Abstract
Cocultures of strains from two Bifidobacterium and two Bacteroides species were performed with exopolysaccharides (EPS) previously purified from bifidobacteria, with inulin, or with glucose as the carbon source. Bifidobacterium longum NB667 and Bifidobacterium breve IPLA20004 grew in glucose but showed poor or no growth in complex carbohydrates (inulin, EPS E44, and EPS R1), whereas Bacteroides grew well in the four carbon sources tested. In the presence of glucose, the growth of Bacteroides thetaiotaomicron DSM-2079 was inhibited by B. breve, whereas it remained unaffected in the presence of B. longum. Ba. fragilis DSM-2151 contributed to a greater survival of B. longum, promoting changes in the synthesis of short-chain fatty acids (SCFA) and organic acids in coculture with respect to monocultures. In complex carbohydrates, cocultures of bifidobacterium strains with Ba. thetaiotaomicron did not modify the behavior of Bacteroides nor improve the poor growth of bifidobacteria. The metabolic activity of Ba. fragilis in coculture with bifidobacteria was not affected by EPS, but greater survival of bifidobacteria at late stages of incubation occurred in cocultures than in monocultures, leading to a higher production of acetic acid than in monocultures. Therefore, cocultures of Bifidobacterium and Bacteroides can behave differently against fermentable carbohydrates as a function of the specific characteristics of the strains from each species. These results stress the importance of considering specific species and strain interactions and not simply higher taxonomic divisions in the relationship among intestinal microbial populations and their different responses to probiotics and prebiotics.
INTRODUCTION
The colon is a complex microbial ecosystem dominated by obligate anaerobes that reach levels up to 1011 cells per gram of intestinal content (1, 2). In spite of the huge diversity of strains, up to 87% of the microbial inhabitants of the human colon belong to only two bacterial phyla, Bacteroidetes and Firmicutes. Actinobacteria and other phyla are present at lower levels (3). Within the group of intestinal Bacteroidetes, Bacteroides spp. account for up to 20% of the human colon microbiota (4). Although a great variety of Bacteroides species has been reported among individuals, Bacteroides thetaiotaomicron always seems to be present (5, 6). This species is considered a human symbiont that stabilizes the colon ecosystem, but the genus also harbors some notorious opportunistic and pathogenic species, as is the case of Bacteroides fragilis (7). Members of the Bacteroides genus are saccharolytic microorganisms producing succinic, acetic, lactic, and propionic acids, but they are also capable of proteolytic fermentation (8). Bifidobacteria account for approximately 3% of the adult human microbiota (9) and are frequently identified as probiotics, based on the implied health-promoting benefits attributed to some strains (10). Bifidobacterium longum is one of the predominant species in adult humans. This species and Bifidobacterium breve are also abundant in the intestine of infants. Bifidobacteria produce lactic and acetic acids as the main metabolic end products of carbohydrate fermentation and smaller amounts of formic acid and ethanol (11). Prebiotics are defined as nondigestible food ingredients that beneficially affect the host by selectively stimulating the growth and/or activity of one or a limited number of bacterial species in the colon, thus improving host health (12). Many of the health-promoting effects attributed to prebiotic substrates are due to their suitability to be fermented by the colonic microbiota producing short-chain fatty acids (SCFA). Bifidobacteria have traditionally been considered the target of prebiotic action, as these substrates can be directly metabolized by these microorganisms; however, some in vitro and in vivo evidences indicate that the effects could also indirectly involve other members of the human colon microbiota through the utilization of these substrates in combination with bifidobacteria. The most well-studied prebiotics to date are inulin-type fructans (13–15). Some researchers have previously demonstrated different degradation mechanisms of oligofructose and inulin-like fructans by Bifidobacterium and Bacteroides species in pure cultures, as well as in cocultures (14–16).
Some bifidobacteria are able to produce exopolysaccharides (EPS), which are complex polymers composed of several units of monosaccharides (17). EPS from bifidobacteria may be released in situ by microorganisms of this species inhabiting the human colon or may be produced by probiotics present as adjunct cultures in fermented dairy products. Although the synthesis of EPS in vivo has not been demonstrated and the amount of polymer released by the producing bacteria would be presumably low, our previous work indicates that bile stimulates the production of EPS by bifidobacteria in in vitro simulated gastrointestinal conditions (17, 18). In addition, EPS could act as fermentable substrates for the human colonic microbiota (19, 20). The fermentation in fecal batch cultures of small amounts of EPS and inulin (0.3%, wt/vol) caused shifts in the synthesis of SCFA related to variations in the levels of some intestinal microbial populations, such as Bacteroides and Bifidobacterium (20). Therefore, in the present work, we selected strains from two species of Bifidobacterium (B. breve and B. longum) and two species of Bacteroides (Ba. thetaiotaomicron and Ba. fragilis) as a model of study, in order to gain an insight into the influence that the presence of EPS and other carbon sources could exert on the interactions between members of these two intestinal microbial groups, by growing them separately and together.
MATERIALS AND METHODS
Bacterial strains.
Two Bacteroides and two Bifidobacterium strains belonging to different species were used in monocultures and cocultures in this study. Ba. thetaiotaomicron DSM-2079 and Ba. fragilis DSM-2151 strains were obtained from the DSMZ bacterial culture collection (Braunschweig, Germany). B. longum NB667 was from the NIZO food research culture collection (Ede, The Netherlands), and B. breve IPLA20004 (also named B. breve BM 12/11) was isolated from breast milk (21) and is held in the IPLA's culture collection. Species identity was confirmed by partial amplification of the 16S rRNA gene using primers plb16 and mlb16 (22) and by sequencing and alignment with sequences from reference strains held in the GenBank database. Strains from frozen stocks were reactivated in Gifu anaerobic medium (GAM) broth (Nissui Pharmaceutical Co, Tokyo, Japan) and in MRS broth (BioKar Diagnostics, Beauvais, France) supplemented with 0.25% (wt/vol) of l-cysteine (Sigma Chemical Co., St. Louis, MO) (named GAMc and MRSc) for Bacteroides and Bifidobacterium, respectively. Strains were incubated overnight at 37°C in an anaerobic cabinet (Mac 1000; Don Whitley Scientific, West Yorkshire, United Kingdom) under a 10% H2, 10% CO2, and 80% N2 atmosphere. To prepare the inoculum stocks, 10 ml of modified carbohydrate-free basal medium (mCFBM; composition specified below) with 1% (wt/vol) glucose was inoculated (1% [vol/vol]) with cultures of Bifidobacterium and Bacteroides strains and incubated for 16 to 18 h, as indicated before. Cultures were then centrifuged at 12,000 × g for 10 min and resuspended in the same volume of mCFBM without a carbon source. Inocula were frozen under liquid N2 and stored at −80°C until use.
EPS isolation.
EPS fractions produced by Bifidobacterium animalis subsp. lactis IPLA R1, a dairy origin strain (23), and by B. longum subsp. longum IPLA E44, a fecal isolate from a healthy adult (24), were isolated and purified from the cellular biomass harvested from agar-MRSc agar plates as specified by Ruas-Madiedo et al. (23).
Batch culture fermentation.
Uncontrolled-pH batch cultures were performed in the nondefined peptone and yeast extract containing CFBM, previously described by Salazar et al. (20). For the present work, it was modified by the addition of vitamin B12 (10 mg liter−1), vitamin K (2 mg liter−1), vitamin B1 (2 mg liter−1), pyridoxal (1 mg liter−1), calcium pantothenate (2 mg liter−1), folic acid (1 mg liter−1), riboflavin (1 mg liter−1), biotin (1 mg liter−1), nicotinic acid (3 mg liter−1), para-aminobenzoic acid (1 mg liter−1), and a solution (2 ml liter−1) of ferrous citrate (25 mM) and trisodium citrate (75 mM) (mCFBM). The final pH of the medium ranged between 6.7 and 7.0.
Pairwise combinations of Bifidobacterium and Bacteroides strains, as well as monocultures of strains, were performed in mCFBM with an added 0.3% (wt/vol) of glucose, inulin, or purified EPS E44 or EPS R1 fractions. The corresponding frozen inocula were added (1% [vol/vol]) to 3.5 ml of the culture medium. Trials of cocultures and the corresponding monocultures in different carbon sources were run in triplicate for a period of 72 h at 37°C under anaerobic conditions. Samples were obtained at fixed times for microbial counts and SCFA and organic acid analyses.
The ability to utilize lactic acid by the two Bacteroides strains considered in this work was tested in mCFBM, with 0.15% lactic acid (vol/vol) added as the carbon source. Additionally, the ability of Bacteroides strains to use the organic nitrogen compounds present in the culture medium was assessed in mCFBM by determining growth and the ability to produce branched-chain fatty acids (BCFA). Cultures were incubated for 72 h in anaerobic conditions as indicated above. At the end of incubation, optical density at 600 nm was determined in cultures, and samples were taken for SCFA and organic acid analyses.
Estimation of bacterial growth by qPCR.
Quantification (cell counts ml−1) of Bifidobacterium and Bacteroides species growing in monoculture and coculture was performed throughout fermentations by quantitative PCR (qPCR) using DNA isolated from batch cultures. Standard curves were obtained by converting 16S rRNA gene copies to cell counts obtained in pure cultures of each strain growing in MRSc in the case of Bifidobacterium and GAMc for Bacteroides. Primers and conditions were those previously described (25).
Analysis of SCFA, organic acids, and glucose.
Cell-free supernatants from cultures were filtered through 0.2-μm-pore-size filters. Identification and quantification of SCFA and BCFA were carried out by gas chromatography-mass spectrometry/flame ionization detector (MS/FID), using a system composed of a 6890N gas chromatograph (Agilent Technologies Inc., Palo Alto, CA, USA) connected with an FID and an MS 5973N detector (Agilent), as described previously (19, 26). A high-performance liquid chromatography (HPLC) system composed of an Alliance 2695 separation module, a photodiode array (PDA) detector (Waters 996), a refractive index detector (Waters 2414), and Empower software (Waters, Milford, MA) was employed. The PDA detector was used for quantification of organic acids at 210 nm, whereas the amount of glucose was analyzed with the refractive index detector. Chromatographic conditions were those indicated previously by Salazar et al. (27). Results of SCFA, BCFA, and organic acids were expressed in millimolar concentrations.
Calculation of carbon recovery.
Carbon recovery (CR), expressed in percentages, was calculated by comparing the total amount of carbon recovered in the metabolites analyzed to the total amount of glucose consumed. For Bacteroides strains, the production of one mole of CO2 for every mole of acetic acid formed (+ 1 × [acetic acid] in the equation below) was considered, as well as the uptake of one mole of CO2 for every mole of succinic acid produced (− 1 × [succinic acid] in the equation) (16, 28–30).
The following equations were used: CR of bifidobacteria ≥ 100 × (3 × [lactic acid] + 2 × [acetic acid] + 1 × [formic acid]/6 × [glucose consumed]) and CR of bacteroides ≥ 100 × (2 × [acetic acid] + 3 × [propionic acid] + 4 × [succinic acid] + 1 × [formic acid] +1 × [acetic acid] − 1 × [succinic acid]/6 × [glucose consumed]).
Statistical analysis.
Statistical analyses were performed using the SPSS-PC software, version 19.0 (SPSS Inc., Chicago, IL, USA). One-way analysis of variance (ANOVA) tests were run in monocultures of Bacteroides and Bifidobacterium for the different SCFA and organic acids. Strains were used as factors, with two categories corresponding to the different species of each genus analyzed. One-way ANOVA was also performed to compare the results of the different parameters by using cocultures versus monocultures and time of incubation as factors. When necessary, a post hoc least significant difference (LSD) comparison test was applied to determine statistical differences between categories.
RESULTS AND DISCUSSION
Behavior of Bifidobacterium species growing in pure culture.
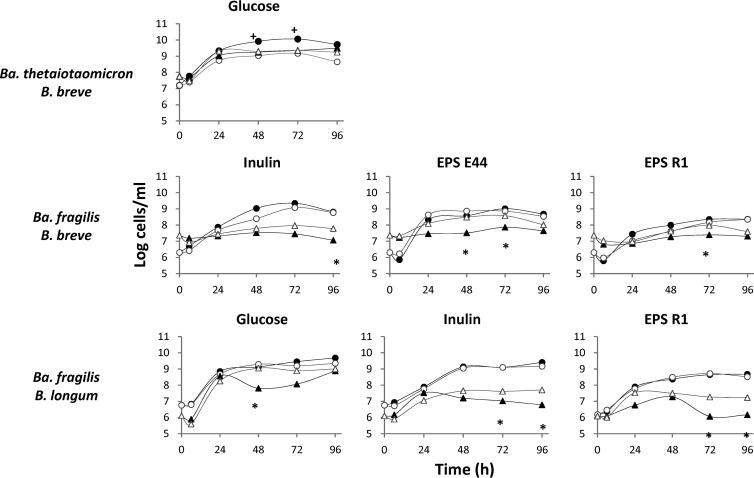
In pure culture, B. longum NB667 was able to grow in glucose, inulin, and EPS (P < 0.05), whereas B. breve IPLA20004 displayed significant growth only in cultures with glucose but not with EPS or inulin (Fig. 1). In monocultures of bifidobacteria, the pH decreased by about 2.5 units throughout fermentation when glucose was the carbon source, whereas it showed little variation in complex carbohydrates (data not shown).
Fig 1.
Growth (mean of log cells ml−1) in single culture and in coculture of Ba. thetaiotaomicron DSMZ 2079 or Ba. fragilis DSMZ 2151 with B. longum NB 667 or B. breve IPLA 20004 in a basal medium supplemented with 0.3% glucose, inulin, EPS E44, or EPS R1 as a carbon source. ●, Bacteroides strain growing in single culture; ○, Bacteroides strain growing in coculture; ▲, Bifidobacterium strain growing in single culture; △, Bifidobacterium strain growing in coculture. The coefficient of variation (standard deviation × 100/mean) of data obtained from the three replicates was about 4.2 to 5.5%. +, significant differences (P < 0.05) of Bacteroides counts reached in coculture compared to the corresponding monoculture; *, significant differences (P < 0.05) of Bifidobacterium counts reached in coculture compared to the corresponding monoculture. Among all the possible culture combinations of Bacteroides and Bifidobacterium strains, the sole combination showing significant variation in the growth of Bacteroides and those enhancing significantly the survival of Bifidobacterium (P < 0.05) are presented.
Glucose was consumed almost completely after 72 h of fermentation in mCFBM cultures of both strains, with a carbon recovery above 90%. Although acetic acid was the most abundant metabolite formed, clear differences between the metabolic profiles of both Bifidobacterium strains were found (Table 1). Thus, whereas B. longum NB667 formed considerable amounts of lactic acid and smaller amounts of formic acid, B. breve IPLA20004 produced more formic than lactic acid.
Table 1.
SCFA and organic acid concentrations and glucose consumption obtained in uncontrolled-pH monocultures of Bifidobacterium and Bacteroides species at 72 h of incubation with glucose, inulin, EPS E44, and EPS R1 as carbon sourcesa
| Carbon source | Species | Glucose consumption (mM) | Concn (mM) |
Carbon recovery (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Acetic acid | Propionic acid | Lactic acid | Succinic acid | Formic acid | ||||
| Glucose | Control (0 h) | 0 | 3.77 ± 1.29 | 0.48 ± 0.01 | 0.08 ± 0.13 | 0.09 ± 0.01 | 0.05 ± 0.07 | NA |
| Ba. thetaiotaomicron | 8.91 ± 1.41 | 10.94 ± 1.81 | 3.74 ± 0.50* | – | 6.18 ± 0.73* | 1.83 ± 0.31 | 97 ± 12 | |
| Ba. fragilis | 7.08 ± 1.24 | 12.58 ± 0.77 | 10.28 ± 1.00 | – | 3.24 ± 0.30 | 1.33 ± 0.20 | 132 ± 27 | |
| B. breve | 11.29 ± 1.22 | 29.24 ± 4.65 | – | 1.96 ± 0.55* | – | 9.50 ± 1.26* | 97 ± 14 | |
| B. longum | 12.88 ± 2.38 | 27.21 ± 4.31 | – | 10.15 ± 0.85 | – | 1.75 ± 0.13 | 102 ± 10 | |
| Inulin | Control (0 h) | NA | 2.02 ± 0.49 | 0.46 ± 0.01 | – | 0.09 ± 0.02 | – | NA |
| Ba. thetaiotaomicron | NA | 4.86 ± 0.80* | 2.25 ± 0.40* | – | 1.26 ± 0.12 | – | NA | |
| Ba. fragilis | NA | 9.55 ± 1.44 | 9.66 ± 1.47 | – | 1.22 ± 0.21 | – | NA | |
| B. breve | NA | 2.95 ± 0.51 | – | – | – | – | NA | |
| B. longum | NA | 3.10 ± 0.76 | – | – | – | – | NA | |
| EPS E44 | Control (0 h) | NA | 1.29 ± 0.14 | 0.45 ± 0.00 | 0.10 ± 0.02 | – | NA | |
| Ba. thetaiotaomicron | NA | 7.40 ± 1.59 | 6.29 ± 1.92 | 1.79 ± 0.32 | – | NA | ||
| Ba. fragilis | NA | 6.12 ± 1.09 | 6.50 ± 0.32 | 1.72 ± 0.30 | – | NA | ||
| B. breve | NA | 3.74 ± 1.06* | – | – | – | – | NA | |
| B. longum | NA | 1.97 ± 0.18 | – | – | – | – | NA | |
| EPS R1 | Control (0 h) | NA | 1.33 ± 0.22 | 0.45 ± 0.00 | 0.10 ± 0.03 | – | NA | |
| Ba. thetaiotaomicron | NA | 7.02 ± 1.74* | 5.58 ± 1.96 | 1.45 ± 0.79 | – | NA | ||
| Ba. fragilis | NA | 3.71 ± 0.51 | 4.09 ± 0.93 | 1.45 ± 0.24 | – | NA | ||
| B. breve | NA | 3.08 ± 0.61* | – | – | – | – | NA | |
| B. longum | NA | 1.68 ± 0.23 | – | – | – | – | NA | |
Initial glucose level in the culture medium was 12.04 ± 1.38 mM. *, significant differences between strains from the same genus (P < 0.05); –, no detection or detection below the quantification limit; NA, not applicable.
Glucomannans from yeast extract present in the culture medium interfere with the detection and quantification of EPS and inulin (19), and, therefore, the calculation of the polymer consumed for cultures with added EPS was not possible in our experimental conditions. The fermentation pattern in cultures with complex carbohydrates by Bifidobacterium differed from those obtained with glucose and led mainly to the formation of small amounts of acetic acid in cultures of both strains (Table 1). Several authors have previously demonstrated a metabolic shift in the glycolytic pathway toward more acetic and formic acids and ethanol production at the expense of lactic acid in bifidobacteria when growth slows (31–33). In this way, less readily fermentable energy sources lead to more ATP formed per mole of sugar consumed (14, 15, 34). The predominant acetic acid production together with the limited or no increase of B. breve and B. longum population levels in cultures with EPS and inulin support the conditions of limited access to energy from these carbohydrates.
Behavior of Bacteroides species growing in pure culture.
Ba. thetaiotaomicron DSM-2079 and Ba. fragilis DSM-2151 grew well in the carbon sources tested (Fig. 1). The pH during incubation decreased more in glucose (1.5 to 1.9 units) than in cultures with EPS (0.3 to 0.7 units). In inulin as the carbohydrate source, Ba. fragilis was able to promote a more pronounced pH decrease than Ba. thetaiotaomicron (1 pH unit compared to 0.1), which is in line with the higher SCFA production by Ba. fragilis than by Ba. thetaiotaomicron (Table 1). About 60 to 70% of the glucose was consumed after 72 h of fermentation in pure cultures of both strains (Table 1). In spite of this, CR at this time was nearly 100% in cultures of Ba. thetaiotaomicron and higher than this value in cultures of Ba. fragilis. A reason for this may be the fermentation of carbohydrates different from glucose and/or organic nitrogen compounds present in the culture medium. In this respect, it is known that members of the genus Bacteroides can ferment proteins and amino acids producing BCFA (35–37). We corroborated that Ba. thetaiotaomicron DSM-2079 and Ba. fragilis DSM-2151 grew slowly in mCFBM, producing BCFA (mainly isobutyric and isovaleric acids) and SCFA (experimental data not shown). This provides a rationale for the high CR values obtained in pure cultures of Ba. thetaiotaomicron and Ba. fragilis.
Different fermentation patterns were evidenced between Ba. thetaiotaomicron DSM-2079 and Ba. fragilis DSM-2151 regarding the production of SCFA (Table 1). Although acetic acid was the most abundant metabolite produced from glucose, in cultures of Ba. thetaiotaomicron, it was followed in abundance by succinic and then propionic acids, whereas Ba. fragilis produced clearly more propionic than succinic acid (Table 1). In complex carbon sources (inulin and EPS), the metabolic profile also differed between Bacteroides strains (Table 1). Propionic acid was the most abundant metabolite produced by Ba. fragilis, followed by acetic acid, whereas Ba. thetaiotaomicron produced more acetic than propionic acid. Previous studies by other authors indicated that the fermentation product profile from carbohydrates by Bacteroides greatly differed depending on the substrates. Succinic acid was generally the main metabolite produced at short generation times, whereas the proportions of acetic and propionic acids increased at long generation times or with less readily fermentable carbohydrates (28, 30, 38). Our results confirm these observations, as the proportion of propionic to succinic acid was higher in complex carbon sources than in glucose.
Interaction of Bacteroides and Bifidobacterium species in coculture.
Decreases in pH paralleled the increases in SCFA concentrations in cocultures of Bifidobacterium and Bacteroides. In inulin, cocultures with Ba. fragilis reached higher concentrations of SCFA and succinic acid than cocultures with Ba. thetaiotaomicron (Table 2), thus leading to more pronounced pH decreases in the former.
Table 2.
SCFA and organic acid concentrations in uncontrolled-pH cocultures of Bifidobacterium with Bacteroides strains at 72 h of incubation with glucose (initial levels of 12.04 ± 1.38 mM), inulin, EPS E44, or EPS R1 as carbon sourcesa
| Carbon source | Sugar consumption or SCFA and organic acid formation | Concn (mM) of glucose consumed or SCFA and organic acids formed |
||||
|---|---|---|---|---|---|---|
| Control (0 h) |
Ba. thetaiotaomicron |
Ba. fragilis |
||||
| B. breve | B. longum | B. breve | B. longum | |||
| Glucose | Glucose consumption | 0 | 11.20 ± 1.43 | 12.91 ± 1.83 | 7.55 ± 0.71↓B | 11.33 ± 0.81↑Ba |
| Acetic acid | 3.77 ± 1.29 | 24.16 ± 3.15↑Ba | 18.20 ± 4.63↓B | 19.61 ± 2.24↑Ba↓B | 20.88 ± 2.92↑Ba↓B | |
| Propionic acid | 0.48 ± 0.01 | 0.83 ± 0.26↓Ba | 3.89 ± 0.50 | 6.37 ± 0.62↓Ba | 5.83 ± 0.47↓Ba | |
| Lactic acid | 0.08 ± 0.13 | 0.27 ± 0.10 | 3.05 ± 1.94↓B | 0.00 ± 0.00↓B | 5.71 ± 1.18↓B | |
| Formic acid | 0.05 ± 0.07 | 8.83 ± 1.27↑Ba | 2.48 ± 0.19↑Ba↑B | 4.75 ± 0.66↑Ba↓B | 1.86 ± 0.21↑Ba | |
| Succinic acid | 0.09 ± 0.01 | 1.04 ± 0.68↓Ba | 6.88 ± 0.74 | 2.95 ± 0.29 | 2.45 ± 0.30↓Ba | |
| Inulin | Acetic acid | 2.02 ± 0.49 | 3.79 ± 0.34 | 4.81 ± 0.92 | 8.21 ± 1.40↑B | 8.48 ± 1.10↑B |
| Propionic acid | 0.46 ± 0.01 | 1.41 ± 0.06 | 2.55 ± 0.46 | 5.34 ± 0.42↓Ba | 9.31 ± 1.10 | |
| Succinic acid | 0.09 ± 0.02 | 0.95 ± 0.15 | 1.25 ± 0.14 | 1.05 ± 0.18 | 1.37 ± 0.04 | |
| EPS E44 | Acetic acid | 1.29 ± 0.14 | 8.52 ± 2.16↑B | 8.47 ± 2.47↑B | 9.03 ± 1.66↑Ba↑B | 6.96 ± 0.99↑Ba↑B |
| Propionic acid | 0.45 ± 0.00 | 4.78 ± 1.97 | 7.24 ± 1.81 | 6.07 ± 0.85 | 5.96 ± 1.94 | |
| Succinic acid | 0.10 ± 0.02 | 1.44 ± 0.06↓Ba | 2.12 ± 0.22 | 1.55 ± 0.14 | 1.52 ± 0.32 | |
| EPS R1 | Acetic acid | 1.33 ± 0.22 | 7.49 ± 1.99 | 7.78 ± 1.67↑B | 4.81 ± 0.47↑Ba↑B | 4.91 ± 0.76↑Ba↑B |
| Propionic acid | 0.45 ± 0.00 | 3.51 ± 0.94 | 6.57 ± 1.00 | 3.39 ± 0.49 | 4.07 ± 0.77 | |
| Succinic acid | 0.10 ± 0.03 | 0.96 ± 0.80 | 1.99 ± 0.17 | 0.96 ± 0.35 | 1.44 ± 0.22 | |
↑Ba and ↓Ba indicate significantly higher or lower levels (P < 0.05), respectively, of a given metabolite in coculture than in the corresponding monoculture of the Bacteroides strain. ↑B and ↓B indicate significantly higher or lower levels (P < 0.05), respectively, of a given metabolite in coculture than in the corresponding monoculture of the Bifidobacterium strain. Glucose consumption is indicated for cocultures with this sugar as the carbon source.
Bacterial levels and metabolite production by pairwise combinations of Bifidobacterium and Bacteroides strains incubated with the different carbon sources were compared with the results obtained from pure cultures of the corresponding strains. In general, Bacteroides reached higher population levels than Bifidobacterium in cocultures, and the presence of bifidobacteria seems not to affect the growth of Bacteroides (data not shown). The only exception to this was the delayed growth at prolonged incubation times of Ba. thetaiotaomicron cocultured with B. breve when glucose was used as the carbon source (Fig. 1). A possible explanation for this inhibition could be the production under such conditions of antimicrobial compounds by B. breve (39) or the outcompetition at prolonged incubation times of bifidobacteria by using carbon sources still available in the culture medium and not consumed by Ba. thetaiotaomicron. Relating to this, we have previously reported on the inhibition by B. longum of other Gram-positive bacteria growing in combined culture (40, 41). Coculture with Ba. thetaiotaomicron did not improve the poor growth displayed by bifidobacteria in pure cultures with complex carbon sources (data not shown). In contrast, the survival of Bifidobacterium increased in the presence of Ba. fragilis in most carbohydrate sources so that cocultivation of both microorganisms resulted in higher population levels of B. breve and B. longum at late stages of incubation than those obtained in the corresponding monocultures (Fig. 1).
Both Bifidobacterium and Bacteroides species can produce acetic and formic acids. Bacteroides is able to form succinic acid, whereas this compound is not synthesized or is produced in very small amounts by bifidobacteria in any condition. Finally, while Bacteroides is a propionic acid producer, the metabolic pathway for the synthesis of propionic acid is not present in bifidobacteria (15, 28, 29, 32). Therefore, the metabolic contribution of microorganisms in coculture was inferred from the levels of propionic and succinic acids produced by Bacteroides, as well as from the levels of other common metabolites from carbohydrate fermentation (SCFA and organic acids) synthesized by both bacteria. With glucose as the carbon source, acetic acid reached levels in cocultures of Ba. thetaiotaomicron and B. breve similar to those in pure cultures of the bifidobacteria, whereas considerably smaller amounts of propionic and succinic acids were obtained from cocultures than in the monocultures of Bacteroides (Table 2). This suggests an impairment of the metabolic activity of Ba. thetaiotaomicron in the presence of B. breve as a consequence of its growth inhibition. In the remaining Bacteroides and Bifidobacterium combinations, acetic acid attained intermediate levels between the lower concentration reached by the monocultures of Bacteroides and the higher level of the monocultures of bifidobacteria. Specifically, in cocultures of Ba. thetaiotaomicron DSM-2079 and B. longum NB667, the production of propionic and succinic acids was similar to that in the monoculture of Bacteroides, suggesting that the metabolic activity of Ba. thetaiotaomicron probably remained unaffected under such conditions; however, in combined cultures of Ba. fragilis and bifidobacteria, lower propionic concentrations, and similar or lower levels of succinic acid than in Bacteroides monocultures, were obtained. This pointed to a probable slowdown of the metabolic activity of Ba. fragilis when bifidobacteria were present. On the other hand, lower lactic acid levels were obtained in most cocultures in glucose with respect to the monocultures of the corresponding Bifidobacterium strain, as was previously reported in coculture fermentations of bifidobacteria and bacteroides with inulin-type fructans (16). Ba. thetaiotaomicron DSM-2079 and Ba. fragilis DSM-2151 growing alone contributed scarcely both to the consumption of lactic acid present in the culture medium (14 to 16%) and to the production of formic acid (experimental data not shown). These findings, together with the increase of formic acid in cocultures of Ba. thetaiotaomicron and B. longum with respect to the corresponding monocultures, point to shifts in the metabolism of lactic and formic acids by one or both microorganisms when they are growing together.
With complex carbon sources, acetic acid was generally the most abundant metabolite produced in cocultures, followed by propionic acid and smaller amounts of succinic acid. Propionic and succinic acid levels in cocultures were similar to levels attained in the corresponding monocultures of Bacteroides for most pairwise combinations of strains, indicating that the metabolism of Bacteroides was probably not affected by the presence of bifidobacteria. In the presence of EPS, cocultures with Ba. thetaiotaomicron displayed levels of acetic acid close to the concentrations reached by monocultures of Bacteroides. In contrast, higher production of acetic acid was obtained in cocultures of bifidobacteria and Ba. fragilis DSM-2151 with EPS as the carbon source than in the corresponding monocultures of Bifidobacterium and Bacteroides, thus indicating an enhancement of the production of this acid in cocultures with Ba. fragilis. Therefore, the behavior of bifidobacterium strains in the same substrate appears to be influenced by the growth and metabolic characteristics of the Bacteroides strain present in the same environment. Falony et al. (16) indicated that the capacity of several Bifidobacterium strains to compete with Ba. thetaiotaomicron for the use of inulin-type fructans was dependent on the ability of the bifidobacteria to degrade fructose and oligofructose in addition to inulin. Using germfree mice colonized with Ba. thetaiotaomicron and B. longum, an expansion in the diversity of polysaccharides targeted for degradation by B. thetaiotaomicron has been observed in the presence of the bifidobacteria, demonstrating an adaptation for substrate utilization by both species in response to one another (42).
In short, differences in growth and metabolic characteristics of Bifidobacterium and Bacteroides strains can influence their joint behavior against EPS and other fermentable carbohydrate sources available in the growth environment. The results presented here stress the importance of considering specific species and strains, and not simply high taxonomic divisions, in the relationship among intestinal microbial populations. Variations at the level of species or strain composition among individuals or human population groups could condition a different response of their intestinal microbiota to specific diets or probiotic and prebiotic interventions.
ACKNOWLEDGMENTS
This work was financially supported by the project AGL2010-16525 from the Spanish Ministry of Economy and Competitiveness (MINECO). David Rios-Covian was funded by a predoctoral fellowship from MINECO (Formación de Personal Investigador Program [FPI]), and Silvia Arboleya was supported by a predoctoral Junta de Ampliación de Estudios Program (JAE) fellowship from CSIC, Spain.
The excellent technical assistance of María Fernández-García and Lidia Aláez is greatly appreciated.
Footnotes
Published ahead of print 27 September 2013
REFERENCES
- 1.Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920 [DOI] [PubMed] [Google Scholar]
- 2.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. 2006. Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl. Acad. Sci. U. S. A. 106:5859–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rigottier-Gois L, Bourhis AG, Gramet G, Rochet V, Dore J. 2003. Fluorescent hybridisation combined with flow cytometry and hybridisation of total RNA to analyse the composition of microbial communities in human faeces using 16S rRNA probes. FEMS Microbiol. Ecol. 43:237–245 [DOI] [PubMed] [Google Scholar]
- 5.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848 [DOI] [PubMed] [Google Scholar]
- 7.Wu S, Rhee KJ, Zhang M, Franco A, Sears CL. 2007. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J. Cell Sci. 120:1944–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacFarlane GT, Cummings JH. 1991. The colonic flora, fermentation and large bowel digestive function, p 51–92 In Phillips SF, Pemberton JH, Shorter RG. (ed), The large intestine: physiology, patho-physiology and disease. Raven Press Ltd., New York, NY [Google Scholar]
- 9.Matsuki T, Watanabe K, Fujimoto J, Miyamoto Y, Takada T, Matsumoto K, Oyaizu H, Tanaka R. 2002. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masco L, Huys G, de Brandt E, Temmerman R, Swings J. 2005. Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int. J. Food Microbiol. 102:221–230 [DOI] [PubMed] [Google Scholar]
- 11.Van der Meulen R, Adriany T, Verbrugghe K, de Vuyst L. 2006. Kinetic analysis of bifidobacterial metabolism reveals a minor role for succinic acid in the regeneration of NAD+ through its growth-associated production. Appl. Environ. Microbiol. 72:5204–5210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401–1412 [DOI] [PubMed] [Google Scholar]
- 13.Duncan SH, Scott KP, Ramsay AG, Harmsen HJ, Welling GW, Stewart CS, Flint HJ. 2003. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl. Environ. Microbiol. 69:1136–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falony G, Lazidou K, Verschaeren A, Weckx S, Maes D, de Vuyst L. 2009. In vitro kinetic analysis of fermentation of prebiotic inulin-type fructans by Bifidobacterium species reveals four different phenotypes. Appl. Environ. Microbiol. 75:454–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van der Meulen R, Makras L, Verbrugghe K, Adriany T, de Vuyst L. 2006. In vitro kinetic analysis of oligofructose consumption by Bacteroides and Bifidobacterium spp. indicates different degradation mechanisms. Appl. Environ. Microbiol. 72:1006–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falony G, Calmeyn T, Leroy F, de Vuyst L. 2009. Coculture fermentations of Bifidobacterium species and Bacteroides thetaiotaomicron reveal a mechanistic insight into the prebiotic effect of inulin-type fructans. Appl. Environ. Microbiol. 75:2312–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruas-Madiedo P, Moreno JA, Salazar N, Delgado S, Mayo B, Margolles A, de los Reyes-Gavilan CG. 2007. Screening of exopolysaccharide-producing Lactobacillus and Bifidobacterium strains isolated from the human intestinal microbiota. Appl. Environ. Microbiol. 73:4385–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruas-Madiedo P, Gueimonde M, Arigoni F, de los Reyes-Gavilán CG, Margolles A. 2009. Bile affects the synthesis of exopolysaccharides by Bifidobacterium animalis. Appl. Environ. Microbiol. 75:1204–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salazar N, Gueimonde M, Hernandez-Barranco AM, Ruas-Madiedo P, de los Reyes-Gavilan CG. 2008. Exopolysaccharides produced by intestinal Bifidobacterium strains act as fermentable substrates for human intestinal bacteria. Appl. Environ. Microbiol. 74:4737–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar N, Ruas-Madiedo P, Kolida S, Collins M, Rastall R, Gibson G, de los Reyes-Gavilan CG. 2009. Exopolysaccharides produced by Bifidobacterium longum IPLA E44 and Bifidobacterium animalis subsp. lactis IPLA R1 modify the composition and metabolic activity of human faecal microbiota in pH-controlled batch cultures. Int. J. Food Microbiol. 135:260–267 [DOI] [PubMed] [Google Scholar]
- 21.Solís G, de los Reyes-Gavilan CG, Fernández N, Margolles A, Gueimonde M. 2010. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 16:307–310 [DOI] [PubMed] [Google Scholar]
- 22.Kullen MJ, Sanozky-Dawes RB, Crowell DC, Klaenhamer TR. 2000. Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J. Appl. Microbiol. 89:511–516 [DOI] [PubMed] [Google Scholar]
- 23.Ruas-Madiedo P, Gueimonde M, Margolles A, de Los Reyes-Gavilan CG, Salminen S. 2006. Exopolysaccharides produced by probiotic strains modify the adhesion of probiotics and enteropathogens to human intestinal mucus. J. Food Prot. 69:2011–2015 [DOI] [PubMed] [Google Scholar]
- 24.Delgado S, Suárez A, Mayo B. 2006. Bifidobacterial diversity determined by culturing and by 16S rDNA sequence analysis in feces and mucosa from ten healthy Spanish adults. Dig. Dis. Sci. 51:1878–1885 [DOI] [PubMed] [Google Scholar]
- 25.Arboleya S, Binetti A, Salazar N, Fernández N, Solís G, Hernández-Barranco A, Margolles A, de los Reyes-Gavilán CG, Gueimonde M. 2012. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol. Ecol. 79:763–772 [DOI] [PubMed] [Google Scholar]
- 26.Salazar N, Binetti A, Gueimonde M, Alonso A, Garrido P, González del Rey C, González C, Ruas-Madiedo P, de los Reyes-Gavilán CG. 2011. Safety and intestinal microbiota modulation by the exopolysaccharide-producing strains Bifidobacterium animalis IPLA R1 and Bifidobacterium longum IPLA E44 orally administered to Wistar rats. Int. J. Food Microbiol. 144:342–351 [DOI] [PubMed] [Google Scholar]
- 27.Salazar N, Prieto A, Leal JA, Bada-Gancedo JC, de los Reyes-Gavilan CG, Ruas-Madiedo P. 2009. Production of exopolysaccharides by Lactobacillus and Bifidobacterium strains of human origin, and metabolic activity of the producing bacteria in milk. J. Dairy Sci. 92:4158–4168 [DOI] [PubMed] [Google Scholar]
- 28.Macy JM, Ljungdahl LG, Gottschalk G. 1978. Pathway of succinate and propionate formation in Bacteroides fragilis. J. Bacteriol. 134:84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller TL. 1978. Pathway of formation of acetate and succinate from pyruvate by Bacteroides succinogenes. Arch. Microbiol. 117:145–152 [DOI] [PubMed] [Google Scholar]
- 30.Salyers AA. 1984. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 38:293–313 [DOI] [PubMed] [Google Scholar]
- 31.Ruas-Madiedo P, Hernandez-Barranco A, Margolles A, de los Reyes-Gavilán CG. 2005. A bile salt-resistant derivative of Bifidobacterium animalis has an altered fermentation pattern when grown on glucose and maltose. Appl. Environ. Microbiol. 71:6564–6570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sánchez B, Champomier-Verges MC, Anglade P, Baraige F, de los Reyes-Gavilán CG, Margolles A, Zagorec M. 2005. Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187:5799–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sánchez B, Champomier-Vergès MC, Stuer-Lauridsen B, Ruas-Madiedo P, Anglade P, Baraige F, de los Reyes-Gavilan CG, Johansen E, Zagorec M, Margolles A. 2007. Adaptation and response of Bifidobacterium animalis subsp. lactis to bile: a proteomic and physiological approach. Appl. Environ. Microbiol. 73:6757–6767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van der Meulen R, Avonts L, de Vuyst L. 2004. Short fractions of oligofructose are preferentially metabolized by Bifidobacterium animalis DN-173010. Appl. Environ. Microbiol. 70:1923–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith EA, Macfarlane GT. 1998. Enumeration of amino acid fermenting bacteria in the human large intestine: effects of pH and starch on peptide metabolism and dissimilation of amino acids. FEMS Microbiol. Ecol. 25:355–368 [Google Scholar]
- 36.Macfarlane GT, Gibson GR, Beatty E, Cummings JH. 1992. Estimation of short-chain fatty acid production from protein by human intestinal bacteria based on branched-chain fatty-acid measurements. FEMS Microbiol. Ecol. 101:81–88 [Google Scholar]
- 37.Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. 2013. The influence of diet on the gut microbiota. Pharmacol. Res. 69:52–60 [DOI] [PubMed] [Google Scholar]
- 38.Kotarski SF, Salyers AA. 1981. Effect of long generation times on growth of Bacteroides thetaiotaomicron in carbohydrate-limited continuous culture. J. Bacteriol. 146:853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Riordan K, Fitzgerald GF. 1998. Evaluation of bifidobacteria for the production of antimicrobial compounds and assessment of performance in cottage cheese at refrigeration temperature. J. Appl. Microbiol. 85:103–114 [DOI] [PubMed] [Google Scholar]
- 40.Ruiz L, Sánchez B, de los Reyes-Gavilan CG, Gueimonde M, Margolles A. 2009. Coculture of Bifidobacterium longum and Bifidobacterium breve alters their protein expression profiles and enzymatic activities. Int. J. Food Microbiol. 133:148–153 [DOI] [PubMed] [Google Scholar]
- 41.Sánchez B, Burns P, Ruiz L, Binetti A, Vinderola G, Reinheimer J, Margolles A, Ruas-Madiedo P, de los Reyes-Gavilan CG. 23 January 2013. Co-culture affects protein profile and heat tolerance of Lactobacillus delbrueckii subsp. lactis and Bifidobacterium longum. Food Res. Int. [Epub ahead of print.] 10.1016/j.foodres.2013.01.026. [DOI] [Google Scholar]
- 42.Sonnenburg JL, Chen CTL, Gordon JI. 2006. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 4:2213–2226. 10.1371/journal.pbio.0040413 [DOI] [PMC free article] [PubMed] [Google Scholar]