Abstract
Pepper mild mottle virus (PMMoV) is a plant virus that has been recently proposed as a potential indicator of human fecal contamination of environmental waters; however, information on its geographical occurrence in surface water is still limited. We aimed to determine the seasonal and geographic occurrence of PMMoV in drinking water sources all over Japan. Between July 2008 and February 2011, 184 source water samples were collected from 30 drinking water treatment plants (DWTPs); viruses from 1 to 2 liters of each sample were concentrated by using an electronegative membrane, followed by RNA extraction and reverse transcription. Using quantitative PCR, PMMoV was detected in 140 (76%) samples, with a concentration ranging from 2.03 × 103 to 2.90 × 106 copies/liter. At least one of the samples from 27 DWTPs (n = 4 or 8) was positive for PMMoV; samples from 10 of these DWTPs were always contaminated. There was a significant difference in the occurrence of PMMoV among geographical regions but not a seasonal difference. PMMoV was frequently detected in samples that were negative for human enteric virus or Escherichia coli. A phylogenetic analysis based on the partial nucleotide sequences of the PMMoV coat protein gene in 12 water samples from 9 DWTPs indicated that there are genetically diverse PMMoV strains present in drinking water sources in Japan. To our knowledge, this is the first study to demonstrate the occurrence of PMMoV in environmental waters across wide geographical regions.
INTRODUCTION
Pepper mild mottle virus (PMMoV), which was first identified in Italy in 1984 (1), is a nonenveloped, rod-shaped, positive-sense, single-stranded RNA virus belonging to the genus Tobamovirus in the family Virgaviridae (2). PMMoV infects pepper species (Capsicum spp.), causing various symptoms such as mild chlorosis, stunting, fruit mottling, and malformation (2). Processed food products containing peppers are therefore known to contain PMMoV, with a concentration of up to 107 copies/ml (3, 4).
A recent metagenomic analysis showed that PMMoV was the most abundant RNA virus in 3 fecal samples from healthy adults in the United States, comprising 75.7 to 99.4% of all RNA viral sequences identified (4). PMMoV was subsequently detected by reverse transcription (RT)-PCR or by RT-quantitative PCR (RT-qPCR) in 3 (50%) of 6 samples in the United States, 6 (67%) of 9 samples in Singapore, and 19 (95%) of 20 samples in Germany and in the feces of 22 (7%) of 304 adult individuals in France, with a concentration ranging from 105 to 1010 copies/g (3–5). In contrast, PMMoV has not been detected in fecal samples of most animals, such as horses, sheep, ducks, pigs, and dogs (5, 6). Although fecal samples from cows, geese, seagulls, and chickens were sometimes positive for PMMoV, virus concentrations in these samples were much lower than those in human feces (5, 6).
Despite the reported presence of PMMoV in human feces, only a few studies have been conducted to determine the occurrence of this virus in water samples such as wastewater, river water, and seawater (5, 6). PMMoV was detected by RT-qPCR in all raw sewage and treated wastewater samples collected from a wastewater treatment plant (WWTP) in Germany (n = 24) (5) and from 12 WWTPs across the United States (n = 34; 2 samples each from 11 WWTPs and 12 samples from 1 WWTP) (6). The concentrations of PMMoV were reported to be 108 to 1010 and 105 to 1010 copies/liter for raw sewage and treated wastewater, respectively (5, 6). Accordingly, PMMoV was detected in a high concentration in river water and seawater that receive effluents from WWTPs (5, 6).
Interestingly, PMMoV was much more abundant in these water samples than human adenoviruses (HuAdVs) and human polyomaviruses (HuPyVs), which had been considered potential viral indicators of human fecal contamination in environmental waters (7–11). In addition, PMMoV is more persistent in water than other viruses, including HuAdVs and HuPyVs (5). These results may confirm the potential of PMMoV as an ideal viral indicator; however, the limited number of studies conducted to date necessitates more research to substantiate this hypothesis.
This study aimed to evaluate seasonal and geographical differences in the occurrence of PMMoV in source water samples of 30 drinking water treatment plants (DWTPs) in Japan. The samples were tested for 3 types of well-studied human enteric viruses (i.e., norovirus genogroup I [NoV GI], NoV GII, and HuAdV serotypes 40 and 41 [HuAdV 40/41]) as well as Escherichia coli, to determine their relationship to PMMoV. In addition, nucleotide sequence analysis was performed to reveal the genetic diversity of PMMoV in the tested samples.
MATERIALS AND METHODS
Collection of water samples.
In total, 30 DWTPs (DWTPs 1 to 30) were selected from 7 geographical regions (Hokkaido, Tohoku, Kanto, Chubu, Kinki, Chugoku-Shikoku, and Kyushu-Okinawa regions, with 3 to 5 DWTPs for each region) in Japan, where these DWTPs used surface water as the source of drinking water. Source waters of most of these DWTPs are suspected to receive effluent from WWTPs and septic tanks upstream as well as wastewater from livestock production facilities at several DWTPs. Water sampling was conducted 8 times between July 2008 and February 2011: all 30 DWTPs were surveyed in June, October, and December 2010 as well as in February 2011, of which 16 were surveyed in July and December of both 2008 and 2009 as well (12). At each sampling time, source water samples (>2.2 liters each) were collected from these DWTPs and transported by a courier company to the laboratory within 2 days of sample collection. The samples were kept at <10°C during transportation and processed for virus concentration and E. coli detection immediately after delivery.
Concentration of viruses.
An adsorption-elution method using an electronegative membrane was utilized to concentrate viruses in source water samples (13). Briefly, 20 ml of 2.5 mol/liter MgCl2 was added to a 2-liter water sample, which was filtered through a mixed cellulose ester membrane (pore size of 0.45 μm and diameter of 90 mm; Merck Millipore, Billerica, MA, USA) attached to a glass membrane holder (Advantec, Tokyo, Japan). Because of membrane clogging, the filtered volume was sometimes <2 liters (1.0 to 1.8 liters). Subsequently, 200 ml of 0.5 mmol/liter H2SO4 (pH 3) was passed through the membrane, followed by 10 ml of 1 mmol/liter NaOH (pH 11) to elute the virus. The filtrate was recovered in an outer vessel of a Centriprep YM-50 device (Merck Millipore) containing 50 μl of 0.1 mol/liter H2SO4 (pH 1) and was further concentrated via centrifugation according to the manufacturer's protocol to obtain a viral concentrate with a volume of 0.62 ± 0.06 ml.
Quantification of PMMoV.
Viral RNA was extracted from 140 μl of the viral concentrate by using a QIAamp viral RNA minikit (Qiagen, Valencia, CA, USA) followed by RT using a High Capacity cDNA reverse transcription kit (Life Technologies, Carlsbad, CA, USA), according to the manufacturers' instructions. Subsequently, a 2.5-μl aliquot of cDNA was mixed with 22.5 μl of a qPCR mixture containing 12.5 μl of Premix Ex Taq (Probe qPCR) (TaKaRa Bio, Otsu, Japan), 10 pmol each forward and reverse primers (PMMV-FP1-rev and PMMV-RP1), and 5 pmol TaqMan minor-groove binder (MGB) probe (PMMV-Probe1). As shown in Table 1, PMMV-RP1 and PMMV-Probe1 were originally developed in a previous study (4), whereas PMMV-FP1-rev was modified from the original primer PMMV-FP1 (GAGTGGTTTGACCTTAACGTTGA) by adding a thymine nucleotide so that the primer sequence perfectly matched target sequences of multiple PMMoV isolates (data not shown).
Table 1.
Oligonucleotide sequences used for detection of PMMoV
| PCR | Function | Primer | Sequence (5′–3′)a | Positionsb | Reference |
|---|---|---|---|---|---|
| qPCR | Forward primer | PMMV-FP1-rev | GAGTGGTTTGACCTTAACGTTTGA | 1878–1901 | This study |
| Reverse primer | PMMV-RP1 | TTGTCGGTTGCAATGCAAGT | 1945–1926 | 4 | |
| TaqMan MGB probe | PMMV-Probe1 | FAM-CCTACCGAAGCAAATG-MGB-NFQ | 1906–1921 | 4 | |
| Single-round PCR | Forward primer | CP/s | ATGGCATACACAGTTACCAGT | 5685–5705 | 14 |
| Reverse primer | CP/a | TTAAGGAGTTGTAGCCCACGTA | 6158–6137 | 14 |
FAM, 6-carboxyfluorescein; MGB, minor groove binder; NFQ, nonfluorescent quencher.
Corresponding nucleotide position of PMMoV isolate S (GenBank accession number M81413).
qPCR amplification was performed by using a Thermal Cycler Dice TP800 real-time system (TaKaRa Bio) programmed as follows: 95°C for 30 s and 45 cycles of 95°C for 5 s and 60°C for 60 s. Serial dilutions of plasmid DNA containing the 68-bp qPCR target sequence for PMMoV isolate S (GenBank accession number M81413) were used as standard samples. A negative control was prepared by using 2.5 μl of PCR-grade water as a template. All water and standard samples and negative controls were analyzed in duplicate.
Threshold cycle (CT) values were determined as the cycle number at which fluorescence intensity exceeded the threshold value. A standard curve was generated from the linear relationship between the log initial concentration of the plasmid DNA and the CT value (r = −0.990 to −0.998, varying between runs). The qPCR assay was able to quantify as few as 5 copies per reaction. The slope (S) of the standard curve ranged from −2.86 to −2.98, and the efficiency (E) of the qPCR amplification was calculated to be 1.17 to 1.24 based on the formula E = (10−1/S − 1).
Since RNA standards were not used to make a standard curve, the efficiency of RT was not taken into account when determining the concentration of PMMoV genomes. Moreover, the PMMoV concentration in the original water sample was calculated from that in the PCR tube, assuming no loss of virus during the detection procedure. Therefore, the viral concentrations may have been underestimated, which means that the actual concentrations in the original water samples could have been higher than our estimation.
Nucleotide sequence analysis of PMMoV.
Selected qPCR-positive samples were further subjected to qualitative single-round PCR, followed by nucleotide sequence analysis. In brief, single-round PCR was performed in a reaction volume of 25 μl containing 12.5 μl of Premix Ex Taq hot-start version (TaKaRa Bio), 7.5 pmol each primer (CP/s and CP/a) (14) (Table 1), and 1 to 2.5 μl of cDNA. Thermal conditions were as follows: 94°C for 2 min; 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and 72°C for 7 min.
The PCR product was separated by using 2% agarose gel electrophoresis, stained with ethidium bromide, and visualized under a UV lamp. The expected length of the PCR product (∼470 bp) was excised from the gel and was purified by using a QIAquick gel extraction kit (Qiagen). Both strands of the product were sequenced by using an Applied Biosystems 3730xl DNA analyzer (Life Technologies), and the sequences were assembled into a single sequence. Genetyx software version 9.1.0 (Genetyx, Tokyo, Japan) was used to perform multiple-sequence alignment of our sequences and reference sequences corresponding to the amplified region excluding the primer sequences (431 nucleotides [nt]) obtained from the GenBank database, with the manufacturer's standard multiple-sequence-alignment algorithm. This software was further used to generate a phylogenetic tree using the neighbor-joining method with bootstrap values of 1,000 replicates.
Quantification of human enteric viruses and E. coli.
Together with PMMoV, NoV GI, NoV GII, and HuAdV 40/41 were measured by RT-qPCR or qPCR as representatives of human enteric viruses. Water samples collected in 2008 and 2009 (n = 64) were tested for these viruses in our previous study, where NoV GI, NoV GII, and HuAdV 40/41 were detected in 8 (13%), 1 (2%), and 25 (39%) samples, respectively (12). Similarly, 120 water samples collected between June 2010 and February 2011 were also used to detect these viruses. Five microliters of each cDNA (for NoV GI and NoV GII) or DNA (for HuAdV 40/41) was used as a template, and qPCR amplification was performed by using a LightCycler 480 System II instrument (Roche Applied Science, Penzberg, Germany). Primers and TaqMan probes used here were designed from conserved nucleotide sequences of each target virus: the junction of open reading frames (ORFs) 1 and 2 for NoV GI and NoV GII (15) and the fiber gene region for HuAdV 40/41 (16).
E. coli, considered a traditional indicator of fecal contamination, was measured using a single-agar-layer method with Chromocult coliform agar (Merck Millipore). Blue colonies were considered those of E. coli after incubation for 24 h at 37°C. Triplicate experiments using 5 ml each were performed for E. coli detection in water samples collected in 2010 and 2011, resulting in a limit of detection of 0.07 CFU/ml. Water samples collected in 2008 and 2009 were also previously analyzed for E. coli (12); however, these data were not included in our analysis because the tested sample volumes (2 ml each) were considerably lower than those of the 2010-2011 samples.
Statistical analyses.
The χ2 test was used to determine whether there was a significant difference in the positive ratios of PMMoV among the different classification categories. Bartlett's test was used to determine if the PMMoV concentrations in different classification categories have equal variances. One-way analysis of variance (ANOVA) followed by Tukey's multiple-comparison test was used if the variances were determined to be equal; otherwise, a nonparametric Kruskal-Wallis test followed by Scheffe's multiple-comparison test was used to determine whether there was a significant difference in the concentrations of PMMoV among the different classification categories. Statistical analyses were performed by using Excel Statistics 2010 (Social Survey Research Information, Tokyo, Japan), and the significance level (P value) was set at 0.05. If PMMoV was not detected in a sample, to calculate a geometric mean concentration of tested samples, the sample was assigned a virus concentration of 100 copies/liter, which corresponded to about one-fifth of the limit of quantification (551 ± 63 copies/liter, depending on the filtered sample volume and the volume of virus concentrate).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers AB828365 to AB828376.
RESULTS AND DISCUSSION
Occurrence of PMMoV in drinking water sources.
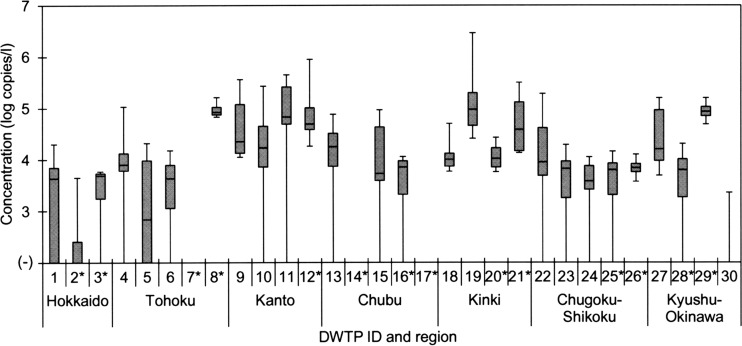
In this study, 184 source water samples collected from 30 Japanese DWTPs between July 2008 and February 2011 were subjected to virus concentration, followed by RNA extraction and RT-qPCR using the primers and the TaqMan MGB probe specific for PMMoV (4) (Table 1). PMMoV was detected in 140 (76%) of 184 samples tested, and its concentrations were quite high, as shown in Fig. 1. The concentration of PMMoV in a PCR tube ranged from 2.00 × 101 to 2.77 × 104 copies/reaction, corresponding to 2.03 × 103 to 2.90 × 106 copies/liter in the original water sample. These concentrations were in agreement with those in German river water samples (3.0 × 103 to 1.1 × 106 copies/liter) (5). Among 30 DWTPs studied, there were only 3 DWTPs (DWTPs 7, 14, and 17) for which PMMoV was not detected in any of the tested samples. On the other hand, PMMoV was present in all the water samples collected from 10 DWTPs (DWTPs 8, 9, 12, 18 to 21, 26, 27, and 29).
Fig 1.
Concentration of PMMoV in source water samples at 30 DWTPs in Japan. Lines within the boxes represent median values; the upper and lower lines of the boxes represent 25th and 75th percentiles, respectively, and the upper and lower bars outside the boxes represent the maximum and minimum values, respectively. Source water samples were collected 4 and 8 times from each DWTP, indicated with and without an asterisk, respectively.
Considering the limited knowledge on the natural occurrence of PMMoV in water, we performed statistical analyses to determine the seasonality and geographic distribution of this virus in our samples. For the seasonality analysis, all the samples were divided into 3 season groups (summer, autumn, and winter) based on the month of sample collection. As summarized in Table 2, PMMoV was detected in 74% (46/62), 67% (20/30), and 80% (74/92) of samples collected in summer, autumn, and winter, respectively, without showing any significant difference (P > 0.05). The concentrations of PMMoV were not significantly different among the 3 seasons (P > 0.05): mean concentrations were 4.59 × 103, 2.66 × 103, and 7.75 × 103 copies/liter for summer, autumn, and winter, respectively. A weaker seasonality in water is one of the essential criteria for appropriate viral indicators (11). In previous studies in the United States and Germany, PMMoV did not show any specific temporal variation in its concentration in wastewater and river water during a short period of 2 weeks or even over a 1-year period (5, 6), which agreed with the results obtained in this study. Conversely, many types of human enteric viruses, such as NoVs or human sapoviruses, are known to be more abundant in water in a certain time of the year, corresponding to the increase in the number of infected individuals during their epidemic period (17–19). The difference in the seasonality between PMMoV and human enteric viruses is probably because PMMoV in water is of dietary origin and is excreted constantly throughout the year in human feces (3, 4).
Table 2.
Comparison of the occurrences of PMMoV in source water samples among seasons
| Season (studied mo) | % of samples positive for PMMoV (no. of positive samples/no. of tested samples) | Significant grouping for positive samplesa | Mean concn of PMMoV (log copies/liter) | Significant grouping for concn of PMMoVa |
|---|---|---|---|---|
| Summer (June–August) | 74 (46/62) | a | 3.66 | a |
| Autumn (October) | 67 (20/30) | a | 3.43 | a |
| Winter (December and February) | 80 (74/92) | a | 3.89 | a |
| Total | 76 (140/184) | 3.74 |
The same letter indicates that there was no significant difference in positive ratios or concentrations of PMMoV (P > 0.05).
As shown in Table 3, there was a significant difference in the occurrence of PMMoV among 7 geographical regions in Japan (P < 0.05). The positive ratio of PMMoV was significantly higher in the Kanto and Kinki regions than in other regions: the virus was detected in 93% (26/28) and 100% (24/24) of the samples collected from the Kanto and Kinki regions, with mean concentrations of 3.38 × 104 and 3.23 × 104 copies/liter, respectively. Both regions are highly urbanized, encompassing a total of about half the population of Japan. Therefore, these results were considered to be attributable to different levels of fecal contamination, and this conclusion was further supported when E. coli was used as an indicator of fecal contamination (see below).
Table 3.
Comparison of the occurrence of PMMoV in source water samples among geographical regions
| Region | % of samples positive for PMMoV (no. of positive samples/no. of tested samples) | Significant grouping for positive samplesa | Mean concn of PMMoV (log copies/liter) | Significant grouping for concn of PMMoVa |
|---|---|---|---|---|
| Hokkaido | 56 (9/16) | a | 3.01 | a |
| Tohoku | 66 (21/32) | a | 3.42 | a |
| Kanto | 93 (26/28) | b | 4.53 | b |
| Chubu | 61 (17/28) | a | 3.31 | a |
| Kinki | 100 (24/24) | b | 4.51 | b, c |
| Chugoku-Shikoku | 84 (27/32) | a | 3.63 | a, c |
| Kyushu-Okinawa | 67 (16/24) | a | 3.60 | a, b |
| Total | 76 (140/184) | 3.74 |
The same letter means that there was no significant difference in positive ratios or concentrations of PMMoV (P > 0.05).
Genetic diversity of PMMoV in drinking water sources.
To determine the geographical genetic diversity of PMMoV in drinking water sources in Japan, at least 1 sample was selected from each of 27 DWTPs, where PMMoV was detected at least once by qPCR, and subjected to single-round PCR targeting the coat protein gene (14). Among 71 tested samples, 15 (21%) samples tested positive for PMMoV.
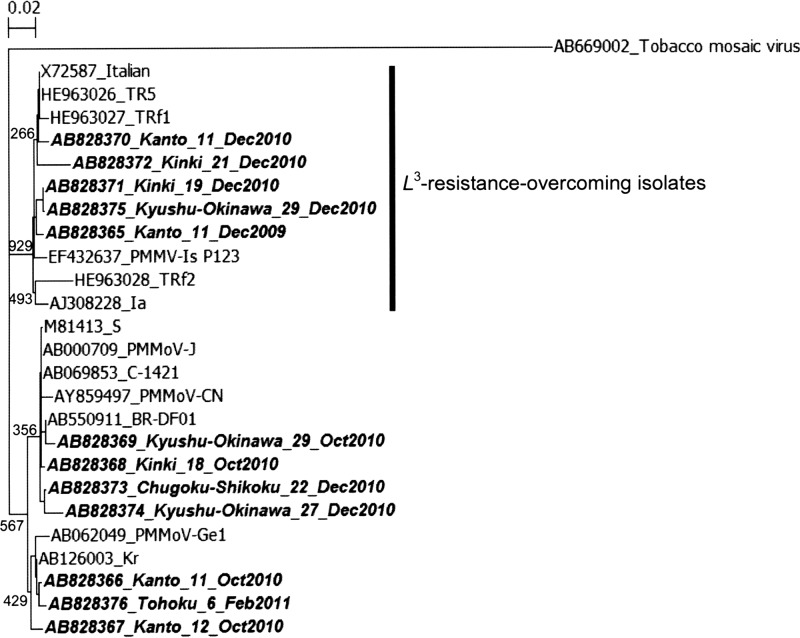
Subsequently, partial coat protein sequences were directly sequenced from the 15 samples positive for single-round PCR. Full-length PCR products (431 nt) were obtained from 6 samples, and partial-length sequences ranging from 228 to 410 nt in length were obtained from 6 samples. On the other hand, no sequence of significant length was obtained from 3 samples, which yielded very faint bands. As shown in the phylogenetic tree in Fig. 2, the 12 sequences originated from 9 DWTPs located in 5 regions (1 DWTP each from the Tohoku and Chugoku-Shikoku regions, 2 DWTPs each from the Kanto and Kyushu-Okinawa regions, and 3 DWTPs from the Kinki region). These sequences shared nucleotide similarities of 85.8 to 100.0% (mean, 95.3%) with each other and 91.2 to 100.0% (mean, 95.9%) with the GenBank sequences shown in Fig. 2. Interestingly, multiple sequences obtained from the same DWTP (DWTP 11 or 29) at different times were classified into different genetic clusters. These results confirm the genetic diversity of PMMoV in drinking water sources in Japan, although more nucleotide sequences need to be analyzed.
Fig 2.
Phylogenetic analysis of PMMoV sequences identified in source water samples. The tree was generated by using the neighbor-joining method with 1,000 bootstrap replicates based on the 431-nt coat protein sequences. The sequence of tobacco mosaic virus was used as an outgroup. The numbers on each branch indicate the bootstrap values, and the scale bar represents the number of nucleotide substitutions per position. PMMoV sequences obtained in this study are labeled with boldface type and italics, and they indicate the GenBank accession number, region name, DWTP identification, and month and year of sample collection. Reference sequences are indicated by the GenBank accession number and strain name.
The single-round PCR assay used in this study further enabled us to determine whether PMMoV has overcome the L3 resistance gene in pepper plants, as PMMoV isolates with the L3 resistance-breaking gene have an amino acid sequence substitution from Met to Asn at position 139 of the coat protein gene of PMMoV isolate TR5 (GenBank accession number HE963026) (14). Based on this, 5 of our samples were classified into a large cluster of L3 resistance-overcoming isolates (Fig. 2). Analysis of surface water samples might be a useful approach to evaluate the spread of PMMoV with the L3 resistance-breaking gene, which is more pathogenic to peppers.
Relationship between PMMoV and human enteric viruses or E. coli.
In addition to the samples collected in 2008 and 2009 (12), those collected in 2010 and 2011 were tested for the presence of NoV GI, NoV GII, and HuAdV 40/41. Of the 184 samples tested, NoV GI, NoV GII, and HuAdV 40/41 were detected in 27 (15%), 37 (20%), and 47 (26%) samples, with maximum concentrations of 3.3 × 104, 2.9 × 103, and 1.7 × 104 copies/liter, respectively. The positive ratio and concentration of these viruses were much lower than those of PMMoV (Fig. 1), which agreed with the results of a previous study, although the types of viruses tested were different (5). As shown in Table 4, at least one of the three virus types was detected in 89 (48%) samples. PMMoV was present in a significantly higher concentration in water samples contaminated with multiple virus types (P < 0.05).
Table 4.
Relationship between occurrences of PMMoV and human enteric viruses in source water samples
| No. of types of human enteric viruses detected by RT-qPCR | % of samples positive for PMMoV (no. of positive samples/no. of tested samples) | Significant grouping for positive samplesa | Mean concn of PMMoV (log copies/liter) | Significant grouping for concn of PMMoVa |
|---|---|---|---|---|
| 0 | 68 (65/95) | a | 3.46 | a |
| 1 | 81 (57/70) | a | 3.85 | b |
| 2 | 94 (15/16) | a | 4.64 | c |
| 3 | 100 (3/3) | NA | 5.02 | NA |
| Total | 76 (140/184) | 3.74 |
The same letter means that there was no significant difference in positive ratios or concentrations of PMMoV (P > 0.05). NA, not analyzed for statistical differences because of the small number of tested samples.
Water samples collected between June 2010 and February 2011 were divided into 3 groups based on the concentration of E. coli, and the difference in the occurrence of PMMoV was examined. As with human enteric viruses, PMMoV was detected with the highest positive ratio and concentration in the group with the highest E. coli concentration, >0.5 CFU/ml (Table 5). In this group, 6 (30%) samples were collected from the Kanto and Kinki regions, where extremely high levels of PMMoV contamination were observed (Fig. 2 and Table 3). Different levels of fecal contamination may have affected the occurrences of PMMoV in water samples from different regions.
Table 5.
Relationship between occurrences of PMMoV and E. coli in source water samples
| Concn of E. coli (CFU/ml) | % of samples positive for PMMoV (no. of positive samples/no. of tested samples) | Significant grouping for positive samplesa | Mean concn of PMMoV (log copies/liter) | Significant grouping for concn of PMMoVa |
|---|---|---|---|---|
| <0.07 | 64 (27/42) | a | 3.47 | a |
| 0.07–0.5 | 76 (48/63) | a | 3.74 | a, b |
| >0.5 | 100 (15/15) | b | 4.38 | b |
| Total | 75 (90/120) | 3.71 |
The same letter means that there was no significant difference in positive ratios or concentrations of PMMoV (P > 0.05).
Notably, PMMoV was detected in 68% (65/95) and 64% (27/42) of samples that were negative for human enteric viruses and E. coli, respectively (Tables 4 and 5). PMMoV has been proposed as a potential indicator of human fecal contamination in water, based on the facts that PMMoV was abundant in human feces, whereas it was rarely found in animal feces (3–6), and that it was absent in a seawater sample that was not polluted by wastewater effluent (6). However, considering the extremely high abundance of PMMoV observed in this study, our samples may have received considerable levels of PMMoV contamination from point and/or nonpoint sources not impacted by sewage, such as wastewater from food processing facilities and runoff water from pepper farms. Our future studies will focus on the occurrence of PMMoV in these non-sewage-impacted water samples.
The detection level of PMMoV in human feces varies greatly, from 7 to 95%, depending on the studied regions (3–5) and also between adults and children, even in the same region (3). Therefore, it is impractical from the viewpoint of required time and cost to analyze individual fecal samples for understanding the incidence of PMMoV. Alternatively, analysis of sewage samples of a WWTP will be able to provide significant information about the incidence of this virus, because viruses shed in feces from infected individuals are mostly transported to the WWTP. Surface water samples, which are highly contaminated by effluents from WWTPs, are also considered suitable for determining the incidence of viruses, although viruses derived from other sources can be present in the samples, as mentioned above. One of the reasons for the more frequent occurrence of PMMoV in water samples tested in this study is likely because viruses in the genus Tobamovirus, including PMMoV, have an extremely high level of resistance against environmental conditions (1, 20). Another reason is the self-evident high concentration of PMMoV in human feces and raw sewage (4–6). Interestingly, PMMoV remains infectious to host pepper plants even after passage through the human gut (4). Further studies are encouraged in order to understand the role of PMMoV in humans.
The adsorption-elution method used in this study (13) has been used previously for recovery tests of human enteric viruses from artificially contaminated water samples: the rates of recovery of poliovirus were 23% ± 19% (n = 20), 65% ± 28% (n = 24), and 82% ± 12% (n = 2) for raw sewage, treated wastewater, and river water, respectively, and that of NoV GII from river water was 15% ± 5% (n = 2) (18, 21). Unfortunately, data on the recovery of PMMoV from water samples are not available. PMMoV, with a 312-nm rod shape (1), is morphologically quite different from icosahedral human enteric viruses and has a lower isoelectric point (3.9 to 4.9) than most of these viruses (2, 22). Therefore, it is unclear whether PMMoV can be adsorbed to and eluted from a 0.45-μm electronegative membrane as effectively as these viruses. PMMoV may exist in drinking water sources with much higher concentrations than those estimated in this study. Further studies should be conducted to evaluate the recovery of PMMoV from various types of water samples.
To our knowledge, this is the first study that demonstrated the seasonal occurrence of PMMoV in environmental waters across wide geographical regions. Since PMMoV was identified in high concentrations in drinking water sources, it remains to be determined if potable treatment facilities efficiently remove the virus from drinking water. If PMMoV is not removed through water treatment, ingestion of drinking water could also contribute to the high concentration of PMMoV observed for human feces. Therefore, our future studies will focus on the evaluation of the efficiency of removal of PMMoV during drinking water treatment processes and its occurrence in treated drinking water. Meanwhile, more environmental water samples will be tested for PMMoV detection to determine the suitability of this virus as a viral indicator of fecal contamination.
ACKNOWLEDGMENTS
This study was partially supported by Environmental Research in Japan, Evaluation and Control of Health Risk from Human-Animal Pollution Sources in Public Water Bodies, from the Ministry of Environment, Japan.
We thank the workers of the DWTPs for their kind cooperation in providing the water samples.
Footnotes
Published ahead of print 20 September 2013
REFERENCES
- 1.Wetter C, Conti M, Altschuh D, Tabillion R, van Regenmortel MHV. 1984. Pepper mild mottle virus, a tobamovirus infecting pepper cultivars in Sicily. Phytopathology 74:405–410 [Google Scholar]
- 2.King AMQ, Lefkowitz E, Adams MJ, Carstens EB. (ed). 2011. Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, Amsterdam, The Netherlands [Google Scholar]
- 3.Colson P, Richet H, Desnues C, Balique F, Moal V, Grob JJ, Berbis P, Lecoq H, Harlé JR, Berland Y, Raoult D. 2010. Pepper mild mottle virus, a plant virus associated with specific immune responses, fever, abdominal pains, and pruritus in humans. PLoS One 5:e10041. 10.1371/journal.pone.0010041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang T, Breitbart M, Lee WH, Rum JQ, Wei CL, Soh SW, Hibberd ML, Liu ET, Rohwer F, Ruan Y. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4:e3. 10.1371/journal.pbio.0040003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamza IA, Jurzik L, Überla K, Wilhelm M. 2011. Evaluation of pepper mild mottle virus, human picobirnavirus and Torque teno virus as indicators of fecal contamination in river water. Water Res. 45:1358–1368 [DOI] [PubMed] [Google Scholar]
- 6.Rosario K, Symonds EM, Sinigalliano C, Stewart J, Breitbart M. 2009. Pepper mild mottle virus as an indicator of fecal pollution. Appl. Environ. Microbiol. 75:7261–7267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bofill-Mas S, Albinana-Gimenez N, Clemente-Casares P, Hundesa A, Rodriguez-Manzano J, Allard A, Calvo M, Girones R. 2006. Quantification and stability of human adenoviruses and polyomavirus JCPyV in wastewater matrices. Appl. Environ. Microbiol. 72:7894–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calgua B, Fumian T, Rusiñol M, Rodriguez-Manzano J, Mbayed VA, Bofill-Mas S, Miagostovich M, Girones R. 2013. Detection and quantification of classic and emerging viruses by skimmed-milk flocculation and PCR in river water from two geographical areas. Water Res. 47:2797–2810 [DOI] [PubMed] [Google Scholar]
- 9.Hamza IA, Jurzik L, Stang A, Sure K, Überla K, Wilhelm M. 2009. Detection of human viruses in rivers of a densely-populated area in Germany using a virus adsorption elution method optimized for PCR analyses. Water Res. 43:2657–2668 [DOI] [PubMed] [Google Scholar]
- 10.McQuaig SM, Scott TM, Lukasik JO, Paul JH, Harwood VJ. 2009. Quantification of human polyomaviruses JC virus and BK virus by TaqMan quantitative PCR and comparison to other water quality indicators in water and fecal samples. Appl. Environ. Microbiol. 75:3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong K, Fong TT, Bibby K, Molina M. 2012. Application of enteric viruses for fecal pollution source tracking in environmental waters. Environ. Int. 45:151–164 [DOI] [PubMed] [Google Scholar]
- 12.Haramoto E, Kitajima M, Kishida N, Katayama H, Asami M, Akiba M. 2012. Occurrence of viruses and protozoa in drinking water sources of Japan and their relationship to indicator microorganisms. Food Environ. Virol. 4:93–101 [DOI] [PubMed] [Google Scholar]
- 13.Katayama H, Shimasaki A, Ohgaki S. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 68:1033–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Çağlar BK, Fidan H, Elbeaino T. 2013. Detection and molecular characterization of Pepper mild mottle virus from Turkey. J. Phytopathol. 161:434–438 [Google Scholar]
- 15.Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko G, Jothikumar N, Hill VR, Sobsey MD. 2005. Rapid detection of infectious adenoviruses by mRNA real-time RT-PCR. J. Virol. Methods 127:148–153 [DOI] [PubMed] [Google Scholar]
- 17.Haramoto E, Katayama H, Phanuwan C, Ohgaki S. 2008. Quantitative detection of sapoviruses in wastewater and river water in Japan. Lett. Appl. Microbiol. 46:408–413 [DOI] [PubMed] [Google Scholar]
- 18.Katayama H, Haramoto E, Oguma K, Yamashita H, Tajima A, Nakajima H, Ohgaki S. 2008. One-year monthly quantitative survey of noroviruses, enteroviruses and adenoviruses in wastewater collected from six plants in Japan. Water Res. 42:1441–1448 [DOI] [PubMed] [Google Scholar]
- 19.Sano D, Pérez-Sautu U, Guix S, Pintó RM, Miura T, Okabe S, Bosch A. 2011. Quantification and genotyping of human sapoviruses in the Llobregat River catchment, Spain. Appl. Environ. Microbiol. 77:1111–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogers SO, Starmer WT, Castello JD. 2004. Recycling of pathogenic microbes through survival in ice. Med. Hypotheses 63:773–777 [DOI] [PubMed] [Google Scholar]
- 21.Haramoto E, Katayama H, Utagawa E, Ohgaki S. 2009. Recovery of human norovirus from water by virus concentration methods. J. Virol. Methods 160:206–209 [DOI] [PubMed] [Google Scholar]
- 22.Michen B, Graule T. 2010. Isoelectric points of viruses. J. Appl. Microbiol. 109:388–397 [DOI] [PubMed] [Google Scholar]