Abstract
Many insects possess symbiotic bacteria that affect the biology of the host. The level of the symbiont population in the host is a pivotal factor that modulates the biological outcome of the symbiotic association. Hence, the symbiont population should be maintained at a proper level by the host's control mechanisms. Several mechanisms for controlling intracellular symbionts of insects have been reported, while mechanisms for controlling extracellular gut symbionts of insects are poorly understood. The bean bug Riptortus pedestris harbors a betaproteobacterial extracellular symbiont of the genus Burkholderia in the midgut symbiotic organ designated the M4 region. We found that the M4B region, which is directly connected to the M4 region, also harbors Burkholderia symbiont cells, but the symbionts therein are mostly dead. A series of experiments demonstrated that the M4B region exhibits antimicrobial activity, and the antimicrobial activity is specifically potent against the Burkholderia symbiont but not the cultured Burkholderia and other bacteria. The antimicrobial activity of the M4B region was detected in symbiotic host insects, reaching its highest point at the fifth instar, but not in aposymbiotic host insects, which suggests the possibility of symbiont-mediated induction of the antimicrobial activity. This antimicrobial activity was not associated with upregulation of antimicrobial peptides of the host. Based on these results, we propose that the M4B region is a specialized gut region of R. pedestris that plays a critical role in controlling the population of the Burkholderia gut symbiont. The molecular basis of the antimicrobial activity is of great interest and deserves future study.
INTRODUCTION
Many insects possess symbiotic bacteria within their cells, tissues, and guts (1). These symbiotic associations have been established for a long time and are known to affect the biology of insects in various ways. Some symbionts play indispensable roles, such as providing essential nutrients (2). Other symbionts play conditionally beneficial roles, such as providing defense against natural enemies and adaptation to specific ecological conditions (3). Others may have parasitic or pathogenic effects on their hosts, causing attenuated host fitness and reproductive aberrations (4).
These symbiotic effects on the host's biology tend to be related to the symbiont population within the host. The level of the symbiont population affects the host's fitness, the fidelity of vertical transmission, and the intensity of the reproductive aberrations (5–8). Therefore, host insects are expected to develop systems to control and maintain the symbiont populations within an optimal range. Previous studies have reported or suggested several mechanisms for controlling symbiont populations, including mechanisms mediated by lysozymes for controlling Buchnera in aphids (9, 10), reactive oxygen species production for controlling Wolbachia in mosquitoes (11), and antimicrobial peptides (AMPs) for controlling primary endosymbionts in Sitophilus weevils (12). However, these studies have mostly focused on the intracellular symbionts of insects, while the mechanisms for controlling the extracellular gut symbionts of insects are poorly understood.
The bean bug Riptortus pedestris harbors a beneficial and specific bacterium of the genus Burkholderia in a specialized region of the posterior midgut (13). This symbiont, which is orally acquired by host nymphs from the environment every generation, is easily cultivable and genetically manipulable (14–16). Hence, the Riptortus-Burkholderia symbiotic system has been recognized as a promising model to study insect-microbe symbioses at the molecular level. Using this model system, we report a previously unrecognized insect organ with antimicrobial activity, which is presumably specialized for controlling the gut symbiont population.
MATERIALS AND METHODS
Insect rearing.
R. pedestris bean bugs were reared in our insect laboratory at 28°C under a long-day regime of 16 h light and 8 h dark as described previously (17). The insects were originally collected from fields of the soybean Glycine max at Tsukuba, Ibaraki, Japan, from which a laboratory strain, TKS-1, has been established (14). Approximately 200 nymphal insects were reared in each of the clean plastic containers (34 cm long by 19.5 cm wide and 27.5 cm high) and supplied with soybean seeds and distilled water containing 0.05% ascorbic acid (DWA). The containers were cleaned every day, and the soybean seeds and DWA were replaced every 2 days. Upon reaching adulthood, the insects were transferred to larger containers (35 cm long by 35 cm wide and 40 cm high) in which soybean plant pots were provided for food and cotton pads were attached to the walls for egg laying. Eggs were collected daily and transferred to new cages for hatching.
Burkholderia symbiont inoculation.
The Burkholderia symbiont strain RPE75, which is a spontaneous rifampin-resistant mutant derived from the strain RPE64 (15, 16), was cultured at 30°C in YG-RIF medium (YG medium [0.5% yeast extract, 0.4% glucose, and 0.1% NaCl] containing 30 μg/ml rifampin). The inoculum solution was prepared by suspending mid-log-phase cultured Burkholderia cells in DWA at a concentration of 107 cells/ml. Newly molted second-instar nymphs were provided with wet cotton balls soaked with the inoculum solution. After the insects were fed with the inoculum solution for 2 days, fresh, sterile DWA was provided to the insects instead of the inoculum solution (18).
Quantitative PCR.
Quantitative PCR (qPCR) for estimating the titers of the Burkholderia symbiont was performed as described previously (17). Dissected midgut samples were subjected to DNA extraction with the QIAamp DNA minikit (Qiagen). The DNA samples were mixed with a master PCR solution containing 2× qPCR premix from the QuantiMix SYBR Kit (PhileKorea) and the primers BSdnaA-F (5′-AGC GCG AGA TCA GAC GGT CGT CGA T-3′) and BSdnaA-R (5′-TCC GGC AAG TCG CGC ACG CA-3′), which target a 0.15-kb region of the dnaA gene of the Burkholderia symbiont (15). The dnaA gene encodes the chromosomal replication initiator protein DnaA (GenBank protein accession number BAN21755) and is a single-copy gene in the Burkholderia symbiont genome (19). The PCR temperature profile was 40 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 15 s. The threshold cycles (CT) of the midgut samples were applied to a standard curve generated with standard DNA samples containing known dnaA copy numbers to estimate the bacterial titers. To generate the standard curve of the dnaA gene copies, Burkholderia symbiont cells were cultured up to mid-log phase and serially diluted to obtain samples containing 103, 104, 105, 106, 107, 108, and 109 cells per 100 μl. The cell numbers were verified by a CFU assay. Each cell sample was subjected to the same procedures for DNA extraction and PCR analysis described above.
CFU assay for the symbiont population.
Dissected midgut samples were collected in 50 μl of 10 mM phosphate buffer (PB), pH 7.0, and homogenized with a pestle. The homogenized samples were serially diluted and spread on 1.5% agar plates of YG-RIF medium. After 2 days of incubation at 30°C, the colonies on the plates were counted and the numbers of symbiont cells in the samples were calculated as CFU times the dilution factor.
FISH.
Fluorescence in situ hybridization (FISH) analysis of symbiotic midgut regions was performed as described previously (16). Two fluorochrome-labeled probes, Alsym16S (5′-ACA CTC AAA GCC TGC CAG T-3′) and BURK129 (5′-CCA CTA CAG GAC ACG TTC-3′), whose 5′ ends were labeled with Alexa Fluor 555 (16), were simultaneously used to target different regions of the 16S rRNA of the Burkholderia symbiont. The symbiotic midgut regions dissected from third-instar nymphs were fixed with 4% paraformaldehyde in PB for 1 h at room temperature. After the fixation, the samples were treated with 0.1% Triton X-100 for 5 min and washed with phosphate-buffered saline (PBS) (137 mM NaCl, 8.1 mM Na2HPO4, 2.7 mM KCl, 1.5 mM KH2PO4, pH 7.5). The samples were incubated overnight in hybridization buffer (20 mM Tris-HCl, pH 8.0, 0.9 M NaCl, 0.01% SDS, 30% formamide) containing 50 nM (each) the probes at room temperature. After being washed with PBS, the samples were mounted on glass slides with 30% glycerol and observed under a fluorescence microscope (AX70; Olympus).
CFU assay for measuring antimicrobial activity.
The following bacterial cells were prepared: the symbiotic Burkholderia strain RPE75 freshly isolated from the M4 region as previously described (18), mid-log-phase Burkholderia RPE75 cultured at 30°C in YG-RIF medium, mid-log-phase Escherichia coli K-12 cultured at 37°C in LB medium (1% tryptone, 0.5% yeast extract, and 0.5% NaCl), and mid-log-phase Staphylococcus aureus RN4220 cultured at 37°C in LB medium. These bacterial cells were washed and diluted with PB to 500 to 1,000 CFU per 50 μl. M4B whole lysate was prepared by homogenizing M4B samples dissected from 10 fifth-instar nymphs in 100 μl PB, and the lysate was serially diluted with PB. Each sample, consisting of 50 μl of the lysate and 50 μl of the bacterial suspension, was incubated at room temperature for 15 min before being spread onto YG-RIF agar plates to count the CFU.
For comparing the M4B lysates between symbiotic and aposymbiotic insects, the M4B whole lysates were centrifuged at 20,000 × g for 15 min, and the supernatant was subjected to protein quantification by the Bradford assay (Bio-Rad). After serial dilution, 50 μl of the lysates at different concentrations (μg protein/ml) was incubated with 50 μl of the Burkholderia symbiont cell suspension. After 15 min of incubation at room temperature, the samples were spread onto YG-RIF agar plates, cultured for 2 days, and subjected to colony counting.
To examine the sensitivity to heat, the M4B lysate (0.2 μg of total protein/ml in PB) was incubated on ice or at 27°C, 37°C, 45°C, or 65°C for 1 h or heated at 100°C for 15 min prior to incubation with the Burkholderia symbiont cell suspension. The treated lysates (50 μl each) were incubated with the cell suspensions of the Burkholderia symbiont (50 μl each), and after 15 min of incubation at room temperature, the samples were spread onto YG-RIF agar plates, cultured for 2 days, and subjected to colony counting.
Reverse transcription-quantitative PCR.
The M4B regions were dissected from fifth-instar nymphs and subjected to total RNA extraction using RiboEx (GeneAll, South Korea). The RNA samples were reverse transcribed using Topscript RT DryMix containing oligo(dT) primers (Enzynomics, South Korea) to synthesize the cDNA. The cDNA was subjected to real-time quantitative PCR after being mixed with a Topreal qPCR 2× PreMix with SYBR green (Enzynomics, South Korea) and 0.25 μM (each) the primers listed in Table 1. The PCR temperature profile was set to 95°C for 10 min, followed by 40 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 20 s, using the CFX96 real-time PCR system (Bio-Rad). The comparative CT (ΔΔCT) method was used to calculate the relative gene expression levels with the elongation factor 1 alpha gene (EF1α) of R. pedestris (GenBank accession number AB591382) as an endogenous control gene.
Table 1.
Primers used for quantitative PCR
| Target gene | Sense primer | Antisense primer |
|---|---|---|
| Defensin-like peptide | TCGGTCGGACTGAGACTGAA | TTGCCGCCTTTGTATCCCTT |
| Pyrrhocoricin-like peptide | TCCGAAGCTGAGGGTCTTCCCG | TCCGCATCCAAGTTCGCGTCC |
| Thanatin-like peptide | GTCTGCCTTCGTTGAAGACG | ATTCGCTTGCAAACGCCG |
| Elongation factor 1 alpha | CCTGCATCCGTTGCTTTTGT | GGCATCGAGGGCTTCAATAA |
RESULTS AND DISCUSSION
A particular midgut region contains dead Burkholderia symbionts.
We measured Burkholderia symbiont populations in the symbiotic organs of R. pedestris, the midgut fourth section (M4) and the bulbous region prior to M4 (M4B) (Fig. 1A). The insects were infected with the Burkholderia symbiont strain RPE75 at early second instar, and when they reached the middle fifth instar, the symbiotic-organ samples, either M4 alone, M4B alone, or M4B plus M4, were dissected from the insects. The symbiont populations in these samples were measured by qPCR assay and CFU assay (Table 2). Unexpectedly, the qPCR results and CFU results exhibited striking discrepancies. Of particular interest was the difference in the symbiont populations in the M4B-alone samples. The qPCR assay detected more than 106 dnaA copies per M4B, while the CFU assay detected less than 10 bacterial cells per M4B. These results suggested that most dnaA copies detected by qPCR may be derived from dead symbiont cells. FISH of M4B and M4 corroborated this idea: while M4 exhibited strong fluorescent signals, M4B was scarcely stained, probably because of degradation of the symbionts' 16S rRNA in M4B (Fig. 1B). Another interesting observation was the difference in the symbiont populations between the M4-alone samples and the M4B-plus-M4 samples. In the CFU assay, the symbiont populations in the M4B-plus-M4 samples were approximately 1,700-fold smaller than those in the M4-alone samples, suggesting that M4B has a strong negative effect on the symbiont population (Table 2). On the basis of these results, we hypothesized that M4B has antimicrobial activity against the Burkholderia symbiont.
Fig 1.
(A) Midgut morphology of R. pedestris. The dissected intestine of a third-instar nymph shows the morphologically distinct midgut regions (M1, M2, M3, M4B, and M4), among which the M4B and M4 regions are symbiotic organs. The yellow dashed circle indicates the midgut regions subjected to FISH analysis. (B) FISH analysis of the dissected M4B and M4 regions. The yellow dashed lines indicate the outlines of the M4B and M4 regions. The red signal in the fluorescence image indicates 16S rRNA of the Burkholderia symbiont.
Table 2.
Symbiont populations in the symbiotic organs, the M4 and M4B midgut regions, in R. pedestris
| Assay | Samplea | Symbiont titerb |
|---|---|---|
| qPCR | M4B | 1.88 × 106 ± 1.18 × 105 |
| M4 | 2.93 × 108 ± 8.22 × 107 | |
| M4 plus M4B | 1.56 × 108 ± 4.71 × 107 | |
| CFU | M4B | 6.00 ± 2.00 |
| M4 | 1.26 × 107 ± 1.70 × 106 | |
| M4 plus M4B | 7.42 × 103 ± 1.77 × 103 |
Dissected from fifth-instar nymphs of R. pedestris.
dnaA copies for qPCR and CFUs for the CFU assay (means ± standard deviations [n = 3]).
The antimicrobial activity of the M4B midgut region is specifically potent against the symbiotic Burkholderia strain.
To investigate the antimicrobial activity of the M4B midgut region, we applied the whole lysate of the M4B samples to the following bacterial cells: symbiotic Burkholderia RPE75, cultured Burkholderia RPE75, E. coli K-12, and S. aureus RN4220. As shown in Fig. 2, the CFUs of the cultured Burkholderia and E. coli were not affected by treatment with the M4B lysate, whereas the CFUs of S. aureus were significantly reduced by treatment with the M4B lysate at relatively high concentrations (1:10, 1:5, and 1:1 dilutions). Strikingly, the CFUs of the symbiotic Burkholderia strain were significantly reduced by treatment with the M4B lysate, even at a low concentration (1:100 dilution), which was equivalent to half of an M4B/ml. These results confirmed that M4B has significant antimicrobial activity and revealed that the antimicrobial activity exhibits high potency specific for the symbiotic Burkholderia strain.
Fig 2.
The specific antimicrobial activity of M4B against the symbiotic Burkholderia strain. Cell suspensions of the Burkholderia symbiont, cultured Burkholderia, E. coli, and S. aureus were subjected to CFU assays after incubation with M4B lysate samples. PB, phosphate buffer without lysate; M4B WL, whole lysate of the M4B midgut region. For concentrations of M4 lysates, M4B WL (1:1) is equivalent to 50 dissected M4B regions/ml, and M4B WL (1:50) is equivalent to a dissected M4B/ml. Means and standard deviations are shown (n = 3). The asterisks indicate statistically significant differences compared to PB-treated bacteria (unpaired t test: *, P < 0.05; **, P < 0.01; ***, P < 0.005).
The antimicrobial activity of the M4B region is a symbiosis-related feature.
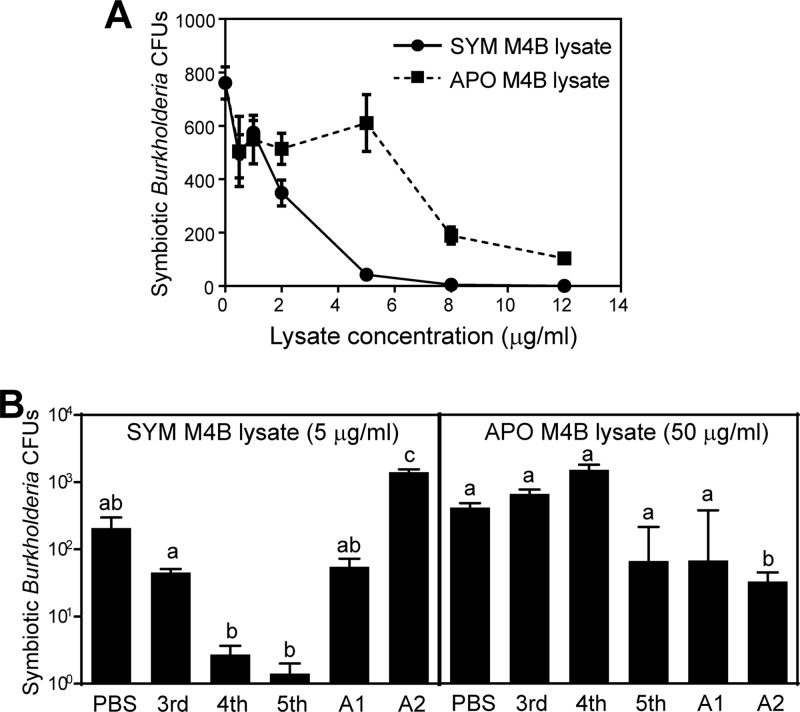
We prepared M4B samples from symbiotic and aposymbiotic fifth-instar insects and measured their antimicrobial activities against the symbiotic Burkholderia strain. The M4B lysates of the symbiotic insects exhibited approximately 6-fold-higher antimicrobial activities (concentration producing 50% inhibition of cell viability [IC50], 1.7 μg/ml) than those of the aposymbiotic insects (IC50, 10.0 μg/ml) (Fig. 3A). This result indicated that the antimicrobial activity against the symbiotic Burkholderia strain is associated with the M4B region of the symbiotic host insect and suggested the possibility that the presence of the Burkholderia symbiont may induce the antimicrobial activity against the symbiont in M4B.
Fig 3.
Symbiosis-related antimicrobial activity of M4B. (A) Comparison of the M4B antimicrobial activities between symbiotic (SYM) insects and aposymbiotic (APO) insects. Symbiotic Burkholderia cells isolated from M4 were incubated with different concentrations of SYM M4B lysates or APO M4B lysates and subjected to CFU assay. Means and standard errors are plotted (n = 3). (B) M4B antimicrobial activities at different developmental stages. The symbiotic Burkholderia cells were incubated with SYM M4B lysate (left) or with APO M4B lysate (right) prepared from different instar stages. 3rd, third instar; 4th, fourth instar; 5th, fifth instar; A1, early adult (within 3 days after adult molting); A2, late adult (approximately 10 days after adult molting). Means and standard errors are plotted (n = 3 to 6). Different letters (a, b, and c) indicate statistically significant differences (unpaired t test with Bonferroni's correction; P < 0.05).
We further surveyed the antimicrobial activities of the M4B lysates from the symbiotic and aposymbiotic insects at different developmental stages. At a protein concentration of 5 μg/ml, the M4B lysates of the symbiotic insects exhibited remarkably different levels of antimicrobial activity against the symbiotic Burkholderia strain: fifth instar > fourth instar > third instar = early adult > late adult (Fig. 3B, left). This pattern may reflect the symbiont populations within the host insects: a previous study reported the order of symbiont populations as fifth instar > fourth instar > third instar (15), and the symbiont populations declined during adult aging (J. K. Kim, unpublished data). On the other hand, even at a protein concentration as high as 50 μg/ml, the M4B lysates of the aposymbiotic insects consistently exhibited little antimicrobial activity (Fig. 3B, right). These results confirmed that the antimicrobial activity against the symbiotic Burkholderia strain is certainly associated with the M4B of the symbiotic host insect and demonstrated that the antimicrobial activity is regulated in a development-dependent manner, with the highest activity at the fifth-instar stage.
The antimicrobial activity of the M4B region against the symbiont is not associated with expression of antimicrobial peptides.
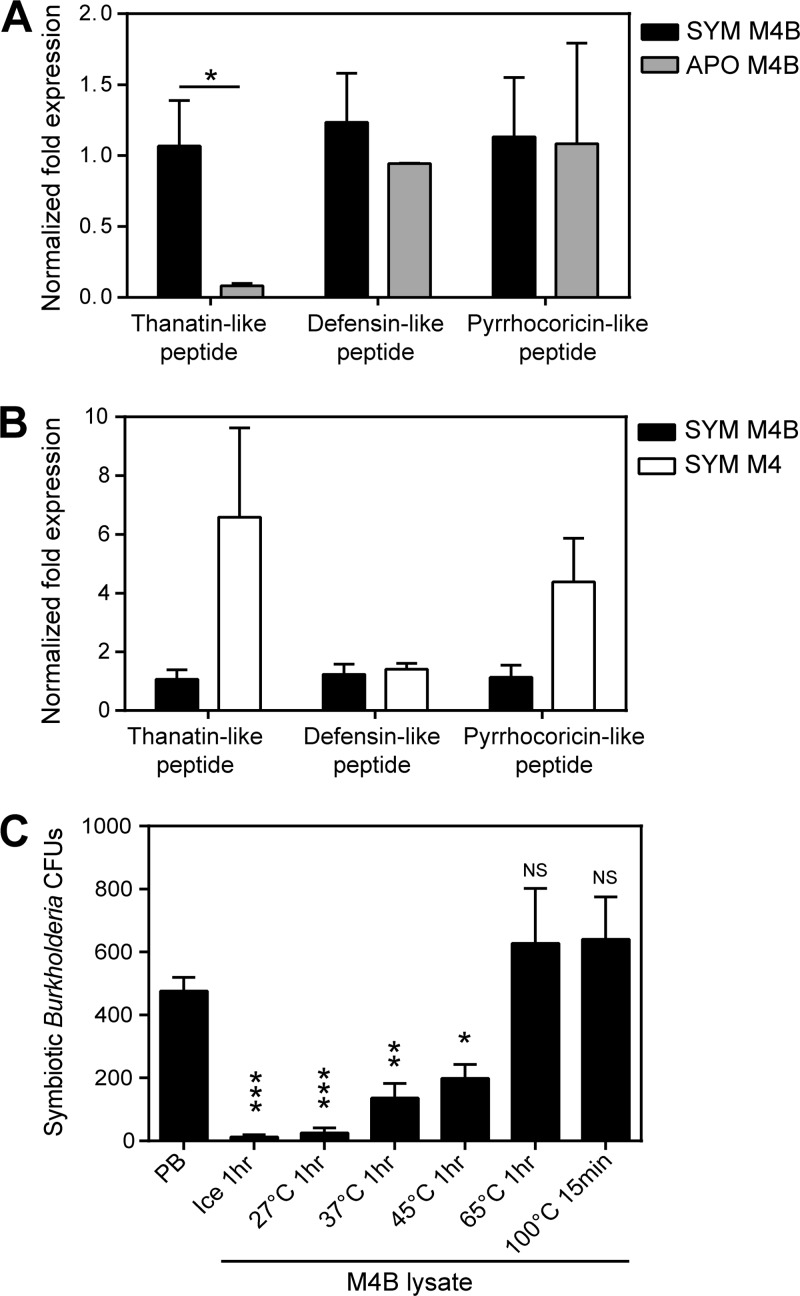
In R. pedestris, the cDNA sequences of three AMPs have been identified: defensin-like peptide (GenBank accession no. AK416895), thanatin-like peptide (AB842298), and pyrrhocoricin-like peptide (AB842297) (20). To address whether these AMPs are responsible for the antimicrobial activity of the M4B region, the expression levels of the AMP genes in the M4B regions of the symbiotic insects and the aposymbiotic insects were compared. As shown in Fig. 4A, the expression levels of the defensin-like peptide gene and the pyrrhocoricin-like peptide gene were similar in the symbiotic insects and the aposymbiotic insects, whereas the expression levels of the thanatin-like peptide gene in M4B were significantly higher in the symbiotic insects than in the aposymbiotic insects. Meanwhile, comparing the M4B and M4 regions of the symbiotic insects, the expression levels of the AMP genes were either similar to each other (for the defensin-like peptide gene) or somewhat higher in the M4 region than in the M4B region (for the pyrrhocoricin-like peptide gene and the thanatin-like peptide gene) (Fig. 4B). These results indicate that these AMPs are not responsible for the antimicrobial activity of the M4B region. When the M4B lysates were treated at different temperatures and subsequently tested for their antimicrobial activities against the symbiotic Burkholderia strain, the activities were reduced by treatments at 37°C and 42°C and completely eliminated by treatment at 65°C (Fig. 4C). Since insect AMPs are generally heat resistant (21), these results corroborate the idea that the antimicrobial activity of the M4B region is not associated with AMP expression.
Fig 4.
(A and B) Expression levels of the AMP genes in symbiotic organs of fifth-instar nymphs of R. pedestris. AMP expression levels were compared between M4B regions from symbiotic and aposymbiotic insects (A) and between the M4B region and the M4 region of symbiotic insects (B). To calculate the normalized fold expression (2−ΔΔCT), EF1α was used as a reference gene, and the expression levels were normalized to the expression of the thanatin-like peptide gene in symbiotic insects, set as 1. Means and standard deviations are shown (n = 3). The asterisk indicates a statistically significant difference (unpaired t test; *, P < 0.05). (C) Antimicrobial activity test of M4B lysates treated at different temperatures. M4B lysate (0.2 μg/ml in PB) was temperature treated prior to testing the antimicrobial activity against the symbiotic Burkholderia strain. Statistically significant differences between the bacterial CFU before (PB) and after incubation with M4B lysate are indicated by asterisks (unpaired t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; n = 3). NS, not significant.
Conclusions and perspective.
In this study, we identified a previously unrecognized insect organ with significant antimicrobial activity, the M4B midgut region, in R. pedestris. Interestingly, the antimicrobial activity of M4B was preferentially detected in the Burkholderia-harboring symbiotic insects (Fig. 3), suggesting the possibility of symbiont-mediated induction of antimicrobial activity. Also of interest is the fact that the antimicrobial activity of M4B selectively acts on the symbiotic Burkholderia strain (Fig. 2), highlighting a highly specific aspect of the host-symbiont interactions.
Although speculative, several major biological roles of the symbiont-killing action of M4B that are not mutually exclusive are conceivable: (i) regulation of the symbiont population, (ii) recruitment of symbiont biomass for host growth, and (iii) prevention of colonization of nonsymbiotic microbial contaminants. Morphologically, the direct connection of M4B to the main symbiotic region, M4 (Fig. 1A and B), appears to be suitable for performing these tasks; overgrown Burkholderia symbiont cells in M4 may continuously flow back to the neighboring M4B, where the bacterial cells may be killed, digested, and absorbed. The highest antimicrobial activity in M4B of the symbiotic fifth-instar insects (Fig. 3B, left) seems to make sense biologically, because fifth-instar nymphs require substantial resources for constructing a thick cuticle, wings, and gonads for the subsequent adult molting. Considering that not only symbiotic Burkholderia, but also S. aureus, was suppressed by the M4B lysate (Fig. 2), M4B may play a suppressive role against nonsymbiotic microbial contaminants acquired orally through host feeding.
Future studies will focus on identification of the molecule(s) involved in the symbiotic Burkholderia-specific antimicrobial activity in the M4B region of R. pedestris. These antimicrobial molecules are sensitive to heat (Fig. 4) and thus are unlikely to be antimicrobial peptides. A recent expression sequence tag analysis of symbiotic and nonsymbiotic midgut regions of R. pedestris identified genes for defense-related proteins, such as lysozymes and cathepsin proteases (20). Our ongoing RNA-sequencing analysis using next-generation sequencers will provide additional candidate genes from which the antimicrobial substance(s) of interest may be identified. Alternatively, the antimicrobial activity may be attributable to a low-molecular-mass secondary metabolite(s) for which metabolomic approaches are needed. We expect that transcriptomic, proteomic, metabolomic, and functional analyses of M4B will uncover pivotal molecular aspects underpinning the regulation of the sophisticated Riptortus-Burkholderia gut symbiotic association.
ACKNOWLEDGMENT
This work was supported by a Global Research Laboratory Grant from the National Research Foundation of Korea (grant number 2011-0021535) to T.F. and B.L.L.
Footnotes
Published ahead of print 13 September 2013
REFERENCES
- 1.Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. Interscience Publishers, New York, NY [Google Scholar]
- 2.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165–190 [DOI] [PubMed] [Google Scholar]
- 3.Oliver KM, Degnan PH, Burke GR, Moran NA. 2010. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55:247–266 [DOI] [PubMed] [Google Scholar]
- 4.Werren JH, Baldo L, Clark ME. 2008. Wolbachia: master manipulators of invertebrate biology. Nat. Rev. Microbiol. 6:741–751 [DOI] [PubMed] [Google Scholar]
- 5.McGraw EA, Merritt DJ, Droller JN, O'Neill SL. 2002. Wolbachia density and virulence attenuation after transfer into a novel host. Proc. Natl. Acad. Sci. U. S. A. 99:2918–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koga R, Tsuchida T, Fukatsu T. 2003. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc. Biol. Sci. 270:2543–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakurai M, Koga R, Tsuchida T, Meng XY, Fukatsu T. 2005. Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl. Environ. Microbiol. 71:4069–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mouton L, Dedeine F, Henri H, Bouletreau M, Profizi N, Vavre F. 2004. Virulence, multiple infections and regulation of symbiotic population in the Wolbachia-Asobara tabida symbiosis. Genetics 168:181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakabachi A, Shigenobu S, Sakazume N, Shiraki T, Hayashizaki Y, Carninci P, Ishikawa H, Kudo T, Fukatsu T. 2005. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc. Natl. Acad. Sci. U. S. A. 102:5477–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishikori K, Morioka K, Kubo T, Morioka M. 2009. Age- and morph-dependent activation of the lysosomal system and Buchnera degradation in aphid endosymbiosis. J. Insect Physiol. 55:351–357 [DOI] [PubMed] [Google Scholar]
- 11.Brennan LJ, Keddie BA, Braig HR, Harris HL. 2008. The endosymbiont Wolbachia pipientis induces the expression of host antioxidant. PLoS One 3:e2083. 10.1371/journal.pone.0002083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Login FH, Balmand S, Vallier A, Vincent-Monegat C, Vigneron A, Weiss-Gayet M, Rochat D, Heddi A. 2011. Antimicrobial peptides keep insect endosymbionts under control. Science 334:362–365 [DOI] [PubMed] [Google Scholar]
- 13.Kikuchi Y, Meng XY, Fukatsu T. 2005. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl. Environ. Microbiol. 71:4035–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kikuchi Y, Hosokawa T, Fukatsu T. 2007. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 73:4308–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kikuchi Y, Hosokawa T, Fukatsu T. 2011. Specific developmental window for establishment of an insect-microbe gut symbiosis. Appl. Environ. Microbiol. 77:4075–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kikuchi Y, Hosokawa T, Fukatsu T. 2011. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 5:446–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JK, Lee HJ, Kikuchi Y, Kitagawa W, Nikoh N, Fukatsu T, Lee BL. 2013. The bacterial cell wall synthesis gene uppP is required for Burkholderia colonization of the stinkbug gut. Appl. Environ. Microbiol. 79:4879–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JK, Won YJ, Nikoh N, Nakayama H, Han SH, Kikuchi Y, Rhee YH, Park HY, Kwon JY, Kurokawa K, Dohmae N, Fukatsu T, Lee BL. 2013. Polyester synthesis genes associated with stress resistance are involved in an insect-bacterium symbiosis. Proc. Natl. Acad. Sci. U. S. A. 110:E2381–E2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata TF, Maeda T, Nikoh N, Yamaguchi K, Oshima K, Hattori M, Nishiyama T, Hasebe M, Fukatsu T, Kikuchi Y, Shigenobu S. 2013. Complete genome sequence of Burkholderia sp. strain RPE64, bacterial symbiont of the bean bug Riptortus pedestris. Genome Announc. 1:e00441–13. 10.1128/genomeA.00441-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Futahashi R, Tanaka K, Tanahashi M, Nikoh N, Kikuchi Y, Lee BL, Fukatsu T. 2013. Gene expression in gut symbiotic organ of stinkbug affected by extracellular bacterial symbiont. PLoS One 8:e64557. 10.1371/journal.pone.0064557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulet P, Hetru C, Dimarcq JL, Hoffmann D. 1999. Antimicrobial peptides in insects; structure and function. Dev. Comp. Immunol. 23:329–344 [DOI] [PubMed] [Google Scholar]