Abstract
Marine macroalgae are rich in bioactive compounds that can, when consumed, impart beneficial effects on animal and human health. The red seaweed Chondrus crispus has been reported to have a wide range of health-promoting activities, such as antitumor and antiviral activities. Using a Caenorhabditis elegans infection model, we show that C. crispus water extract (CCWE) enhances host immunity and suppresses the expression of quorum sensing (QS) and the virulence factors of Pseudomonas aeruginosa (strain PA14). Supplementation of nematode growth medium with CCWE induced the expression of C. elegans innate immune genes, such as irg-1, irg-2, F49F1.6, hsf-1, K05D8.5, F56D6.2, C29F3.7, F28D1.3, F38A1.5 ZK6.7, lys-1, spp-1, and abf-1, by more than 2-fold, while T20G5.7 was not affected. Additionally, CCWE suppressed the expression of PA14 QS genes and virulence factors, although it did not affect the growth of the bacteria. These effects correlated with a 28% reduction in the PA14-inflicted killing of C. elegans. Kappa-carrageenan (K-CGN), a major component of CCWE, was shown to play an important role in the enhancement of host immunity. Using C. elegans mutants, we identified that pmk-1, daf-2/daf-16, and skn-1 are essential in the K-CGN-induced host immune response. In view of the conservation of innate immune pathways between C. elegans and humans, the results of this study suggest that water-soluble components of C. crispus may also play a health-promoting role in higher animals and humans.
INTRODUCTION
Seaweeds are rich in bioactive compounds, such as proteins, peptides, amino acids, lipids, fibers, pigments, polyphenols, and polysaccharides (1, 2), that are responsible for imparting various health benefits. For example, β-carotene and lutein were identified as antimutagenic substances in edible red algae, which indicated their potential anticancer activity (3). Furthermore, studies on mice/rats and humans demonstrated that dietary supplementation with various extracts of a variety of seaweeds correlated with a decreased risk of breast cancer (4, 5). Polysaccharides, proteins, peptides, and amino acids from a number of seaweeds showed beneficial activity against diabetes, cancer, AIDS, and vascular diseases (2). The red seaweed Chondrus crispus (Rhodophyta) is widely distributed in the northern Atlantic. The work presented here was undertaken using a proprietary strain of C. crispus which was cultivated on land in Nova Scotia, Canada, for the Asian food market by Acadian Seaplants Limited. Besides high contents of total proteins, oligopeptides, and pigments, this red alga is rich in the water-soluble polysaccharide carrageenan (CGN) (6), which has been reported to have antiviral (7, 8) and antitumor (9, 10) activities.
CGNs are linear polymers of digalactose residues and can be extracted from some species of red seaweed. CGNs are widely used in the food industry as thickeners, stabilizers, and emulsifiers. C. crispus produces three types of CGN at different stages of its life cycle. In the diploid sporophyte phase, it produces lambda-CGN, while the haploid gametophyte produces predominantly kappa-CGN (K-CGN) with some iota-CGN. The gametophyte also makes the precursor types of kappa- and iota-CGNs, mu- and nu-CGNs. The mu- and nu-CGNs are more sulfated than the kappa- and iota-CGN types, and they are of nongelling forms. These precursors are more similar to lambda-CGN with respect to sulfation levels and solubility properties (J. S. Craigie, personal communication). The water extract used in the present work contained multiple compounds, with K-CGN being the main type of CGN.
The nematode (Caenorhabditis elegans) model has been used to study fundamental biological processes and also for the rapid screening of chemical compounds for various health effects. In the present work, the C. elegans model of infection with Pseudomonas aeruginosa was used to investigate the immune-enhancing activity of a cultivated C. crispus water extract (CCWE). The ubiquitous bacterium P. aeruginosa is an emerging opportunistic human pathogen which infects immunodeficient or immunocompromised patients (11). It also causes lethal infection of the nematode (C. elegans) (12). The pathogenesis of P. aeruginosa is mediated by secreted virulence factors, which include toxins, such as pyocyanin, pyoverdine, siderophores, and hydrogen cyanide (12, 13), as well as by bacterial enzymes, such as elastase and alkaline protease (14). Moreover, biofilm formation protects the bacteria from adverse environmental factors and increases their antibiotic resistance and pathogenesis (15). Interestingly, the virulence factors and biofilm formation were found to be regulated by quorum sensing (QS), the cell-to-cell communication system of the bacteria. The two QS systems include the lasR-lasI and the rhlR-rhlI systems, where lasR and rhlR are transcription activators and lasI and rhlI are the synthases of small interactive autoinducer molecules (14, 16).
The immune response of C. elegans to P. aeruginosa is mediated through the p38 mitogen-activated protein kinase (PMK-1), transforming growth factor β (TGF-β), and the DAF-2/DAF-16 insulin-like and ZIP-2 pathways (17–20). Recently, an extract of the brown seaweed Ascophyllum nodosum was demonstrated to protect C. elegans from infection by P. aeruginosa strain PA14 (21). In the present study, we tested the effects of a water extract from the cultivated red seaweed C. crispus on host immunity and PA14 pathogenicity using the C. elegans infection model with PA14. We further examined the effect of pure K-CGN, the predominant water-soluble polysaccharide present in C. crispus, on host immunity. Moreover, we utilized a number of mutant lines of C. elegans to determine the role of various signaling pathways in the K-CGN-elicited immune response.
MATERIALS AND METHODS
Preparation of seaweed extracts.
The on-land-cultivated proprietary strain of C. crispus used in this study was obtained from Acadian Seaplants Limited (Dartmouth, NS, Canada). The dried seaweed (250 g) was extracted with water (two times with 1.5 liters each time) by sonication for 1 h at room temperature. The aqueous fraction was concentrated under reduced pressure and freeze-dried overnight, yielding water extract (CCWE; 67.1 g). The CCWE was stored at −20°C. Research-grade K-CGN powder was obtained from Cargill Texturant Systems, France. A stock solution of CCWE or K-CGN was prepared by dissolving the powder in distilled water to a concentration of 25 mg/ml or 10 mg/ml, respectively, and stored at 4°C. The stock solutions were prepared no longer than 1 week before use in order to avoid degradation of bioactive compounds due to prolonged storage.
Bacterial strain and culture.
A clinical isolate of P. aeruginosa, strain PA14, was a kind gift from Eric Déziel (INRS, Institute Armand-Frappier-Microbiologieet Biotechnologie, Laval, QC, Canada). PA14 was cultured and maintained in King's B (KB) medium or on KB medium agar unless otherwise stated. PA14 bacterial stocks kept at −80°C were thawed and cultured in 3 ml of KB medium at 37°C for 6 h with constant shaking, followed by streaking the culture onto a KB agar plate. The plate was incubated at 37°C overnight, and then a single colony was picked from the KB agar plate, inoculated into 10 ml of KB medium, and cultured overnight at 37°C with constant gentle shaking (150 rpm). The culture was diluted to an optical density at 600 nm (OD600) of 1.0 with fresh KB medium, stored at 4°C, and used within 1 week.
C. elegans strains, culture, and PA14 killing assay.
Wild-type C. elegans strain N2 Bristol, mutant strains of the worm with mutations in the daf-16 (mutant mu86), daf-2 (mutant e1370), skn-1 (mutant zu67), and pmk-1 (mutant km25) genes, and the standard laboratory bacterial food source Escherichia coli strain OP50 were obtained from the Caenorhabditis Genetics Center, University of Minnesota, Minneapolis, MN. Cultures of E. coli OP50 were grown overnight in Luria-Bertani (LB) broth, concentrated by centrifugation, and stored at 4°C until use. The worms were maintained at 20°C on solid nematode growth medium (NGM), which had been seeded with live E. coli OP50, according to standard procedures. For the PA14 killing assay, synchronized worms were grown on NGM with 0, 250, 500, or 750 μg/ml of CCWE or 200 μg/ml of K-CGN in the medium from early L1 stage to young adult stage. The day 1 adults were treated with 75 μM fluorodeoxyuridine (FUdR; Sigma) for 3 h before being transferred onto modified NGM plates (22) with a preestablished PA14 lawn. Ten microliters of fresh PA14 culture was used to establish a PA14 bacterial lawn in the presence of 0, 250, 500, or 750 μg/ml of CCWE or 200 μg/ml of K-CGN. FUdR was present at the same concentration in each plate to prevent egg hatching. After 8 h of incubation at 37°C to allow the establishment of a PA14 bacterial lawn, open plates and lids were kept in a biosafety hood at room temperature for 20 to 30 min to remove excessive moisture. This step was crucial to prevent the worms from avoiding the pathogen. Approximately 30 worms were transferred to each plate and counted. PA14 bacteria with or without the various concentrations of CCWE were spread onto the central area (diameter, 2.5 cm) of the 3.5-cm plates to ensure effective exposure of worms to the pathogen and also to prevent the worms from crawling onto the sides of the plates. Worms which were transferred to new plates with an established OP50 lawn served as a control for the environmental condition. All plates were kept at 25°C and observed under a dissection microscope to count the number of live and dead worms every 24 h until all worms in the control plates were killed. Worms were scored as dead when they failed to respond to a gentle touch by a platinum worm picker. Worms which died due to being stuck on the side of plates were excluded from analysis. The protection effect of CCWE was tested with the PA14 killing assay under three experimental conditions: (i) CCWE was used as a food supplement only, where CCWE was added to the worms as a supplement during their developmental stage before being subjected to PA14 infection; (ii) CCWE was used as a PA14 pathogenicity inhibitor only, where worms were raised without CCWE supplementation and were then exposed to the PA14 lawn which was preestablished in the presence of CCWE; and (iii) CCWE was used as both a food supplement and a PA14 pathogenicity inhibitor, where CCWE was present during the worm developmental stage and the PA14 infection stage. The third condition was designed to test for any synergistic effects. All experiments were performed in three biological replicates.
Infection experiments and gene expression analysis of C. elegans and PA14.
Pathogen infection experiments were performed as described above for the killing assay, except that approximately 100 worms were transferred to each plate and harvested at 3 h, 6 h, 24 h, or 48 h postexposure to the PA14 lawn. The worms were washed 3 times in M9 buffer to eliminate excess bacteria. The tubes with worms were briefly chilled on ice to acquire stacked worms, and the buffer was removed by pipetting. Prior to the gene expression analysis of PA14, bacteria with an initial OD600 of 0.02 were cultured at 37°C in KB broth in the presence (treatment) or absence (control) of 500 μg/ml CCWE with constant shaking. The overnight culture was centrifuged for 10 min at 1,500 × g to pellet the bacteria. Total RNA was extracted from stacked worms or bacterial pellets with TRIzol reagent (Invitrogen) and an RNeasy RNA kit (Qiagen) following the manufacturer's protocol. The integrity and quantity of the RNA were assessed by agarose gel electrophoresis and with a NanoDrop ND-2000 spectrophotometer (NanoDrop Technologies Wilmington, DE), respectively. For gene expression analysis, RNA samples derived from three biological replicates of each treatment were pooled. From 2 μg of total RNA, cDNA was synthesized using a High Capacity cDNA reverse transcription kit (Applied Biosystems). Real-time quantitative PCR (qPCR) was performed on a StepOne real-time PCR system (Applied Biosystems) using Promega GoTaq SYBR green reagent (Roche Diagnostics, Mississauga, ON, Canada) with 0.2 μM each gene-specific primer and 10 ng of cDNA as the template. Each pooled sample, representing three biological replicates, was run in a reaction mixture with a final volume of 10 μl in triplicate, following the manufacturer's instructions. For gene expression analysis of C. elegans, ama-1 or nhr-23, whichever was more stably expressed in the specific sample sets, was used as an endogenous control. For PA14 gene expression, 16S rRNA served as an endogenous control. The sequences of the primers are listed in Table S2 in the supplemental material.
Total protease assay.
PA14 was cultured in the presence (treatment) or absence (control) of 500 μg/ml of CCWE, and protease activity was determined spectrophotometrically at 600 nm by measurement of the skim milk hydrolysis efficacy of the protease secreted in the culture. The protease activity was expressed as mg of protein hydrolyzed per ml culture per hour, using a standard curve generated from a serial dilution of skim milk (skim milk powder for microbiology; BDH) (23).
Alkaline protease assay.
The alkaline protease activity of the supernatant of PA14, cultured in the presence (treatment) or absence (control) of 500 μg/ml of CCWE, was assayed using Hide-Remazol brilliant blue fibrous powder (Sigma). The alkaline protease activity was expressed as units where 1 unit was defined as an increase of 1.0 in the OD595 per ml per hour (24).
Elastase assay.
The elastase activity of PA14, cultured in the presence (treatment) or absence (control) of 500 μg/ml of CCWE, was determined using elastin-Congo red powder (Sigma). Elastase activity was expressed as the increase in the OD495 per ml of PA14 filtrate (25).
HCN assay.
Hydrogen cyanide (HCN) production by PA14 in the presence (treatment) or absence (control) of 500 μg/ml of CCWE was visualized by the changing color of sodium picrate due to the reduction activity of HCN (26). Filter paper disks were saturated with sodium picrate and attached onto the lower side of the lid of culture plates which were seeded with PA14. The plates were sealed and incubated prior to elution, and the brownish yellow compound on the discs was spectrophotometrically quantified. The quantity of the corresponding HCN was expressed as the OD625 (27).
Pyocyanin assay.
Pyocyanin in 3 ml of filtrate of PA14, cultured in the presence or absence of 500 μg/ml of CCWE, was extracted with chloroform-HCl and quantified by determination of the absorbance at 520 nm, as previously described (28), with minor modifications.
Siderophore assay.
The PA14 siderophore in a 20-ml culture filtrate with (treatment) or without (control) 500 μg/ml of CCWE was extracted in ethyl acetate, dried with a nitrogen evaporator (N-EVAP; Efamol Research Inc., Canada), dissolved in ethanol, mixed with Hathway's reagent, and subjected to reading of the absorbance at 700 nm. The quantity of siderophore was expressed as the OD700 of the culture filtrate (29).
Microtiter plate biofilm assay.
The relative quantity of biofilm was determined by reading the OD590 on a microplate reader (BioTek) (30). The PA14 biofilm which was formed in the presence (treatment) or absence (control) of 500 μg/ml of CCWE was stained with crystal violet and washed, after which the absorbance of the ethanol-eluted biofilm-conjugated stain was determined. The quantity of biofilm was expressed as the OD590 per ml of the filtrate.
Statistical analyses.
The statistical analyses were performed using SPSS software, version 15.0. The comparison of survival curve data was carried out using the log-rank test of the Kaplan-Meier survival function, while an independent t test was applied to the analysis of the other data. Differences were considered significant when P was <0.05.
RESULTS
CCWE and K-CGN protect C. elegans from PA14 infection.
Wild-type C. elegans N2 worms were cultured with CCWE as a food supplement from the early L1 stage. On day 1 of adulthood, the worms were exposed to PA14 infection in the presence or absence of CCWE in the culture medium. The survival rate of the worms was recorded every 24 h. Worms cultured without CCWE supplementation, or plain worms, followed by plain PA14 (PA14 cultured without CCWE) exposure, served as the control. It was found that CCWE treatment resulted in survival rates higher than those for the control when CCWE was used either as a worm food supplement (P < 0.0001) (see Fig. S1A and Table S1 in the supplemental material) or as an inhibitor of PA14 pathogenicity in the culture medium (P < 0.0001) (see Fig. S1B and Table S1 in the supplemental material). Interestingly, the combination of the two treatments protected a larger fraction of the worms from bacterial infection (Fig. 1 and Table 1). Specifically, all untreated worms died at 120 h, while 500 μg/ml of CCWE protected up to 28% of the worms against the lethal effect of PA14 infection (P < 0.0001). Although the treatment with 750 μg/ml of CCWE as a food supplement resulted in a better survival rate than treatment with 500 μg/ml of CCWE as a food supplement (see Fig. S1A in the supplemental material), 250 or 750 μg/ml of CCWE showed less efficacy for protection in both the inhibition of PA14 pathogenicity (see Fig. S1B in the supplemental material) and the synergism (Fig. 1) experiments. CCWE at concentrations between 250 and 750 μg/ml did not negatively affect the growth or development of the animal (data not shown). On the basis of these observations, 500 μg/ml was identified as the optimum CCWE concentration for protection of the worms from the lethal effect of PA14 infection; therefore, this dosage was used in all further assays.
Fig 1.
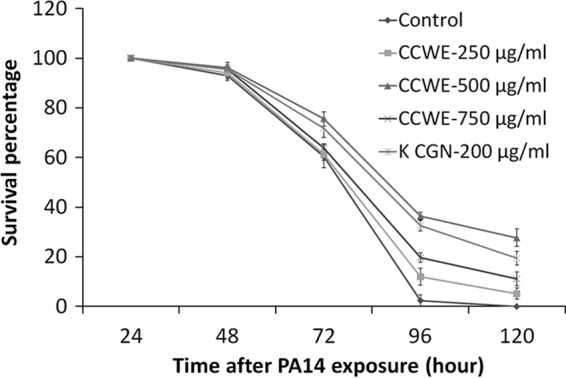
CCWE- and K-CGN-induced survival of C. elegans wild-type worms against P. aeruginosa PA14 infection. Various concentrations of CCWE and K-CGN were tested in comparison with a negative control (water). Day 1 adult N2 worms raised with CCWE or K-CGN as a food supplement were exposed to a PA14 bacterial lawn which was established in the presence of CCWE or K-CGN. Data are presented as the mean ± SD. See Table 1 for statistical data.
Table 1.
CCWE/K-CGN protects wild-type C. elegans against P. aeruginosa PA14 infection
| Treatmenta | No. of C. elegans worms |
Mean ± SE survival time (h) | P value vs controlb | |
|---|---|---|---|---|
| Total | Censored | |||
| Control | 93 | 0 | 85.2 ± 1.66 | |
| CCWE250 | 98 | 6 | 89.1 ± 1.78 | 0.063 |
| CCWE500 | 109 | 31 | 98.4 ± 1.96 | <0.0001 |
| CCWE750 | 111 | 13 | 91.5 ± 1.83 | 0.004 |
| K-CGN200 | 92 | 18 | 96.0 ± 2.17 | <0.0001 |
CCWE250, CCWE500, and CCWE750, 250, 500, or 750 μg/ml of Chondrus crispus water extract, respectively; K-CGN200, 200 μg/ml of kappa-carrageenan.
P values were determined by the log-rank test.
The components of CCWE were investigated for their effects on host immunity induction. On the basis of the previously reported immune-modulating effects of CGN and the fact that CGN constituted 40% of the weight of our CCWE preparation (31), we reasoned that the predominant water-soluble polysaccharide in CCWE, K-CGN, might have played a role in the observed protection of C. elegans against PA14 infection. Consequently, the effect of pure K-CGN was tested using the C. elegans infection model. The purity of the K-CGN sample used in the present work was confirmed by proton nuclear magnetic resonance on a Bruker 700-MHz spectrometer (see Fig. S2 in the supplemental material). To match the optimal concentration of CCWE, 200 μg/ml of K-CGN was used. The protective effects of K-CGN observed were similar to the effects previously observed with CCWE (Fig. 1 and Table 1), especially during the period from 24 h to 96 h, with a 20% protection rate at 120 h (P < 0.0001). Thus, the K-CGN component in CCWE could have played an important role in the induction of host immunity in the presence of a PA14 challenge. To rule out the possibility that the observed effects are a result of a hormetic effect of CCWE and K-CGN, we tested the toxicity of CCWE and K-CGN. Results suggest that they were not toxic to the nematode, as both CCWE and K-CGN extended the life span, decreased the aging, increased the reproduction, and improved the overall physiology of the worms (see Fig. S3 and S4 in the supplemental material). Taken together, the data suggest a general health-promoting effect of CCWE and K-CGN in C. elegans under noninfection conditions and imply a potential immune modulation effect in response to PA14 infection.
The daf-2/daf-16, pmk-1, and skn-1 signaling pathways are essential for K-CGN-induced protection against PA14 infection.
Mutants with one of four loss-of-function mutations, i.e., mutants which were defective in one of the daf-2, daf-16, pmk-1, or skn-1 immune response pathways, were tested to investigate which immune pathway was important for the K-CGN-induced protection against lethal infection with PA14. As shown in Fig. 2, K-CGN treatment did not protect any of the mutants, while it protected N2 worms, which suggested that functional daf-2, daf-16, pmk-1, and skn-1 are essential in K-CGN-mediated immunity against PA14.
Fig 2.
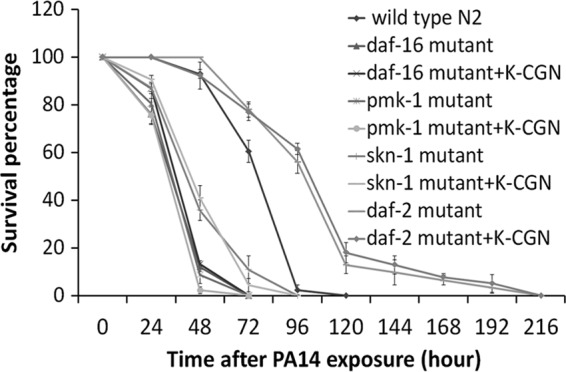
K-CGN-induced survival was impaired in daf-16, daf-2, pmk-1, and skn-1 mutants. K-CGN was supplemented to the food source of worms from early L1 stage to adulthood. Day 1 adult worms were then transferred to a P. aeruginosa PA14 bacterial lawn which was preestablished in the presence of K-CGN. Worms raised without K-CGN supplement exposed to a PA14 lawn established in the absence of K-CGN served as a control. P was >0.05 for each mutant versus the same mutant plus K-CGN. Data are presented as the mean ± SD.
CCWE and K-CGN induce immune response genes in C. elegans under both PA14 infection and noninfection conditions.
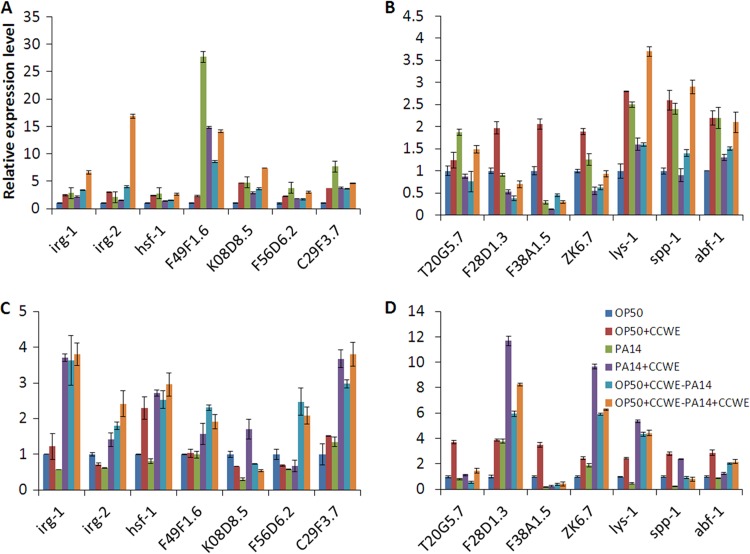
Immune response gene regulation may be responsible for the CCWE- and K-CGN-induced protection of C. elegans against PA14 infection. The independent effects of CCWE and K-CGN on the expression of 14 immune response genes in PA14-infected and noninfected worms were analyzed by qPCR. The selected genes included 7 early response genes, i.e., irg-1 (infection response gene 1; C07G3.2), irg-2 (infection response gene 2; C49G7.5), F49F1.6 (ShK domain-like, secreted surface protein), hsf-1 (heat shock factor 1), K08D8.5 (CUB-like domain), F56D6.2 (C-type lectin), and C29F3.7 (CUB-like domain) (20, 32), and 7 late response genes, i.e., T20G5.7 (matridin SK domain protein), F28D1.3 (thaumatin-like protein), F38A1.5 (lectin family protein), ZK6.7 (lypase), lys-1 (lysozyme-like protein), spp-1 (saponin-like protein), and abf-1 (antibacterial protein) (21, 33). Worms fed E. coli OP50, the standard laboratory food, were used as a noninfection control, while worms fed OP50 prior to exposure to PA14 served as the control for infection conditions. Under the noninfection conditions, at 6 h of adulthood, all tested immune genes except T20G5.7 were upregulated by 2- to 5-fold by CCWE compared to the level of regulation for the control (P < 0.0001) (Fig. 3A and B). At 24 h of adulthood, half of the genes, such as the early response genes hsf-1 and C29F3.7 (Fig. 3C) and the late response genes T20G5.7, F28D1.3, F38A1.5, ZK6.7, lys-1, spp-1, and abf-1 (Fig. 3D), were significantly upregulated compared to the level of regulation for the control (P < 0.05) (Fig. 3C and D). Thus, when the host was not challenged by the PA14 infection, food supplementation with CCWE resulted in elevated immunity of the host. However, immune genes were found to be expressed differentially at various time points postinfection. At 6 h after infection, the majority of the immune genes were significantly activated by PA14 infection; on the other hand, the genes were not further activated by CCWE, except for irg-1, irg-2, and lys-1 (Fig. 3A and B). On the contrary, at 24 h postinfection, except for C29F3.7, F28D1.3, and ZK6.7, all genes were suppressed in PA14-infected worms compared to their expression in the control worms fed OP50. Notably, at this time point, CCWE treatment of either worms or PA14, or both worms and PA14, resulted in a significant activation of all 14 immune genes in PA14-infected worms. The CCWE-activated genes with more than 2-fold changes under infection conditions were as follows: irg-1, irg-2, F49F1.6, hsf-1, F56D6.2, C29F3.7, F28D1.3, ZK6.7, lys-1, spp-1, and abf-1. The expression of K05D8.5, T20G5.7, and F38A1.5 was apparently less affected by CCWE (P < 0.01) (Fig. 3C and D). Overall, CCWE enhanced immunity in C. elegans. K-CGN activated most of the immune genes tested under both infection and noninfection conditions at both time points (6 h and 24 h) (see Fig. S5 in the supplemental material), in a pattern similar to that of CCWE treatment.
Fig 3.
CCWE-induced immune responses in wild-type C. elegans. N2 worms were grown with or without dietary supplementation with CCWE, and expression of immune genes was analyzed by qPCR at 6 h (A and B) or 24 h (C and D) of adulthood or postexposure to P. aeruginosa PA14 that was preestablished with or without CCWE. Worms fed Escherichia coli strain OP50, the standard laboratory food source, were used as a control for noninfection conditions, and OP50-fed PA14-infected worms were used as a control for infection conditions. OP50, worms fed OP50; OP50 + CCWE, worms fed OP50 with CCWE as food supplement; PA14, worms grown with OP50 and exposed to PA14; PA14 + CCWE, worms grown with OP50 and exposed to PA14 which was cultured in the presence of CCWE; OP50 + CCWE-PA14, worms raised on OP50 with CCWE as a food supplement and exposed to PA14; OP50 + CCWE-PA14 + CCWE, worms raised on OP50 with CCWE as food supplement exposed to PA14 which was cultured in the presence of CCWE. Data are presented as the mean ± SD. P was <0.0001 for OP50 versus OP50 + CCWE for all genes except T20G5.7 (A and B); P was <0.0001 for OP50 versus OP50 + CCWE for all late immune response genes (D); P was < 0.05 for OP50 versus OP50 + CCWE for hsf-1 and C29F3.7 (C); P was <0.0001 for PA14 versus PA14 + CCWE, OP50 + CCWE-PA14, or OP50 + CCWE-PA14 + CCWE for irg-1, irg-2, and K08D8.5 (A) and for F38A1.5, lys-1, and spp-1 (B); P was <0.01 for PA14 versus PA14 + CCWE, OP50 + CCWE-PA14, or OP50 + CCWE-PA14 + CCWE for all genes (C and D).
CCWE suppresses expression of quorum-sensing and virulence factor genes of PA14.
Besides the enhanced immunity of the host, another mechanism which may act to attenuate the death of worms following PA14 exposure is the inhibition of virulence factors by CCWE treatment. Therefore, the effect of CCWE on the expression of the quorum-sensing genes and the virulence factor genes of PA14 was investigated. CCWE repressed the expression of a number of QS and virulence factor genes in PA14, without affecting its housekeeping genes (Fig. 4). The QS genes, namely, lasI, lasR, rhlI, and rhlR, were repressed by 4- to 20-fold (P < 0.0001). Likewise, virulence factor genes, including hcnC, aroE, rpoN, sbe, and sodB, were downregulated by approximately 2- to 50-fold with treatment with 500 μg/ml of CCWE in the culture medium (P < 0.01).
Fig 4.
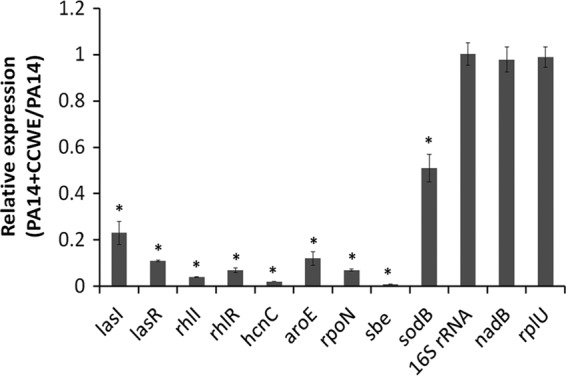
CCWE suppressed the quorum-sensing and virulence factor genes of P. aeruginosa PA14 but not the housekeeping genes. Gene expression of PA14 cultured in the presence of CCWE was quantified by qPCR and compared with that of PA14. Data are presented as the mean ± SD. *, P < 0.01 for PA14 versus PA14 plus CCWE.
Biochemical evidence for inhibition of the virulence factors of PA14 by CCWE.
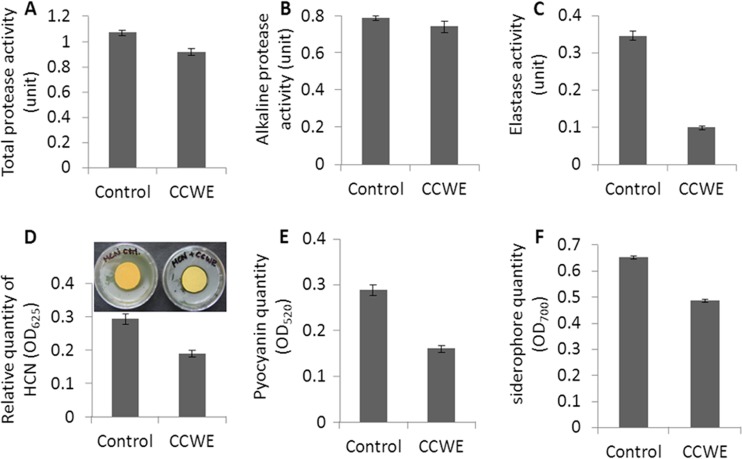
CCWE significantly downregulated PA14 virulence factor genes; therefore, quantification of PA14 virulence factors was undertaken. Biochemical assays demonstrated an overall reduction of the virulence factors by CCWE treatment (P < 0.01) (Fig. 5). The activity of the pathogen-secreted enzyme total protease was significantly inhibited by CCWE treatment (P = 0.001), while the alkaline protease activity was not apparently affected (P = 0.07) (Fig. 5A and B). Another PA14-secreted enzyme, elastase, showed a 3-fold decrease in activity with CCWE treatment (P < 0.0001) (Fig. 5C). Additionally, all 4 PA14-secreted toxins were significantly reduced (P < 0.01) (Fig. 5D to F). Specifically, in the presence of CCWE in the culture medium, PA14 produced 30% less HCN than the control, which was visualized by the lighter color of the sodium picrate-saturated filter paper in the CCWE-treated PA14 plates (P = 0.001) (Fig. 5D). In addition, pyocyanin production was reduced by as much as 45% in CCWE-treated PA14 (P < 0.0001) (Fig. 5E). Similarly, PA14 secretion of siderophore was reduced by 25% by CCWE treatment (P < 0.0001) (Fig. 5F).
Fig 5.
Biochemical assays of PA14-secreted virulence factors. (A) Effect of CCWE on the total protease activity of PA14; (B) effect of CCWE on the alkaline protease activity of PA14; (C) effect of CCWE on the elastase activity of PA14; (D) effect of CCWE on HCN production in PA14; (E) effect of CCWE on pyocyanin production in PA14; (F) effect of CCWE on siderophore production in PA14. Control, no CCWE was added to the PA14 culture; CCWE, PA14 was cultured with CCWE. The experiments were conducted with three biological replicates and three technical replicates. Data are presented as the mean ± SD. P values, 0.001, 0.07, <0.0001, 0.001, <0.0001, and <0.0001 (for the control versus CCWE) for panels A to F, respectively.
CCWE inhibits biofilm formation of PA14.
The presence of biofilms affects the pathogenicity of PA14. The effect of CCWE on biofilm formation was studied by crystal violet staining. When 500 μg/ml of CCWE was present in the culture medium, PA14 formed 0.09 unit of biofilm, which was a 3-fold decrease compared to the 0.29 unit formed by the control (P = 0.001).
DISCUSSION
The clinical isolate P. aeruginosa PA14 is pathogenic to both Arabidopsis thaliana and severely burned mice, with PA14 infection in the latter case being lethal (34). The pathogenicity in both the plant and animal models was mediated by a number of virulence factors, including toxA, plcS, and gacA. Interestingly, PA14 and another strain of P. aeruginosa, PAO1, were demonstrated to kill the soil nematode C. elegans in either hours or days, depending on the type of growth medium used in the killing assay (12, 35). In the present study, we utilized the C. elegans infection model to investigate the effects of a water extract from a commercially cultivated strain of the red seaweed C. crispus on both host immunity and P. aeruginosa pathogenicity. We observed that 500 μg/ml of CCWE exerted the best protection against PA14 killing for the worms, while higher or lower concentrations were less efficient. This result was consistent with what had previously been observed for an extract from the commercially harvested brown seaweed, A. nodosum, in a similar model system (21).
Thriving in coastal waters, seaweeds have evolved robust defense mechanisms, for example, inhibition of QS, against pathogens, such as the ubiquitous bacterium P. aeruginosa. Strikingly, CCWE presented properties as an inhibitor of QS in pathogenic strain PA14, which suggested that the red seaweed C. crispus per se can be of benefit for applications broader than just a functional food alone. In the growth curve assays, CCWE and its major component, K-CGN, did not abolish the growth of PA14 (see Fig. S6 in the supplemental material). In line with this, in a disk diffusion test, CCWE showed no direct antimicrobial activity (see Fig. S7 in the supplemental material). However, CCWE repressed the expression of PA14 QS genes and reduced the levels of PA14 virulence factors secreted, without affecting its housekeeping genes (Fig. 4 and 5). Interestingly, in another study which screened 30 species of green, brown, and red seaweeds, quorum-sensing inhibitors were shown to be present only in the red seaweed Asparagopsis taxiformis (36); another species of red alga, Delisea pulchra, was the first seaweed reported to contain compounds with anti-QS activity (37).
Another aspect of the C. elegans infection model was that the endpoint of PA14-inflicted killing for the control N2 worms was no earlier than 120 h postexposure, which was later than that described in previous reports (12, 38). An explanation for the slower killing can be the use of FUdR in the present study. FUdR blocks the midproliferation stage of the embryonic development of C. elegans (39), thus preventing eggs from hatching, which helped to avoid the confounding effects of progeny production. Immunity suppression in C. elegans was shown to be correlated with reproduction, or, more precisely, normal embryonic development. It was reported that sterile mutants of C. elegans, when placed under PA14 infection conditions, survived longer than wild-type N2 worms, and in accordance with this, FUdR-treated worms also survived longer (38).
C. elegans exhibited differential immune responses to PA14 at early (i.e., 6 h) and late (i.e., 24 h) stages of infection in this study. At 6 h postinfection, the majority of the tested immune genes were upregulated, while at 24 h postexposure to PA14, most genes were repressed. This finding was largely consistent with previous reports. For example, 4 h of exposure of C. elegans to PA14 resulted in activation of immune response genes, such as irg-1, irg-2, F49F1.6, K08D8.5, F56D6.2, and others (20). In addition, lys-1 and another two lysozyme genes, as well as the gene for a lipase, ZK6.7, were demonstrated to be inducible in C. elegans by the pathogenic bacterium Serratia marcescens (40). Similarly, infection of C. elegans by the Gram-positive bacterium Microbacterium nematophilus was correlated with the upregulation of a set of immune effectors, including, but not restricted to, F49F1.6, F56D6.2, and lysozymes (Lys3 and Lys7) (41). Despite the abundance of studies supporting immune induction by infection with a pathogenic organism, there is evidence showing immune suppression at a later time point of pathogen exposure. For instance, at 12 h of exposure to PA14, immune response genes such as spp-1, lys-7, and thn-2 were suppressed in C. elegans by activation of the DAF-2 insulin-like signaling pathway (33). Host immune suppression may represent one of the many strategies that the pathogen has evolved to counteract host immune defenses through a yet unknown mechanism (33, 42). However, research has shed light on both sides of the pathogen-host interaction: mounting of immune defense by the host and suppression of host immunity by the pathogen(s). It is possible that one side overcomes the other at certain time points during infection (32, 40, 41) and that the outcome of the battle can be regulated by external intervention. Indeed, in this study it was demonstrated at 24 h postexposure that a water extract of the commercially cultivated strain of the red seaweed C. crispus (CCWE) blocked the immune repression involving a majority of the tested immune response genes in C. elegans induced by PA14. To better understand the function of these early and late immune response genes, we also examined their expression at 3 h and 48 h postexposure (see Fig. S8 in the supplemental material) and showed overall activation of the early immune response genes to PA14 infection and less activation of the late immune response genes. Strikingly, it is evident that CCWE enhances the immune activation or counteracts the immune suppression induced by pathogenic bacteria at various time points during the course of infection (Fig. 3; see Fig. S8 in the supplemental material). This is also associated with the suppression of QS and virulence genes, which contributes to an elevated rate of survival of the host. Surprisingly, K-CGN, a water-soluble polysaccharide present in Chondrus, induced a very similar pattern of host immune response in the N2 worms, which implies an important role for K-CGN in the CCWE-induced immunity. However, we did not observe a protective effect of K-CGN in the mutants, but a protective effect of CCWE cannot be ruled out, as it contains other bioactive components, albeit in less abundance.
Notably, in our present work, the selected immune genes were regulated by at least four distinct signaling pathways. For example, the infection response genes irg-1 and irg-2 are mediated by the zip-2 pathway, while the ShK domain-like secreted surface protein F49F1.6 and the C-type lectin F56D6.2 are regulated by the PMK-1 pathway (20). Additionally, the lysozyme-like protein Lys-1 is regulated by both TGF-β and PMK-1 signaling pathways, whereas the CUB-like protein K08D8.5 is under the regulation of both the PMK-1 and DAF-2/DAF-16 insulin-like pathways (43). Moreover, the sapsonin-like protein Spp-1 was thought to be regulated by the DAF-2/DAF-16 pathway, together with an as yet unknown pathway (33). Thus, the regulation of immune response genes by various known and yet unknown signaling pathways may further account for the differential expression of the immune response genes.
Despite the differential expression of immune genes in C. elegans worms with PA14 infection, under noninfection conditions, CCWE and K-CGN were observed to enhance the expression of the tested immune response genes at both 6 h (for the early immune response genes) and 24 h (for the late immune response genes) of adult worms, again suggesting an important role of CCWE and K-CGN in the positive modulation of immunity in the animals. The immune modulation effect of CCWE may have resulted from a variety of the bioactive compounds that it contains, among which K-CGN predominates. Although low-molecular-weight CGN has been related to a higher incidence of tumors in animals treated with chemical carcinogens (44), a large amount of research supports the beneficial effects of native or undegraded CNGs (6, 8, 45). The controversial effect of CGN might be attributed to the following factors: first, studies are usually undertaken with commercially available products with mixtures of CGNs of various sources and various degrees of purity and/or fractionation. In line with this, a purified iota-CGN, but not lambda-CGN, was previously observed to protect Arabidopsis thaliana from infection with a pathogenic organism (46). Second, animals of different ages may have diverse responses to various routes of administration and dosages (44). The work presented here, using the model animal C. elegans, supports a beneficial effect of CCWE and K-CGN in host immune modulation. Further studies are required to elucidate the mechanism(s) of the immune modulation effect of CCWE and CGNs.
Supplementary Material
ACKNOWLEDGMENTS
The research group of B.P. is supported by the Natural Sciences Engineering Council of Canada (NSERC), Acadian Seaplants Limited, and the Atlantic Canada Opportunities Agency (ACOA)—Atlantic Innovation Fund (AIF). We are most grateful for the NSERC Industrial Research Development Fellowship (IRDF) awarded to J.L.
Footnotes
Published ahead of print 20 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01927-13.
REFERENCES
- 1.Kumar CS, Ganesan P, Suresh PV, Bhaskar N. 2008. Seaweeds as a source of nutritionally beneficial compounds—a review. J. Food Sci. Technol. 45:1–13 [Google Scholar]
- 2.Holdt SL, Kraan S. 2011. Bioactive compounds in seaweed: functional food applications and legislation. J. Appl. Phycol. 23:543–597 [Google Scholar]
- 3.Okai Y, Higashi-Okai K, Yano Y, Otani S. 1996. Identification of anti-mutagenic substances in an extract of edible red alga, Porphyra tenera (Asakusa-nori). Cancer Lett. 100:235–240 [DOI] [PubMed] [Google Scholar]
- 4.Maruyama H, Watanabe K, Yamamoto I. 1991. Effect of dietary kelp on lipid peroxidation and glutathione per-oxidase activity in livers of rats given breast carcinogen DMBA. Nutr. Cancer 15:221–228 [DOI] [PubMed] [Google Scholar]
- 5.Yang YJ, Nam S-J, Kong G, Kim MK. 2010. A case-control study on seaweed consumption and the risk of breast cancer. Br. J. Nutr. 103:1345–1353 [DOI] [PubMed] [Google Scholar]
- 6.Chopin T, Gallant T, Davison I. 1995. Phosphorus and nitrogen nutrition in Chondrus crispus (Rhodophyta): effects on total phosphorus and nitrogen content, carrageenan production, and photosynthetic pigments and metabolism. J. Phycol. 31:283–293 [Google Scholar]
- 7.Skoler-Karpoff S, Ramjee G, Ahmed K, Altini L, Plagianos MG, Friedland B, Govender S, Kock AD, Cassim N, Palanee T. 2008. Efficacy of Carraguard for prevention of HIV infection in women in South Africa: a randomised, double-blind, placebo-controlled trial. Lancet 372:1977–1987 [DOI] [PubMed] [Google Scholar]
- 8.Luescher-Mattli M. 2003. Algae, a possible source for new drugs in the treatment of HIV and other viral diseases. Curr. Med. Chem. 2:219–225 [Google Scholar]
- 9.Zhou G, Sheng W, Yao W, Wang C. 2006. Effect of low molecular lambda-carrageenan from Chondrus ocellatus on anti-tumor H-22 activity of 5-Fu. Pharmacol. Res. 53:129–134 [DOI] [PubMed] [Google Scholar]
- 10.Shiau S-Y, Chang GW. 1986. Effects of certain dietary fibers on apparent permeability of the rat intestine. J. Nutr. 116:223–232 [DOI] [PubMed] [Google Scholar]
- 11.Wood RE. 1976. Pseudomonas: the compromised host. Hosp. Pract. 11:91–100 [DOI] [PubMed] [Google Scholar]
- 12.Tan MW, Rahme LG, Sternberg J, Tompkins RG, Ausubel FM. 1999. Pseudomonas aeruginosa killing of Caenorhabditis elegans used to identify P. aeruginosa virulence factors. Proc. Natl. Acad. Sci. U. S. A. 96:2408–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J. Bacteriol. 21:6454–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith RS, Iglewski BH. 2003. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6:56–60 [DOI] [PubMed] [Google Scholar]
- 15.Gander S. 1996. Bacterial biofilms: resistance to anti-microbial agents. J. Antimicrob. Chemother. 37:1047–1050 [DOI] [PubMed] [Google Scholar]
- 16.Juhas M, Eberl L, Tummler B. 2005. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ. Microbiol. 7:459–471 [DOI] [PubMed] [Google Scholar]
- 17.Ewbank JJ. 23 January 2006. Signaling in the immune response. In The C. elegans Research Community (ed), WormBook. 10.1895/wormbook.1.83.1 http://www.wormbook.org/chapters/www_signalingimmuneresponse/signalingimmuneresponse.html. [DOI] [PMC free article] [PubMed]
- 18.Mochii M, Yoshida S, Morita K, Kohara Y, Ueno N. 1999. Identification of transforming growth factor-β-regulated genes in Caenorhabditis elegans by differential hybridization of arrayed cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:15020–15025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gravato-Nobre MJ, Hodgkin J. 2005. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell. Microbiol. 7:741–751 [DOI] [PubMed] [Google Scholar]
- 20.Estes KA, Dunbar TL, Powell JR, Ausubel FM, Troemel ER. 2010. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 107:2153–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kandasamy S, Khan W, Evans F, Critchley AT, Prithiviraj B. 2012. Tasco®: a product of Ascophyllum nodosum enhances immune response of Caenorhabditis elegans against Pseudomonas aeruginosa infection. Mar. Drugs 10:84–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sulston J, Hodgkin J. 1988. Methods, p 587 In Wood WB. (ed), The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 23.Dow JM, Clarke BR, Milligan DE, Tang JL, Daniels MJ. 1990. Extracellular proteases from Xanthomonas campestris pv. campestris, the black rot pathogen. Appl. Environ. Microbiol. 56:2994–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howe TR, Iglewski BH. 1984. Isolation and characterization of alkaline protease-deficient mutants of Pseudomonas aeruginosa in vitro and in a mouse eye model. Infect. Immun. 43:1058–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohman DE, Cryz SJ, Iglewski BH. 1980. Isolation and characterization of Pseudomonas aeruginosa PAO mutant that produces altered elastase. J. Bacteriol. 142:836–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadasivam S, Manickam A. 1996. Biochemical methods, 1st ed. New Age International Ltd., New Delhi, India [Google Scholar]
- 27.Miller RL, Higgins VJ. 1970. Association of cyanide with infection of birdfoot trefoil Stemphylium loti. Phytopathology 60:104–110 [Google Scholar]
- 28.Denervaud V, Tuquoc P, Blanc D, Favre-Bonte S, Krishnapillai V, Remmann C, Heas D, Delden CV. 2004. Characterization of cell-to-cell signaling-deficient Pseudomonas aeruginosa strains colonizing intubated patients. J. Clin. Microbiol. 42:554–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeves M, Pine L, Neilands JB, Bullows A. 1983. Absence of siderophore activity in Legionella species grown in iron-deficient media. J. Bacteriol. 154:324–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shanks RMQ, Donegan NP, Graber ML, Buckingham SE, Zegans ME, Cheung AL, O'Toole GA. 2005. Heparin stimulates Staphylococcus aureus biofilm formation. Infect. Immun. 73:4596–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Craigie JS, Leigh C. 1978. Carrageenans and agars, p 109–131 In Hellebust JA, Craigie JS. (ed), Handbook of phycological methods: physiological and biochemical methods. Cambridge University Press, New York, NY [Google Scholar]
- 32.Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM. 2006. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2:e183. 10.1371/journal.pgen.0020183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans EA, Kawli T, Tan MW. 2008. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog. 4:e1000175. 10.1371/journal.ppat.1000175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahme LG, Stevens EJ, Wolfort SF, Shao J, Tompkins RG, Ausubel FM. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899–1902 [DOI] [PubMed] [Google Scholar]
- 35.Darby C, Cosma CL, Thomas JH, Manoil C. 1999. Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:15202–15207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jha B, Kavita K, Westphal J, Hartmann A, Schmitt-Kopplin P. 2013. Quorum sensing inhibition by Asparagopsis taxiformis, a marine macroalga: separation of the compound that interrupts bacterial communication. Mar. Drugs 11:253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Givskov M, DeNys R, Manefield M, Gram L, Maximilien R, Eberl L, Molin S, Steinberg P, Kjelleberg S. 1996. Eukaryotic interference with homoserine lactone-mediated prokaryotic signaling. J. Bacteriol. 178:6618–6622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyata S, Begun J, Troemel ER, Ausubel FM. 2008. DAF-16-dependent suppression of immunity during reproduction in Caenorhabditis elegans. Genetics 178:903–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stroeher VL, Kennedy BP, Millen KJ, Schroeder DF, Hawkins MG. 1994. DNA-protein interactions in the Caenorhabditis elegans embryo: oocyte and embryonic factors that bind to the promoter of the gut-specific ges-1 gene. Dev. Biol. 163:367–380 [DOI] [PubMed] [Google Scholar]
- 40.Mallo GV, Kurz CL, Couillault C, Pujol N, Granjeaud S. 2002. Inducible antibacterial defense system in C. elegans. Curr. Biol. 12:1209–1214 [DOI] [PubMed] [Google Scholar]
- 41.O'Rourke D, Baban D, Demidova M, Mott R, Hodgkin J. 2006. Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 16:1005–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Apidianakis Y, Mindrinos MN, Xiao W, Lau GW, Baldini RL. 2005. Profiling early infection responses: Pseudomonas aeruginosa eludes host defenses by suppressing antimicrobial peptide gene expression. Proc. Natl. Acad. Sci. U. S. A. 102:2573–2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA. 2007. Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol. Cell. Biol. 27:5544–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tobacman JK. 2001. Review of harmful gastrointestinal effects of carrageenan in animal experiments. Environ. Health Perspect. 109:983–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiner ML, Nuber D, Blakemore WR, Harriman JF, Cohen SM. 2007. A 90-day dietary study on kappa carrageenan with emphasis on the gastrointestinal tract. Food Chem. Toxicol. 45:98–106 [DOI] [PubMed] [Google Scholar]
- 46.Sangha JS, Ravichandran S, Prithiviraj K, Critchley AT, Prithiviraj B. 2010. Sulfated macroalgal polysaccharide iota-carrageenan induces resistance against Sclerotinia sclerotiorum in Arabidopsis thaliana, whereas lambda-carrageenan enhances susceptibility. Physiol. Mol. Plant Pathol. 75:38–45 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.