Abstract
Cyanophages are important components of aquatic ecosystems, but their genetic diversity has been little investigated in freshwaters. A yearlong survey was conducted in surface waters of the two largest natural perialpine lakes in France (Lake Annecy and Lake Bourget) to investigate part of this cyanophage diversity through the analysis of both structural (e.g., g20) and functional (e.g., psbA) genes. We found that these cyanophage signature genes were prevalent throughout the year but that the community compositions of g20 cyanomyoviruses were significantly different between the two lakes. In contrast, psbA-containing cyanophages seemed to be more similar between the two ecosystems. We also found that a large proportion of g20 sequences grouped with cyanomyophage isolates. psbA sequences, belonging to phages of Synechococcus spp., were characterized by distinct triplet motifs (with a novel viral triplet motif, EFE). Thus, our results show that cyanophages (i) are a diverse viral community in alpine lakes and (ii) are clearly distinct from some other freshwater and marine environments, suggesting the influence of unique biogeographic factors.
INTRODUCTION
Viruses infecting cyanobacteria (i.e., cyanophages) have been recognized as an important player in aquatic ecosystems (1). They have been shown to influence host population mortality as well as population succession and community structure and thus are a major driving force behind biogeochemical cycles (2–4). To the best of our knowledge, most of the characterized cyanophages (largely of marine origin) are tailed double-stranded DNA viruses and belong to three families (1, 5–7): the Myoviridae (T4-like, TIM5-like), the Podoviridae (T7-like), and the Siphoviridae (lambda-like). The ratio of these three groups of cyanophages varies depending on both biological hosts and environmental conditions (1, 8). In surface waters of the vast open ocean, the relative proportion of cyanomyoviruses/cyanopodoviruses/cyanosiphoviruses is about 20:10:1, as revealed by BLAST searching against the Global Ocean Sampling (GOS) metagenome database using the available 29 cyanophage genomes and their TerL amino acid sequences as queries (9).
The T4-like cyanomyoviruses generally have a broad host range (10) and are abundant in various environments. The genetic diversity of this group has been examined using the T4-like portal protein-encoding gene g20 as a molecular marker in different locations of both marine and freshwater ecosystems (11–24) and even in paddy soils (25). Distinct cyanomyoviruses could be detected in freshwater (12, 18, 21, 26) and paddy soil (25) ecosystems in comparison to their marine counterpart. Also, the diversity, abundance, or structure of cyanomyoviruses can vary between stations of the same lake (21) or ocean (15, 18, 27). When sampled from a single location, it has also been shown that abundance and community structure and/or diversity of these cyanophages can vary with time (11, 12, 16, 20, 23, 28, 29) and depth (13, 27). Together, these studies reflect distinct temporal, vertical, and/or geographical distribution patterns of cyanomyoviruses across aquatic environments. Comparatively, the cyanopodoviruses and cyanosiphoviruses are much more host specific (9, 12, 30). The occurrence and diversity of cyanopodoviruses (5, 8, 31) were investigated only in marine ecosystems, but no report has been provided so far for freshwaters. Such a lack of data is also true for cyanosiphoviruses even if several genomes of these cyanophages have been made available recently from the Mediterranean Sea, Atlantic Ocean, and Chesapeake Bay (9, 26, 32).
Among remarkable features discovered in cyanophages are the host-derived photosynthesis reaction genes (e.g., mainly psbA and psbD) present in their genomes. These auxiliary metabolic genes are assumed to benefit the host through their expression during infection, supplying substantial energy and carbon for phage production (33–36). The psbA gene encodes the photosystem II D1 protein, and it has been found in approximately 2/3 of both cyanomyovirus and cyanopodovirus isolates (30) but not in cyanosiphoviruses (5, 9, 26). As the psbA gene may not be specific to a single cyanophage family, it represents an additional genetic marker to explore diversity and evolutionary history of cyanophages in aquatic environments. Furthermore, Sharon et al. (37) found that viral psbA sequences can be very abundant in the environment, and 62% of identifiable psbA sequences issued from the GOS metagenome database have been assigned to virus origin. PCR-based studies have also revealed that psbA gene sequences are diverse in both marine waters (36–39) and freshwaters (38, 40, 41).
During the last decade, previous studies have partially identified the diversity, dynamics, or role (as mortality agents of picocyanobacteria) of cyano(myo)phages in French perialpine lakes (12, 17, 23, 42, 43). This body of work has suggested that the cyano(myo)phage community is diverse, displays marked seasonal variations, and is concentrated mainly in surface waters down to 20 m deep. Furthermore, the cyanophage community could, at certain periods of the year (e.g., mainly in spring and only occasionally in summer or autumn), be an important mortality factor for the picocyanobacterial community. However, we have not yet provided a detailed description of the cyanophage genetic composition, and more generally such an investigation is absent from large and deep lakes. Thus, we conducted a complete annual survey of Lake Annecy and Lake Bourget in the 0- to 20-m upper lit layer with the goal of investigating cyanophage community composition. Such sampling strategy was chosen to respond to the potential high temporal, vertical, and geographical variability of cyanophages as described previously, so as to unveil a global and comprehensive image of cyanophages in perialpine lakes. Specifically, we employed two genetic markers, using PCR-denaturing gradient gel electrophoresis (DGGE) and cloning sequencing approaches, to examine part of the genetic diversity of T4-like cyanomyoviruses (g20 gene) and psbA-containing cyanophages (psbA gene) in two contrasting ecosystems: an oligotrophic (Annecy) and an oligo-mesotrophic lake (Bourget). To the best of our knowledge, this is the first study for freshwater ecosystems with such a temporal and taxonomic resolution.
MATERIALS AND METHODS
Sample collection and processing.
Water samples were collected once or twice each month between January and November 2011 at reference stations of Lake Annecy and Lake Bourget as detailed elsewhere (23). A few hours following sampling, 20-liter samples were first filtered through a 60-μm-pore-size mesh and then filtered through 1-μm-pore-size filters (Millipore, Bedford, MA). The final filtrate (i.e., <1-μm fraction) was concentrated to a final volume of 200 to 250 ml by using a 30,000-Da-molecular-weight-cutoff, spiral-wound, Millipore ultrafiltration cartridge (regenerated cellulose, PLTK Prep/Scale TFF, 1 ft2; Millipore). To ensure that all remaining small free-living bacteria were removed, we filtered the <1-μm concentrated fraction through 0.45-μm-pore-size filters twice (Millipore). The absence of cellular contamination was verified using flow cytometry (not shown). This <0.45-μm viral concentrate (VC) was stored in the dark at −20°C until further processing.
PCR amplification and DGGE.
The PCR was conducted in two stages prior to the DGGE analysis (17, 23, 30). The first PCR stage on the VC, the template, was conducted with the primer set in the absence of the GC clamp. The second PCR was then performed on the product of the first-stage PCR, using the GC clamp-containing primer set (i.e., with a 40-nucleotide [nt] GC clamp attached to the 5′ end of the forward primer). PCRs were performed using the DNA thermal cycler T-Professional (Biometra), whereby we amplified the T4-like portal protein-encoding g20 gene using the primer set CPS1.1/8.1 (44) and the photosystem II D1 protein-encoding psbA gene using the primer set Pro-psbA-1F/1R (36). Briefly, for all primer sets, 25 μl of reaction mix contained 1× PCR buffer, 4 mM MgCl2, 200 μM each deoxynucleoside triphosphate (dNTP), 0.4 μM each primer, 0.5 U of Platinum Taq DNA polymerase (Invitrogen), and 1 μl of VC. Prior to PCR with the psbA gene, VC was treated with DNase (0.002 units of DNase I for 1 μl of VC; Sigma) to remove environmental free DNA and to avoid the potential amplification of psbA sequences from free DNA released from cellular hosts. The program for the first-stage PCR was a 15-min virion lysing and denaturation at 95°C, followed by 34 cycles of denaturation at 95°C for 30 s, annealing for 30 s, extension at 72°C for 45 s, and a final extension at 72°C for 5 min. The program for the second-stage PCR was 5 min of denaturation at 95°C, followed by 24 cycles of denaturation at 95°C for 30 s, annealing for 30 s, extension at 72°C for 45 s, and a final extension at 72°C for 5 min. After various tests of optimization (not shown), the best annealing temperatures were 46°C and 50°C for CPS1.1/8.1 and Pro-psbA-1F/1R, respectively.
The DGGE was conducted as described previously (23). The linear denaturing gradient was at 25 to 50% and 40 to 60% for amplicons of CPS1.1/8.1 and Pro-psbA-1F/1R, respectively. Twenty microliters of PCR products was loaded into wells with 5 μl of 5× loading buffer (12.5% Ficoll, 25 mM Tris, 5 mM EDTA [pH 8.0], 0.5% SDS, 0.1% [wt/vol] xylene cyanol, and 0.1% [wt/vol] bromophenol blue). Electrophoresis was carried out for 16 h in 1× TAE buffer (pH 7.4; 40 mM Tris base, 20 mM sodium acetate, 1 mM EDTA) at 120 V and a constant temperature of 60°C using the CBS-DGGE 2000 system (C.B.S. Scientific Co., Inc.). Due to the limited gel size (i.e., the number of wells) accepted by this device, we ran samples in two gels (one for Lake Annecy and the other for Lake Bourget for each marker gene) put together in the same tank. To standardize the banding pattern in each gel, a pooled sample (corresponding to equal volumes of all obtained samples from the two lakes mixed together) was loaded in the middle and at each side of the gels. To identify the DGGE bands shared by the two lakes, in addition to the pooled sample, the samples of the other lake (those possessing typical distinct banding patterns) were also loaded at the same time in the DGGE migration to serve as an extra reference. Gels were stained in a 2× SYBR green I (Molecular Probes, Invitrogen) solution for 45 min and were visualized on a UV transilluminator (Tex-35 M; Bioblock Scientific) and photographed with GelDoc (Bio-Rad).
DNA purification, cloning, and sequencing.
Gel slices of 47 and 39 visibly different DGGE band representatives were excised from the g20 and psbA DGGE gels, respectively. DNA of each DGGE band representative was eluted from the excised gel slice by adding 100 μl of 1× sterile TAE buffer and heated for 15 min at 95°C. Three microliters of eluted DNA served as the template in a 22-μl PCR mixture using the corresponding primer set. PCRs were performed under the same conditions of the first-stage PCR, as described above. The amplicons were first verified by electrophoresis in a 1.5% agarose gel, purified by the Illustra GFX PCR DNA and gel band purification kit (GE Healthcare), and finally cloned into pCR4-TOPO vectors by using the TOPO TA cloning kit (Invitrogen). Respectively, 45 and 29 bands of g20 and psbA resulted in clones, and the other bands (mainly DGGE bands with weak luminosity or fluorescent density) failed, probably due to the low DNA concentration. Eight randomly selected clones for each band representative were then sent for sequencing to GATC Biotech. After cleaning and correcting sequences by using BioEdit 7.0.5.3 (45), we obtained 110 and 62 different nonredundant sequences for g20 and psbA, respectively.
Phylogenetic analysis.
For the analysis of g20, all sequences we obtained were translated into amino acids and aligned using MAFFT version 7 (46), using reference sequences of both environmental and culture representative cyanophages (Table 1). Multiple alignments were then cured using Gblocks (47) with a stringent option by not allowing a contiguous unconserved position inside the final blocks. Phylogenies were reconstructed using Bayesian inference and maximum likelihood methods. Bayesian inference was carried out using MrBayes 3.2.1 (48) with two runs, four chains, 1.5 million generations, sampling every 1,000 generations, mixed models of amino acid substitution, and a burn-in value of 25%. The maximum likelihood phylogeny was constructed using PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/) (49) with 100 bootstrap replicates, the rtREV (general reverse transcriptase) model, and gamma-distributed rate heterogeneity among amino acid substitutions (i.e., the best model for this aligned g20 sequence data set determined by using MEGA5) (50).
Table 1.
Origins of g20 environmental sequences used for the phylogenetic tree shown in Fig. 2
| Environment | Prefix of sequence label | Location(s) | No. of sequences obtained | GenBank accession no. | Reference |
|---|---|---|---|---|---|
| Freshwater | LAB_g20 | Lake Annecy and Bourget (HS, France) | 110 | KC626330 to KC626439 | This study |
| LB | Lake Bourget (HS, France) | 47 | AY426128 to AY426174 | 12 | |
| LAC95 | Lake Constance (Germany) | 6 | AY705091 to AY705145 | 18 | |
| CUL | Cultus Lake (BC, Canada) | 9 | AY705091 to AY705145 | 18 | |
| CHL | Chilliwack Lake (BC, Canada) | 1 | AY705091 to AY705145 | 18 | |
| CAT | Catfish Pond | 1 | AY705091 to AY705145 | 18 | |
| SPM | Shore pond mat (Arctic mat) | 8 | AY705091 to AY705145 | 18 | |
| LE | Lake Erie (North America) | 45 | DQ318388 to DQ318432 | 21 | |
| PFW | Paddy field floodwater (Japan) | 77 | AB471562 to AB471638 | 20 | |
| Marine | BES | Beaufort Sea (Arctic Ocean) | 9 | AY705091 to AY705145 | 18 |
| CHS | Chuckchi Sea (Arctic Ocean) | 3 | AY705091 to AY705145 | 18 | |
| ANT | Antarctic Peninsula (Southern Ocean) | 4 | AY705091 to AY705145 | 18 | |
| GOM | Gulf of Mexico (NW Atlantic Ocean) | 5 | AY705091 to AY705145 | 18 | |
| MAL | Malaspina Inlet (NE Pacific Ocean) | 1 | AY705091 to AY705145 | 18 | |
| SAI | Salmon Inlet (NE Pacific Ocean) | 2 | AY705091 to AY705145 | 18 | |
| PES | Pendrell Sound (NE Pacific Ocean) | 2 | AY705091 to AY705145 | 18 | |
| COL | Coast of Colombia (SE Pacific Ocean) | 1 | AY705091 to AY705145 | 18 | |
| CHI | Coast of Chile (SE Pacific Ocean) | 1 | AY705091 to AY705145 | 18 | |
| SE | Skidaway Estuary (Savannah, GA) | 29 | AY027985 to AY028013 | 24 | |
| GS | Gulf Stream (along edge of Sargasso Sea) | 36 | AY027938 to AY027973 | 24 | |
| SS | Sargasso Sea (NW Atlantic Ocean) | 65 | AY028014 to AY028078 | 24 | |
| CB | Chesapeake Bay (United States) | 15 | AY152732 to AY152746 | 19 | |
| Soil | AnCf | Paddy soil of Anjo (Japan) | 25 | AB560191 to AB560215 | 25 |
| KuCf | Paddy soil of Kuroishi (Japan) | 26 | AB560146 to AB560171 | 25 | |
| OmCf | Paddy soil of Omagari (Japan) | 19 | AB560172 to AB560190 | 25 |
Since a high level of conservation exists among psbA sequences, DNA sequences were used for phylogenetic reconstruction instead of the deduced amino acid sequences as for g20 (30, 51, 52). All obtained psbA nucleotide sequences were also aligned using MAFFT with culture representatives of cyanophages, cyanobacteria, and eukaryotic microalgae, as well as environmental sequences from other studies (25, 36–39). The multiple alignments were again cured by using Gblocks, and phylogenies were reconstructed as described above. The general time-reversible (GTR) model and gamma-distributed rate heterogeneity among nucleotide substitutions was for the maximum likelihood phylogeny. The Bayesian phylogeny employed the codon model of nucleotide substitution and allowed different rates for transitions and transversions.
Statistical analysis.
To evaluate whether the (g20 or psbA) cyanophage community composition of Lake Annecy differed from that of Lake Bourget, we carried out statistical analyses using the UniFrac distance metric statistical tools available at http://bmf.colorado.edu/unifrac/ (53, 54). We used the unweighted UniFrac option in order to compare community composition based on presence/absence importance (i.e., on qualitative data). This tool measures the distance between two communities by calculating the fraction of the branch length in a phylogenetic tree (54). In brief, we used the Bayesian phylogenetic tree and a file mapping sequence labels to their habitats as input for each analysis (g20 or psbA). The P test was conducted based on the UniFrac distance matrix generated for each cyanophage assemblage.
Nucleotide sequence accession numbers.
Sequences were deposited in GenBank under the accession numbers KC626330 to KC626439 for g20 and KC626440 to KC626501 for psbA.
RESULTS
Photosynthetic gene psbA sequences.
From the 29 visibly different representative bands excised, we obtained 62 nonredundant sequences for both lakes, with nucleotide similarity varying between 72.9% and 99.9% and GC content varying between 43.3% and 48.1% (Fig. 1). Among the eight sequenced clones analyzed for each band, between 1 and 6 different sequences were obtained, with nucleotide dissimilarity up to 21.7%. Eleven out of the 29 psbA bands contained a single sequence.
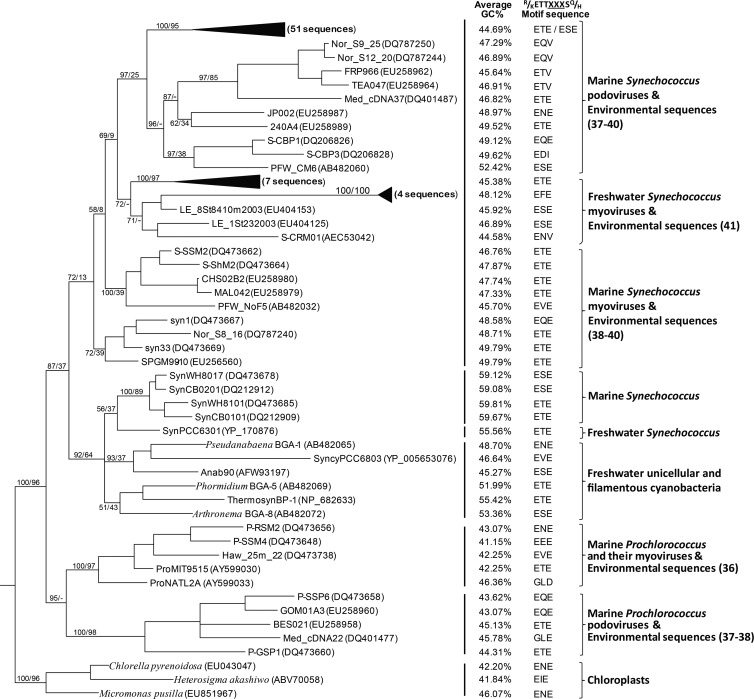
Fig 1.
Phylogenetic tree, GC contents, and triplet sequences from D1 protein motifs of a partial psbA gene from the culture representatives of cyanophages, cyanobacteria, and eukaryotic microalgae and from the environmental samples of this study and others (36–41). A Bayesian phylogenetic tree was constructed based on the alignment of 611 homologous nucleotides of the psbA gene. Values at nodes as shown on the main branches are the Bayesian inference (BI) posterior probability and the maximum likelihood (ML) bootstrap value in the order of BI/ML. With the clade support value of both BI and ML of ≥90%, the sequences of Lake Annecy and Bourget are grouped into black triangles. The number of sequences is given in parentheses and written in bold.
The triplet peptides associated with the D1 protein motif (R/K)ETTXXXS(Q/H) were also analyzed, and we detected three triplets, referred to as ETE, ESE, and EFE, which accounted for 75.8%, 17.7%, and 6.5% of obtained sequences, respectively. The phylogenetic analysis of these sequences with phytoplankton and cyanophage culture representatives showed that all obtained psbA sequences were grouped together with viruses infecting Synechococcus spp. (Fig. 1), of which 82.3% were more closely related to cyanopodoviruses (S-CBP1 and S-CBP3) rather than to cyanomyoviruses. The other 17.7% (11 sequences) had the closest homologue to the freshwater Synechococcus myovirus S-CRM01.
The UniFrac analysis failed to detect any significant differences between Lake Annecy and Lake Bourget psbA sequences (P = 0.15).
Capsid assembly gene g20 sequences.
From the 45 visibly different representative bands excised, we obtained 110 nonredundant sequences, with nucleotide similarity varying between 66.3% and 98.9%. Among the eight sequenced clones analyzed for each band, between 1 and 6 different sequences were obtained, with nucleotide dissimilarity up to 33.7%. Fourteen out of the 45 g20 bands contained a single sequence.
The phylogenetic analysis of these sequences with both cyanomyovirus culture representatives and other environmental sequences (Table 1) allowed us to discriminate 17 clades (Fig. 2). Five of them were initially identified by Sullivan et al. (44) as the culture-containing clusters I, II, and III, cluster 3 (P-SSM9/11/12 cluster), and the environmental sequence-only cluster 2. Another one was the cluster A previously defined by Zhong et al. (24). We redefined the other 11 clusters as environmental sequence-only clusters Env 1 to 10 and cluster 4. Overall, our global clustering pattern agreed with the analysis of Sullivan et al. (44). However, the previously defined environmental sequence-only clusters (like cluster 1 but not cluster 2) were reorganized when sequences of a paddy field (20, 25) and alpine lakes (this study) were introduced, for which sequences were distributed in cluster Env 1 and 3 to 9.
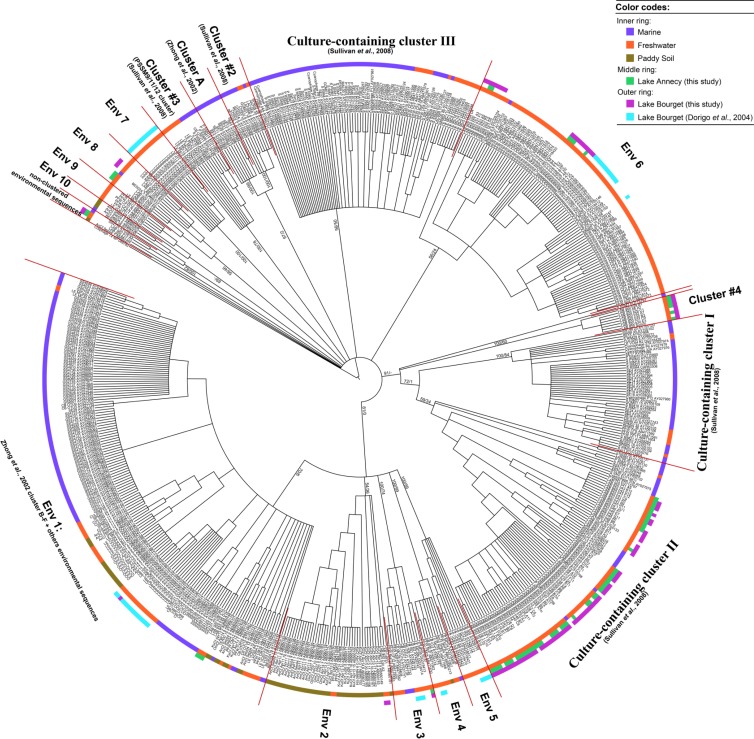
Fig 2.
Bayesian phylogenetic tree based on the alignment of 112 homologous amino acid positions of the T4-like portal protein-encoding gene g20 from 616 available g20 sequences. Sequences are from Lake Annecy and Lake Bourget, cyanomyovirus isolates (18, 24, 28, 44, 58, 68), and other different environments as described in Table 1. Values at nodes as shown on the main branches are the Bayesian inference (BI) posterior probability and the maximum likelihood (ML) bootstrap value in the order of BI/ML. The inner ring constituted with color strips aims at distinguishing among g20 sequences of the marine field (violet), freshwater (orange), and the paddy soil (antique bronze). The middle ring, with color strips in green, corresponds to sequences from Lake Annecy. In the outer ring, sequences labeled with color strips are from Lake Bourget, of which those in purple were obtained by this study and those in cyan were obtained by Dorigo et al. (12).
In the color ring in the phylogenetic tree of Fig. 2, cluster A contains exclusively g20 sequences from the marine field, whereas only freshwater-originated sequences could be detected in clusters Env 6, 7, and 9. No cluster contained exclusively sequences from the paddy field soil, which were clustered with other water-originated sequences in clusters Env 1, 2, 4, and 10. Sixty-three percent of the sequences obtained from Lake Annecy and Lake Bourget were grouped into culture-containing cluster II, whereas the others were in different environmental sequence-only clades, like Env 1, 2, 3, 6, and 8 and cluster 4. No sequences were in the culture-containing clusters I and III, clusters 2 and 3, or cluster A (Fig. 2).
When the g20 sequences were pooled over the year, the UniFrac analysis revealed significant differences between Lake Annecy and Lake Bourget (P = 0.03).
DISCUSSION
Methodological considerations.
DGGE allows fingerprinting communities based on the fractionation of amplicon sequences (55). In our study, we observed that the cyanophage community composition (either from g20 or psbA) varied throughout the year (23) (data not shown), so considering a sample(s) at a single time point or at selected periods of the year was likely to miss some bands (i.e., subpopulations) occurring at other time points or periods. Taking g20 cyanomyoviruses as an example, all visibly different g20 band types throughout the year were excised for cloning sequencing, of which 96% (45 of 47 bands) resulted in reliable sequences. When considering a single sample from Lake Bourget, only between 7 (February) and 17 (November) visible bands could be detected, corresponding to less than half of the total band types obtained in this lake throughout 2011 (41 bands) and less than one-third of the total band types obtained from the two lakes (47 bands). Therefore, our study completed over the year allowed us to obtain a global picture of cyanomyovirus communities in these ecosystems.
We are aware, however, that DGGE-based community composition studies present important drawbacks. First of all, they are limited to major taxa. Minor populations may produce no visible bands or bands insufficiently visible to be cut from gels. Also, when cut, low-density bands can result in too-low concentrations of purified DNAs to be cloned. This was observed for psbA: 10 out of 39 band types failed to produce clones. Second, a single band type can contain multiple sequences whose composition may vary with time. As we selected only one or two bands for each band type for sequencing, it is possible we missed some sequences (also typical of the same band type) that could occur during periods other than the one analyzed.
All in all, our DGGE-based approach was likely to provide a rough minimum estimate of cyanophage community composition. Considering these viral populations followed the seed bank model (i.e., only a few members of viruses were actively abundant at any given time, and most of them were rather rare and/or inactive) (29, 51, 56), this 1-year investigation could at least reveal some of these annually actively abundant populations. Hence, our approach provided a solution to identify rapidly and at reduced cost the actively abundant viral fraction occurring at some periods of the year in response to environmental and/or biological factors.
Cyanophage g20 diversity.
Our analysis, using PCR-DGGE with primers CPS1.1 and CPS8.1 to target the g20 gene in Lake Annecy and Lake Bourget, first revealed the prevalence of cyanomyoviruses in these perialpine lakes. Cyanomyoviruses of Lake Bourget have been previously investigated by Dorigo et al. (12) by using a PCR-DGGE-based approach between September 2002 and January 2003 and by Roux et al. (57) through a metagenomic-based approach in July 2008. When we blasted against the virome using our g20 sequences as queries (E value of ≤0.01), only 0.004% (24 read sequences) of reads were detected to have similarity to g20. After tentative assembly of these reads to contigs, only two of them overlapped with our g20 sequences and could be recruited for the phylogenetic analysis. This earlier metagenomic study generated relatively short reads (on average 427 bp in length), and the sequencing depth was likely not sufficient to allow the construction of longer contigs of rare sequences (cyanomyoviruses accounted for <0.1% of total viruses as revealed by flow cytometry and qPCR) (23). However, the two g20 contig sequences obtained from the virome were detected to be different from those obtained by Dorigo et al. (12) and from the present study. One clustered in the culture-containing cluster II and the other was in cluster Env 7. A significant difference of g20 sequences in Lake Bourget was also detected between this study and the one of Dorigo et al. (12) (P = 0), and only one nucleotide sequence was identical. Our sequences were located mainly in the culture-containing cluster II, whereas no sequence obtained by Dorigo et al. (12) clustered with cyanomyovirus isolates. Most of them were in clusters Env 1, 3, 4, 5, and 7, where our sequences were rarely present (Fig. 2). Such a difference between these three studies, for the same lake, may be due to the technique employed (PCR-DGGE versus metagenomic), the sampling strategy (water at 5 m versus integrated water at 0 to 20 m), the period of investigation (July versus September to January versus January to November, moreover, in different years), or the difference in primer specificities (primers CPS1/8 used by Dorigo et al. [12] versus CPS1.1/8.1 in our study). It is noteworthy here that Sullivan et al. (44) tested CPS1/8 and CPS1.1/8.1 over a wide range of marine cyanomyovirus isolates and revealed that CPS1/8 could amplify only 56% of cyanomyoviruses, whereas such a percentage reached 100% when using CPS1.1/8.1. This could explain the observation that numerous g20 sequences were closely related to cyanomyovirus isolates in the present study compared to the relatively few reported by Dorigo et al. (12). Sixty-three percent of g20 sequences (69 out of 110) obtained from Lake Annecy and Lake Bourget were in the culture-containing cluster II, which is in agreement with the distribution pattern in the GOS metagenome database (i.e., 65.6%) (44). However, we did not have sequences in the culture-containing clusters I and III, while 12.7% (65 out of 512) of sequences in GOS field samples were in these clusters. This was the first application of the revised primers CPS1.1 and CPS8.1 for environmental samples, and our results seem to support their improvement in targeting the g20 gene of cyanomyoviruses.
The g20 homologue is ubiquitous among T4-like myoviruses, with hosts ranging from proteobacteria to cyanobacteria (14, 44). While using the primer set CPS4/G20-2, which targets the same region of the g20 gene, Short and Suttle (18) obtained sequences (BES02B-27 and BES02B-28) from waters at a 3,246-m-deep station of the Arctic Beaufort Sea, where cyanobacteria and their phages are not supposed to be found. The two authors speculated that the environmental g20 sequences out of clusters containing cyanomyovirus isolates were not of cyanophage origin. The g20 sequence of the clone BES02B-27 grouped with sequences obtained from alpine lakes (this study) (12) and also with those from a paddy field floodwater (20) in the environmental sequence-only cluster Env 6 (Fig. 2). In contrast, the BES02B-28 clustered with g20 sequences obtained from paddy soil (25) in cluster Env 10. Although the g20 sequences of BES02B-27 and BES02B-28 clones were generated by another primer set and the CPS1.1/8.1 could not amplify some noncyanomyovirus isolates (44), we might not exclude the possibility that CPS1.1/8.1 could amplify g20 sequences of T4-like viruses other than cyanomyoviruses for which the sequences of the environmental sequence-only cluster Env 6 may be suspect (see above). This is a common issue of diversity studies through the culture-independent approach by using PCR, since the validity of the primers depends on a limited number of isolates. Recently, Clasen et al. (11) employed the isolation-based/culture-dependent approach to examine marine cyanomyoviruses infecting Synechococcus sp. WH7803 and revealed important cooccurring cyanomyovirus genetic diversity. Such an approach could be used for cyanomyoviruses (because they can have a wide host range), and it displays advantages, compared to the culture-independent primer assays, for a robust evaluation of host-virus dynamics in response to environmental factors and changes. However, the isolation-based/culture-dependent approach is likely to explore only a small portion of the cyanomyoviruses and has limitations in terms of gaining access to nonviable viruses as well as those that cannot cross-infect Synechococcus sp. WH7803.
Cyanophage psbA diversity.
Lake Annecy and Lake Bourget are two perialpine lakes located in the same ecoregion and are both dominated by Synechococcus spp. (43). It is thus not surprising that all psbA sequences obtained from these ecosystems were grouped with viruses infecting Synechococcus spp. in the phylogenetic reconstruction, since the viral psbA gene was initially acquired from a host, and its transfer was largely limited by host range (36). Viruses infecting Synechococcus can be discriminated based on their families and environmental origins in psbA phylogenetic trees as clades containing marine podoviruses, freshwater myoviruses, or marine myoviruses (Fig. 1). It is noteworthy that such clustering still deserves validation because of the lack of freshwater podoviruses and the insufficient representation of freshwater myoviruses and marine podoviruses in phylogenetic analyses that have been proposed until now. Moreover, the intragenic recombination of the psbA gene within cyanophages as well as between hosts and cyanophages has been reported as a common event (36), which could possibly cause phylogenetic confusion if viral psbA genes of different cyanobacterial genera and/or different phage families within the same cyanobacterial genus are involved in genetic exchanges. For example, S-ShM1, S-SSM1, and P-SSM1 cluster differently with the myoviruses of the same cyanobacterial genus, and they have been suspected of intragenic psbA gene exchanges between marine myoviruses of Synechococcus and Prochlorococcus (36). The phage-to-phage intragenic exchange of the psbA gene among cyanophage families via host intermediates during infection might also be possible in the case of broad-host-range myoviruses (e.g., S-CBM2), which may share the same host (i.e., Synechococcus CB0101) with podoviruses (e.g., S-CBP1 and S-CBP3).
The GC content of the psbA nucleotide sequences and triplet peptides associated with the D1 protein motif (R/K)ETTXXXS(Q/H) (where XXX marks the position of the triplet peptides) have been proposed or used to distinguish viral psbA sequences from the host's in marine ecosystems (36, 38, 39, 52). Although this approach has not been widely tested for freshwater systems, we noticed that the GC content of Synechococcus viruslike psbA sequences of Lake Annecy and Lake Bourget ranged between 43.3% and 48.2%. In fact, we found that 90% of these psbA sequences (56 out of 62) had a GC content below 46% and were thus out of the percentage range (46 to 51%) found in marine Synechococcus phages (36). Our results echo the other freshwater studies (11, 41), where low GC content has also been detected in some Synechococcus-virus-like psbA sequences and in the only known characterized freshwater Synechococcus phage, S-CRM01 (44.6%), at the same 729-nt region. Together, these results suggest that freshwater Synechococcus phages might have a lower GC content in psbA sequences than their marine counterparts, and this may be a result of a lower GC content in their Synechococcus hosts. However, one may argue that this conservative psbA fragment (729 bp) is too short to predict for the complete psbA-coding sequence (approximately 1.1 kb) and to support the above-given assumption. This result will deserve further investigation with increasing available sequences.
Only one virus-specific triplet (EDV) could be detected in alpine lakes and was obtained from the virome of Lake Bourget. The psbA sequence to which it belongs is closely related to the freshwater cyanomyovirus S-CRM01 (data not shown). Our study revealed the existence of three triplets (ETE, ESE, and EFE) possessed by the psbA-containing cyanophages from Lake Annecy and Lake Bourget, and EFE has never been described before. The psbA sequences containing EFE triplets belonged to a single DGGE band and were also closely related to the freshwater cyanomyovirus S-CRM01 (Fig. 2). These sequences occurred in spring in Lake Bourget, when a previous study reported important viral lysis induced by cyanophages (30). ESE and ETE triplets were comparatively more frequently observed in both viruses and hosts (Fig. 1) (37). The ETE triplet was detected in 20 out of 33 currently known marine psbA-containing Synechococcus myoviruses, in one Synechococcus podovirus (syn26), in 3 Prochlorococcus myoviruses (P-SSM1/9/12), and one podovirus (P-GSP1). Approximately 17% of viruslike psbA sequences obtained from the GOS metagenomic data set (37) contained this ETE triplet, and only 5.3% were found in psbA-containing cyanophages from Lake Erie (41). Interestingly, the majority of Synechococcus viruslike psbA sequences (75.3%) with ETE triplets were from Lake Annecy and Lake Bourget, which may reflect the unique features of their viral psbA sequences or their common Synechococcus hosts in alpine lakes. The ESE triplet signature was rarely detected in viruslike psbA sequences from marine environments (37, 38). It was initially thought to be issued from cyanobacteria exclusively (37). However, the ESE triplet could be found in two freshwater cyanophages obtained from Lake Erie (MC15 and MC17) (41) and three marine cyanophages: S-RSM4 (58) and S-PWM2 and S-PWM3 (38). Our analysis revealed that 17.7% of Synechococcus viruslike psbA sequences from alpine lakes, 25% from paddy field floodwater (40), and 39.5% from Lake Erie were with ESE. Therefore, it seems that the ESE triplet was preferentially observed in freshwaters, suggesting also perhaps a unique community of potential hosts.
The ecological relevance of the existence of a particular motif found in perialpine lakes is in question. The D1 protein motif (R/K)ETTXXXS(Q/H) is a variable region within the PEST-like domain located in the loop between transmembrane helices D and E. It is implicated as the site of initial cleavage of the D1 protein and initiates the protein turnover from one isoform to another (37, 59, 60). There are two isoforms for D1 protein detected in cyanobacterial hosts (61). Isoform I is constitutively expressed, whereas isoform II is upregulated in response to high light and UV stress (36, 62–64). Sharon et al. (37) analyzed the triplet signatures of D1 protein motifs from the GOS metagenomic database and found that some viruses contain unique viral triplets (virus-specific triplet), while some others share the same motif as the host (host-like triplet). They hypothesized that the unique viral triplets may make D1 protein less susceptible to photodamage and turnover than the host D1, by conferring to the phage a feature similar to isoform II of host D1 protein, which could be important for life support during short periods of phage morphogenesis. Since viral psbA sequences were initially acquired from hosts through horizontal gene transfer, the interpretation for the difference of these unique viral triplets could be that, throughout their evolution, viruses modified the D1 protein motif in order to (i) facilitate adaptation to harsh light conditions or (ii) alter its role for their selfish benefits (e.g., by replacing the host light-sensitive D1 protein by the more stable viral D1 protein during infection with the goal to supply continuous energy for viral replication) (37, 60). Interestingly, as described above for our lakes, most viral D1 proteins possess host-like triples (e.g., ETE/ESE, which are detected in 94% of psbA sequences, most of which are related to Synechococcus podoviruses) and only a few contained virus-specific triplets (e.g., EDV and/or EFE, which are of Synechococcus myovirus origin and detected only occasionally in Lake Bourget). These results could thus suggest that the modification of the D1 protein motif by viruses in response to high light and/or stress conditions has not been a frequent event in perialpine lakes, compared to in the ocean, where virus-specific triplets account for up to 30% of GOS viral D1 proteins. This agrees with the finding that D1 proteins of most freshwater Synechococcus strains are more resistant to high light and stress (37, 62, 65), so the modification of the D1 protein for their cooccurring viruses may not be required. The sporadic occurrence of unique viral triplet-containing cyanomyophages could thus indicate (i) the existence of only a few Synechococcus spp. containing light-sensitive D1 proteins, so their viruses needed to produce their own stable D1 to replace the host's during replication in Lake Bourget, (ii) underwater light intensity in Lake Bourget could be reduced compared to that in Lake Annecy because of the trophic state and higher biomass levels in surface waters of the former (66). All above-given assumptions need a deeper analysis, experimental tests, and verifications to examine how the triplet divergence of viruses affects D1 protein structure and thereby photosynthesis, the difference between the D1 proteins of virus and host in terms of structure and function and between freshwater and marine environments, and if virus-specific D1 is restricted to specific depths or displays a narrow vertical distribution pattern in Lake Bourget.
The majority of psbA sequences (51 out of 62) obtained in our study had phylogenetic proximity to estuarine Synechococcus podoviruses. These results may indicate that these psbA sequences could be of Synechococcus podovirus origin and that the perialpine lakes may thus sustain diverse psbA-containing cyanopodoviruses. The cyanopodoviruses were also investigated for the same samples by targeting the DNA polymerase gene polA, using the primers designed by Chen et al. (8). However, the obtained sequences did not seem to be polA sequences and did not belong to cyanopodoviruses (data not shown). When blasted against the virome of Lake Bourget (57) using polA sequences of currently available cyanopodovirus isolates (P60, S-CBP1, S-CBP2, S-CBP3, S-CBP42, P-SSP7, syn5, Pf-WMP3, and Pf-WMP4) as queries, no hits were obtained, even when setting the E value at ≤0.1. These results suggest that in perialpine lakes, (i) the polA sequences of cyanopodoviruses may be very different from known cyanopodovirus isolates and (ii) the primers used might not be suitable for freshwater cyanopodoviruses, as they were designed based on marine cyanophages. To examine freshwater cyanopodovirus diversity, it is now crucial to obtain sequences from cyanophage isolates since, to the best of our knowledge, no Synechococcus podovirus genome has been reported from continental waters so far.
Contrasted cyanophage community compositions among environments.
For g20 sequences, significant differences were detected between Lake Annecy and Lake Bourget (P = 0.03) from the pooled 1-year data. This difference could reflect the difference of trophic states between the two lakes resulting in population shifts of the potential hosts in response to environmental changes and/or biological impact, i.e., grazing, predation, or the viral lysis by itself (23). In contrast, however, the psbA-containing cyanophages did not shown significant composition differences between the two lakes (P = 0.15) when considered over the year. This could be explained by the fact that (i) the psbA-containing cyanophages contain both cyanomyoviruses and cyanopodoviruses, the majority of which being podoviruses, and (ii) not every cyanophage possesses the host-derived psbA gene (5, 30). These results are consistent with a parallel analysis where we observed that psbA-containing cyanophages display seasonal dynamic patterns in community structure in both lakes, while it was not the case for g20 cyanomyoviruses (X. Zhong, F. Rimet, and S. Jacquet, unpublished data). The detection of similar psbA-containing phages in Lake Annecy and Lake Bourget may suggest similar annual host populations in these ecosystems, since cyanopodoviruses are generally host specific. Beyond this local aspect, it is of note that possessing psbA in cyanophage genomes may be beneficial for the hosts, the photosynthesis productivity being indifferently maintained or enhanced in Lake Annecy and Lake Bourget (34, 67). The selection pressure might in priority accommodate the limited nutrients for hosts of psbA-containing phages to sustain an annually constant population for which the viral lysis could help in nutrient recycling. This will need further investigation.
At last, we examined whether cyanomyoviruses of perialpine lakes differed from other freshwater and marine environments. Previous studies used the UniFrac analysis (25, 44) to statistically infer community composition dissimilarity of g20 cyanomyoviruses between environments. However, because the sequences recruited for phylogenetic reconstruction of each environment (Table 1) were from different studies for which the sample(s) was either from a single time point or period or from water issued from different depths and treated using different primers (CPS1/8, CPS1.1/8.1, and CPS4/G20-2) and with different sequencing depths of coverage, the method could not be used. Indeed, such an unbalanced data set to compare environmental traits of g20 sequences using Unifrac would generate here an erroneous view, particularly for Chilliwack Lake, Catfish Pond, Malaspina Inlet, Pendrell Sound, Coast of Colombia, and Coast of Chile, for which only one sequence was available. Nevertheless, our phylogenetic reconstruction was sufficient to show that g20 cyanomyoviruses are clearly different between the subalpine lakes and the other environments, especially the freshwaters. Among the four clusters containing cyanomyovirus isolates (i.e., culture-containing clusters I, II, and III and cluster 3), the culture-containing cluster II had no freshwater g20 sequences other than ours. In contrast, the other three clusters contained g20 sequences from other freshwaters (Lake Erie, Lake Constance, Cultus Lake, paddy field floodwater, and shore pond mat) but none from our lakes (Fig. 2). This could reflect the unique environment of perialpine lakes, where a distinct viral community exists. This is not surprising when one knows that both Lake Annecy and Lake Bourget have the same geological and water origin and also a unique host community (66).
Conclusion.
As in other investigated ecosystems, cyanophages seem very diverse in (peri)alpine lakes, and we found new sequences that seem to be unique to these ecosystems. In contrast to the only available and comparable study for Lake Bourget conducted a few years ago (12), a large proportion of g20 sequences obtained in 2011 were grouped with cyanomyovirus culture isolates. This result was likely due to the improvement of primers CPS1.1/8.1 on targeting cyanomyoviruses. For the first time, psbA-containing cyanophages were also investigated, and our results revealed that (i) they were all of Synechococcus phage origin, most of which were likely podoviruses, and that (ii) freshwater psbA sequences displayed distinct viral triplet D1 protein motifs compared to their marine counterparts, and a novel one was discovered (e.g., EFE). The cyanomyovirus community compositions were significantly different between the two lakes, while it was not the case for psbA-containing cyanophages, suggesting that this last group differed from the former and that all cyanophages could respond differently to their surrounding biology and/or environment within each lake.
ACKNOWLEDGMENTS
We thank Yves Desdevises for his critical reading of the manuscript and Irene Gregory-Eaves for English corrections. We particularly thank three anonymous reviewers who helped us build this article.
This work was supported by a fellowship from the region of Rhône-Alpes (France) awarded to X.Z.
Footnotes
Published ahead of print 13 September 2013
REFERENCES
- 1.Suttle CA. 2000. Ecological, evolutionary, and geochemical consequences of viral infection of cyanobacteria and eukaryotic algae, p 247–296 In Hurst C. (ed), Viral ecology. Academic Press, San Diego, CA [Google Scholar]
- 2.Fuhrman JA. 1999. Marine viruses and their biogeochemical and ecological effects. Nature 399:541–548 [DOI] [PubMed] [Google Scholar]
- 3.Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801–812 [DOI] [PubMed] [Google Scholar]
- 4.Wilhelm SW, Suttle CA. 1999. Viruses and nutrient cycles in the sea. Bioscience 49:781–788 [Google Scholar]
- 5.Dekel-Bird NP, Avrani S, Sabehi G, Pekarsky I, Marston MF, Kirzner S, Lindell D. 2013. Diversity and evolutionary relationships of T7-like podoviruses infecting marine cyanobacteria. Environ. Microbiol. 15:1476–1491 [DOI] [PubMed] [Google Scholar]
- 6.Mann NH. 2003. Phages of the marine cyanobacterial picophytoplankton. FEMS Microbiol. Rev. 27:17–34 [DOI] [PubMed] [Google Scholar]
- 7.Sabehi G, Shaulov L, Silver DH, Yanai I, Harel A, Lindell D. 2012. A novel lineage of myoviruses infecting cyanobacteria is widespread in the oceans. Proc. Natl. Acad. Sci. U. S. A. 109:2037–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen F, Huang SJ, Cai HY, Zhao MR, Jiao NZ, Wommack KE. 2009. Diverse and dynamic populations of cyanobacterial podoviruses in the Chesapeake Bay unveiled through DNA polymerase gene sequences. Environ. Microbiol. 11:2884–2892 [DOI] [PubMed] [Google Scholar]
- 9.Huang S, Wang K, Jiao N, Chen F. 2012. Genome sequences of siphoviruses infecting marine Synechococcus unveil a diverse cyanophage group and extensive phage-host genetic exchanges. Environ. Microbiol. 14:540–558 [DOI] [PubMed] [Google Scholar]
- 10.Sullivan MB, Waterbury JB, Chisholm SW. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047–1051 [DOI] [PubMed] [Google Scholar]
- 11.Clasen JL, Hanson CA, Ibrahim Y, Weihe C, Marston MF, Martiny JBH. 2013. Diversity and temporal dynamics of Southern California coastal marine cyanophage isolates. Aquat. Microb. Ecol. 69:17–31 [Google Scholar]
- 12.Dorigo U, Jacquet S, Humbert JF. 2004. Cyanophage diversity, inferred from g20 gene analyses, in the largest natural lake in France, Lake Bourget. Appl. Environ. Microbiol. 70:1017–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frederickson CM, Short SM, Suttle CA. 2003. The physical environment affects cyanophage communities in British Columbia inlets. Microb. Ecol. 46:348–357 [DOI] [PubMed] [Google Scholar]
- 14.Fuller NJ, Wilson WH, Joint IR, Mann NH. 1998. Occurrence of a sequence in marine cyanophages similar to that of T4 g20 and its application to PCR-based detection and quantification techniques. Appl. Environ. Microbiol. 64:2051–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jameson E, Mann NH, Joint I, Sambles C, Mühling M. 2011. The diversity of cyanomyovirus populations along a North–South Atlantic Ocean transect. ISME J. 5:1713–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mühling M, Fuller NJ, Millard A, Somerfield PJ, Marie D, Wilson WH, Scanlan DJ, Post AF, Joint I, Mann NH. 2005. Genetic diversity of marine Synechococcus and co-occurring cyanophage communities: evidence for viral control of phytoplankton. Environ. Microbiol. 7:499–508 [DOI] [PubMed] [Google Scholar]
- 17.Parvathi A, Zhong X, Jacquet S. 2012. Dynamics of various viral groups infecting autotrophic plankton in Lake Geneva. Adv. Oceanogr. Limnol. 3:171–191 [Google Scholar]
- 18.Short CM, Suttle CA. 2005. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl. Environ. Microbiol. 71:480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Chen F. 2004. Genetic diversity and population dynamics of cyanophage communities in the Chesapeake Bay. Aquat. Microb. Ecol. 34:105–116 [Google Scholar]
- 20.Wang G, Murase J, Asakawa S, Kimura M. 2010. Unique viral capsid assembly protein gene (g20) of cyanophages in the floodwater of a Japanese paddy field. Biol. Fertil. Soils 46:93–102 [Google Scholar]
- 21.Wilhelm SW, Carberry MJ, Eldridge ML, Poorvin L, Saxton MA, Doblin MA. 2006. Marine and freshwater cyanophages in a Laurentian Great Lake: evidence from infectivity assays and molecular analyses of g20 genes. Appl. Environ. Microbiol. 72:4957–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson WH, Fuller NJ, Joint IR, Mann NH. 1999. Analysis of cyanophage diversity and population structure in a south-north transect of the Atlantic Ocean. Bull. Inst. Oceanogr. 1999:209–216 [Google Scholar]
- 23.Zhong X, Berdjeb L, Jacquet S. 16 June 2013. Temporal dynamics and structure of picocyanobacteria and cyanomyoviruses in two large and deep peri-alpine lakes. FEMS Microbiol. Ecol. [Epub ahead of print.] 10.1111/1574-6941.12166. [DOI] [PubMed] [Google Scholar]
- 24.Zhong Y, Chen F, Wilhelm SW, Poorvin L, Hodson RE. 2002. Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl. Environ. Microbiol. 68:1576–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang G, Asakawa S, Kimura M. 2011. Spatial and temporal changes of cyanophage communities in paddy field soils as revealed by the capsid assembly protein gene g20. FEMS Microbiol. Ecol. 76:352–359 [DOI] [PubMed] [Google Scholar]
- 26.Sullivan MB, Krastins B, Hughes JL, Kelly L, Chase M, Sarracino D, Chistolm SW. 2009. The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial mobilome. Environ. Microbiol. 11:2935–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matteson AR, Loar SN, Bourbonniere RA, Wilhelm SW. 2011. Molecular enumeration of an ecologically important cyanophage in a Laurentian Great Lake. Appl. Environ. Microbiol. 77:6772–6779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marston MF, Sallee JL. 2003. Genetic diversity and temporal variation in the cyanophage community infecting marine Synechococcus species in Rhode Island's coastal waters. Appl. Environ. Microbiol. 69:4639–4647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandaa R-A, Larsen A. 2006. Seasonal variations in virus-host populations in Norwegian coastal waters: focusing on the cyanophage community infecting marine Synechococcus spp. Appl. Environ. Microbiol. 72:4610–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K, Chen F. 2008. Prevalence of highly host-specific cyanophages in the estuarine environment. Environ. Microbiol. 10:300–312 [DOI] [PubMed] [Google Scholar]
- 31.Huang S, Wilhelm SW, Jiao N, Chen F. 2010. Ubiquitous cyanobacterial podoviruses in the global oceans unveiled through viral DNA polymerase gene sequences. ISME J. 4:1243–1251 [DOI] [PubMed] [Google Scholar]
- 32.Mizuno CM, Rodriguez-Valera F, Garcia-Heredia I, Martin-Cuadrado A-B, Ghai R. 2013. Reconstruction of novel cyanobacterial siphovirus genomes from Mediterranean metagenomic fosmids. Appl. Environ. Microbiol. 79:688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clokie MRJ, Mann NH. 2006. Marine cyanophages and light. Environ. Microbiol. 8:2074–2082 [DOI] [PubMed] [Google Scholar]
- 34.Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. 2005. Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438:86–89 [DOI] [PubMed] [Google Scholar]
- 35.Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T, Kettler G, Sullivan MB, Steen R, Hess WR, Church GM, Chisholm SW. 2007. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449:83–86 [DOI] [PubMed] [Google Scholar]
- 36.Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW. 2006. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 4:e234. 10.1371/journal.pbio.0040234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharon I, Tzahor S, Williamson S, Shmoish M, Man-Aharonovich D, Rusch DB, Yooseph S, Zeidner G, Gilden SS, Mackey SR, Adir N, Weingart U, Horn D, Venter JC, Mandel-Gutfreund Y, Béjà O. 2007. Viral photosynthetic reaction center genes and transcripts in the marine environment. ISME J. 1:492–501 [DOI] [PubMed] [Google Scholar]
- 38.Chénard C, Suttle CA. 2008. Phylogenetic diversity of sequences of cyanophage photosynthetic gene psbA in marine and freshwaters. Appl. Environ. Microbiol. 74:5317–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandaa R-A, Clokie M, Mann NH. 2008. Photosynthetic genes in viral populations with a large genomic size range from Norwegian coastal waters. FEMS Microbiol. Ecol. 63:2–11 [DOI] [PubMed] [Google Scholar]
- 40.Wang G, Murase J, Asakawa S, Kimura M. 2009. Novel cyanophage photosynthetic gene psbA in the floodwater of a Japanese rice field. FEMS Microbiol. Ecol. 70:79–86 [DOI] [PubMed] [Google Scholar]
- 41.Wilhelm SW, Matteson AR. 2008. Freshwater and marine virioplankton: a brief overview of commonalities and differences. Freshw. Biol. 53:1076–1089 [Google Scholar]
- 42.Parvathi A, Zhong X, Pradeep-Ram AS, Jacquet S. 2013. Dynamics of auto- and heterotrophic picoplankton and associated viruses in Lake Geneva. Hydrol. Earth Syst. Sci. Dis. 10:8715–8746 [Google Scholar]
- 43.Personnic S, Domaizon I, Dorigo U, Berdjeb L, Jacquet S. 2009. Seasonal and spatial variability of virio, bacterio- and picophytoplanktonic abundances in three peri-alpine lakes. Hydrobiology 627:99–111 [Google Scholar]
- 44.Sullivan MB, Coleman ML, Quinlivan V, Rosenkrantz JE, DeFrancesco AS, Tan G, Fu R, Lee JA, Waterbury JB, Bielawski JP, Chisholm SW. 2008. Portal protein diversity and phage ecology. Environ. Microbiol. 10:2810–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 46.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552 [DOI] [PubMed] [Google Scholar]
- 48.Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 50.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Short CM, Rusanova O, Short SM. 2011. Quantification of virus genes provides evidence for seed-bank populations of phycodnaviruses in Lake Ontario, Canada. ISME J. 5:810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeidner G, Bielawski JP, Shmoish M, Scanlan DJ, Sabehi G, Béjà O. 2005. Potential photosynthesis gene recombination between Prochlorococcus and Synechococcus via viral intermediates. Environ. Microbiol. 7:1505–1513 [DOI] [PubMed] [Google Scholar]
- 53.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lozupone CA, Hamady M, Kelley ST, Knight R. 2007. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73:1576–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muyzer G. 1999. DGGE/TGGE a method for identifying genes from natural ecosystems. Curr. Opin. Microbiol. 2:317–322 [DOI] [PubMed] [Google Scholar]
- 56.Breitbart M, Rohwer F. 2005. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 13:278–284 [DOI] [PubMed] [Google Scholar]
- 57.Roux S, Enault F, Robin A, Ravet V, Personnic S, Theil S, Colombet J, Sime-Ngando T, Debroas D. 2012. Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS One 7:e33641. 10.1371/journal.pone.0033641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Millard AD, Zwirglmaier K, Downey MJ, Mann NH, Scanlan DJ. 2009. Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: implications for mechanisms of cyanophage evolution. Environ. Microbiol. 11:2370–2387 [DOI] [PubMed] [Google Scholar]
- 59.Greenberg BM, Gaba V, Mattoo AK, Edelman M. 1987. Identification of a primary in vivo degradation product of the rapidly-turning-over 32 kd protein of photosystem II. EMBO J. 6:2865–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mulo P, Sicora C, Aro E-M. 2009. Cyanobacterial psbA gene family: optimization of oxygenic photosynthesis. Cell. Mol. Life Sci. 66:3697–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Golden SS, Brusslan J, Haselkorn R. 1986. Expression of a family of psbA genes encoding a photosystem II polypeptide in the cyanobacterium Anacystis nidulans R2. EMBO J. 5:2789–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clarke AK, Soitamo A, Gustafsson P, Oquist G. 1993. Rapid interchange between two distinct forms of cyanobacterial photosystem II reaction center protein D1 in response to photoinhibition. Proc. Natl. Acad. Sci. U. S. A. 90:9973–9977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garczarek L, Dufresne A, Blot N, Cockshutt AM, Peyrat A, Campbell DA, Joubin L, Six C. 2008. Function and evolution of the psbA gene family in marine Synechococcus: Synechococcus sp. WH7803 as a case study. ISME J. 2:937–953 [DOI] [PubMed] [Google Scholar]
- 64.Sicora CI, Appleton SE, Brown CM, Chung J, Chandler J, Cockshutt AM, Vass I, Campbell DA. 2006. Cyanobacterial psbA families in Anabaena and Synechocystis encode trace, constitutive and UVB-induced D1 isoforms. Biochim. Biophys. Acta 1757:47–56 [DOI] [PubMed] [Google Scholar]
- 65.Schaefer MR, Golden SS. 1989. Light availability influences the ratio of two forms of D1 in cyanobacterial thylakoids. J. Biol. Chem. 264:7412–7417 [PubMed] [Google Scholar]
- 66.Jacquet S, Domaizon I, Anneville O. 2012. Evolution de paramètres clés indicateurs de la qualité des eaux et du fonctionnement écologique des grands lacs péri-alpins (Léman, Annecy, Bourget): etude comparative de trajectoires de restauration post-eutrophisation. Arch. Sci. 65:225–242 [Google Scholar]
- 67.Bragg JG, Chisholm SW. 2008. Modeling the fitness consequences of a cyanophage-encoded photosynthesis gene. PLoS One 3:e3550. 10.1371/journal.pone.0003550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dreher TW, Brown N, Bozarth CS, Schwartz AD, Riscoe E, Thrash C, Bennett SE, Tzeng SC, Maler CS. 2011. A freshwater cyanophage whose genome indicates close relationships to photosynthetic marine cyanomyophages. Environ. Microbiol. 13:1858–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]