments. The mole percentage of Gly is also known to be a good indicator of the early stages of DOM degradation (45). Kaiser and Benner (5) reported the opposite result. However, their conclusion was based on the data from deepwater samples from the Hawaii Ocean Time-Series (HOT), where the fraction of bacterially derived carbon was lower in deep water than in surface water. Since DOM from deep water at HOT is the oldest DOM in the ocean, intensive degradation of bacterial biomarkers could occur, resulting in lower bacterial contribution to a DOM pool. If this is true, bacterially derived carbon is more resistant to microbial degradation than bulk carbon, and the accumulation of recalcitrant DOM could be due to the accumulation of bacterially derived DOM.
Implication for carbon cycling in Lake Kasumigaura.
High bacterial production and bacterial contribution to DOM found in this study suggest that bacteria are the major sources of organic matter and the microbial loop is an important carbon and energy transfer mechanism for higher trophic levels in Lake Kasumigaura. Recently, increasing trends of chemical oxygen demands (COD) have been reported for many lakes in Japan, whereas biological oxygen demands (BOD), standing for easily biodegradable organic matter, have been decreasing (22), indicating that recalcitrant DOM is accumulating in lake waters.
Due to long-term regulation of DOM source origins in a drainage basin, the loading of DOM from the drainage basin appears to have gradually decreased recently. Therefore, DOM from a drainage basin may not be a main cause of increasing recalcitrant DOM. In Lake Kasumigaura, a major shift of cyanobacteria from Microcystis to Planktothrix has been observed, although a dominant phytoplankton group consists of diatoms in Lake Kasumigaura (46, 47). This shift could cause changes in the quality of DOM released from phytoplankton and a subsequent increase in recalcitrant DOM. However, Imai and Matsushige (46) conducted the degradation experiments using exudates from both Microcystis and Planktothrix. They found that the degradation rate of DOC from Microcystis was much lower than that for Planktothrix (50% versus 90%), indicating that it is unlikely that the shift of cyanobacterial species could result in the increase in recalcitrant DOM. On the other hand, bacteria have been reported to produce recalcitrant DOM (48, 49). Imai et al. (50) suggested that the increase in recalcitrant DOM in Lake Kasumigaura could be due to an increase in the discharge of sewage treatment plant effluent (STPE). The STPE is discharged directly into Lake Kasumigaura, with a flow rate of 76,323 m3 per day (Tsuchiura city home page [http://www.city.tsuchiura.lg.jp/index.php?code=523]), and the discharge has been increasing due to an increase in population. Since the STPE is water intensively treated by bacteria, DOM from STPE is expected to be highly degraded and thus recalcitrant. The major contribution of DOM from bacteria suggests that the increase in recalcitrant DOM in Japanese lakes could result from increasing contributions of bacterially derived DOM.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Grant-in-Aid for Scientific Research grants (no. 19710019, 21241108, and 22710019) from the Japan Society for the Promotion of Science and by the Environment Research and Technology Development Fund of the Ministry of the Environment, Japan (5-1304). Sampling was supported by the GEMS/Water Trend Monitoring Project at Lake Kasumigaura.
We thank Ronald Benner for his comments and suggestions for greatly improving our manuscript. We also thank the members of the project for their cooperation.
Footnotes
Published ahead of print 13 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01504-13.
REFERENCES
- 1.Carlson CA, Ducklow HW, Sleeter TD. 1996. Stocks and dynamics of bacterioplankton in the northwestern Sargasso Sea. Deep Sea Res. II 43:491–515 [Google Scholar]
- 2.Cole JJ, Findlay S, Pace M. 1988. Bacterial production in fresh and saltwater ecosystems—a cross system overview. Mar. Ecol. Prog. Ser. 43:1–10 [Google Scholar]
- 3.del Giorgio PA, Cole JJ. 1998. Bacterial growth efficiency in natural aquatic systems. Annu. Rev. Ecol. Syst. 29:503–541 [Google Scholar]
- 4.McCarthy MD, Hedges JI, Benner R. 1998. Major bacterial contribution to marine dissolved organic nitrogen. Science 281:231–234 [DOI] [PubMed] [Google Scholar]
- 5.Kaiser K, Benner R. 2008. Major bacterial contribution to the ocean reservoir of detrital organic carbon and nitrogen. Limnol. Oceanogr. 53:99–112 [Google Scholar]
- 6.Kawasaki N, Sohrin R, Ogawa H, Nagata T, Benner R. 2011. Bacterial carbon content and the living and detrital bacterial contributions to suspended particulate organic carbon in the North Pacific Ocean. Aquat. Microb. Ecol. 62:165–176 [Google Scholar]
- 7.Mazzotti G, Rizzoli R, Galanzi A, Papa S, Vitale M, Falconi M, Neri LM, Zini N, Maraldi NM. 1990. High-resolution detection of newly synthesized DNA by anti-bromo-deoxyuridine antibodies identifies specific chromatin domains. J. Histochem. Cytochem. 38:13–22 [DOI] [PubMed] [Google Scholar]
- 8.Steward GF, Azam F. 1999. Bromodeoxyuridine as an alternative to 3H-thymidine for measuring bacterial productivity in aquatic samples. Aquat. Microb. Ecol. 19:57–66 [Google Scholar]
- 9.Hamasaki K, Long RA, Azam F. 2004. Individual cell growth rates of marine bacteria, measured by bromodeoxyuridine incorporation. Aquat. Microb. Ecol. 35:217–227 [Google Scholar]
- 10.Nelson CE, Carlson CA. 2005. A nonradioactive assay of bacterial productivity optimized for oligotrophic pelagic environments. Limnol. Oceanogr. Methods 3:211–220 [Google Scholar]
- 11.Benner R, Kaiser K. 2003. Abundance of amino sugars and peptidoglycan in marine particulate and dissolved organic matter. Limnol. Oceanogr. 48:118–128 [Google Scholar]
- 12.Tanoue E, Nishiyama S, Kamo M, Tsugita A. 1995. Bacterial membranes: possible source of a major dissolved protein in seawater. Geochim. Cosmochim. Acta 59:2643–2648 [Google Scholar]
- 13.Wakeham SG, Pease TK, Benner R. 2003. Hydroxy fatty acids in marine dissolved organic matter as indicators of bacterial membrane material. Org. Geochem. 34:857–868 [Google Scholar]
- 14.Fukushima T, Takahashi M, Matsushita B, Okanishi Y. 2007. Land use/cover change and its drivers: a case in the watershed of Lake Kasumigaura, Japan. Landsc. Ecol. Eng. 3:21–31 [Google Scholar]
- 15.Porter KG, Feig YS. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943–948 [Google Scholar]
- 16.Hamasaki K. 2006. Comparison of bromodeoxyuridine immunoassay with tritiated thymidine radioassay for measuring bacterial productivity in oceanic waters. J. Oceanogr. 62:793–799 [Google Scholar]
- 17.Benner R, Strom M. 1993. A critical evaluation of the analytical blank associated with DOC measurements by high-temperature catalytic oxidation. Mar. Chem. 41:153–160 [Google Scholar]
- 18.Kaiser K, Benner R. 2005. Hydrolysis-induced racemization of amino acids. Limnol. Oceanogr. Methods 3:318–325 [Google Scholar]
- 19.Shinohara A, Imai A, Komatsu K, Matsushige K, Nara F. 2008. Application of HPLC-PAD to highly sensitive analysis of dissolved carbohydrates and their composition in lake water and extracellular organic matter derived from algae. J. Jpn. Soc. Water Environ. 31:447–454 (In Japanese.) [Google Scholar]
- 20.Dauwe B, Middelburg JJ, Herman PMJ, Heip CHR. 1999. Linking diagenetic alteration of amino acids and bulk organic matter reactivity. Limnol. Oceanogr. 44:1809–1814 [Google Scholar]
- 21.Kaiser K, Benner R. 2009. Biochemical composition and size distribution of organic matter at the Pacific and Atlantic time-series stations. Mar. Chem. 113:63–77 [Google Scholar]
- 22.Imai A, Matsushige K. 2004. Studies on mass balance of dissolved organic matter in lake and its functions and effects on lacustrine ecosystems and water quality. Report of Special Research from the National Institute for Environmental Studies, Japan, SR-62-2004. (In Japanese.) National Institute for Environmental Studies, Tsukuba, Japan [Google Scholar]
- 23.Ducklow HW. 2000. Bacterial production and biomass in the oceans, p 85–120 In Kirchman DL. (ed), Microbial ecology of the oceans. John Wiley, New York, NY [Google Scholar]
- 24.Takamura N, Iwakuma T, Yasuno M. 1984. The biomass and production of phytoplankton in Lake Kasumigaura during 1981-83. Res. Rep. Natl. Inst. Environ. Stud. 51:11–56 [Google Scholar]
- 25.Takamura N, Aizaki M. 1991. Change in primary production in Lake Kasumigaura (1986–1989) accompanied by transition of dominant species. Jpn. J. Limnol. 52:173–187 [Google Scholar]
- 26.Obernosterer I, Kawasaki N, Benner R. 2003. P-limitation of respiration in the Sargasso Sea and uncoupling of bacteria from P-regeneration in size-fractionation experiments. Aquat. Microb. Ecol. 32:229–237 [Google Scholar]
- 27.Proctor LM, Fuhrman JA. 1990. Viral mortality of marine bacteria and cyanobacteria. Nature 343:60–62 [Google Scholar]
- 28.Søndergaard M, Middelboe M. 1995. A cross-system analysis of labile dissolved organic carbon (LDOC). Mar. Ecol. Prog. Ser. 118:283–294 [Google Scholar]
- 29.Weiss M, Simon M. 1999. Consumption of labile dissolved organic matter by limnetic bacterioplankton: the relative significance of amino acids and carbohydrates. Aquat. Microb. Ecol. 17:1–12 [Google Scholar]
- 30.Benner R. 2002. Chemical composition and reactivity, p 59–90 In Hansell DA, Carlson CA. (ed), Biogeochemistry of marine dissolved organic matter. Academic Press, New York, NY [Google Scholar]
- 31.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. 2011. A guide to the natural history of freshwater lake bacteria. Microbiol. Mol. Biol. Rev. 75:14–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zwart G, Crump BC, Agterveld MPK, Hagen F, Han SK. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141–155 [Google Scholar]
- 33.Gonzalez JM, Moran MA. 1997. Numerical dominance of a group of marine bacteria in the α-subclass of the class Proteobacteria in coastal seawater. Appl. Environ. Microbiol. 63:4237–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe K, Komatsu N, Kitamura T, Ishii Y, Park HD, Miyata R, Noda N, Sekiguchi Y, Satou T, Watanabe M, Yamamura S, Imai A, Hayashi S. 2012. Ecological niche separation in the Polynucleobacter subclusters linked to quality of dissolved organic matter: a demonstration using a high sensitivity cultivation-based approach. Environ. Microbiol. 10.1111/j.1462-2920.2012.02815.x [DOI] [PubMed] [Google Scholar]
- 35.Hobbie JE. 1988. A comparison of the ecology of planktonic bacteria in fresh and salt water. Limnol. Oceanogr. 33:750–764 [Google Scholar]
- 36.Baldock JA, Masiello CA, Géinas Y, Hedges JI. 2004. Cycling and composition of organic matter in terrestrial and marine ecosystems. Mar. Chem. 92:39–64 [Google Scholar]
- 37.Imai A, Fukushima T, Matsushige K, Kim YH. 2001. Fractionation and characterization of dissolved organic matter in a shallow eutrophic lake, its inflowing rivers, and other organic matter sources. Water Res. 35:4019–4028 [DOI] [PubMed] [Google Scholar]
- 38.Meyers-Schultke J, Hedges JI. 1986. Molecular evidence for a terrestrial component of organic matter dissolved in seawater. Nature 321:61–63 [Google Scholar]
- 39.Opsahl S, Benner R. 1997. Distribution and cycling of terrigenous dissolved organic matter in the ocean. Nature 386:480–482 [Google Scholar]
- 40.Shimotori K, Omori Y, Hama T. 2010. Bacterial production of marine humic-like fluorescent dissolved organic matter and its biogeochemical importance. Aquat. Microb. Ecol. 58:55–66 [Google Scholar]
- 41.Tranvik LJ. 1993. Microbial transformation of labile dissolved organic matter into humic-like matter in seawater. FEMS Microbiol. Ecol. 12:177–183 [Google Scholar]
- 42.Kawasaki N, Benner R. 2006. Bacterial release of dissolved organic matter during cell growth and decline: molecular origin and composition. Limnol. Oceanogr. 51:2170–2180 [Google Scholar]
- 43.Watanabe FN, Imai A, Matsushige K, Komatsu K, Kawasaki N, Shibata Y. 2010. Radiocarbon measurements of dissolved organic carbon in sewage-treatment-plant effluent and domestic sewage. Nucl. Instrum. Methods Phys. Res. B 268:1142–1145 [Google Scholar]
- 44.Williams PM, Druffel ERM. 1987. Radiocarbon in dissolved organic carbon in the central north Pacific Ocean. Nature 330:246–248 [Google Scholar]
- 45.Amon RMW, Fitznar HP, Benner R. 2001. Linkages among the bioreactivity, chemical composition, and diagenetic state of marine dissolved organic matter. Limnol. Oceanogr. 46:287–297 [Google Scholar]
- 46.Imai A, Matsushige K. 2001. Studies on origin and dynamics of recalcitrant organic matter in lake and its effects on lacustrine ecosystems and water quality. Report of Special Research from the National Institute for Environmental Studies, Japan, SR-36-2001. (In Japanese.) National Institute for Environmental Studies, Tsukuba, Japan [Google Scholar]
- 47.Takamura N, Iwakuma T, Yasuno M. 1985. Photosynthesis and primary production of Microcystis aeruginosa Kütz in Lake Kasumigaura. J. Plankton Res. 7:303–312 [Google Scholar]
- 48.Brophy JE, Carlson DJ. 1989. Production of biologically refractory dissolved organic carbon by natural seawater microbial populations. Deep Sea Res. 36:497–507 [Google Scholar]
- 49.Ogawa H, Amagai Y, Koike I, Kaiser K, Benner R. 2001. Production of refractory dissolved organic matter by bacteria. Science 292:917–920 [DOI] [PubMed] [Google Scholar]
- 50.Imai A, Fukushima T, Matsushige K, Kim YH, Choi K. 2002. Characterization of dissolved organic matter in effluents from wastewater treatment plants. Water Res. 36:859–870 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.