Abstract
Chitin, a major component of fungal cell walls and invertebrate cuticles, is an exceedingly abundant polysaccharide, ranking next to cellulose. Industrial demand for chitin and its degradation products as raw materials for fine chemical products is increasing. A bacterium with high chitin-decomposing activity, Paenibacillus sp. strain FPU-7, was isolated from soil by using a screening medium containing α-chitin powder. Although FPU-7 secreted several extracellular chitinases and thoroughly digested the powder, the extracellular fluid alone broke them down incompletely. Based on expression cloning and phylogenetic analysis, at least seven family 18 chitinase genes were found in the FPU-7 genome. Interestingly, the product of only one gene (chiW) was identified as possessing three S-layer homology (SLH) domains and two glycosyl hydrolase family 18 catalytic domains. Since SLH domains are known to function as anchors to the Gram-positive bacterial cell surface, ChiW was suggested to be a novel multimodular surface-expressed enzyme and to play an important role in the complete degradation of chitin. Indeed, the ChiW protein was localized on the cell surface. Each of the seven chitinase genes (chiA to chiF and chiW) was cloned and expressed in Escherichia coli cells for biochemical characterization of their products. In particular, ChiE and ChiW showed high activity for insoluble chitin. The high chitinolytic activity of strain FPU-7 and the chitinases may be useful for environmentally friendly processing of chitin in the manufacture of food and/or medicine.
INTRODUCTION
Chitin is a linear homopolysaccharide of β-1,4-linked N-acetyl-d-glucosamine (GlcNAc) that is widely distributed in fungal cell walls and invertebrate cuticles and represents a biomass that is second in abundance to cellulose (1–4). Although chitin is insoluble and relatively resistant to degradation due to its crystalline structure, some soil and marine bacteria, such as Bacillus circulans WL-12 (5), Serratia marcescens (6), Streptomyces coelicolor A3 (7), Aeromonas caviae (8), Pseudoalteromonas sp. strain S91 (9), and Vibrio harveyi (10), can degrade this biopolymer, transport it, and utilize it as an energy source by using chitinases (EC 3.2.1.14), as well as other chitin-related proteins/enzymes, such as chitin-binding-domain proteins and surface-active CBM33-type lytic polysaccharide monooxygenases (11). Glycosyl hydrolases (GH) are currently classified into 130 families based on amino acid sequence similarities (12). Most chitinases are classified in the GH-18 and GH-19 families and are present in a wide range of organisms, including bacteria, fungi, insects, plants, and animals (13–16), and their functions are diverse. In addition to the different amino acid sequences among families, they differ in three-dimensional structures and catalytic mechanisms (17, 18). GH-19 chitinases, found mainly in plants, share a lysozyme-type α+β fold. In contrast, GH-18 chitinases have a canonical (β/α)8-barrel fold. Recently, chitinase activities were also found in GH-23, GH-20, and GH-48 (19–21). Various benefits, such as elicitor action and anti-tumor activity, have been confirmed for chitin and chitin-derived sugars, which are increasingly exploited for broad use in industry, agriculture, and medicine (1, 4). In particular, the demand for GlcNAc in the health food industry is growing rapidly (2). The conventional enzymatic degradation method for the insoluble chitin in plentiful crustacean shells is, however, inefficient and unsuitable for industrial use. In the present manufacturing process, chitin is hydrolyzed with concentrated hydrochloride on the manufacturing site, which results in an environmental burden and operational risks. Moreover, pretreatment of chitin by powdering, together with purification of the degrading enzymes, adds considerable expense. Thus, further development of enzymatic or bacterial chitinolysis is required (13, 14, 22, 23). On the other hand, chitinolytic bacteria play a critical role in the chitin recycling process (24). In addition to their industrial applications, microbial chitinases are important for biocirculation of carbon and nitrogen sources in nature. Details of their mechanism of chitinolysis and subsequent metabolism of the degradation products (GlcNAc and its oligosaccharides), however, remain to be elucidated.
In order to avoid the difficulties in industrial use and to elucidate the bacterial chitinolytic process, we isolated a soil bacterial strain with high chitinolytic activity using a screening medium containing insoluble crystalline α-chitin. The isolate belonged to the genus Paenibacillus, which is widely distributed in nature (25, 26). Recently, Paenibacillus strains have been found to produce a number of antibiotics and cell wall-degrading enzymes. Hence, their ability to exert biological control of bacteria and fungi (27) has attracted considerable notice. In this study, we identified the chitinase genes, characterized the enzymes, and demonstrated that Paenibacillus sp. strain FPU-7 has an efficient chitinolytic system.
MATERIALS AND METHODS
Materials.
α-Chitin powder from crab shells, Congo red, polypeptone N, and agar were purchased from Wako Pure Chemical Industries, Ltd. (Osaka, Japan). Chitin flakes from crab shells were obtained from Yaegaki Bio-industry, Inc. (Hyogo, Japan). Bonito extract was purchased from Maruha Nichiro Foods, Inc. (Tokyo, Japan). d-Glucosamine hydrochloride (GlcN) and GlcNAc were purchased from Sigma Chemical Co. (St. Louis, MO). N,N′-Diacetyl-β-d-chitobiose [(GlcNAc)2], 4-nitrophenyl N,N′-diacetyl-β-d-chitobioside [pNP-(GlcNAc)2], a chitooligosaccharide mixture containing oligomers from monomer to hexamer (5 mg/vial each) for high-performance liquid chromatography (HPLC), and ethylene glycol chitin were purchased from Seikagaku Biobusiness Co. (Tokyo, Japan). A prestained protein molecular weight marker (Precision Plus protein, all-blue standards) was purchased from Bio-Rad Laboratories Inc. (Hercules, CA). Bacto yeast extract and Bacto tryptone were purchased from Becton, Dickinson (Cockeysville, MD). Restriction enzymes were purchased from TaKaRa Bio Inc. (Shiga, Japan). Ampicillin and isopropyl-β-d-thiogalactopyranoside (IPTG) were obtained from Nippon Gene (Toyama, Japan). All other chemicals were reagent grade or molecular biological grade.
Culture conditions for chitinolytic bacteria.
The enrichment culture medium used to isolate chitinolytic bacteria was a mixture of the following three solutions. Solution I contained 0.1% (wt/vol) sodium citrate · 2H2O, 1.4% (wt/vol) K2HPO4, 0.6% (wt/vol) KH2PO4, and 0.2% (wt/vol) (NH4)2SO4 (100 ml). Solution II was a 0.02% (wt/vol) MgSO4 · 7H2O solution (10 ml), and solution III was a 1.1% (wt/vol) α-chitin powder solution (890 ml). The three solutions were autoclaved separately and then mixed immediately before use. Chitin powder plates were supplemented with 1.0% (wt/vol) α-chitin, 1.0% (wt/vol) bonito extract, and 1.5% (wt/vol) agar. The isolated strain was also grown at 30°C for 5 days with shaking in liquid medium containing 1.0% (wt/vol) bonito extract, 0.5% (wt/vol) NaCl, and/or 5.0% (wt/vol) crab shell chitin flakes, pH 7.5.
A Spizizen minimal medium (28) [per liter: 2 g (NH4)2SO4, 14 g K2HPO4, 6 g KH2PO4, 1 g sodium citrate · 2H2O, 0.2 g MgSO4 · 7H2O] supplemented with or without 1.0% (vol/wt) polypeptone N, 1.0% (vol/wt) chitin flakes, 1.0% (vol/wt) broth, 1.0% (vol/wt) bonito extract, or 1.0% (vol/wt) yeast extract, was used for optimizing FPU-7 growth. To attain efficient cell yield, strain FPU-7 was grown at 30°C with shaking in a medium containing 1.0% polypeptone N and 0.5% NaCl at pH 7.5 (polypeptone medium). For zymogram analysis, FPU-7 cells were grown at 30°C with shaking in polypeptone medium supplemented with various carbon sources, such as 24 mM glucose (Glc), 24 mM GlcN, 24 mM GlcNAc, or 12 mM (GlcNAc)2.
Scanning electron microscopy (SEM).
Strain FPU-7 was grown at 30°C for 24 h with shaking in polypeptone medium supplemented with chitin flakes. Cells were harvested by centrifugation (5,000 × g, 5 min, 4°C) and resuspended in 1 ml of 2% (wt/vol) glutaraldehyde in 30 mM HEPES buffer at 4°C overnight (prefixation). The prefixed cells were rinsed with 30 mM HEPES buffer, postfixed in 2% (wt/vol) osmium tetroxide for 3 h in an ice bath, and dehydrated in graded ethanol (50, 70, 80, 90, 95, and 100% [vol/vol]) for 10 to 15 min, respectively. The specimens were immersed in isoamyl acetate, critical-point dried by using liquefied carbon dioxide (JCPD-5, JEOL Ltd., Tokyo), and coated with a layer of sublimated osmium tetroxide using an osmium plasma coater (NL-OPC80N; JEOL Ltd., Tokyo, Japan). The osmium plasma-coated specimens were viewed by SEM (JSM-6320F; JEOL Ltd., Tokyo, Japan) according to the manufacturer's instructions.
Biological analysis and toxicity test of FPU-7.
Biological analyses for identification of bacteria, including motility, morphology, Gram staining, and enzyme activities, were carried out by TechnoSuruga Laboratory Co., Ltd. (Shizuoka, Japan). The toxicity of strain FPU-7 was tested as follows. Male and female ICR mice weighing 25 to 30 g were purchased from Koatech Ltd. (Pyungtaek, South Korea). All animals were kept in a room maintained under environmentally controlled conditions of 24 ± 1°C and a 12-h light–12-h dark cycle. The animals had free access to water and a standard diet. Mice were deprived of food for 16 to 18 h prior to administration of the test substances. All animal experiments were performed following the Guidelines for Animal Care and Use Committee of the Catholic University of Daegu. A single-dose toxicity test was performed according to the Organization of Economic Cooperation and Development (OECD) guidelines (54). Mice were randomly divided into two groups. The mice in the treatment group were orally given lyophilized powder of the FPU-7 culture (test sample) at a single dose of 2,000 mg/kg body weight. The mice in the control group received the same volume of phosphate-buffered saline (PBS). Body weight, clinical signs, and mortality were observed once a day for 14 days. On day 15, all mice fasted overnight and were sacrificed for necropsy examination. The heart, lungs, liver, kidneys, spleen, and brain were examined. The 50% lethal dose (LD50) was estimated according to the method described by Miller and Tainter (29). Hematological analysis was performed using an automatic hematological analyzer (Beckman Coulter, Indianapolis, IN). Parameters included white blood cell, red blood cell, hemoglobin, hematocrit, and platelet counts. For biochemical analysis, blood was subjected to centrifugation at 1,500 × g for 10 min to obtain serum, which was stored at −20°C, and the following parameters were determined: albumin, alkaline phosphatase, alanine aminotransferase, aspartate aminotransferase, direct bilirubin, total bilirubin, and total protein. Analyses were made using Architect (Abbott, Abbott Park, IL) automation with Boehringer Ingelheim biochemical kits (Ingelheim am Rhein, Germany).
Activity staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Zymogram analysis (30) was performed as follows. FPU-7 was grown in polypeptone medium containing the various carbon sources described above. The FPU-7 culture (0.5 ml) was centrifuged, and the supernatant was mixed with the same volume of 2× SDS-PAGE sample buffer. The mixed solutions were heated at 60°C for 30 min and loaded on an SDS-polyacrylamide gel containing 0.1% ethylene glycol chitin. The gel was washed with 100 ml of 10 mM Tris-HCl buffer (pH 7.5) for 16 h at room temperature and incubated at 37°C for 5 h. The gel was then immersed in 0.5% (wt/vol) Congo red solution for 15 min and washed with 1 M NaCl. An active band appeared as a yellow halo on a red background. The gel was photographed using a Stage-1000BGR system (AMZ System Science, Inc., Osaka, Japan).
Expression cloning of the chitinase genes.
Genomic DNA was extracted from strain FPU-7, purified, and subjected to partial digestion with Sau3AI. The DNA fragments were subcloned into the BamHI site of pBluescript II SK(+), electrotransferred into E. coli DH5α, and plated on LB agar containing colloidal chitin (31). After incubation at room temperature for 1 month, 53 clones were found to have a lytic halo around each colony. Single-colony isolation was repeated, and 29 clones with stable halo-forming ability were selected.
DNA sequencing and sequence data analysis.
Nucleotide sequences were determined by the dideoxy method, with an ABI Prism 3100 genetic analyzer (Applied Biosystems, Foster City, CA). Both strands of each PCR product were directly sequenced using appropriate primers. De novo next-generation sequencing of the FPU-7 genome was carried out using the Roche 454 GS FLX system (Roche Diagnostics, Mannheim, Germany). The nucleotide sequence data were analyzed using Genetyx computer software (Software Development Co. Ltd., Tokyo, Japan). The chromosomal DNA sequence database was searched using BLASTN search algorithms (http://www.ncbi.nlm.nih.gov/BLAST/) and the Apollo phylogenetic analysis system for the identification of microbes at TechnoSuruga Laboratory Co., Ltd. (Shizuoka, Japan). The protein domains and functional sites were annotated using the Pfam server (http://pfam.sanger.ac.uk/) (32).
DNA manipulations.
Plasmid pBluescript II SK(+) (Agilent Technologies Inc., Santa Clara, CA) and pCold I (TaKaRa Bio Inc., Shiga, Japan) were used as the cloning vectors. Electroporation was used to introduce plasmid DNA into E. coli strain DH5α (31). Plasmid DNA was extracted from the recombinant E. coli DH5α using the GenElute plasmid miniprep kit (Sigma Chemical Co., St. Louis, MO). Genomic DNA from strain FPU-7 was prepared essentially as described previously (33). In brief, FPU-7 was grown at 30°C for 24 h, with shaking, in 20 ml of polypeptone medium. The cells from an overnight culture were incubated for 1 h at 37°C in 4.7 ml of lysis solution (composed of 10 U/ml mutanolysin, 2 mg/ml lysozyme, 1 mM EDTA, and 10 mM Tris-HCl [pH 8.0]). After incubation, 0.2 ml of 10% (wt/vol) SDS solution and 0.1 ml of proteinase K solution (20 mg/ml) were added and mixed well, and the suspension was incubated for 3 h at 56°C. Then, 2.0 ml of KCl buffer (composed of 3.9 M NaCl, 0.63 M KCl, and 10 mM Tris-HCl [pH 8.0]) was added. The mixture was treated with phenol (equilibrated with 1 M Tris-HCl [pH 8.0]) and chloroform, followed by centrifugation at 12,000 × g for 10 min. The top phase was collected and mixed with 1 volume of isopropyl alcohol, and the mixture was centrifuged at 6,000 × g for 5 min. The precipitate was rinsed with 80% (vol/vol) ethanol and finally resuspended in TE buffer (10 mM Tris-HCl [pH 8.0] containing 1 mM EDTA).
Subcloning of the chitinase genes.
A Phusion high-fidelity PCR kit was purchased from New England BioLabs Inc. (Ipswich, MA). Enzymes and buffers were used according to suppliers' recommendations. Each gene encoding chitinase (ChiA to ChiF and ChiW) was amplified from chromosomal DNA of FPU-7 by PCR, using specific primer sets. Synthetic oligonucleotide primers used in this study are listed in Table S1 in the supplemental material. The PCR products were digested with restriction enzymes and ligated with pCold I vector for fusion with an N-terminal 6×His tag. These subcloned vectors were introduced into E. coli strain BL21 cells for overproduction of chitinases.
Overproduction and purification of chitinases.
Overnight cultures of E. coli strain BL21 harboring each chitinase gene were diluted 1:20 into fresh 2× YT medium (composed of 1.0% (wt/vol) Bacto yeast extract, 1.6% (wt/vol) Bacto tryptone, and 0.5% (wt/vol) sodium chloride, pH 7.5) and grown at 37°C until the optical density at 660 nm (OD660) reached 0.5. The culture was then induced with 1 mM IPTG at 15°C for 24 h, and the His-tagged chitinases were extracted using BugBuster protein extraction reagent (Merck KGaA, Darmstadt, Germany) as follows. Cells were harvested by centrifugation at 5,000 × g for 10 min at 4°C, washed with PBS, and resuspended in the extraction reagent at room temperature. After protein extraction, insoluble cell debris was removed by centrifugation (12,000 × g, 30 min), and the supernatant was used as the protein source. The protein extracts were dialyzed against 50 mM sodium phosphate buffer, pH 7.4, containing 0.01% (wt/vol) sodium azide (buffer A), at 4°C overnight. After removal of denatured material by centrifugation (10,000 × g, 20 min), the extract was loaded on a TSKgel BioAssist chelating column (Tosoh Co., Tokyo, Japan; 2.4 ml) equilibrated with Ni2SO4 and buffer A containing 20 mM imidazole. To wash out proteins that were nonspecifically bound to the column, 0.1 M sodium phosphate buffer (pH 7.8) containing 3 M NaCl and 0.01% (wt/vol) sodium azide was passed through the column. Elution was performed at a flow rate of 1.0 ml/min at 20°C, and the protein elution profile was obtained by monitoring the absorbance at 280 nm. His-tagged chitinases were recovered by a 20 to 250 mM linear gradient of imidazole in buffer A (30 ml). The chitinase fraction was pooled and dialyzed against 0.1 M citric acid buffer, pH 5.6. The protein content was determined using a commercial Bradford assay kit (rapid protein quantification kit; Dojindo Molecular Technologies, Inc., Kumamoto, Japan). The enzymes (ChiA to -F and ChiW) were purified to homogeneity, as assessed by 10% (wt/vol) SDS-PAGE analysis followed by Coomassie brilliant blue (CBB) R-250 staining (34).
NH2-terminal amino acid sequence.
The purified recombinant ChiW was subjected to SDS-PAGE and then transferred to a Clearblot-P polyvinylidene difluoride (PVDF) membrane (ATTO Co., Tokyo, Japan). The PVDF membrane was stained with CBB R-250, and protein bands were cut out of the membrane. Protein sequencing of the bands was performed by automated Edman degradation with a PE Applied Biosystems model 491 Procise protein sequencing system (Applied Biosystems, Foster City, CA).
Immunoblot analysis of intact ChiW of FPU-7.
Rabbit anti-ChiW antiserum was raised against the C-terminal synthetic peptide (1405-KLLGAIFNELKAPK-1418), which had an additional Cys residue and was conjugated to keyhole limpet hemocyanin. FPU-7 cells were grown at 30°C with shaking in polypeptone medium with or without 24 mM (GlcNAc)2 as an inducer for ChiW. The FPU-7 culture (0.5 ml) was centrifuged and the supernatant was subjected to SDS-PAGE and transferred onto a PVDF membrane by electroblotting. The pelleted cells were washed twice with PBS and boiled at 100°C for 5 min in 0.1 ml of SDS-PAGE sample buffer. The remaining bacterial debris and unlysed cells were removed by centrifugation for 5 min (12,000 × g). The resulting materials were subjected to SDS-PAGE in parallel to the supernatants described above. After transfer, the PVDF membrane was blocked using a Blocking One kit (Nacalai Tesque, Inc., Kyoto, Japan) for 1 h. The membrane was washed with PBS containing 0.05% (vol/vol) Tween 20 (PBST), then anti-ChiW antiserum (diluted 1:5,000 in PBST) was added, and the blot was incubated for 1 h. Next, peroxidase-conjugated AffiniPure goat anti-rabbit IgG (Wako Pure Chemical Industries, Ltd.) was added at a 1:10,000 dilution in PBST and incubated for 1 h, and the bands were visualized by using an ImageQuant LAS 4000 mini-luminescent image analyzer (GE Healthcare Japan, Tokyo, Japan) and an ECL Prime Western blotting detection system (GE Healthcare Japan, Tokyo, Japan).
Enzyme and protein assay.
Chitinase activity was determined using pNP-(GlcNAc)2 or colloidal chitin as the substrate. Reaction mixture A, consisting of 50 μl of 0.1% (wt/vol) pNP-(GlcNAc)2, 40 μl of 0.1 M citric acid buffer (pH 5.6) containing 0.1% bovine serum albumin (BSA), and 10 μl of enzyme solution, was incubated for 30 min at 37°C. Then, 200 μl of 0.2 N NaOH was added, and the released p-nitrophenol was determined by monitoring the increase in absorbance at 405 nm. One unit of activity was defined as the amount of enzyme catalyzing the production of 1 μmol of p-nitrophenol per min. Reaction mixture B, consisting of 240 μl of 1.5% (wt/vol) colloidal chitin, 240 μl of 0.1 M citric acid buffer (pH 5.6) containing 0.1% BSA and 120 μl of enzyme solution, was incubated for 60 min at 37°C. The amounts of the degradation products, GlcNAc, (GlcNAc)2, and tri-N-acetylchitotriose, were estimated with a Tosoh 8020 HPLC system equipped with a TSKgel Amide-80 column (4.6 by 250 mm; Tosoh Co., Tokyo, Japan). The products were eluted with a mobile phase of 70% (vol/vol) CH3CN, and detected at 210 nm. One unit of activity was defined as the amount of enzyme catalyzing the production of 1 μmol of product per min.
Effects of protease treatment on chitinase activity of FPU-7 cells.
Cultured FPU-7 cells in polypeptone medium supplemented with 24 mM (GlcNAc)2 were collected by centrifugation at 6,000 × g and 4°C for 20 min and washed twice with 100 mM Tris-HCl buffer, pH 8.0. Cells (3.2 g [wet weight]) were then resuspended in 10 ml of the same buffer. The suspension was incubated with trypsin (0.0135 to 1.35 μg/ml; Cooper Biomedical, Inc., Palo Alto, CA), pronase-P (1 mg/ml; Kaken Pharmaceutical Co., Ltd., Tokyo, Japan), or proteinase K (1 mg/ml; Wako Pure Chemical Industries, Ltd., Osaka, Japan) at 37°C for 1 h. After centrifugation at 13,860 × g and room temperature for 5 min, each supernatant was subjected to SDS-PAGE, and the remaining precipitate in the tube was resuspended in 100 μl of 0.1 M citric acid buffer, pH 5.6. The residual chitinase activities in the suspension of the cell fraction were measured with 0.05% (wt/vol) pNP-(GlcNAc)2 using the assay method described above.
Nucleotide sequence accession numbers.
Nucleotide sequence data for chiA, chiB, chiC, chiD, chiE, chiF, and chiW have been deposited in the DDBJ/EMBL/GenBank database under accession numbers AB683959, AB683960, AB683961, AB683962, AB683963, AB683964 and AB683965, respectively.
RESULTS
Isolation and characterization of chitinolytic bacteria.
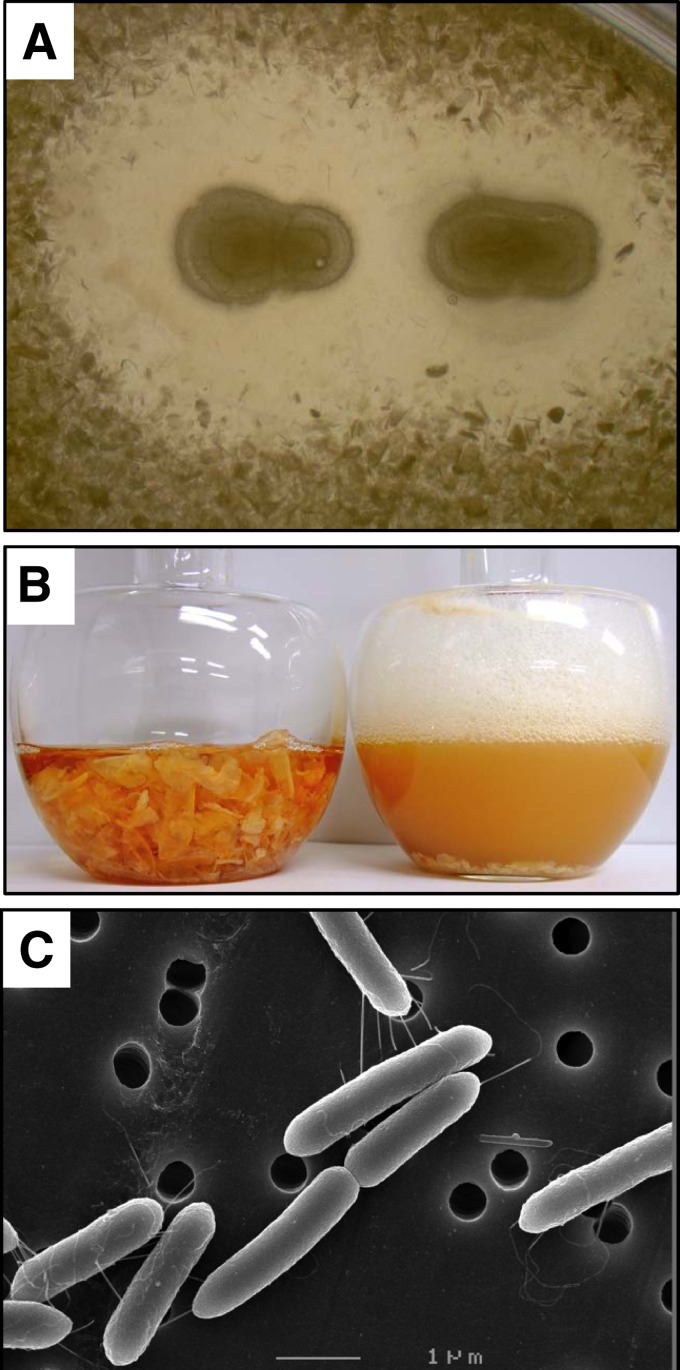
In order to isolate bacteria with high chitinolytic activity, soil samples from Fukui Prefecture were screened using the enrichment culture technique. Each soil specimen was inoculated into selective growth medium containing α-chitin powder as the only source of carbon and was incubated at room temperature for 2 weeks. Since the structure of chitin powder is more resistant to bacterial degradation than that of colloidal chitin, the powder was thought to be suitable for selecting bacteria with high chitinase activity. Then, a portion of each culture was added to a flask containing fresh medium, and each was incubated with shaking. After several cycles of cultivation, 29 candidate bacterial isolates were obtained, and each was streaked on an agar plate containing α-chitin powder and bonito extract, followed by incubation at room temperature. Among several bacterial colonies forming a lytic halo, the strain (FPU-7) with the largest clear halo was isolated (Fig. 1A). This organism grew well in liquid medium and thoroughly hydrolyzed flake chitin from crab shells (Fig. 1B). Morphological examination by SEM revealed structural similarity to Bacillus species, a 0.5- by 2.0-μm rod-shaped Gram-positive spore-forming bacterium with flagella (Fig. 1C). The isolated bacterium hydrolyzed casein and showed catalase activity as well as oxidase function. Bacterial growth proceeded under both aerobic and anaerobic conditions but did not take place above 55°C. These traits correspond to those of Paenibacillus members. As shown in Fig. 2, molecular phylogenetic analysis based on the sequence of 16S rRNA supported this assignment. The sequence of the 16S rRNA gene from FPU-7 was 96.8% identical to that of the standard strain Paenibacillus chinjuensis WN9 (a threshold of 97% has been customarily used to define operational taxonomic units at the species level) (35). Among Paenibacillus members thus far reported (36), species or strains identical to FPU-7 have not been identified. A study comparing the properties of strain FPU-7 with those of Paenibacillus validus, which was phylogenetically most closely related to FPU-7, was then performed (96.4% sequence identity between their 16S rRNA genes). Both FPU-7 and P. validus oxidized glucose, maltose, and sucrose but not galactitol or inulin. They also showed β-galactosidase and arginine dihydrolase activities. In contrast to P. validus (37), however, FPU-7 liquefied gelatin but could not oxidize glycerol, ribose, or xylose. Based on these observations, the newly isolated strain was designated Paenibacillus sp. strain FPU-7 (see Table S2 in the supplemental material). In the single-dose toxicity test, the sample, at a dose of 2 mg/g, did not cause death of mice during 14 days of observation. The mice did not show any signs of intoxication or changes in general behavior or other physiological activities. There were no body weight or other observable differences between the control group and the treatment group. The pathological examination of the internal organs revealed no signs of abnormalities (data not shown). Thus, it can be concluded that the test sample has no significant toxic effects. This result will offer an advantage for industrial applications, e.g., GlcNAc production by Paenibacillus sp. strain FPU-7.
Fig 1.
Chitin degradation and cell morphology of Paenibacillus sp. strain FPU-7. (A) Colonies of strain FPU-7 on an α-chitin powder plate. Cells of strain FPU-7 were smeared on a 1.0% (wt/vol) α-chitin powder plate, and the plate was incubated at 30°C for 1 week. (B) Strain FPU-7 was grown at 30°C for 5 days with shaking, in bonito extract medium containing 5.0% (wt/vol) crab shell chitin flakes. The solid matter (chitin flakes) in the left flask disappeared after 5 days of incubation (right flask). (C) Cell morphology of strain FPU-7 (stationary phase) observed by SEM. Bar, 1 μm.
Fig 2.
Phylogenetic analysis of the 16S rRNA genes of Paenibacillus type strains and FPU-7 (shaded). The numbers at the nodes indicate the percentage bootstrap values for each node based on 1,000 bootstrap resamplings estimated by ClustalW software, using the neighbor-joining method. The tree was rooted with the 16S rRNA gene of Bacillus subtilis as an outgroup. The scale bar indicates 0.01 substitution per nucleotide position.
Effects of carbon source on production of secreted chitinase.
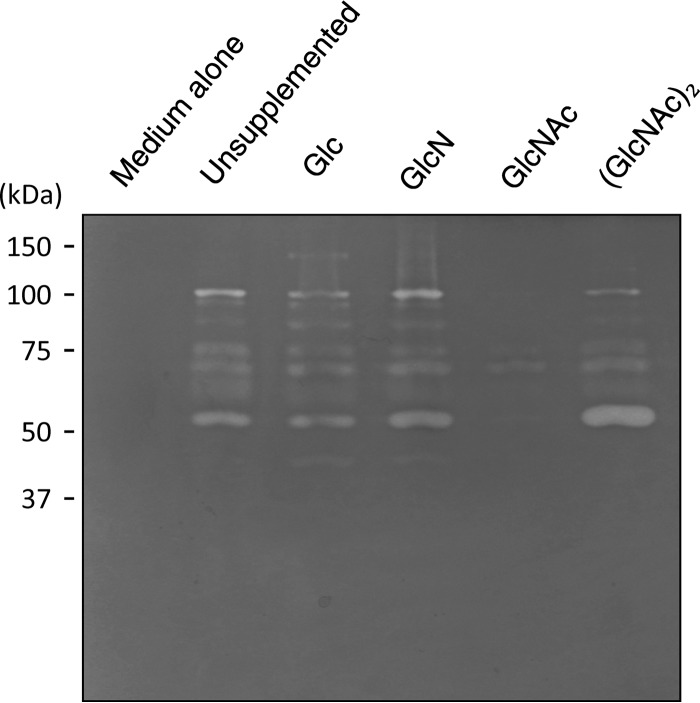
Initially, Spizizen's minimal medium (28) was used to investigate the production of chitinase by FPU-7; however, growth of the strain in the minimal medium was very poor, requiring yeast extract, bonito extract, or peptone. Therefore, a medium containing polypeptone N and NaCl (polypeptone medium) was used in subsequent experiments. Under culture conditions with polypeptone N, FPU-7 secreted several chitinases into the medium. Zymogram analysis revealed that the chitinases in the medium had molecular masses ranging from 40 to 150 kDa (Fig. 3, polypeptone medium lane). Although the presence of Glc, GlcN, or (GlcNAc)2 had no influence on chitinase secretion, GlcNAc exerted a severe inhibitory effect on the synthesis of secreted chitinases (Fig. 3, GlcNAc lane). These results suggest that secretion of the enzymes was constitutive but was subject to catabolite repression by GlcNAc.
Fig 3.
Zymogram analysis of the active chitinases in culture supernatants. Strain FPU-7 was grown at 30°C for 2 days with shaking in polypeptone medium supplemented with Glc, GlcN, GlcNAc, or (GlcNAc)2 as described in Materials and Methods. Aliquots of culture supernatants were loaded onto an SDS-PAGE gel containing 0.1% (wt/vol) ethylene glycol chitin, and activity staining was performed. Medium alone served as a negative control; unsupplemented cultures (polypeptone medium alone) and cultures supplemented with Glc, GlcN, GlcNAc, and (GlcNAc)2 were tested.
Expression cloning of extracellular chitinase gene products from FPU-7.
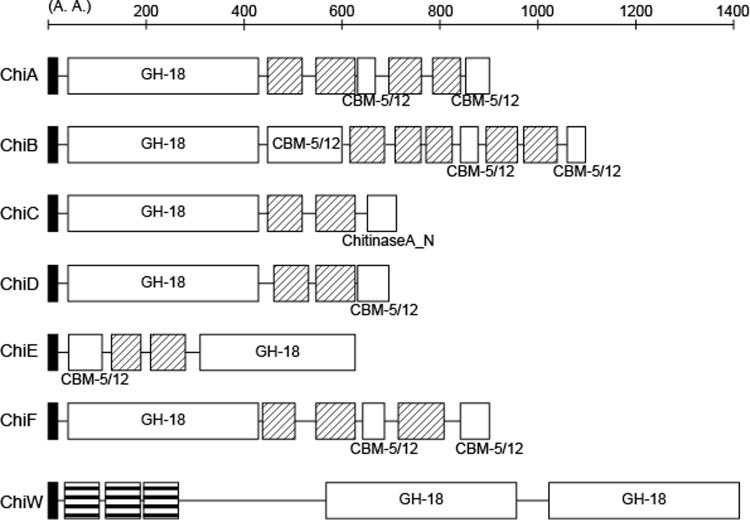
Strain FPU-7 had high chitinolytic activity and produced several extracellular chitinases, described above. Since their isolation and purification from the medium were not easy, shotgun cloning of the chitinase genes was attempted using E. coli, and 29 clones with stable halo-forming ability were selected. Analysis of the nucleotide sequences indicated the presence of 4 genomic regions (I to IV) and six chitinase genes (chiA to chiF) among these clones (Fig. 4; Table 1). Region I contained chiA and chiB in tandem, and clones carrying this sequence formed halos most rapidly. In region II, chiC and chiD were present in tandem, and the rate of halo formation of clones carrying these genes was second to that of region I clones. The halo formation rate of clones carrying region III (chiE) was higher than those with region IV (chiF). A 146-bp spacer was present between chiA and chiB, whereas a 122-bp spacer was present between chiC and chiD. A typical promoter sequence was found upstream of only chiA and chiC, suggesting that each gene pair forms an operon.
Fig 4.
Domain structures of FPU-7 chitinases. The bars and numbers indicate positions of amino acid sequences. Protein sequence was analyzed by using the Pfam server, and the locations of the following domains are indicated: signal peptide (black boxes), GH-18 catalytic domains (GH-18; Pfam ID, PF00704), carbohydrate-binding module (CBM-5/12; Pfam ID, PF02839), chitinase A N-terminal domain (ChitinaseA_N; Pfam ID, PF08329), fibronectin type III domain (hatched boxes; Pfam ID, PF00041), and SLH domains (boxes with horizontal lines; Pfam ID, PF00395).
Table 1.
Properties of the purified FPU-7 extracellular and cell surface chitinases
| Chitinase | No. of amino acids (molecular mass [kDa]) | Predicted domainsa | Activity (U/mg) |
|
|---|---|---|---|---|
| pNP-(GlcNAc)2 | Colloidal chitin | |||
| ChiA | 933 (97) | Signal peptide, GH-18, FnIII × 4, CBM-5/12 × 2 | 0.041 | 2.5 |
| ChiB | 1,162 (122) | Signal peptide, GH-18, FnIII × 5, CBM-5/12 × 3 | 1.3 | 2.0 |
| ChiC | 766 (82) | Signal peptide, GH-18, FnIII × 2, chitinase A N-terminal domain | 0.25 | 2.5 |
| ChiD | 724 (78) | Signal peptide, GH-18, FnIII × 2, CBM-5/12 | 0.085 | 0.58 |
| ChiE | 585 (61) | Signal peptide, GH-18, FnIII × 2, CBM-5/12 | 270 | 4.6 |
| ChiF | 834 (87) | Signal peptide, GH-18, FnIII × 3, CBM-5/12 × 2 | 0.048 | 2.1 |
| ChiW | 1,418 (153) | Signal peptide, SLH × 3, GH-18 × 2 | 0.71 | 5.2 |
Figure 4 shows the domain structures predicted from the nucleotide sequences of chitinases ChiA to -F analyzed by the Pfam server (38, 39). Each cloned chitinase had a glycoside hydrolase family 18 (GH-18) catalytic domain (TIM barrel), a carbohydrate-binding domain/module (CBM-5/12; immunoglobulin-like fold) involved in substrate binding, a chitinase A N-terminal domain also involved in chitin binding (immunoglobulin-like fold), and a fibronectin type III domain of unknown function (immunoglobulin-like fold). In addition, a signal peptide sequence was detected at each N-terminal region, indicating that all of these chitinases were secreted enzymes. The expression and characterization of ChiA to -F are described below.
In silico cloning of the cell surface chitinase gene from FPU-7.
When spent medium from FPU-7 cultures was initially tested for degradation of the chitin flakes, significant chitinolysis did not take place. Similarly, the washed cell suspension alone could not hydrolyze the substrate. Chitin flakes were thoroughly degraded only in growing cultures when the pH of the medium was between 5 and 8. These results indicate that efficient degradation of the insoluble chitin flakes required the collaboration of the living cells with the secreted chitinases, suggesting the involvement of a cell-bound chitinase(s). Indeed, draft genome analysis detected a candidate gene (chiW) for a cell-bound chitinase, an unique multimodular protein composed of 1,418 amino acid residues (Fig. 4). At the N-terminal region, this chitinase ChiW had a signal peptide sequence and three surface layer homology (SLH) domains predicted to be the cell surface signal (40). At the C-terminal region, two sets of GH-18 catalytic domains were also arranged in tandem.
Production of recombinant ChiW.
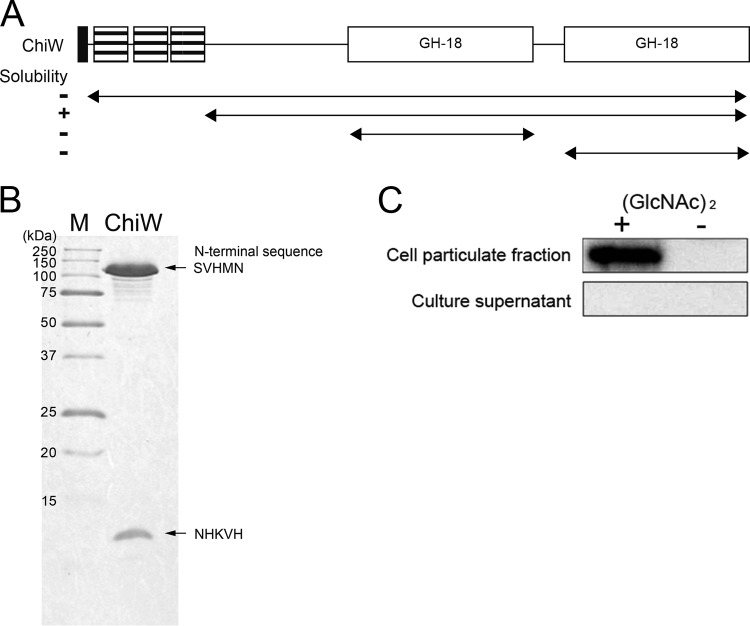
To confirm enzymatic activity of ChiW, a His-tagged recombinant protein was produced in E. coli. The ChiW protein without the putative signal peptide and each GH-18 domain alone each formed insoluble inclusion bodies (Fig. 5A). Truncation of the three SLH domains led to successful production of a soluble protein, which was subsequently purified by affinity chromatography with a nickel-chelate column. Two bands differing in molecular mass were detected by SDS-PAGE analysis of the purified enzyme (Fig. 5B). N-terminal sequence analysis of the two fragments identified the His tag sequence derived from the expression vector (NHKVH) in the smaller fragment, whereas the sequence downstream of Ser283 (SVHMN) was found for the larger band. Thus, limited proteolysis of ChiW occurred within the N-terminal region between Asn282 and Ser283.
Fig 5.
Overproduction and purification of the recombinant His-tagged ChiW protein and localization of native ChiW in FPU-7 cells. (A) Arrangement of domains and deleted regions of ChiW. The solubility of the truncated mutants is indicated on the left. The ChiW with the SLH domain deleted was the only soluble form obtained. (B) SDS-PAGE of purified SLH domain deletion-containing ChiW (right lane). The purified enzyme was subjected to SDS-PAGE (15% gel). Protein bands were stained with CBB R-250. Lane M, molecular weight standards. The N-terminal sequences of the bands were SVHMN and NHKVH, as determined by protein sequencing. The site-specific proteolysis occurred between Asn282 and Ser283 during the purification step. (C) Expression and localization of ChiW in FPU-7. FPU-7 was grown in medium containing 1.0% (wt/vol) polypeptone N and 0.5% NaCl with or without 24 mM (GlcNAc)2 at pH 7.5. ChiW protein was induced by (GlcNAc)2 and was detected in the cell particulate fraction by immunoblotting.
Cellular localization of ChiW in strain FPU-7.
A specific polyclonal antibody raised against the C-terminal sequence of ChiW was used to examine the expression and localization of ChiW in FPU-7 by Western blotting. As seen in Fig. 5C, ChiW was not found in the culture supernatant but was detected in the cell particulate fraction from the (GlcNAc)2-supplemented culture. These results suggest that ChiW is not a secreted enzyme and that it is specifically produced in the presence of (GlcNAc)2, in contrast to the other constitutively secreted chitinases in strain FPU-7 (Fig. 3). Furthermore, proteinase K treatment of whole cells decreased the chitinolytic activity of the cell particulate fraction to 43.3% ± 9.5%. The cell particulate fraction was resistant to other proteases, such as trypsin and pronase, probably due to the complicated cell envelope containing many layers of peptidoglycan and teichoic acids. These results suggest that ChiW is present on the cell surface, most likely anchored by the SLH domain.
Specific activities of the recombinant FPU-7 chitinases for two substrates.
By using a recombinant gene technique, the FPU-7 extracellular chitinases (ChiA to -F) lacking the putative signal peptides were fused with a His tag at the N terminus and were expressed in E. coli as soluble forms, although they were multimodular proteins (Fig. 4A). Then, these enzymes were purified by nickel-chelate column chromatography. Initial characterization of each recombinant chitinase (ChiA to -F and ChiW) was performed using colloidal chitin and pNP-(GlcNAc)2 as the substrates (Table 1). Although all of the chitinases contained GH-18 catalytic domains (Fig. 4), their specific activities varied. ChiE exerted the highest activity for pNP-(GlcNAc)2, whereas ChiA, -D, and -F were less active. The activities of ChiB, ChiC, and ChiW with this synthetic chromogenic substrate were moderate. Activity with colloidal chitin was higher for ChiE and ChiW but somewhat lower for ChiD. ChiA, -B, -C, and -F were rather active on this insoluble substrate. They exhibited maximum activity in the acidic pH range of 4 to 6. The optimal pH for ChiE was 4, which was lower than that for the other FPU-7 chitinases.
DISCUSSION
The seven chitinase genes, chiA to chiF and chiW, were cloned from FPU-7, a strain that efficiently degrades α-chitin powder. Among the gene products, ChiW is a surface-bound enzyme whose production is induced by (GlcNAc)2. It seems probable that degradation of the chitin in fungal cell walls or arthropod shells is the primary role of the extracellular chitinases of the soil bacterium FPU-7. The secreted enzymes allow an attack on scattered substrates, but utilization of the degradation products is somewhat difficult for the bacterial cells. On the other hand, digestion on the cell surface seems to be profitable for the direct incorporation of (GlcNAc)2. In most chitinolytic microbes, chitobiose is intracellularly hydrolyzed to GlcNAc (22). Indeed, a putative gene encoding (GlcNAc)2 hydrolase was identified in the FPU-7 draft genome sequence, although a (GlcNAc)2 transporter gene has not yet been detected. In FPU-7 cells, the resultant intracellular GlcNAc may repress expression of the extracellular chitinases.
ChiW is one of the most complicated and multifunctional chitinases, having N-terminal SLH domains, two catalytic domains, and a limited proteolysis site within the relatively long sequence (1,418 amino acid residues). This multimodular enzyme, however, does not contain any known carbohydrate-binding modules, which are often present in endo-type chitinases (41). Thus far, microbial chitinases with two catalytic domains have been identified in the Chlorella virus CVK2 (42), the hyperthermophilic archaeon Thermococcus kodakaraensis strain KOD1 (43), and the Gram-negative marine bacterium Microbulbifer degradans strain 2-40 (44). In the CVK2 chitinase vChti-1, the N-terminal signal peptide was replaced with a carbohydrate-binding module (42), and the viral lytic function might substitute for the secretory signal. The KOD1 chitinase ChiA may also be a secreted protein, as it has a putative signal peptide and three carbohydrate-binding modules (one between the signal peptide and the GH-18 catalytic domain at the N-terminal end and two between the GH-18 catalytic domains). On the other hand, the Microbulbifer degradans strain 2-40 chitinase ChiB has a lipobox motif (45) adjacent to the N-terminal signal sequence, which may be necessary for anchoring the enzyme to the membrane for cell surface expression. However, the localization of the 2-40 ChiB has not yet been determined. Although ChiW is the fourth microbial enzyme with two GH-18 catalytic domains, this is the first example from a Gram-positive bacterium and unique in being expressed on the cell surface.
Paenibacillus species are widely distributed in nature and have been found to be important for industrial, agricultural, and medical applications. Recently, their biocontrol potential against bacteria and fungi has attracted considerable notice, owing to the production of antimicrobial substances and cell wall-degrading enzymes (27, 46–49). Molecular biological information regarding Paenibacillus chitinases is, however, scarce, and their amino acid sequences mostly remain to be determined. Although the FPU-7 chitinases all belong to the GH-18 family, their substrate specificities for colloidal chitin and pNP-(GlcNAc)2 differ considerably. In particular, ChiE and ChiW hydrolyze insoluble chitin effectively. Moreover, ChiE is extremely active with pNP-(GlcNAc)2. The inference that the pair chiA and chiB, as well as the pair chiC and chiD, formed an operon suggested possible synergism between the products. Mixed incubation of ChiA and ChiB with colloidal chitin or powder chitin, however, did not show a synergistic effect (data not shown). Similarly, synergism was not observed between ChiC and ChiD (data not shown).
Solid biomaterials, such as crustacean shells and fungal cell walls, contain a variety of biopolymers, including glucans, chitosans, and proteins, besides the main component, chitin. They have such complicated structures that other factors in addition to chitinases may be required to degrade them efficiently. In fact, other Paenibacillus species, such as Paenibacillus fukuinensis, produce additional degradation enzymes, including glucanase, chitosanase, and protease, which work cooperatively (30, 50). Moreover, a unique type of enzyme was recently discovered: surface-active CBM33-type lytic polysaccharide monooxygenase, which acts synergistically with chitinase by splitting the glycoside bond of chitin by an oxidative mechanism (51–53). The presence of putative genes encoding these enzymes in the FPU-7 genome has been confirmed, and their expression is now under investigation. Detailed analyses of the structure and function of the FPU-7 chitinases, together with metabolism of the chitinolysis products, will be useful for industrial application of these enzymes. We are now attempting to detect other components required for the complete degradation of fungal cell walls through transcriptomic and proteomic analyses using the FPU-7 genome sequence. Attempts are also in progress to isolate a mutant lacking ChiW.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a grant from FPU Promotion of Community Fund and by the Special Coordination Funds for Promoting Science and Technology of the Ministry of Education, Culture, Sports, Science, and Technology of the Japanese government. This work was also supported in part by a Grant-in-Aid for Young Scientists (B) (to T.I.; grant 25850234) from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print 27 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02483-13.
REFERENCES
- 1.Aam BB, Heggset EB, Norberg AL, Sorlie M, Varum KM, Eijsink VG. 2010. Production of chitooligosaccharides and their potential applications in medicine. Mar. Drugs 8:1482–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen JK, Shen CR, Liu CL. 2010. N-acetylglucosamine: production and applications. Mar. Drugs 8:2493–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park BK, Kim MM. 2010. Applications of chitin and its derivatives in biological medicine. Int. J. Mol. Sci. 11:5152–5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Hadrami A, Adam LR, El Hadrami I, Daayf F. 2010. Chitosan in plant protection. Mar. Drugs 8:968–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe T, Oyanagi W, Suzuki K, Tanaka H. 1990. Chitinase system of Bacillus circulans WL-12 and importance of chitinase A1 in chitin degradation. J. Bacteriol. 172:4017–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuchs RL, McPherson SA, Drahos DJ. 1986. Cloning of a Serratia marcescens gene encoding chitinase. Appl. Environ. Microbiol. 51:504–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saito A, Fujii T, Yoneyama T, Miyashita K. 1998. glkA is involved in glucose repression of chitinase production in Streptomyces lividans. J. Bacteriol. 180:2911–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sitrit Y, Vorgias CE, Chet I, Oppenheim AB. 1995. Cloning and primary structure of the chiA gene from Aeromonas caviae. J. Bacteriol. 177:4187–4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Techkarnjanaruk S, Goodman AE. 1999. Multiple genes involved in chitin degradation from the marine bacterium Pseudoalteromonas sp. strain S91. Microbiology 145:925–934 [DOI] [PubMed] [Google Scholar]
- 10.Montgomery MT, Kirchman DL. 1993. Role of chitin-binding proteins in the specific attachment of the marine bacterium Vibrio harveyi to chitin. Appl. Environ. Microbiol. 59:373–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaaje-Kolstad G, Horn SJ, Sorlie M, Eijsink VG. 2013. The chitinolytic machinery of Serratia marcescens—a model system for enzymatic degradation of recalcitrant polysaccharides. FEBS J. 280:3028–3049 [DOI] [PubMed] [Google Scholar]
- 12.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahiya N, Tewari R, Hoondal GS. 2006. Biotechnological aspects of chitinolytic enzymes: a review. Appl. Microbiol. Biotechnol. 71:773–782 [DOI] [PubMed] [Google Scholar]
- 14.Eijsink V, Hoell I, Vaaje-Kolstada G. 2010. Structure and function of enzymes acting on chitin and chitosan. Biotechnol. Genet. Eng. Rev. 27:331–366 [DOI] [PubMed] [Google Scholar]
- 15.Okazaki K, Yamashita Y, Noda M, Sueyoshi N, Kameshita I, Hayakawa S. 2004. Molecular cloning and expression of the gene encoding family 19 chitinase from Streptomyces sp. J-13-3. Biosci. Biotechnol. Biochem. 68:341–351 [DOI] [PubMed] [Google Scholar]
- 16.Ueda M, Kojima M, Yoshikawa T, Mitsuda N, Araki K, Kawaguchi T, Miyatake K, Arai M, Fukamizo T. 2003. A novel type of family 19 chitinase from Aeromonas sp. no. 10S-24. Cloning, sequence, expression, and the enzymatic properties. Eur. J. Biochem. 270:2513–2520 [DOI] [PubMed] [Google Scholar]
- 17.Beintema JJ. 1994. Structural features of plant chitinases and chitin-binding proteins. FEBS Lett. 350:159–163 [DOI] [PubMed] [Google Scholar]
- 18.Iseli B, Armand S, Boller T, Neuhaus JM, Henrissat B. 1996. Plant chitinases use two different hydrolytic mechanisms. FEBS Lett. 382:186–188 [DOI] [PubMed] [Google Scholar]
- 19.Wohlkonig A, Huet J, Looze Y, Wintjens R. 2010. Structural relationships in the lysozyme superfamily: significant evidence for glycoside hydrolase signature motifs. PLoS One 5:e15388. 10.1371/journal.pone.0015388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu T, Zhang H, Liu F, Chen L, Shen X, Yang Q. 2011. Active-pocket size differentiating insectile from bacterial chitinolytic beta-N-acetyl-d-hexosaminidases. Biochem. J. 438:467–474 [DOI] [PubMed] [Google Scholar]
- 21.Fujita K, Shimomura K, Yamamoto K, Yamashita T, Suzuki K. 2006. A chitinase structurally related to the glycoside hydrolase family 48 is indispensable for the hormonally induced diapause termination in a beetle. Biochem. Biophys. Res. Commun. 345:502–507 [DOI] [PubMed] [Google Scholar]
- 22.Bhattacharya D, Nagpure A, Gupta RK. 2007. Bacterial chitinases: properties and potential. Crit. Rev. Biotechnol. 27:21–28 [DOI] [PubMed] [Google Scholar]
- 23.Patil RS, Ghormade VV, Deshpande MV. 2000. Chitinolytic enzymes: an exploration. Enzyme Microb. Technol. 26:473–483 [DOI] [PubMed] [Google Scholar]
- 24.Gooday GW. 1994. Physiology of microbial degradation of chitin and chitosan. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 25.Slepecky R, Hemphill E. 1992. The genus Bacillus—nonmedical, p 1663–1696 In Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH.The prokaryotes, 2nd ed. Springer, New York, NY [Google Scholar]
- 26.Claus D, Berkeley RCW. 1986. Genus Bacillus Cohn 1872, 174AL, p 1105–1139 In Sneath PHA, Mair NS, Sharpe ME, Holt JG. (ed), Bergey's manual of systematic bacteriology, vol 2, 1st ed. Williams & Wilkins, Baltimore, MD [Google Scholar]
- 27.von der Weid I, Alviano DS, Santos AL, Soares RM, Alviano CS, Seldin L. 2003. Antimicrobial activity of Paenibacillus peoriae strain NRRL BD-62 against a broad spectrum of phytopathogenic bacteria and fungi. J. Appl. Microbiol. 95:1143–1151 [DOI] [PubMed] [Google Scholar]
- 28.Spizizen J. 1958. Transformation of biochemically deficient strains of Bacillus subtilis by deoxyribonucleate. Proc. Natl. Acad. Sci. U. S. A. 44:1072–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller LC, Tainter ML. 1944. Estimation of ED50 and its error by means of logarithmic probit graph paper. Proc. Soc. Exp. Biol. Med. 57:261–264 [Google Scholar]
- 30.Kimoto H, Kusaoke H, Yamamoto I, Fujii Y, Onodera T, Taketo A. 2002. Biochemical and genetic properties of Paenibacillus glycosyl hydrolase having chitosanase activity and discoidin domain. J. Biol. Chem. 277:14695–14702 [DOI] [PubMed] [Google Scholar]
- 31.Kimoto H, Taketo A. 1997. Initial stage of DNA-electrotransfer into E. coli cells. J. Biochem. 122:237–242 [DOI] [PubMed] [Google Scholar]
- 32.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimoto H, Fujii Y, Yokota Y, Taketo A. 2005. Molecular characterization of NADase-streptolysin O operon of hemolytic streptococci. Biochim. Biophys. Acta 1681:134–149 [DOI] [PubMed] [Google Scholar]
- 34.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 35.Hamady M, Knight R. 2009. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 19:1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K. 1997. Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int. J. Syst. Bacteriol. 47:289–298 [DOI] [PubMed] [Google Scholar]
- 37.Heyndrickx M, Vandemeulebroecke K, Scheldeman P, Hoste B, Kersters K, De Vos P, Logan NA, Aziz AM, Ali N, Berkeley RC. 1995. Paenibacillus (formerly Bacillus) gordonae (Pichinoty et al. 1986) Ash et al. 1994 is a later subjective synonym of Paenibacillus (formerly Bacillus) validus (Nakamura 1984) Ash et al. 1994: emended description of P. validus. Int. J. Syst. Bacteriol. 45:661–669 [DOI] [PubMed] [Google Scholar]
- 38.Sonnhammer EL, Eddy SR, Durbin R. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405–420 [DOI] [PubMed] [Google Scholar]
- 39.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukuda M, Watanabe S, Yoshida S, Itoh H, Itoh Y, Kamio Y, Kaneko J. 2010. Cell surface xylanases of the glycoside hydrolase family 10 are essential for xylan utilization by Paenibacillus sp. W-61 as generators of xylo-oligosaccharide inducers for the xylanase genes. J. Bacteriol. 192:2210–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang QS, Xie XL, Liang G, Gong F, Wang Y, Wei XQ, Wang Q, Ji ZL, Chen QX. 2012. The GH18 family of chitinases: their domain architectures, functions and evolutions. Glycobiology 22:23–34 [DOI] [PubMed] [Google Scholar]
- 42.Hiramatsu S, Fujie M, Usami S, Sakai K, Yamada T. 2000. Two catalytic domains of Chlorella virus CVK2 chitinase. J. Biosci. Bioeng. 89:252–257 [DOI] [PubMed] [Google Scholar]
- 43.Tanaka T, Fukui T, Imanaka T. 2001. Different cleavage specificities of the dual catalytic domains in chitinase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Biol. Chem. 276:35629–35635 [DOI] [PubMed] [Google Scholar]
- 44.Howard MB, Ekborg NA, Taylor LE, II, Weiner RM, Hutcheson SW. 2004. Chitinase B of “Microbulbifer degradans” 2-40 contains two catalytic domains with different chitinolytic activities. J. Bacteriol. 186:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Babu MM, Priya ML, Selvan AT, Madera M, Gough J, Aravind L, Sankaran K. 2006. A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J. Bacteriol. 188:2761–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh AK, Ghodke I, Chhatpar HS. 2009. Pesticide tolerance of Paenibacillus sp. D1 and its chitinase. J. Environ. Manage. 91:358–362 [DOI] [PubMed] [Google Scholar]
- 47.Pason P, Kyu KL, Ratanakhanokchai K. 2006. Paenibacillus curdlanolyticus strain B-6 xylanolytic-cellulolytic enzyme system that degrades insoluble polysaccharides. Appl. Environ. Microbiol. 72:2483–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DasGupta SM, Khan N, Nautiyal CS. 2006. Biologic control ability of plant growth-promoting Paenibacillus lentimorbus NRRL B-30488 isolated from milk. Curr. Microbiol. 53:502–505 [DOI] [PubMed] [Google Scholar]
- 49.Aktuganov G, Melentjev A, Galimzianova N, Khalikova E, Korpela T, Susi P. 2008. Wide-range antifungal antagonism of Paenibacillus ehimensis IB-X-b and its dependence on chitinase and beta-1,3-glucanase production. Can. J. Microbiol. 54:577–587 [DOI] [PubMed] [Google Scholar]
- 50.Kimoto H, Akamatsu M, Fujii Y, Tatsumi H, Kusaoke H, Taketo A. 2010. Discoidin domain of chitosanase is required for binding to the fungal cell wall. J. Mol. Microbiol. Biotechnol. 18:14–23 [DOI] [PubMed] [Google Scholar]
- 51.Purushotham P, Arun PV, Prakash JS, Podile AR. 2012. Chitin binding proteins act synergistically with chitinases in Serratia proteamaculans 568. PLoS One 7:e36714. 10.1371/journal.pone.0036714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaaje-Kolstad G, Bohle LA, Gaseidnes S, Dalhus B, Bjoras M, Mathiesen G, Eijsink VG. 2012. Characterization of the chitinolytic machinery of Enterococcus faecalis V583 and high-resolution structure of its oxidative CBM33 enzyme. J. Mol. Biol. 416:239–254 [DOI] [PubMed] [Google Scholar]
- 53.Vaaje-Kolstad G, Westereng B, Horn SJ, Liu Z, Zhai H, Sorlie M, Eijsink VG. 2010. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330:219–222 [DOI] [PubMed] [Google Scholar]
- 54.OECD 2001. Guideline 423. OECD guideline for testing of chemicals. Acute oral toxicity—acute toxic class method. OECD, Paris, France [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.