Abstract
3,5,6-Trichloro-2-pyridinol (TCP) is a widespread pollutant. Some bacteria and fungi have been reported to degrade TCP, but the gene clusters responsible for TCP biodegradation have not been characterized. In this study, a fragment of the reduced flavin adenine dinucleotide (FADH2)-dependent monooxygenase gene tcpA was amplified from the genomic DNA of Ralstonia sp. strain T6 with degenerate primers. The tcpA disruption mutant strain T6-ΔtcpA could not degrade TCP but could degrade the green intermediate metabolite 3,6-dihydroxypyridine-2,5-dione (DHPD), which was generated during TCP biodegradation by strain T6. The flanking sequences of tcpA were obtained by self-formed adaptor PCR. tcpRXA genes constitute a gene cluster. TcpR and TcpX are closely related to the LysR family transcriptional regulator and flavin reductase, respectively. T6-ΔtcpA-com, the complementation strain for the mutant strain T6-ΔtcpA, recovered the ability to degrade TCP, and the strain Escherichia coli DH10B-tcpRXA, which expressed the tcpRXA gene cluster, had the ability to transform TCP to DHPD, indicating that tcpA is a key gene in the initial step of TCP degradation and that TcpA dechlorinates TCP to DHPD. A library of DHPD degradation-deficient mutants of strain T6 was obtained by random transposon mutagenesis. The fragments flanking the Mariner transposon were amplified and sequenced, and the dhpRIJK gene cluster was cloned. DhpJ could transform DHPD to yield an intermediate product, 5-amino-2,4,5-trioxopentanoic acid (ATOPA), which was further degraded by DhpI. DhpR and DhpK are closely related to the AraC family transcriptional regulator and the MFS family transporter, respectively.
INTRODUCTION
TCP (3,5,6-trichloro-2-pyridinol) is the main degradation product of the herbicide triclopyr and the insecticides chlorpyrifos and chlorpyrifos-methyl (1–4), which have been widely used in agriculture because of their high efficiency and moderate toxicity. TCP is classified as persistent and mobile by the U.S. EPA, with a half-life ranging from 65 to 360 days in soil (5), and is more migratory than its parent molecule due to its greater water solubility, which causes widespread contamination of soils and aquatic environments (3, 6, 7). TCP was detected in >95% of the indoor floor dust and hard-floor surface wipe, solid food, and indoor air samples in the everyday environments of preschool children in six North Carolina counties, posing a potential hazard to children (8). TCP has high affinity to the DNA molecule and can bind to the DNA groove, which might cause the metabolite to act as a potential health hazard (9).
Some bacteria, bacterial consortia, and fungi have been reported to degrade TCP and can grow using it as their sole carbon source; these organisms include Alcaligenes faecalis DSP3 (1), Burkholderia sp. strain KR100 (4), Pseudomonas sp. strain ATCC 700113 (7), Paracoccus sp. strain TRP (10), Pseudomonas sp. strains 1 to 4 (11), Agrobacterium sp. strains 5 and 6 (11), Bacillus sp. strain 7 (11), Trichosporon sp. strain TCF (12), Bacillus pumilus C2A1 (13), Pseudomonas strain M285 (14), and Ralstonia sp. strain T6 (15). However, the genes and metabolic pathways responsible for TCP biodegradation have rarely been reported, and the gene clusters affecting TCP biodegradation have not been well characterized. There are only two reports on the genes of TCP biodegradation. A novel gene involved in TCP degradation, tcp3A, was isolated from a cow rumen metagenomic library and was functionally verified in Escherichia coli (16). The genome sequence of the chlorpyrifos and TCP degrader Paracoccus sp. TRP was completed, but no genes related to TCP degradation were identified (17). A reductive dechlorination mechanism was proposed in the TCP degrader Pseudomonas sp. ATCC 700113 (18), which is the well-known metabolic pathway of TCP biodegradation.
Ralstonia sp. T6 degrades TCP more effectively than do other TCP degraders (1, 4, 7, 10, 11, 12, 13, 14, 15), and discovering the genes and metabolic pathway for TCP degradation is valuable. We report here the cloning and functional characterization of two gene clusters and the partial metabolic pathway responsible for TCP degradation in strain T6.
MATERIALS AND METHODS
Materials, bacterial strains, cultures, and plasmids.
TCP (99.3%) was obtained from the Gu'an Enkang Medicine Chemical Raw Material Co. Ltd., Langfang, China. A concentrated stock solution of TCP (10 g/liter) was prepared in water. All solvents were of pure analytical grade, except for chromatographically pure methanol. PCRs were performed with Taq, LA Taq, or PrimeSTAR HS DNA polymerase (TaKaRa Biotechnology, Dalian, China), and the primers were purchased from the Invitrogen Co. (Shanghai, China). Restriction endonucleases were purchased from TaKaRa Biotechnology Co. (Dalian, China). The minimal salts medium (MSM) contained (per liter) 1.5 g K2HPO4, 0.5 g KH2PO4, 1.0 g (NH4)2SO4, 0.03 g MgSO4, and 1.0 g NaCl, pH 7.0. An appropriate concentration of TCP was added to the MSM to prepare the TMM medium. The Luria-Bertani (LB) medium contained (per liter) 10.0 g tryptone, 5.0 g yeast extract, and 10.0 g NaCl, pH 7.0. Solid medium plates were prepared by adding 16 g/liter agar into the above liquid media. The bacterial strains and plasmids used in this study are listed in Table 1. Ralstonia sp. T6 and its mutant derivatives were grown at 30°C in LB medium or on LB agar. All E. coli strains were routinely cultivated at 37°C in LB medium or on LB agar, except E. coli BL21(DE3) and E. coli DH10B, which were cultivated at 30°C when used to express enzyme proteins and to transform TCP to 3,6-dihydroxypyridine-2,5-dione (DHPD), respectively. Ampicillin (Amp), kanamycin (Km), tetracycline (Tc), and gentamicin (Gm) were used at 100, 50, 30, and 30 mg/liter in culture medium, respectively.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Ralstonia sp. | ||
| T6 | Wild type, TCP degrader, Gm− Tcr | Lab stock |
| T6-ΔtcpA | tcpA insertion mutant of T6 | This study |
| T6-ΔtcpA-com | Complementation for the tcpA insertion mutant of T6 | This study |
| Cupriavidus necator JMP134 | Wild type, 2,4,6-trichlorophenol degrader | 24 |
| E. coli | ||
| DH5α | Host strain for cloning vectors | Lab stock |
| DH10B | Host strain for cloning vectors | Lab stock |
| SM10λpir | Conjugation strain | Lab stock |
| BL21(DE3) | Host strain for expressing vectors | Lab stock |
| Plasmids | ||
| pMD19-T | T-A cloning vector, Ampr | TaKaRa |
| pMD19-RXA | Vector for tcpRXA heterologous expression | This study |
| pMD19-RXA-JMP134 | Vector for heterologous expression of tcpRXA from C. necator JMP134 | This study |
| pUC19 | Parent vector of pMD19-T digested with EcoRV | TaKaRa |
| pJQ200SK | Suicide vector, Gmr | 22 |
| pJQ-TY1 | Vector for the tcpA insertion mutant of T6 | This study |
| pRK2013 | Helper plasmid, mob+ tra+ Kmr | Lab stock |
| pSC123 | Vector for transposon-mediated mutagenesis of bacteria, Kmr | 25 |
| pBBR1MCS-2 | Broad-host-range cloning vector, Kmr | 23 |
| pBB-XA | pBBR1MCS-2 derivative carrying tcpX and tcpA genes | This study |
| pET-29a(+) | Expression vector, Kmr | Lab stock |
| pET-I | pET-29a(+) derivative carrying dhpI | This study |
| pET-J | pET-29a(+) derivative carrying dhpJ | This study |
Cloning of tcpA and the tcpRXA gene cluster by SEFA PCR.
The partial tcpA gene was amplified from Ralstonia sp. T6 with the degenerate primers TftD-S and TftD-A (19) (Table 2). The flanking sequences of tcpA were obtained by self-formed adaptor PCR (SEFA PCR) (20), and the primers used are presented in Table 2. The primers TftDR-Sp1, TftDR-Sp2, and TftDR-Sp3 were used to amplify the 3′ flanking sequence of tcpA. The primers TftDF-Sp1, TftDF-Sp2, and TftDF-Sp3 were used to amplify the 5′ flanking sequence of tcpA. The genomic DNA of strain T6 and its mutant was extracted using a high-concentration salt precipitation method (21). All the amplified nucleotide sequences were confirmed by DNA sequencing with an ABI Genetic Analyzer 3730 (Invitrogen Bio Inc., Shanghai, China).
Table 2.
Oligonucleotide primers used in this study
| Primer | Sequence (5′ to 3′)a | Purpose | Position |
|---|---|---|---|
| TftD-S | AGTACCTGGAGTCSCTSAACGAC | To amplify part of sequence of tcpA | 1955–1977 in tcp gene cluster |
| TftD-A | CGGSGTSCCGTTGAATTCTCGAA | To amplify part of sequence of tcpA | 3285–3262 in tcp gene cluster |
| TY-S | CCGTGGGAAACGCAGACTATCG | To amplify the homologous sequence of tcpA | 2290–2311 in tcp gene cluster |
| TY-A | TGCACTGCGGGAATCTTGTTGG | To amplify the homologous sequence of tcpA | 2906–2885 in tcp gene cluster |
| TftDR-Sp1 | CTCACAGGGCGACGAACTGGACG | To amplify the flanking sequence of tcpA | 2676–2698 in tcp gene cluster |
| TftDR-Sp2 | GTCGCACGTGTTCCACCTAGGCA | To amplify the flanking sequence of tcpA | 2733–2755 in tcp gene cluster |
| TftDR-Sp3 | CACCAACAAGATTCCNNNNNNNNNGACTCG | To amplify the flanking sequence of tcpA | 2883–2912 in tcp gene cluster |
| TftDF-Sp1 | CGCACAGTTCAGGTCATGCTGCTTG | To amplify the flanking sequence of tcpA | 2388–2364 in tcp gene cluster |
| TftDF-Sp2 | TCAGCGCCGATAGTCTGCGTTTC | To amplify the flanking sequence of tcpA | 2318–2296 in tcp gene cluster |
| TftDF-Sp3 | CGTCGATGTAGGTCTNNNNNNNNNAGTTGT | To amplify the flanking sequence of tcpA | 2281–2252 in tcp gene cluster |
| TCPXA-S | GGTACCGACGCGAGGTGATTAGCAAA (KpnI) | Complementation for the tcpA insertion mutant | 1132–1151 in tcp gene cluster |
| TCPXA-A | GGGCCCTGCACTCCTTCCTTGGTTCA (ApaI) | Complementation for the tcpA insertion mutant | 3556–3537 in tcp gene cluster |
| TCPRXA-S | GGTACCGACTATAACCGGCGCTCATC (KpnI) | Heterologous expression of the tcpRXA gene cluster of T6 | 145–164 in tcp gene cluster |
| TCPRXA-A | GGGCCCTGCACTCCTTCCTTGGTTCA (ApaI) | Heterologous expression of the tcpRXA gene cluster of T6 | 3556–3537 in tcp gene cluster |
| ARB1 | GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT | To amplify the flanking sequence of Mariner transposon | None |
| ARB2 | GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC | To amplify the flanking sequence of Mariner transposon | None |
| ARB3 | GGCCACGCGTCGACTAGTAC | To amplify the flanking sequence of Mariner transposon | None |
| F(1)-SP1 | AGCCAGGGATGTAACGCACT | To amplify the flanking sequence of Mariner transposon | 198–179 in Mariner transposon |
| F(1)-SP2 | TAACGGCTGACATGGGAATT | To amplify the flanking sequence of Mariner transposon | 128–109 in Mariner transposon |
| F(2)-SP1 | GCAAGAATCCCAGCAATCG | To amplify the flanking sequence of Mariner transposon | 1721–1703 in dhp gene cluster |
| F(2)-SP2 | GACGCCACGGTGACAAGAT | To amplify the flanking sequence of Mariner transposon | 1632–1614 in dhp gene cluster |
| F(3)-SP1 | CAGGCTCTGGTACAAACACG | To amplify the flanking sequence of Mariner transposon | 404–385 in dhp gene cluster |
| F(3)-SP2 | CATGAAGCTGAAGATGCGGT | To amplify the flanking sequence of Mariner transposon | 361–342 in dhp gene cluster |
| R(1)-SP1 | TTGAAGGATCAGATCACGCA | To amplify the flanking sequence of Mariner transposon | 3322–3341 in Mariner transposon |
| R(1)-SP2 | GGTATCGCTCTTGAAGGGAACT | To amplify the flanking sequence of Mariner transposon | 3463–3484 in Mariner transposon |
| R(2)-F | GAACTCGTCGTTGCTATCGG | To amplify the flanking sequence of Mariner transposon | 2610–2629 in dhp gene cluster |
| R(2)-R | CAGTGTCAGGAAACTACCGC | To amplify the flanking sequence of Mariner transposon | 5361–5342 in dhp gene cluster |
| R(3)-SP1 | ACCTTGCTATCGGTCTTCGG | To amplify the flanking sequence of Mariner transposon | 5192–5211 in dhp gene cluster |
| R(3)-SP2 | TGTCGTTGCCTGTGCGTAAG | To amplify the flanking sequence of Mariner transposon | 5247–5266 in dhp gene cluster |
| R(4)-SP1 | CTTCTTCGGCTAAGCGTGC | To amplify the flanking sequence of Mariner transposon | 5491–5509 in dhp gene cluster |
| R(4)-SP2 | CGCATTGTGCCATGGTTG | To amplify the flanking sequence of Mariner transposon | 5575–5592 in dhp gene cluster |
| DHPI-S | CATATGCCTAGAGCCAGTGCGGA (NdeI) | Expression of dhpI | 2340–2359 in dhp gene cluster |
| DHPI-A | CTCGAGGGACTCAGACGATCTCGACC (XhoI) | Expression of dhpI | 3060–3041 in dhp gene cluster |
| DHPJ-S | CATATGAAACCACTCGAACGCA (NdeI) | Expression of dhpJ | 3081–3099 in dhp gene cluster |
| DHPJ-A | CTCGAGTGCACGGATCCGGCTATGA (XhoI) | Expression of dhpJ | 3972–3954 in dhp gene cluster |
Restriction sites of primers are underlined.
RT-PCR analyses of the common promoter for tcpX and tcpA.
The total RNA of strain T6 cells grown in LB medium was extracted with the RNAprep Pure Cell/Bacteria kit (Tiangen Biotech Co., Ltd., Beijing, China). Reverse transcriptase PCR (RT-PCR) analyses were carried out using the GoScript reverse transcription system (Promega Corp., Madison, WI) according to the manufacturer's instructions. Three primer sets were used for RT-PCR. Primer set A (forward, 5′-TCGTCCGCAGTATCCATC-3′; reverse, 5′-ACCATCGCGGATTTGCTG-3′) amplified the internal 356 bp of tcpX. Primer set B (forward, 5′-ACCTGGAATCGCTCAACG-3′; reverse, 5′-GCCGATAGTCTGCGTTTC-3′) amplified the internal 392 bp of tcpA. Primer set C (forward, 5′-GCCATACAGCAAATCCGC-3′; reverse, 5′-GCCGATAGTCTGCGTTTC-3′) amplified 663 bp of the tcpX-tcpA-spanning region. All the amplified fragments were confirmed by DNA sequencing with an ABI Genetic Analyzer 3730 (Invitrogen Bio Inc., Shanghai, China).
Disruption and complementation of tcpA in Ralstonia sp. T6.
Homologous recombination was used to disrupt tcpA. The homologous recombination-directing sequence was amplified with the primers TY-S and TY-A (Table 2), using strain T6 genomic DNA as a template. The purified PCR product was ligated to the pMD19-T vector (TaKaRa Biotechnology, Dalian, China), which was then recovered by digestion with PstI and SacI and ligated into the suicide vector pJQ200SK (22), yielding pJQ-TY. The plasmid pJQ-TY was transformed into Ralstonia sp. T6 by triparental conjugation. The donor, helper, and recipient strains were E. coli SM10λpir containing plasmid pJQ-TY, E. coli pRK2013, and Ralstonia sp. T6, respectively. All of the strains were cultured in LB medium to an optical density at 600 nm (OD600) of 1.0. Then, 1 ml of each culture was collected by centrifugation at 2,500 × g for 5 min and washed three times with fresh LB medium to remove any residual antibiotics. Cells were resuspended in 50 μl LB medium. The mixture was spread onto a nitrocellulose filter (with an average pore size of 0.45 μm) on an LB plate and incubated at 30°C for 24 h. The mixtures were then resuspended in 1 ml of LB medium and spread onto LB plates (200 μl per plate) supplemented with Gm and Tc. After incubation at 30°C for 3 days, colonies with single recombination events were selected on LB plates containing Gm and Tc. The disruption of tcpA was confirmed by the presence/absence of the expected PCR products in the wild-type and mutant strains using the primers TftD-S and TftD-A. One of the selected mutant strains was designated T6-ΔtcpA.
To complement the disrupted tcpA gene of T6-ΔtcpA, a fragment carrying both tcpX and tcpA was amplified from the genomic DNA of strain T6 with the primers TCPXA-S and TCPXA-A (Table 2) because of their common promoter. The fragment (89% identity to tcpXA from the 2,4,6-trichlorophenol degrader Cupriavidus necator JMP134) was inserted into the broad-host-range plasmid pBBR1MCS-2 (23), yielding pBB-XA. The plasmid pBB-XA was transformed into T6-ΔtcpA by triparental conjugation to generate the complementation strain T6-ΔtcpA-com, and the resulting transconjugants were selected on LB plates with Gm, Tc, and Km. Complementation of tcpA was confirmed by extracting the plasmid pBB-XA.
Heterologous expression of the tcpRXA gene cluster in E. coli DH10B.
The gene cluster of tcpRXA (88% identity to tcpRXA from C. necator JMP134) was cloned from the genomic DNA of strain T6 with the primers TCPRXA-S and TCPRXA-A (Table 2). Then, the fragment was ligated into the pMD19-T vector, producing the plasmid pMD19-RXA, which was transformed into E. coli DH10B, yielding the tcpRXA gene cluster expression strain DH10B-tcpRXA. Meanwhile, the strain DH10B-pUC19 containing the plasmid pUC19 (TaKaRa Biotechnology, Dalian, China) was taken as a control, and the heterologous expression strain E. coli DH10B containing tcpRXA from C. necator JMP134 (24) was constructed for comparison with DH10B-tcpRXA.
Screening for green metabolite degradation-deficient mutants.
In the process of TCP degradation by Ralstonia sp. T6, a green intermediate metabolite was detected (15) and was used as a selection marker for strain T6 mutants losing part of their TCP degradation ability. A mutant library of strain T6 was generated by Mariner transposon mutagenesis. E. coli SM10λpir containing the plasmid pSC123 (25) was used as the donor strain. The conjugation procedure was the same as that described in the literature (26), except that the antibiotics used were Km and Tc. After incubation at 30°C for 3 days, conjugants were transferred onto numbered LB plates supplemented with Km and Tc. All of the colonies on numbered LB plates were then transferred into 96-well plates with filter-sterilized MSM containing the green intermediate metabolite, Km, and Tc. Colonies that grew on numbered LB plates but did not make the green color fade were selected and streaked to yield single colonies.
Cloning of the fragments flanking the Mariner transposon.
The sequences flanking the Mariner transposon in the T6 mutant were obtained by thermal asymmetric interlaced PCR (TAIL PCR) (27) and sequence homology to Ralstonia sp. strain H16 with the primers presented in Table 2. The primers ARB1, ARB2, ARB3, F(1)-SP1, and F(1)-SP2 were used for the first round of amplifying the 5′ flanking sequence of the transposon. Besides ARB1, ARB2, and ARB3, the primer pairs F(2)-SP1 and F(2)-SP2 and F(3)-SP1 and F(3)-SP2 were used for the second and third rounds of amplifying the 5′ flanking sequence, respectively. The primers ARB1, ARB2, ARB3, R(1)-SP1, and R(1)-SP2 were used for the first round of amplifying the 3′ flanking sequence of the transposon, while the primers R(2)-S and R(2)-A were used for the second round of amplifying the 3′ flanking sequence, according to sequence homology. Besides ARB1, ARB2, and ARB3, the primer pairs R(3)-SP1 and R(3)-SP2 and R(4)-SP1 and R(4)-SP2 were used for the third and fourth rounds of amplifying the 3′ flanking sequence, respectively. The conditions for PCR with the primers R(2)-S and R(2)-A were as follows: 5 min of denaturation at 94°C, followed by 30 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 2.5 min, and a final extension at 72°C for 10 min.
After purification by agarose gel electrophoresis, PCR fragments were ligated into the pMD19-T vector and transformed into E. coli DH5α for sequencing.
Expression of dhpI and dhpJ in E. coli.
DNA fragments containing dhpI and dhpJ were independently amplified by PCR from genomic DNA of strain T6 using two primer sets, DHPI-S and DHPI-A for dhpI and DHPJ-S and DHPJ-A for dhpJ. The amplified PCR fragments were sequenced and separately inserted into the expression vector pET29a with NdeI and XhoI to produce plasmids pET-I and pET-J. The pET29a constructs were then transformed into E. coli BL21(DE3) for expression with induction of 0.2 mM isopropyl-β-d-thiogalactoside (IPTG) at 30°C for 3.5 h after it was grown in LB with 50 mg/liter kanamycin at 37°C to an optical density at 600 nm (OD600) of 0.5.
The cells were harvested and suspended in 8 ml of 20 mM phosphate-buffered saline (PBS) buffer (pH 7.0) containing 0.5 mM phenylmethylsulfonyl fluoride (PMSF). After the cells were disrupted by ultrasonication (Kutota; Insonator 201 M) on ice, the lysate was centrifuged at 15,000 × g for 15 min, and the supernatant was obtained as the cell extracts. The expressed proteins were identified by standard sodium dodecyl sulfate-polyacrylamide gel electrophoresis (28).
TCP degradation or transformation by Ralstonia sp. T6 and E. coli DH10B derivatives.
The inocula for all of the biodegradation or transformation experiments by Ralstonia sp. T6 and E. coli DH10B derivatives were prepared by growing bacteria in 50 ml LB medium overnight on a rotary shaker (180 rpm) at 30°C and 37°C, respectively. Cells were collected by centrifugation at 3,800 × g for 10 min at room temperature. The pellets of Ralstonia sp. T6 and DH10B derivatives were washed twice and resuspended with MSM to give OD600s of 2.1 and 1.1, respectively. The cells were inoculated at 5% (vol/vol) into a 50-ml flask containing 20 ml TMM (50 mg/liter, pH 7.0) for T6 derivatives and 20 ml TMM (10 mg/liter, pH 7.0) for DH10B derivatives and incubated at 30°C in a rotary shaker (180 rpm). TCP was measured by a previously described method (15). All experiments were performed in triplicate.
TCP and 2,4,6-trichlorophenol conversion by E. coli DH10B derivatives, Ralstonia sp. T6, and C. necator JMP134.
All of the TCP and 2,4,6-trichlorophenol conversion experiments by E. coli DH10B derivatives, Ralstonia sp. T6, and C. necator JMP134 were carried out as mentioned above. Ralstonia sp. T6 and C. necator JMP134 were grown at 30°C, and OD600 was adjusted to 2.1; E. coli DH10B derivatives were grown at 37°C, and OD600 was adjusted to 1.1. The initial concentration of 2,4,6-trichlorophenol was 20 mg/liter, and those of TCP for E. coli DH10B derivatives and C. necator JMP134 were 11 mg/liter and 10 mg/liter, respectively.
Enzyme assay of cell extracts.
The crude cell extract enzyme assay was demonstrated by detecting the depletion of the substrate. To detect DphJ, activity against 3,6-dihydroxypyridine-2,5-dione (DHPD) was determined by monitoring the disappearance of absorbance at 390 nm (UV-2450; Shimadzu). Enzymatic activity was measured in MSM (pH 7.0) containing 0.2 mM DHPD and 0.02 mg protein/ml of DphJ cell extracts. The product formed from complete conversion of DHPD was prepared as the substrate of DhpI. The reaction mixture of DhpI contained 0.03 mg protein/ml of the cell extracts and 0.2 mM substrate. The reaction was followed by monitoring the loss of absorbance at 332 nm. Those reactions were carried out in a 3.05-ml reaction mixture for DhpJ and a 3.1-ml reaction mixture for DhpI, both of which were incubated at 30°C for 40 min until substrates completely vanished in a UV-visible scanning spectrum. All assays were also conducted with strains carrying only vectors as controls. One unit of enzyme activity was defined as the amount required for the disappearance of 1 nmol substrate per min. Specific activities are expressed as units per milligram of protein. All experiments were performed in triplicate.
Analysis of the metabolites during TCP degradation by strain T6.
A preliminary analysis of the green metabolite produced during TCP biodegradation by T6 was previously reported (15). To further identify the green metabolite, it was analyzed by tandem mass spectrometry (MS/MS) (Finnigan TSQ Quantum Ultra AM; Thermal, USA) with direct injection. The green metabolite was purified with a preparative liquid chromatograph equipped with a YMC-Pack Pro C18 column (20 by 150 mm, 5-μm grain size) for nuclear magnetic resonance (NMR) analysis to further verify its structure, and the mobile phase was acetonitrile-water (30:70, vol/vol). 1H NMR measurements were conducted on a Bruker Avance III 400 spectrometer operating at 300.18 MHz with dimethyl sulfoxide (DMSO) inside at room temperature.
Another metabolite in TCP biodegradation by T6 was named Met332 for its UV absorbance at 332 nm. Met332 was analyzed by MS/MS (Agilent 6410; triple quadrupole liquid chromatograph/mass spectrometer [QQQ LC/MS]) and ionized by electrospray with a negative polarity. The metabolite was identified from 15 ml of the reaction mixtures of the DhpJ crude extracts, which were dried by lyophilization and dissolved in 2 ml methanol. The mobile phase was methanol-water (60:40, vol/vol), and the flow rate was 0.5 ml/min.
Nucleotide sequence accession numbers.
The DNA sequences obtained in this study have been deposited in the GenBank database (accession no. KC294622 for the tcpRXA gene cluster and accession no. KC294623 for the dhpRIJK gene cluster).
RESULTS
Disruption of tcpA in Ralstonia sp. T6 and cloning of the tcpRXA gene cluster.
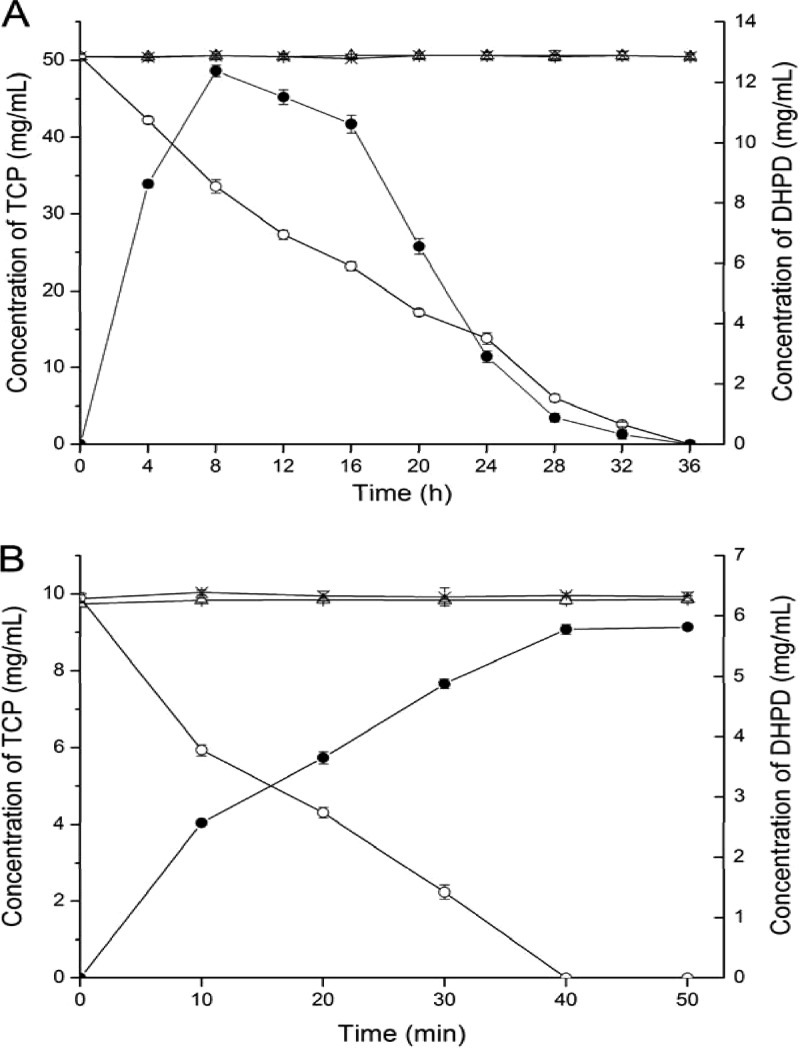
Considering the structural similarity of 3,5,6-trichloropyridinol and 2,4,6-trichlorophenol, we deduced that 2,4,6-trichlorophenol monooxygenase TcpA from C. necator JMP134 might be involved in the degradation of 3,5,6-trichloropyridinol by Ralstonia sp. T6. To test our hypothesis, an approximately 1,330-bp fragment of tcpA was amplified from the genomic DNA of Ralstonia sp. T6, and it showed 91% identity to tcpA of strain JMP134. The tcpA gene of T6 was then disrupted with the suicidal plasmid pJQ-TY, which carried a 617-bp internal fragment of tcpA, to confirm its function in the biodegradation of TCP. The wild-type strain of T6 could turn the medium green and degrade TCP. In contrast, the mutant strain T6-ΔtcpA could not degrade TCP (Fig. 1A) and no green intermediate was produced, but it could degrade the green metabolite DHPD generated by strain T6 (data not shown).
Fig 1.
(A) Degradation of 3,5,6-trichloro-2-pyridinol (TCP) by the derivatives of Ralstonia sp. T6. △, concentration of TCP not inoculated; ×, concentration of TCP inoculated with the mutant T6-ΔtcpA; ○, concentration of TCP inoculated with the complementation strain T6-ΔtcpA-com; ●, concentration of DHPD inoculated with T6-ΔtcpA-com. (B) Transformation of 3,5,6-trichloro-2-pyridinol (TCP) by the derivatives of E. coli DH10B. △, concentration of TCP not inoculated; ×, concentration of TCP inoculated with DH10B-pUC19; ○, concentration of TCP inoculated with DH10B-tcpRXA; ●, concentration of DHPD inoculated with DH10B-tcpRXA.
Disruption of tcpA from strain T6 indicated that tcpA and its flanking genes function in the initiation step of TCP biodegradation. A 6,412-bp fragment was amplified by SEFA PCR and contained three putative genes that constitute the tcpRXA gene cluster (Fig. 2). All tcp genes start with the ATG codon. The genes tcpR, tcpX, and tcpA are 972 bp, 552 bp, and 1,554 bp in length, respectively. Gene tcpR is located approximately 150 bp upstream of tcpX, which is spaced from tcpA by a sequence of 104 bp. RT-PCR analyses were performed to clarify the transcriptional features of these genes. Total RNA extracted from strain T6 was used as the template, and three oligonucleotide sets were used as the PCR primers. RT-PCR with each of the primer sets amplified DNA fragments of the expected sizes (see Fig. S1 in the supplemental material). These results indicated that tcpX and tcpA were transcribed from a common promoter. The best matches with existing database sequences for the tcp genes are shown in Table 3. TcpA and TcpR are closely related to 2,4,6-trichlorophenol monooxygenase and LysR family transcriptional regulator, respectively, and TcpX is closely related to a flavin reductase that can deliver H from NAD(P)H to flavin adenine dinucleotide (FAD) to produce reduced FAD (FADH2). The proteins encoded by orf1 and orf2 show high identity to NADH-dependent maleylacetate reductase TcpD from C. necator JMP134, which is involved in 2-chloromaleylacetate reduction, and the GntR family transcriptional regulator, respectively.
Fig 2.
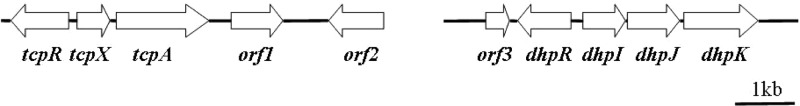
Organization of 3,5,6-trichloro-2-pyridinol catabolic clusters in Ralstonia sp. T6. The primers for generating the tcpRXA and dhpRIJK gene clusters are listed in Table 2. The homology of Ralstonia sp. T6 gene products is shown in Table 3.
Table 3.
BLAST results for deduced amino acid sequences of TCP catabolic gene clusters
| T6 protein | Representative homolog | Source | GenBank accession no. | % identity |
|---|---|---|---|---|
| TcpR | TcpR, LysR family transcriptional regulator | Cupriavidus necator JMP134 | YP_295793 | 89 |
| LysR family transcriptional regulator | Cupriavidus necator N-1 | YP_004687804 | 89 | |
| TcpX | TcpX, putative flavin reductase | Cupriavidus necator JMP134 | YP_295794 | 87 |
| RutF, NAD(P)H-flavin reductase | Cupriavidus necator N-1 | YP_004687805 | 87 | |
| TcpA | TcpA, 2,4,6-trichlorophenol monooxygenase | Cupriavidus necator JMP134 | YP_295795 | 96 |
| 2,4,6-Trichlorophenol monooxygenase | Cupriavidus necator N-1 | YP_004687806 | 96 | |
| Orf1 | TcpD, maleylacetate reductase | Cupriavidus necator JMP134 | YP_295799 | 91 |
| TftE, maleylacetate reductase | Cupriavidus necator N-1 | YP_004687810 | 92 | |
| Orf2 | GntR family transcriptional regulator | Cupriavidus necator JMP134 | YP_295790 | 82 |
| GntR family transcriptional regulator | Cupriavidus necator N-1 | YP_004687751 | 83 | |
| DhpR | AraC family transcriptional regulator | Ralstonia eutropha H16 | YP_728590 | 99 |
| AraC family transcriptional regulator | Burkholderia dolosa AUO158 | ZP_04947452 | 41 | |
| DhpI | Putative fumarylpyruvate hydrolase | Ralstonia eutropha H16 | YP_728591 | 97 |
| Fumarylacetoacetate (FAA) hydrolase family protein 14 | Achromobacter xylosoxidans A8 | YP_003982846 | 70 | |
| DhpJ | Chitooligosaccharide deacetylase | Ralstonia eutropha H16 | YP_728592 | 99 |
| Polysaccharide deacetylase | Methylobacterium radiotolerans JCM 2831 | YP_001757215 | 50 | |
| DhpK | MFS family transporter | Ralstonia eutropha H16 | YP_728593 | 99 |
| Major facilitator transporter | Achromobacter xylosoxidans AXX-A | EGP45397 | 43 | |
| Orf3 | Hypothetical protein | Ralstonia eutropha H16 | YP_728589 | 95 |
| Glutathione-dependent formaldehyde-activating protein | Marinobacter aquaeolei VT8 | YP_960185 | 76 |
Function of tcpA in TCP biodegradation.
To verify the function of tcpA in TCP biodegradation, a DNA fragment of 2,437 bp, including tcpXA with its original promoter, was amplified using the primers TCPXA-S and TCPXA-A. The strain T6-ΔtcpA-com recovered the ability to degrade TCP and showed an accumulation of DHPD comparable to strain T6 (Fig. 1A); the degradation rate was 33.3 mg liter−1 day−1 (22% of strain T6). Additionally, the tcpRXA gene cluster expression strain, DH10B-tcpRXA, could completely transform 10 mg/liter TCP to DHPD in 40 min, but the control strain DH10B-pUC19 could not transform TCP to DHPD (Fig. 1B). Strain DH10B-tcpRXA could not further transform DHPD (data not shown). These data indicate that tcpA is the key gene in TCP degradation by strain T6, the protein of which can dehalogenate TCP to yield DHPD. In addition, in contrast to E. coli DH10B with tcpRXA (DH10B-tcpRXA), E. coli DH10B with tcpA alone could not transform TCP to DHPD (data not shown); it indicated that monooxygenase TcpA needs flavin reductase TcpX to supply FADH2 when transforming TCP.
Cloning and sequence analysis of the dhpRIJK gene cluster from T6.
A library of DHPD degradation mutants of T6 was obtained by random transposon mutagenesis. Four clones that could not degrade DHPD (data not shown) were selected from more than 6,000 mutants, and one of them was designated DHPD-RM-1 and used to amplify the flanking sequences of the Mariner transposon.
The genomic DNA of the mutant DHPD-RM-1 was extracted for PCR amplification. Subsequently, a 5,942-bp fragment containing the mutated gene and its flanking sequences was obtained after several rounds of genome walking by PCR. Sequence analysis showed that the Mariner transposon inserted in a putative AraC family transcriptional regulator gene. Four putative genes were identified and annotated as the dhpRIJK gene cluster, on the basis of BLAST analysis (Fig. 2). Most dhp genes start with an ATG codon, except for dhpK, which starts with a GTG codon. The genes dhpR, dhpI, dhpJ, and dhpK are 900 bp, 717 bp, 879 bp, and 1,251 bp in length, respectively. The spacing between dhpR and dhpI is 211 bp, the spacing between dhpI and dhpJ is 24 bp, and the spacing between dhpJ and dhpK is 62 bp. The best database matches with existing sequences for the dhp genes are shown in Table 3. DhpR, DhpI, DhpJ, and DhpK are highly similar to AraC family transcriptional regulator, putative fumarylpyruvate hydrolase, chitooligosaccharide deacetylase, and MFS family transporter of the Ralstonia eutropha H16 genome (accession number AM260480), respectively.
Functional analysis of dhpI and dhpJ in TCP biodegradation.
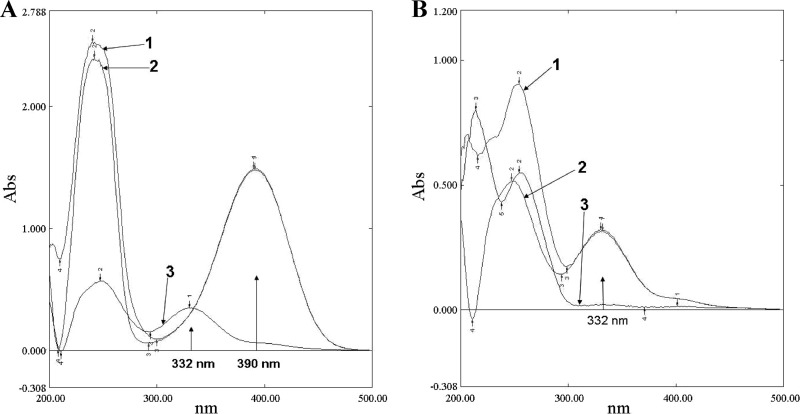
IPTG-induced expression of DhpI and DhpJ was confirmed by SDS-PAGE (see Fig. S2 in the supplemental material). The molecular mass of DhpI is approximately 30 kDa, and that of DhpJ is approximately 37 kDa, which agree well with the molecular masses deduced from their amino acid sequences. Cell extracts of E. coli BL21-pETJ were found to contain DhpJ with a specific activity of 250 U/mg. The characteristic absorption peak of DHPD at 390 nm vanished completely in UV scanning, and a new absorption peak at 332 nm appeared, which indicated that a new metabolite was produced (Fig. 3A). The metabolite was named Met332 according to its absorption at 332 nm. Neither DHPD consumption nor Met322 release was detected in the negative control. This result indicated that dhpJ is the key gene in the biodegradation of DHPD.
Fig 3.
(A) UV-visible spectral changes in DHPD conversion. Line 1, added with cell extracts of BL21-pET29a; line 2, added with MSM; line 3, added with cell extracts of BL21-pET-J. (B) UV-visible spectral changes in ATOPA conversion. Line 1, added with cell extracts of BL21-pET29a; line 2, added with MSM; line 3, added with cell extracts of BL21-pET-I.
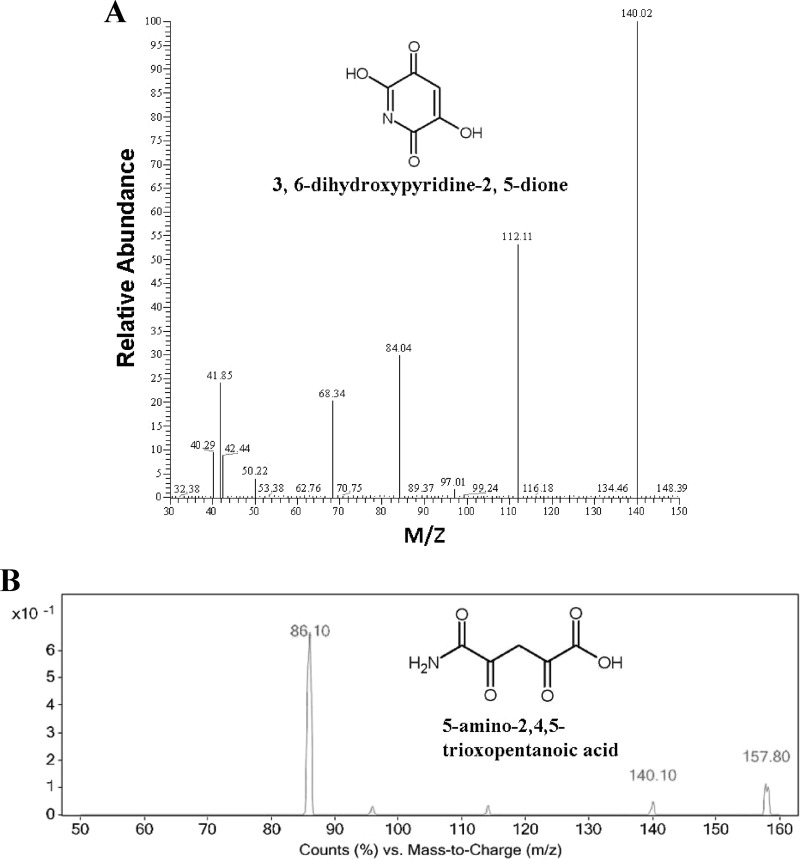
The metabolite Met332 generated a deprotonated prominent molecular ion at m/z = 158 [M-H]− in the first-order MS that is 18 Da more than its parent molecule of DHPD. This finding indicates that Met332 is the product generated by the hydrolysis of DHPD. DhpJ is highly similar to chitooligosaccharide deacetylase that hydrolyzes the amide bond and has a structure similar to the amide bond in DHPD. Met332 is preliminarily identified as 5-amino-2,4,5-trioxopentanoic acid (ATOPA), which shows characteristic fragment ion peaks of the second-order MS at m/z = 140 [M-18]−, 86 [M-72]−, indicating the results of losing H2O, CO + CO + NH2 (Fig. 4B).
Fig 4.
Mass spectra of the intermediate metabolites in 3,5,6-trichloro-2-pyridinol degradation by Ralstonia sp. T6. (A) Negative-ion mass spectra of the metabolite 3,6-dihydroxypyridine-2,5-dione; (B) negative-ion mass spectra of the metabolite 5-amino-2,4,5-trioxopentanoic acid.
The hydrolysis product of DHPD, ATOPA, was used as the substrate for DhpI. The characteristic absorption peak at 332 nm of ATOPA vanished after incubation with DhpI for 40 min at 30°C (Fig. 3B). ATOPA was transformed completely by DhpI, which showed a specific activity of 167 U/mg. These results indicated that DhpI is the key enzyme in the degradation of ATOPA.
DISCUSSION
The environmental fate of 3,5,6-trichloro-2-pyridinol (TCP) has been widely investigated because of its potential toxicity to the environment (29–33). The complete degradation of TCP in soil is believed to be microbially mediated (7), but the bacteria that degrade TCP are difficult to screen for because of the antibiotic activity of TCP (6). Most TCP degraders isolated degraded TCP with a relatively low efficiency; 3 to 12 days was required to degrade less than 100 mg/liter of TCP with the concentration of 108 cells/ml, and some of them lost the ability when the TCP concentration was higher than 200 mg/liter (11). Strain T6 was a highly effective TCP degrader and could degrade 100 mg/liter TCP in 12 h by equivalent numbers of cells and even retain the degradative ability at high concentrations of TCP up to 700 mg/liter (15). Strain T6 is a good model system for researching the molecular mechanism of TCP biodegradation.
The tcpRXA gene cluster was cloned from strain T6 and showed significant identity with the tcpRXA gene cluster involved in 2,4,6-trichlorophenol degradation from C. necator JMP134. tcpB (quinone reductase gene), tcpC (6-chlorohydroxy-quinol 1,2-dioxygenase gene), tcpY (unknown protein gene), and tcpD (maleylacetate reductase gene) were downstream of tcpRXA in C. necator JMP134, but they were all missed in Ralstonia sp. T6; however, tcpD showed significant identity to orf1 (34). The insertion mutation strain T6-ΔtcpA could not degrade TCP but could degrade the green metabolite DHPD, and the complementation strain T6-ΔtcpA-com recovered the ability to degrade TCP. The tcpRXA gene cluster expression strain, E. coli DH10B-tcpRXA, had the ability to transform TCP to DHPD. These results indicated that tcpA is the key gene for the initiation step of TCP degradation by strain T6.
TcpA of strain T6 was completely different from Tcp3A derived from the cow rumen metagenomic library (16). It has been reported that E. coli DH5α harboring tcp3A could use TCP as the sole source of carbon for its growth, but the reaction catalyzed by Tcp3A has not been identified (16). Strain T6 could also use trichlorophenol as the substrate, but transformation activity of C. necator JMP134 was not observed when TCP was used as the substrate (see Fig. S3 in the supplemental material). However, when the tcpRXA genes from C. necator JMP134 and Ralstonia sp. T6 were expressed in E. coli, both of them could use TCP and trichlorophenol as the substrates (see Fig. S4 in the supplemental material). This outcome implied the lack of transporter of TCP in C. necator JMP134. In addition, it is reasonable to speculate that TcpA from Ralstonia sp. T6 confers a reaction mechanism similar to that proposed for TcpA from C. necator JMP134. TcpA of strain JMP134 catalyzes sequential dechlorinations of 2,4,6-trichorophenol by oxidative and hydrolytic reactions (19, 35, 36). TcpA of strain T6 performs a function toward dehalogenase from the pyridine ring similar to that of TcpA of strain JMP134. We failed to detect any dehalogenation intermediates during the degradation of TCP by strain T6, and the 3 chlorides were released simultaneously, as determined by chloride titration during the degradation process (15). The green intermediate was preliminarily identified as 3,6-dihydroxypyridine-2,5-dione (15). To further clarify its structure, the green metabolite, with a molecular ion at m/z 140 [M]− in the first-order MS, was analyzed by MS/MS. The green metabolite showed characteristic fragment ion peaks of the second-order MS at m/z = 112 [M-28]−, 84 [M-56]−, 68 [M-72]−, 42 [M-98]−, indicating that CO, CO + CO, CO + CO + O, and CO + CO + CH COH were lost (Fig. 4A). We further verified its structure with NMR. In the NMR spectrum of DHPD (see Fig. S5 in the supplemental material), only one hydrogen atom on the pyridine ring (7.30 ppm) could be detected. The hydrogens of the hydroxyl group were too active to be detected. According to the MS and NMR results, the structure of DHPD was proved to be right. These results indicated that 2,4,6-trichlorophenol monooxygenase TcpA of strain T6 might have adopted novel dehalogenation mechanisms for the degradation of TCP.
COH were lost (Fig. 4A). We further verified its structure with NMR. In the NMR spectrum of DHPD (see Fig. S5 in the supplemental material), only one hydrogen atom on the pyridine ring (7.30 ppm) could be detected. The hydrogens of the hydroxyl group were too active to be detected. According to the MS and NMR results, the structure of DHPD was proved to be right. These results indicated that 2,4,6-trichlorophenol monooxygenase TcpA of strain T6 might have adopted novel dehalogenation mechanisms for the degradation of TCP.
The products of tcpRXA from strain T6 could not further utilize DHPD. It can be expected that diverse genes are involved in the degradation of TCP. The dhpRIJK gene cluster was cloned after random transposon insertion, which is closely related to a conceptually annotated gene cluster from R. eutropha H16 on the basis of BLAST analysis. This 5,341-bp (bp 271 to 5611) fragment showed 98% similarity to the genome sequence of R. eutropha H16, and a 265-bp sequence in the 5′ terminus was similar to a transposase from Ralstonia oxalatica transposon Tn437. No significant similar sequences were identified for the 331-bp sequence at the 3′ terminus. Such an organized structure implies that strain T6 perhaps acquired this genomic region through bilateral transfer. DhpJ (H16_B0428) from strain H16 is a putative chitooligosaccharide deacetylase, which catalyzes the hydrolysis of carbon-nitrogen bonds. DHPD also has a similar bond structure. Based on the structure of the product, 5-amino-2,4,5-trioxopentanoic acid (ATOPA), we deduced that DhpJ of strain T6 could cleave the pyridine ring of DHPD by hydrolysis. This is different from the ring fission of other hydroxylated pyridines (37). Dioxygenase and monooxygenase were proposed to be involved in the transformation of hydroxylated pyridines (37). ATOPA could be degraded further by DhpI. 3,6-Dihydroxypyridine-2,5-dione hydrolase DhpJ and 5-amino-2,4,5-trioxopentanoic acid hydrolase DhpI could collaboratively degrade DHPD. Additionally, it was deduced that DhpR and DhpK play a regulatory role and a transporter role, respectively, in the dhpRIJK gene cluster by analysis of protein sequence homology. Insertion mutations in dhpR or dhpJ caused strain T6 to be deficient in DHPD degradation.
A proposed pathway of TCP biodegradation by strain T6 is shown in Fig. 5. 2,4,6-Trichlorophenol monooxygenase TcpA of strain T6 transforms TCP to DHPD first, which is then cleaved by the hydrolysis reaction of 3,6-dihydroxypyridine-2,5-dione hydrolase DhpJ to produce ATOPA. ATOPA is further degraded by 5-amino-2,4,5-trioxopentanoic acid hydrolase DhpI. One well-known report on the TCP biodegradation metabolic pathway addresses the combination of photolytic and microbiological degradation by Pseudomonas sp. ATCC 700113. The resting cells of Pseudomonas sp. ATCC 700113 degraded the reductive dechlorination products of TCP generated by photolysis, suggesting that Pseudomonas sp. ATCC 700113 utilizes a reductive dechlorination pathway in degrading TCP (18). However, we propose that TCP is dechlorinated by oxidative and hydrolytic reactions in strain T6, to generate DHPD. The metabolic pathway of TCP biodegradation in strain T6 has never been reported before.
Fig 5.
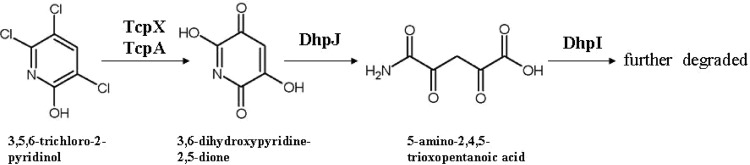
Proposed metabolic pathway for 3,5,6-trichloro-2-pyridinol in Ralstonia sp. T6.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Natural Science Foundation of Jiangsu Province, China (BK2012029), the Natural Science Foundation of China (31270095), and the National Science and Technology Support Program (2012BAD14B02) and grant 2011-Z21 from the Ministry of Agriculture of China.
We thank Luying Xun for the helpful discussion on the catalytic mechanism of TcpA on 3,5,6-trichloropyridinol.
Footnotes
Published ahead of print 20 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01817-13.
REFERENCES
- 1.Yang L, Zhao Y, Zhang B, Yang C, Zhang X. 2005. Isolation and characterization of a chlorpyrifos and 3,5,6-trichloro-2-pyridinol degrading bacterium. FEMS Microbiol. Lett. 251:67–73 [DOI] [PubMed] [Google Scholar]
- 2.Chapman RA, Harris CR. 1980. Persistence of chlorpyrifos in a mineral and an organic soil. J. Environ. Sci. Health B 15:39–46 [DOI] [PubMed] [Google Scholar]
- 3.Racke KD, Coats JR, Titus KR. 1988. Degradation of chlorpyrifos and its hydrolysis product, 3,5,6-trichloro-2-pyridinol, in soil. J. Environ. Sci. Health B 23:527–539 [Google Scholar]
- 4.Kim JR, Ahn YJ. 2009. Identification and characterization of chlorpyrifos-methyl and 3,5,6-trichloro-2-pyridinol degrading Burkholderia sp. strain KR100. Biodegradation 20:487–497 [DOI] [PubMed] [Google Scholar]
- 5.Armbrust KL. 2001. Chlorothalonil and chlorpyrifos degradation products in golf course leachate. Pest Manag. Sci. 57:797–802 [DOI] [PubMed] [Google Scholar]
- 6.Racke KD, Robbins ST. 1991. Factors affecting the degradation of 3,5,6-trichloro-2-pyridinol in soil. ACS Symp. Ser. Am. Chem. Soc. 459:93–107 [Google Scholar]
- 7.Feng Y, Racke KD, Bollag JM. 1997. Isolation and characterization of a chlorinated-pyridinol-degrading bacterium. Appl. Environ. Microbiol. 63:4096–4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan MK, Sheldon LS, Croghan CW, Jones PA, Robertson GL, Chuang JC, Wilson NK, Lyu CW. 2005. Exposures of preschool children to chlorpyrifos and its degradation product 3,5,6-trichloro-2-pyridinol in their everyday environments. J. Expo. Anal. Environ. Epidemiol. 15:297–309 [DOI] [PubMed] [Google Scholar]
- 9.Kashanian S, Shariati Z, Roshanfekr H, Ghobadi S. 2012. DNA binding studies of 3,5,6-trichloro-2-pyridinol pesticide metabolite. DNA Cell Biol. 31:1341–1348 [DOI] [PubMed] [Google Scholar]
- 10.Xu G, Zheng W, Li Y, Wang S, Zhang J, Yan Y. 2008. Biodegradation of chlorpyrifos and 3,5,6-trichloro-2-pyridinol by a newly isolated Paracoccus sp. strain TRP. Int. Biodeterior. Biodegradation 62:51–56 [Google Scholar]
- 11.Maya K, Singh RS, Upadhyay SN, Dubey SK. 2011. Kinetic analysis reveals bacterial efficacy for biodegradation of chlorpyrifos and its hydrolyzing metabolite TCP. Process Biochem. 46:2130–2136 [Google Scholar]
- 12.Xu G, Li Y, Zheng W, Peng X, Li W, Yan Y. 2007. Mineralization of chlorpyrifos by co-culture of Serratia and Trichosporon spp. Biotechnol. Lett. 29:1469–1473 [DOI] [PubMed] [Google Scholar]
- 13.Anwar S, Liaquat F, Khan QM, Khalid ZM, Iqbal S. 2009. Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. J. Hazard. Mater. 168:400–405 [DOI] [PubMed] [Google Scholar]
- 14.Feng Y, Racke KD, Bollag JM. 1997. Use of immobilized bacteria to treat industrial wastewater containing a chlorinated pyridinol. Appl. Microbiol. Biotechnol. 47:73–77 [DOI] [PubMed] [Google Scholar]
- 15.Li J, Liu J, Shen W, Zhao X, Hou Y, Cao H, Cui Z. 2010. Isolation and characterization of 3, 5, 6-trichloro-2-pyridinol-degrading Ralstonia sp. strain T6. Bioresour. Technol. 101:7479–7483 [DOI] [PubMed] [Google Scholar]
- 16.Math RK, Asraful Islam SM, Cho KM, Hong SJ, Kim JM, Yun MG, Cho JJ, Heo JY, Lee YH, Kim H, Yun HD. 2010. Isolation of a novel gene encoding a 3,5,6-trichloro-2-pyridinol degrading enzyme from a cow rumen metagenomic library. Biodegradation 21:565–573 [DOI] [PubMed] [Google Scholar]
- 17.Li K, Wang S, Shi Y, Qu J, Zhai Y, Xu L, Xu Y, Song J, Liu L, Rahman MA, Yan Y. 2011. Genome sequence of Paracoccus sp. strain TRP, a chlorpyrifos biodegrader. J. Bacteriol. 193:1786–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng Y, Minard RD, Bollag JM. 1998. Photolytic and microbial degradation of 3,5,6-trichloro-2-pyridinol. Environ. Toxicol. Chem. 17:814–819 [Google Scholar]
- 19.Louie TM, Webster CM, Xun L. 2002. Genetic and biochemical characterization of a 2,4,6-trichlorophenol degradation pathway in Ralstonia eutropha JMP134. J. Bacteriol. 184:3492–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S, He J, Cui Z, Li S. 2007. Self-formed adaptor PCR: a simple and efficient method for chromosome walking. Appl. Environ. Microbiol. 73:5048–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller SA, Dykes DD, Polesky HF. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quandt J, Hynes MF. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15–21 [DOI] [PubMed] [Google Scholar]
- 23.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 24.Yin Y, Xiao Y, Liu HZ, Hao F, Rayner S, Tang H, Zhou NY. 2010. Characterization of catabolic meta-nitrophenol nitroreductase from Cupriavidus necator JMP134. Appl. Microbiol. Biotechnol. 87:2077–2085 [DOI] [PubMed] [Google Scholar]
- 25.Chiang SL, Mekalanos JJ. 2000. Construction of a Vibrio cholerae vaccine candidate using transposon delivery and FLP recombinase-mediated excision. Infect. Immun. 68:6391–6397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu J, Ma Y, Chen L, Wu L, Wen Y, Liu W. 2011. A sirA-like gene, sirA2, is essential for 3-succinoyl-pyridine metabolism in the newly isolated nicotine-degrading Pseudomonas sp. HZN6 strain. Appl. Microbiol. Biotechnol. 92:1023–1032 [DOI] [PubMed] [Google Scholar]
- 27.Liu YG, Whittier RF. 1995. Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from Pl and YAC clones for chromosome walking. Genomics 25:674–681 [DOI] [PubMed] [Google Scholar]
- 28.Shapiro AL, Viñuela E, Maizel JV. 1967. Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem. Biophys. Res. Commun. 28:815–820 [DOI] [PubMed] [Google Scholar]
- 29.Hanley TR, Carney EW, Johnson EW. 2000. Developmental toxicity studies in rats and rabbits with 3,5,6-trichloro-2-pyridinol, the major metabolite of chlorpyrifos. Toxicol. Sci. 53:100–108 [DOI] [PubMed] [Google Scholar]
- 30.Baskaran S, Kookana RS, Naidu R. 2003. Contrasting behaviour of chlorpyrifos and its primary metabolite, TCP (3,5,6-trichloro-2-pyridinol), with depth in soil profiles. Aust. J. Soil Res. 41:749–760 [Google Scholar]
- 31.Randhawa MA, Anjum FM, Ahmed A, Randhawa MS. 2007. Field incurred chlorpyrifos and 3,5,6-trichloro-2-pyridinol residues in fresh and processed vegetables. Food Chem. 103:1016–1023 [Google Scholar]
- 32.Cáceres T, He W, Naidu R, Megharaj M. 2007. Toxicity of chlorpyrifos and TCP alone and in combination to Daphnia carinata: the influence of microbial degradation in natural water. Water Res. 41:4497–4503 [DOI] [PubMed] [Google Scholar]
- 33.Ling Y, Wang H, Yong W, Zhang F, Sun L, Yang M, Wu Y, Chu X. 2011. The effects of washing and cooking on chlorpyrifos and its toxic metabolites in vegetables. Food Control 22:54–58 [Google Scholar]
- 34.Sánchez MA, González B. 2007. Genetic characterization of 2,4,6-trichlorophenol degradation in Cupriavidus necator JMP134. Appl. Environ. Microbiol. 73:2769–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matus V, Sánchez MA, Martínez M, González B. 2003. Efficient degradation of 2,4,6-trichlorophenol requires a set of catabolic genes related to tcp genes from Ralstonia eutropha JMP134(pJP4). Appl. Environ. Microbiol. 69:7108–7115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xun L, Webster CM. 2004. A monooxygenase catalyzes sequential dechlorinations of 2,4,6-trichlorophenol by oxidative and hydrolytic reactions. J. Biol. Chem. 279:6696–6700 [DOI] [PubMed] [Google Scholar]
- 37.Kaiser JP, Feng Y, Bollag JM. 1996. Microbial metabolism of pyridine, quinoline, acridine, and their derivatives under aerobic and anaerobic conditions. Microbiol. Rev. 60:483–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.