Abstract
Bartonella henselae (Rhizobiales: Bartonellaceae) is a Gram-negative fastidious bacterium of veterinary and zoonotic importance. The cat flea Ctenocephalides felis (Siphonaptera: Pulicidae) is the main recognized vector of B. henselae, and transmission among cats and humans occurs mainly through infected flea feces. The present study documents the use of a quantitative molecular approach to follow the daily kinetics of B. henselae within the cat flea and its excreted feces after exposure to infected blood for 48 h in an artificial membrane system. B. henselae DNA was detected in both fleas and feces for the entire life span of the fleas (i.e., 12 days) starting from 24 h after initiation of the blood meal.
INTRODUCTION
Bartonella henselae is a Gram-negative facultative intracellular bacterium of veterinary and zoonotic importance distributed worldwide (1). At present, its major competent vector is the cat flea, Ctenocephalides felis (2). Viable B. henselae or its DNA has also been detected in several other blood-feeding arthropods, such as ticks (Dermacentor spp., Ixodes spp.) (3–6) and biting flies (Haematobia spp., Stomoxys spp.) (7); however, no evidence of the role of these insects as competent vectors exists. Cats, particularly kittens, represent the major reservoir of B. henselae (8). Infected cats are usually asymptomatic but experience chronic recurring bacteremia (9). Clinical signs observed after experimental infection of cats include febrile illness, transient anemia, neurological dysfunction, and endocarditis (10–12).
In humans, B. henselae is the causative agent of cat scratch disease (CSD), a syndrome characterized by a persistent regional lymphadenopathy that is usually self-limiting within 2 to 4 months in immunocompetent patients (13). However, infected immunocompromised individuals (such as those with AIDS or organ transplant recipients) can develop severe vasoproliferative tumors known as bacillary angiomatosis and bacillary peliosis (13, 14, 15).
Although the prevalence of B. henselae infection in cats fluctuates significantly, the highest levels of infection occur in temperate regions where conditions are most favorable for the development of C. felis (1, 16, 17). The seroprevalence of antibodies against B. henselae in healthy cat populations ranges from 25 to 45% throughout the world (17). Nonetheless, in North America, where C. felis is endemic, seroprevalence was reported to reach up to more than 90% in some cat colonies (18). Fleas acquire B. henselae during their blood meal on highly bacteremic cats (16). Once in the arthropod vector, the bacterium seems to replicate within the gut and is then excreted in the feces (19, 20). While transmission by flea saliva still requires further investigation (21), the exposure to infected flea feces appears to be the main route of infection for cats and, accidentally, humans (22–24). B. henselae can also be inoculated into the skin of a naive host via scratching or biting by a flea-contaminated carrier animal (16, 20, 25).
Knowledge of the kinetics of B. henselae in C. felis and its feces throughout the life span of the cat flea is an essential prerequisite for devising efficient control strategies against this arthropod-borne microorganism. To date, however, among the limited number of studies carried out in this respect, only one monitored the presence of B. henselae in the flea gut and excretion in feces, yet monitoring was for a limited time (i.e., up to 9 days) and was by means of qualitative immunofluorescence (19).
Therefore, the aim of this study was to assess, using quantitative molecular approaches, the earliest detectability of B. henselae in artificially fed C. felis fleas and their excreted feces and to monitor the persistence of B. henselae, on a daily basis, for the whole life span of the fleas.
The findings will help address future culture-based investigations aiming to assess the occurrence of possible extragut routes of dissemination or replication of B. henselae in C. felis.
MATERIALS AND METHODS
Ethics statement.
Animals were handled in strict accordance with good animal practice as defined by the relevant European standards of welfare for animals in research. Work performed on animals at the École Nationale Vétérinaire de Toulouse (ENVT), Toulouse, France, was reviewed and approved by the Institut National de la Recherche Agronomique (INRA) Toulouse/ENVT Ethics Committees (agreement no. MP/01/22/06/09).
Bacterial strain.
The reference strain B. henselae Houston-1 ATCC 49882 (26) was used to infect the blood on which fleas were fed.
Medium and growth conditions.
The B. henselae strain was grown on sheep blood agar (Columbia blood agar [CBA] base) medium (bioMérieux, Craponne, France) in a humidified atmosphere at 35°C in a 5% carbon dioxide atmosphere. For flea infection assays, the bacteria were collected after 5 days of growth on CBA plates and suspended in sterile phosphate-buffered saline (PBS) buffer. The bacterial suspension was diluted with PBS to obtain approximately 2 × 108 bacteria per ml.
Flea maintenance and supply.
The C. felis strain employed was obtained from a laboratory-reared colony originating from a wild strain that was harvested from a cat and that has been maintained on cats under laboratory conditions at ENVT since 1990. Prior to use for this experiment, fleas were ascertained to be quantitative PCR (qPCR) negative for Bartonella species infection (see below).
Feeding of C. felis with B. henselae-infected blood.
Canine blood used in all experiments was obtained from three healthy 6-year-old beagle dogs (weight, 13 kg) from ENVT. The dogs had been vaccinated every year with the vaccine DHPPi/L, which protects dogs against canine distemper virus, canine adenovirus type 2, canine parvovirus, canine parainfluenza virus, and Leptospira interrogans. Blood smears and the microhematocrit of the three dogs were checked before their inclusion in the study, and none of the dogs showed any abnormalities. The absence of Bartonella spp. in dog blood was also confirmed by qPCR. Blood obtained by venipuncture was placed in lithium heparin-coated Vacutainer tubes (Venosafe; Terumo Europe, Heverlee, Belgium). Blood functional complement was deactivated by storing blood samples at room temperature for 1 h after the blood was drawn and before storage at 4°C. Blood samples were kept at 4°C for no longer than 48 h.
One thousand unfed fleas were placed in a Plexiglas box in contact with a glass feeder closed at the bottom with a thin Parafilm membrane (Parafilm3 M; Pechiney Plastic Packaging, IL). The fleas had never received any blood meal before the start of the study. To stimulate the fleas to feed on blood, a constant temperature of 38.5°C, mimicking the host's body temperature, was maintained by a water jacket system that circulated water through the glass feeder. Fleas were fed on the glass feeder until no more live fleas were retrieved in the Plexiglas box. For the first 48 h of feeding, fleas were fed 5 ml of blood mixed with 500 μl of B. henselae in suspension in PBS at a concentration of approximately 2 × 108 bacteria per ml. Uninfected blood was then placed in the feeder until the end of each trial. Every 24 h, the glass feeder was cleaned with a quaternary ammonium disinfectant (Anios, D.D.S.H.; Laboratoires Anios, Lille-Hellemmes, France) and then with distilled water. A new Parafilm membrane was stretched and blood was changed. At the same time, 20 mg of the feces excreted by the flea colony daily and 20 live engorged fleas were collected. All removed fleas were identified as having fed successfully by observation of distension of the abdomen with the naked eye. About 1 ml of 70% ethanol was added to the fleas and the feces that were collected. Samples were then stored at −20°C until PCR analyses.
Simultaneously, 1,000 negative-control fleas were fed on uninfected canine blood and were sampled at the same time points. They were followed until no more live fleas were retrieved from the box.
Three trials of infected fleas and two trials of uninfected fleas were conducted at the same time.
DNA extraction.
DNA was extracted from all samples (including negative and positive controls) by using a NucleoSpin tissue kit according to the manufacturer's recommendations (Macherey-Nagel, Hoerdt, France). The daily quantity of biological material used for independent DNA extraction was a pool of 20 fleas (230 ng/μl of DNA) and 20 mg of flea feces (6.7 ng/μl of DNA). Fleas and feces were incubated overnight to 56°C for the proteinase K lysis step. For all samples, the final elution volume was 100 μl.
qPCR amplification.
First, for qPCR amplification, the DNA of C. felis was detected by amplification of a fragment of C. felis 18S ribosomal DNA (rDNA) derived from a partial sequence of C. felis 18S rDNA available in GenBank (accession number AF136859). The primers and TaqMan probe were designed with online GenScript real-time PCR (TaqMan) Primer Design software (accessed 12 June 2012) to generate a 122-bp amplicon. The sequences of the primer set were 5′-AGCGGAACCGTTTACAAGTC-3′ for forward primer 18SF and 5′-GGAACTCGAACGCTCATACA-3′ for reverse primer 18SR. The TaqMan probe was labeled with 6-carboxyfluorescein (FAM) at the 5′ end and N,N,N′,N′-tetramethyl-6-carboxytetramethylrhodamine (TAMRA) at the 3′ end. The sequence of the probe, named 18ST, was 5′-6-FAM-CCGTATCTCCCGACGGCGTC-TAMRA-3′. This amplification was used as a reference for the efficiency of DNA extraction from fleas and fecal samples. Second, amplification of B. henselae DNA was performed by targeting a conserved region on the citrate synthase gene (gltA), as described by Molia et al. (27). The sequences of the primers and probe were 5′-GTGCTAATCCATTTGCATGTATT-3′ for forward primer Bart.738f, 5′-GTAACATTTTTAGGCATGCTTCATTA-3′ for reverse primer Bart.831r, and 5′-6-FAM-AGCTGGTCCCCAAAGGCATGCAA-TAMRA-3′ for the fluorescent TaqMan probe Bart.772p, labeled as described above for the 18ST probe with FAM at the 5′ end and TAMRA at the 3′ end. The size of the gltA fragment targeted was 96 bp (27).
Each reaction was carried out in a final volume of 25 μl. For the B. henselae qPCR, 400 nM each primer and 80 nM fluorogenic TaqMan probe were added to TaqMan buffer containing carboxy-X-rhodamine (ROX) as a passive reference (TaqMan Universal PCR master mix; Life Technologie SAS, Saint-Aubin, France). For the C. felis qPCR, the concentrations were optimized to 100 nM each primer and 50 nM probe. Five microliters of DNA template was added to each reaction mixture, and reactions with each template were conducted in triplicate. Amplifications were run using a Stratagene MX3005P thermal cycler (Agilent Technologies, La Jolla, CA).
Data acquisition and analysis were performed using MxPro QPCR (version 4.10) software (Agilent Technologies, La Jolla, CA). Cycling conditions were as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 15 s at 95°C and 60 s at 60°C, as previously reported (27). The signal of FAM as the reporter dye was measured against the signal of ROX, used as an internal reference dye, in order to normalize for the non-PCR-related fluorescence fluctuations between wells. The B. henselae DNA concentration was determined using a Genova spectrophotometer (Jenway, United Kingdom).
For C. felis qPCR amplification, ultrapure water (Simplicity UV; Millipore) and a sample of 1 ng/μl of B. henselae DNA, corresponding to 2.5 × 106 bacteria, were used as negative controls. DNA extracted from fleas engorged on noninfected blood was used as a positive control. For the B. henselae qPCR amplification, ultrapure water (Simplicity UV; Millipore) and a solution of DNA extracted from fleas engorged on uninfected canine blood were used as negative controls. A solution of extracted B. henselae DNA at a concentration of 1 ng/μl was used as a positive control. Negative and positive controls were included in each reaction plate.
Standard curve and reproducibility of the detection of B. henselae by qPCR.
To generate a standard curve and to assess the detection limit of the qPCR using the primers and probe described by Molia et al. (27), a B. henselae suspension was serially diluted 10-fold in canine blood to achieve levels of B. henselae bacteremia ranging from 2 × 106 to 2 × 101 total bacteria. The standard curve obtained with these primers and the related linear equation were previously generated by Molia et al. (27), with cycle threshold (CT) values (y axis) being plotted against B. henselae plasmid concentrations (x axis). However, the standard curve was determined again in the present study in order to express the data on the x axis as the log of the number of bacteria. The amplification efficiency (E) of the qPCR was calculated with the formula E = (10(−1/S)) − 1, where S is the slope of the standard curve. To determine the intra-assay reproducibility, assays with eight replicates of 10-fold serial dilutions of B. henselae DNA (from 2 × 106 to 2 × 101 bacteria) were performed. The interassay reproducibility was assessed on independently repeated amplifications performed over six consecutive days with three replicates per day.
qPCR assay specificity.
Both the C. felis and B. henselae qPCRs were tested for inhibition and potential cross-hybridization between probes and nontarget DNA. The B. henselae qPCR was tested against DNA extracted from C. felis, and the C. felis qPCR was tested against B. henselae DNA. Both PCRs were tested against DNA extracted from uninfected canine blood sampled from each of the three dogs used in the study.
Data analysis.
To correct each sample size according to the quality of DNA extraction, the B. henselae CT values were modified by computing the difference between the daily and the average flea CT values for all trials. The result of this difference was added to the associated B. henselae CT values for each sample. The corrected CT value was then transformed into the number of bacteria according to the equation of the linear regression of the standard curve.
The means, standard deviations (SDs), and coefficients of variation (CVs) of the ranges of CT values of flea DNA obtained daily from both 20 individual C. felis fleas and 20 mg of flea feces were computed with Microsoft Excel 2010 software (Microsoft Corporation). The CVs of flea DNA and B. henselae from the standard curve were calculated by dividing the SD of the CT values of replicates by the mean CT value of replicates. Analysis of variance (ANOVA) was used to compare the means between the CT values for fleas and their feces and was carried out to compare the data between the three trials using the software package StatXact, release 3.1 (Cytel Software Corporation).
RESULTS
Standard curve, sensitivity, and reproducibility of the B. henselae qPCR assay.
A linear correlation was obtained for both intra- and interassay standard curves, with R2 equal to 0.99 and efficiency equal to 97% for the intra-assay standard curve and R2 equal to 0.98 and efficiency equal to 93% for the interassay standard curve. The efficiency (E) of the qPCR by use of the combination of the intra- and interassay standard curves was 92%, with a strong linear correlation (R2 = 0.9816), a y-intercept value of 44.027, and a slope of −3.5507. The detection limit determined from the standard curve was 20 bacteria, corresponding to a CT value of 39. This value was calculated on the basis of the mean CT value at which all replicates of the highest 10-fold dilution of B. henselae DNA provided amplification plots among the intra- and interassay standard curves. Amplification of lower numbers of bacteria (<20) provided inconstant and nonreproducible CT values even when an increased number of cycles was used (data not shown).
The intra- and interassay results were combined to assess the reproducibility of the B. henselae qPCR. The coefficient of variation ranged from 2.59% for 2 × 102 bacteria to 3.86% for 2 × 106 bacteria, showing strong reproducibility. Inhibition of the PCR was not recorded in the presence of the highest B. henselae load (2 × 106 bacteria). Cross-hybridization between B. henselae probes and C. felis DNA and between C. felis probes and B. henselae DNA was not recorded.
Reproducibility of the C. felis batch and flea excrement qPCR.
The reproducibility of qPCR assays based on the same number of fleas (20) and the same quantity of flea feces (20 mg) was determined by computation of CVs for each of the three trials. In the three trials carried out for fleas and flea excrement, the CVs of the CT values ranged from 4.23% to 5.24% and from 8.49% to 12.81%, respectively (Tables 1 and 2). As they were all lower than 15%, these CV values were consistent with previously published data and were highly reproducible (28). The mean CT values did not significantly differ between the three trials of batches of 20 C. felis fleas or between the three trials of 20 mg of flea feces. The P values determined by ANOVA were 0.09 and 0.13, respectively. Then, the mean CT values of 13.23 for batches of 20 fleas (Table 1) and of 18.48 for 20 mg of flea feces (Table 2) were used as corrections for the CT values for B. henselae for each trial.
Table 1.
Distribution of CT values obtained by qPCR from batches of 20 Ctenocephalides felis fleas for each replicatea
| Trial | CT value |
CV | |||
|---|---|---|---|---|---|
| Minimum | Maximum | Mean | SD | ||
| 1 | 12.26 | 14.35 | 13.3 | 0.7 | 5.24 |
| 2 | 11.62 | 14.37 | 12.92 | 0.64 | 4.98 |
| 3 | 12.7 | 14.42 | 13.5 | 0.57 | 4.23 |
| All | 11.62 | 14.42 | 13.23 | 0.67 | 5.06 |
Fleas were fed on blood containing B. henselae. CT, cycle threshold.
Table 2.
Distribution of CT values obtained by qPCR from 20 mg of Ctenocephalides felis feces for each replicatea
| Trial | CT value |
CV | |||
|---|---|---|---|---|---|
| Minimum | Maximum | Mean | SD | ||
| 1 | 15.39 | 23.79 | 18.23 | 2.18 | 11.98 |
| 2 | 15.35 | 20.77 | 18.46 | 1.57 | 8.49 |
| 3 | 15.46 | 24.57 | 18.76 | 2.4 | 12.81 |
| All | 15.35 | 24.57 | 18.48 | 2.03 | 11 |
Fleas were fed on blood containing B. henselae. CT, cycle threshold.
Persistence of B. henselae within fleas.
The two replicates of control fleas lived for 10 and 14 days, respectively, on the artificial membrane feeding system. Both colonies were qPCR negative for B. henselae throughout the study. The infected fleas from trials 1 and 2 lived for 13 days, whereas the infected fleas from trial 3 lived for 12 days.
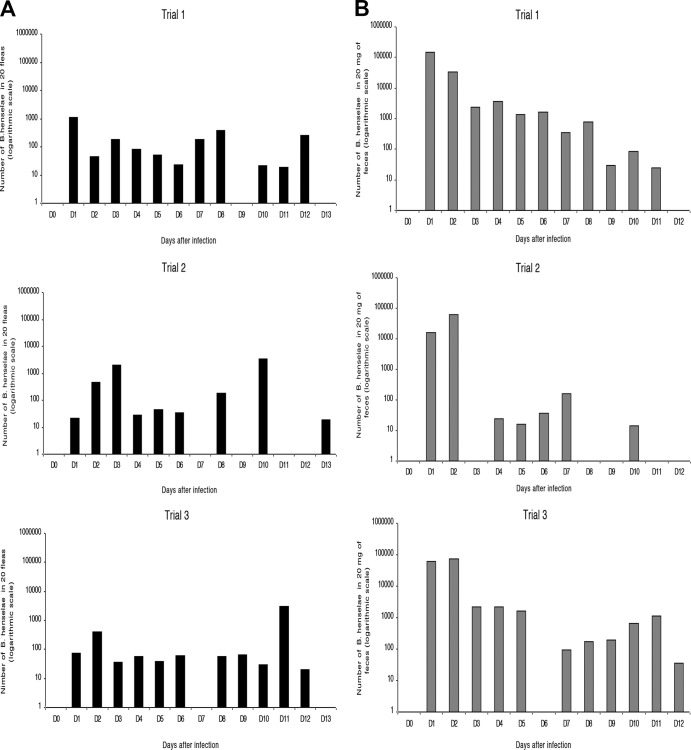
From day 1 to day 13 after infection, a total of 760 fleas were sampled and their DNA was extracted to estimate the number of bacteria according to the equation of the standard linear regression curve. From days 1 to 6, the fluctuation in the quantity of bacteria followed a similar pattern in all three trials of fleas sampled: a peak concentration was recorded on either day 1, 2, or 3 and a rather steady smaller amount was found until day 6 (Fig. 1A). Afterwards, the concentration of B. henselae in flea pools decreased, remaining rather steady at less than 102 bacteria between days 4 and 6 for trials 1 and 2 and days 3 and 6 for trial 3. A final increase in the B. henselae load was then seen on days 8 (trials 1 and 2), 10 (trial 2), 11 (trial 3), and 12 (trial 1). No B. henselae DNA was detected on days 9 and 13 for trial 1, on days 7, 9, 11, and 12 for trial 2, and on day 7 for trial 3 (Fig. 1A).
Fig 1.
Dynamics of B. henselae in fleas (A) and excreted feces (B). Fleas were fed blood infected with B. henselae from day 0 to day 2 and with uninfected blood from day 2 onwards.
Persistence of B. henselae within flea feces.
The feces of control fleas were sampled daily and analyzed, and B. henselae DNA was not detected throughout the study. Sampling could not be performed on day 3 for trial 2, on day 6 for trial 3, and on day 12 for trials 1 and 2 (Fig. 1B) because the amount of excreted feces was too small.
For trial 1, the bacterial load in the excreted feces remained above 1 × 103 bacteria from day 1 until day 6, with an average of 9 × 104 bacteria being excreted within the first 2 days. Then, the amount of bacteria decreased until day 11. No bacteria were detected on day 12 (Fig. 1B). For trial 2, the amount of bacteria in feces gradually increased until day 2, reaching a bacterial load of 6 × 104 bacteria and decreasing dramatically on day 3, when DNA was not detected. An increase was then recorded between days 4 and 7. No quantification could be obtained on days 8 and 9 or on days 11 and 12 due to the absence of B. henselae DNA detection. Bacteria were, however, still excreted in feces on day 10, with the lowest value of approximately 20 bacteria being obtained for this replicate (Fig. 1B). For trial 3, feces were infected with B. henselae on days 1 and 2, with an average load of 6.7 × 104 bacteria. The bacterial load remained above 1 × 103 bacteria until day 5. B. henselae kept being rather constantly excreted from day 7 until the last sampling on day 12, with a bacterial load of 35 (Fig. 1B).
DISCUSSION
This study investigated, using a quantitative molecular approach, the daily kinetics of B. henselae within the cat flea and cat flea feces after exposure to infected blood for 48 h. The qPCR employed for both C. felis and B. henselae primers and probes showed good reproducibility, specificity, and sensitivity, consistent with the results of similar studies carried out previously (29). In addition, this approach can also be used to detect the presence of Bartonella spp. directly from patient tissues (30–32). The artificial membrane feeding system allowing experimental infection has also been used to investigate several vector-pathogen interactions, like those involving Xenopsylla cheopis and Yersinia pestis, Pediculus humanus and B. quintana, and Ixodes ricinus and B. henselae (5, 33, 34). In the case of B. henselae, it is known that its acquisition by C. felis can already occur from 3 h after the start of a contaminated blood meal (19). In this study, a time lapse of 48 h from the initiation of feeding on contaminated blood was used in order to allow the largest but undetermined number of fleas to engorge on infected blood. In both the infected and the control groups of fleas, an important but not quantified mortality was observed daily throughout the study among nonengorged specimens. This can be attributable to several causes, including the natural death of fleas, the reluctance of some fleas to feed on an artificial membrane system, and their daily handling for sampling purposes. Nonetheless, the maximum life span recorded for infected fleas (i.e., 12 to 13 days) was not shortened in comparison with that of control fleas (i.e., 10 to 14 days). This is in accordance with the findings of a previous study, where Xenopsylla ramesis fleas infected with Bartonella spp. did not show a shorter life span than uninfected fleas (35).
Although no clear replication pattern was reported throughout the three trials in the current investigation, B. henselae DNA was not cleared from the fleas after contamination but instead was maintained in the fleas until 10 to 13 days. All trials differed in the mean initial B. henselae load, with trial 1 showing the highest bacterial load and trial 2 showing the lowest. On days 7, 9, 11, and 12 for trial 2 and on day 7 for trial 3, however, no DNA could be detected in fleas, suggesting that the amount of bacteria was below the detection threshold of the optimized qPCR. For trial 2, the absence of B. henselae in all fleas on day 9 was anticipated and accompanied by the absence of bacteria in their feces on days 8 and 9. Similarly, in a previous study, no B. quintana DNA was detected in louse feces on either day 5, 7, 9, or 11 after infection (34).
For all three trials, a general increase in the amount of bacteria in the fleas was observed after day 7. These findings suggest the possible replication of B. henselae in the cat flea at this time of its life span. This is in accordance with the findings of previous studies showing the persistence and qualitative increase in bacteria in flea guts 9 days after an infected blood meal (19) and the replication of B. henselae within adult fleas at 6 to 8 days after feeding (20). The first decrease in bacterial DNA in the fleas was observed 2 days after the infected blood meal and may have been due to fleas clearing themselves of an excess of bacteria, as was suggested for C. felis and X. cheopis after a highly septicemic blood meal with Rickettsia felis (36) and Y. pestis, respectively (37, 38). Then, the persistence of Bartonella DNA until the end of the study could be explained by several assumptions requiring further investigation: the formation of aggregates in the flea gut; attachment to an extracellular matrix, as shown for Y. pestis and B. schoenbuchensis, respectively (33, 39); or sequestration in other flea tissues. According to the first hypothesis, bacteria would then be continuously released from the gut biofilm and excreted in feces. In our study, the excretion of the bacteria was indeed continuous, although the bacterial load decreased gradually in the second part of the flea life span, starting on day 7. However, the reduction in the amount of bacteria in the feces was not correlated with the decrease in the amount of bacteria in all fleas at the end of the study, which, conversely, increased on days 12 (trial 1), 10 (trial 2), and 11 (trial 3). This could suggest the second hypothesis of migration of B. henselae from the digestive tract to the body cavity and/or other organs (for example, reproductive tissues, hemolymph, Malpighian tubules) in the second part of the flea life span. Similarly, R. felis has recently been shown to replicate in the digestive tract of C. felis in the first stage of infection, and then to migrate to the hemolymph, disseminating to the excretory system (Malpighian tubules, hindgut, and rectal ampulla) and reproductive tissues (36). At present, the survival of B. henselae in excreted feces is estimated to last for at least 3 days in the environment (20) and could be possible because of the aggregation of the bacteria in a biofilm on the surface of the feces, as suggested for B. quintana on louse feces (34). Nevertheless, the dispersion of B. henselae within the cat flea and B. henselae excretion have not yet been fully investigated. This is of epidemiological importance, as flea feces are indeed the principal infectious material for the transmission of B. henselae (24) and can be cultured to produce viable colonies (19, 20). The viability of B. henselae in flea feces should be addressed by mRNA amplification, as DNA-based qPCR alone does not allow differentiation between live and dead bacteria. Such an approach may help assess the actual infectivity of feces harvested from the fur of flea-infested cats as a key indicator of the risk for intraspecies and zoonotic transmission of B. henselae.
To conclude, the persistence of B. henselae DNA occurred in both the arthropod vector and its feces for the whole life span of the fleas on the artificial system. This is epidemiologically relevant, as the persistence of B. henselae in both fleas and feces enhances contamination risks, especially in the presence of a high density of reservoir hosts.
Further studies are required to determine whether the replication of B. henselae could occur also in extragut tissues. In these regards, microdissection of fleas could possibly help address this issue with the aim of identifying the organs (e.g., flea gut, digestive epithelial cells, as well as other tissues) in which B. henselae replication and dissemination take place in its most competent arthropod host and distinguishing viable from nonviable bacteria.
ACKNOWLEDGMENTS
We are grateful to Francis Biville (Institut Pasteur, Paris, France) for his thoughtful comments on this work; Vincenzo Lorusso (University of Edinburgh, Edinburgh, United Kingdom) for his valuable suggestions on the manuscript and his contribution to the English of the text; Vincent Bourret (ENVT, Toulouse, France) for his help with data analysis; MaFeng Liu, Yann Ferrandez, and Martine Monteil (ANSES, Maison-Alfort, France) for their technical assistance; Solange Vermot, Martine Roques, and Sonia Gounaud (ENVT, Toulouse, France) for their help with dog restraint and flea colony maintenance; and Christelle Grisez, Francoise Prévot, and Quentin Pagé (ENVT, Toulouse, France) for their technical assistance with qPCR.
Footnotes
Published ahead of print 20 September 2013
REFERENCES
- 1.Chomel BB, Kasten RW. 2010. Bartonellosis, an increasingly recognized zoonosis. J. Appl. Microbiol. 109:743–750 [DOI] [PubMed] [Google Scholar]
- 2.Chomel BB, Kasten RW, Floyd-Hawkins K, Chi B, Yamamoto K, Roberts-Wilson J, Gurfield AN, Abbott RC, Pedersen NC, Koehler JE. 1996. Experimental transmission of Bartonella henselae by the cat flea. J. Clin. Microbiol. 34:1952–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsai YL, Chang CC, Chuang ST, Chomel BB. 2011. Bartonella species and their ectoparasites: selective host adaptation or strain selection between the vector and the mammalian host? Comp. Immunol. Microbiol. Infect. Dis. 34:299–314 [DOI] [PubMed] [Google Scholar]
- 4.Podsiadly E, Chmielewski T, Sochon E, Tylewska-Wierzbanowska S. 2007. Bartonella henselae in Ixodes ricinus ticks removed from dogs. Vector Borne Zoonotic Dis. 7:189–192 [DOI] [PubMed] [Google Scholar]
- 5.Cotté V, Bonnet S, Le Rhun D, Le Naour E, Chauvin A, Boulouis HJ, Lecuelle B, Lilin T, Vayssier-Taussat M. 2008. Transmission of Bartonella henselae by Ixodes ricinus. Emerg. Infect. Dis. 14:1074–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietrich F, Schmidgen T, Maggi RG, Richter D, Matuschka FR, Vonthein R, Breitschwerdt EB, Kempf VA. 2010. Prevalence of Bartonella henselae and Borrelia burgdorferi sensu lato DNA in Ixodes ricinus ticks in Europe. Appl. Environ. Microbiol. 76:1395–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung CY, Kasten RW, Paff SM, Van Horn BA, Vayssier-Taussat M, Boulouis HJ, Chomel BB. 2004. Bartonella spp. DNA associated with biting flies from California. Emerg. Infect. Dis. 10:1311–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomel BB, Boulouis HJ, Maruyama S, Breitschwerdt EB. 2006. Bartonella spp. in pets and effects on human health. Emerg. Infect. Dis. 12:389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breitschwerdt EB, Kordick DL. 1995. Bartonellosis. J. Am. Vet. Med. Assoc. 206:1928–1931 [PubMed] [Google Scholar]
- 10.Guptill L, Slater L, Wu CC, Lin TL, Glickman LT, Welch DF, Hogenesch H. 1997. Experimental infection of young specific pathogen-free cats with Bartonella henselae. J. Infect. Dis. 176:206–216 [DOI] [PubMed] [Google Scholar]
- 11.Kordick DL, Brown TT, Shin K, Breitschwerdt EB. 1999. Clinical and pathologic evaluation of chronic Bartonella henselae or Bartonella clarridgeiae infection in cats. J. Clin. Microbiol. 37:1536–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breitschwerdt EB, Kordick DL. 2000. Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin. Microbiol. Rev. 13:428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boulouis HJ, Chang CC, Henn JB, Kasten RW, Chomel BB. 2005. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet. Res. 36:383–410 [DOI] [PubMed] [Google Scholar]
- 14.Klotz SA, Ianas V, Elliott SP. 2011. Cat-scratch disease. Am. Fam. Physician 83:152–155 [PubMed] [Google Scholar]
- 15.Breitschwerdt EB. 2008. Feline bartonellosis and cat scratch disease. Vet. Immunol. Immunopathol. 123:167–171 [DOI] [PubMed] [Google Scholar]
- 16.Guptill L. 2010. Bartonellosis. Vet. Microbiol. 140:347–359 [DOI] [PubMed] [Google Scholar]
- 17.Lappin MR, Griffin B, Brunt J, Riley A, Burney D, Hawley J, Brewer MM, Jensen WA. 2006. Prevalence of Bartonella species, Haemoplasma species, Ehrlichia species, Anaplasma phagocytophilum, and Neorickettsia risticii DNA in the blood of cats and their fleas in the United States. J. Feline Med. Surg. 8:85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jameson P, Greene C, Regnery R, Dryden M, Marks A, Brown J, Cooper J, Glaus B, Greene R. 1995. Prevalence of Bartonella henselae antibodies in pet cats throughout regions of North America. J. Infect. Dis. 172:1145–1149 [DOI] [PubMed] [Google Scholar]
- 19.Higgins JA, Radulovic S, Jaworski DC, Azad AF. 1996. Acquisition of the cat scratch disease agent Bartonella henselae by cat fleas (Siphonaptera:Pulicidae). J. Med. Entomol. 33:490–495 [DOI] [PubMed] [Google Scholar]
- 20.Finkelstein JL, Brown TP, O'Reilly KL, Wedincamp J, Jr, Foil LD. 2002. Studies on the growth of Bartonella henselae in the cat flea (Siphonaptera: Pulicidae). J. Med. Entomol. 39:915–919 [DOI] [PubMed] [Google Scholar]
- 21.Bouhsira E, Ferrandez Y, Liu M, Franc M, Boulouis H-J, Biville F. 2013. Ctenocephalides felis an in vitro potential vector for five Bartonella species. Comp. Immunol. Microbiol. Infect. Dis. 36:105–111 [DOI] [PubMed] [Google Scholar]
- 22.Abbot RC, Chomel BB, Kasten RW, Floyd-Hawkins KA, Kikuchi Y, Koehler JE, Pedersen NC. 1997. Experimental and natural infection with Bartonella henselae in domestic cats. Comp. Immunol. Microbiol. Infect. Dis. 20:41–51 [DOI] [PubMed] [Google Scholar]
- 23.Foil L, Andress E, Freeland RL, Roy AF, Rutledge R, Triche PC, O'Reilly KL. 1998. Experimental infection of domestic cats with Bartonella henselae by inoculation of Ctenocephalides felis (Siphonaptera: Pulicidae) feces. J. Med. Entomol. 35:625–628 [DOI] [PubMed] [Google Scholar]
- 24.Telford SR, Wormser GP. 2010. Bartonella spp. transmission by ticks not established. Emerg. Infect. Dis. 16:379–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mosbacher M, Elliott SP, Shehab Z, Pinnas JL, Klotz JH, Klotz SA. 2010. Cat scratch disease and arthropod vectors: more to it than a scratch? J. Am. Board Fam. Med. 23:685–686 [DOI] [PubMed] [Google Scholar]
- 26.Sölder B, Allerberger F, Covi B, Maurer K, Scheminzky C, Kreczy A, Schön G, Dierich MP. 1995. Cat scratch disease caused by Bartonella henselae. Immun. Infekt. 23:228–231 [PubMed] [Google Scholar]
- 27.Molia S, Chomel BB, Kasten RW, Leutenegger CM, Steele BR, Marker L, Martenson JS, Keet DF, Bengis RG, Peterson RP, Munson L, O'Brien SJ. 2004. Prevalence of Bartonella infection in wild African lions (Panthera leo) and cheetahs (Acinonyx jubatus). Vet. Microbiol. 100:31–41 [DOI] [PubMed] [Google Scholar]
- 28.Zemanick ET, Wagner BD, Sagel SD, Stevens MJ, Accurso FJ, Harris JK. 2010. Reliability of quantitative real-time PCR for bacterial detection in cystic fibrosis airway specimens. PLoS One 5:e15101. 10.1371/journal.pone.0015101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robinson MT, Morgan ER, Woods D, Shaw SE. 2010. Real-time and multiplex real-time polymerase chain reactions for the detection of Bartonella henselae within cat flea, Ctenocephalides felis, samples. Med. Vet. Entomol. 24:449–455 [DOI] [PubMed] [Google Scholar]
- 30.Ciervo A, Mastroianni CM, Ajassa C, Pinto A, Ciceroni L. 2005. Rapid identification of Bartonella henselae by real-time polymerase chain reaction in a patient with cat scratch disease. Diagn. Microbiol. Infect. Dis. 53:75–77 [DOI] [PubMed] [Google Scholar]
- 31.Bergmans AM, Rossen JW. 2013. Detection of Bartonella spp. DNA in clinical specimens using an internally controlled real-time PCR assay. Methods Mol. Biol. 943:217–228 [DOI] [PubMed] [Google Scholar]
- 32.Liberto MC, Matera G, Lamberti AG, Quirino A, Barreca GS, Marascio N, Baudi F, Caroleo B, Staltari O, Focà A. 2013. Diagnosis and follow up of Bartonella henselae infection in the spleen of an immunocompetent patient by real-time quantitative polymerase chain reaction. J. Med. Microbiol. 62:1081–1085 [DOI] [PubMed] [Google Scholar]
- 33.Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, Whitney AR, Kobayashi SD, DeLeo FR, Hinnebusch BJ. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783–792 [DOI] [PubMed] [Google Scholar]
- 34.Seki N, Kasai S, Saito N, Komagata O, Mihara M, Sasaki T, Tomita T, Sasaki T, Kobayashi M. 2007. Quantitative analysis of proliferation and excretion of Bartonella quintana in body lice, Pediculus humanus. Am. J. Trop. Med. Hyg. 77:562–566 [PubMed] [Google Scholar]
- 35.Morick D, Krasnov BR, Khokhlova IS, Gutierrez R, Fielden LJ, Gottlieb Y, Harrus S. 2013. Effects of Bartonella spp. on flea feeding and reproductive performance. Appl. Environ. Microbiol. 79:3438–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thepparit C, Hirunkanokpun S, Popov VL, Foil LD, Macaluso KR. 2013. Dissemination of bloodmeal acquired Rickettsia felis in cat fleas, Ctenocephalides felis. Parasit. Vectors 6:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinnebusch BJ, Perry RD, Schwan TG. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367–370 [DOI] [PubMed] [Google Scholar]
- 38.Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, Forsberg A. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296:733–735 [DOI] [PubMed] [Google Scholar]
- 39.Dehio C, Sauder U, Hiestand R. 2004. Isolation of Bartonella schoenbuchensis from Lipoptena cervi, a blood-sucking arthropod causing deer ked dermatitis. J. Clin. Microbiol. 42:5320–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]