Abstract
In aquatic ecosystems, carbon (C) availability strongly influences nitrogen (N) dynamics. One manifestation of this linkage is the importance in the dissolved organic matter (DOM) pool of dissolved organic nitrogen (DON), which can serve as both a C and an N source, yet our knowledge of how specific properties of DOM influence N dynamics are limited. To empirically examine the impact of labile DOM on the responses of bacteria to DON and dissolved inorganic nitrogen (DIN), bacterial abundance and community composition were examined in controlled laboratory microcosms subjected to various combinations of dissolved organic carbon (DOC), DON, and DIN treatments. Bacterial communities that had colonized glass beads incubated in a stream were treated with various glucose concentrations and combinations of inorganic and organic N (derived from algal exudate, bacterial protein, and humic matter). The results revealed a strong influence of C availability on bacterial utilization of DON and DIN, with preferential uptake of DON under low C concentrations. Bacterial DON uptake was affected by the concentration and by its chemical nature (labile versus recalcitrant). Labile organic N sources (algal exudate and bacterial protein) were utilized equally well as DIN as an N source, but this was not the case for the recalcitrant humic matter DON treatment. Clear differences in bacterial community composition among treatments were observed based on terminal restriction fragment length polymorphisms (T-RFLP) of 16S rRNA genes. C, DIN, and DON treatments likely drove changes in bacterial community composition that in turn affected the rates of DON and DIN utilization under various C concentrations.
INTRODUCTION
Dissolved organic matter (DOM) is the largest component of the organic matter pool in lotic ecosystems (1, 2). The concentrations and compositions of DOM vary spatially and temporally (3–5), which in turn affects the productivity of stream food webs (6, 7). Heterotrophic bacteria are major consumers of DOM (8, 9), and thus, DOM influences bacterial community composition (i.e., structure) and abundance (10–12).
The processing and fate of organic carbon (C) is fundamentally and synergistically linked to the nitrogen (N) cycle (13, 14). Despite this linkage between the C and N cycles, many studies focus solely on C dynamics (15–17) or N dynamics (18–20) rather than their interrelationship. In streams, specifically, C and N are tightly linked (21), and C availability strongly influences N dynamics (22, 23), yet our knowledge of how specific properties of DOM influence N dynamics is limited.
Nitrogen is an important component of DOM (24), and a major fraction (often more than 50%) of the total dissolved N pool is dissolved organic nitrogen (DON) (25–27). The DON pool is composed of a continuum of compounds ranging from high-molecular-weight polymers, like polypeptides, to low-molecular-weight monomers, like amino acids and urea (28, 29). DON is also derived from different sources (e.g., allochthonous versus autochthonous), which influence the composition and lability (30, 31).
Allochthonous sources contribute the majority of refractory DON to streams (32), whereas alga-derived DON is more labile (33, 34). DON utilization by heterotrophic bacteria varies; the labile fraction is readily utilized (35–38), while the recalcitrant fraction is utilized with the aid of extracellular enzymes (39).
Reliance on organic versus inorganic forms of N depends on N availability. The ability to take up dissolved inorganic nitrogen (DIN) in the form of nitrate or ammonia is widespread among bacteria; ammonia uptake is energetically favorable, but often nitrate is more available (40, 41). Although DON serves as a potential N (and C) source for microbial communities (28, 29, 42), bacterial metabolism of DON is influenced by inorganic N concentrations (43). High DIN concentrations inhibit the production of enzymes that scavenge N from DON (44) and reduce extracellular hydrolysis of refractory DON (44, 45).
DOM serves as a substrate for bacterial growth (46), which in turn increases the demand for assimilation of nitrogen (21). DON serves, potentially, as both a C and an N source. The ability of DON to meet the metabolic demand for organic C likely influences the assimilative demand for N and whether, energetically, that demand is best met by N from DON or DIN. To empirically examine the impact of labile DOM on the responses of bacteria to DON and DIN, bacterial abundance and community composition (based on terminal restriction fragment length polymorphisms [T-RFLP] of 16S rRNA genes) (47) were examined in controlled laboratory microcosms subjected to various dissolved organic carbon (DOC), DON, and DIN treatments. Bacterial communities that had colonized glass beads incubated in a stream were treated with three labile C (glucose) concentrations and four combinations of inorganic and organic N. Because DON is diverse and compounds vary in bioavailability, three DON sources were examined: humic matter, bacterial protein, and algal exudate. Additional energy expenditure is required for utilization of humic matter and protein because extracellular enzyme production is required for their assimilation (44, 48, 49). In contrast, algal exudates are typically low molecular weight, more labile, and readily utilized by bacteria (50–52).
MATERIALS AND METHODS
The bacterial communities used in the experiments were grown on etched soda lime glass beads (4 mm in diameter; Fisher Scientific) that were incubated in the West Branch of the Mahoning River, Portage County, OH, USA (12, 53). The beads were autoclaved, packed in mesh bags (80-μm mesh size; WildCo Wild Life Supply, Yulee, FL), and incubated for 45 days in a riffle in the stream.
Water temperature, pH, conductivity, and dissolved oxygen (DO) were measured using a Qd/IntelliCAL Rugged Field Kit (Hach Company, Loveland, CO), and turbidity was measured with a Hach turbidometer, model 2100P, when substrates were deployed and retrieved. DOC and total nitrogen (TN) concentrations were determined using a Shimadzu TOC5000 analyzer with TNM-1 (Shimadzu Corporation, Columbia, MD).
Upon retrieval, beads from bags were pooled and divided into subsamples. A portion of the beads were frozen at −80°C for DNA extraction from the initial bacterial community, another portion were preserved with 8% paraformaldehyde in phosphate-buffered saline (pH 7.2) for bacterial enumeration, and the remainder were used in experiments as described below.
In the laboratory, each treatment and control was carried out in triplicate. Experiments were performed in flasks containing 10 g of beads in 150 ml of artificial stream water (ASW) (composition per liter, 12 mg NaHCO3, 7.5 mg CaSO4 · 2H2O, 7.5 mg MgSO4, 0.5 mg KCl, 10 mg CaCO3, 10 mg K2HPO4, pH 6.4). Treatments consisted of different N sources (and concentrations) and different organic C concentrations (Table 1). The organic N sources were bacterial protein, algal exudate, and humic matter (obtained as described below), and the source of inorganic N was NaNO3. For each N treatment, there were three glucose concentrations (low, medium, and high). N and DOC concentrations were selected based on the average concentrations in the study stream (12). Positive controls (without N but amended with low, medium, or high concentrations of glucose) were used to examine the effect of glucose amendment on bacterial communities. Negative controls (without C and N amendments) were also set up. The flasks were incubated in the dark, to prevent photochemical degradation, with shaking at 25°C and were sampled after 5 or 10 days.
Table 1.
Carbon and nitrogen treatments used in the experimenta
| Carbon treatment | Nitrogen treatment | Glucose (mg C/liter) | Organic nitrogen (mg N/liter) | Inorganic nitrogen (mg N/liter) |
|---|---|---|---|---|
| C-low | OH + IL | 0.5 | 1 | 0.5 |
| OL + IH | 0.5 | 0.5 | 1 | |
| OL + IL | 0.5 | 0.5 | 0.5 | |
| OH + IH | 0.5 | 1 | 1 | |
| C-medium | OH + IL | 10 | 1 | 0.5 |
| OL + IH | 10 | 0.5 | 1 | |
| OL + IL | 10 | 0.5 | 0.5 | |
| OH + IH | 10 | 1 | 1 | |
| C-high | OH + IL | 25 | 1 | 0.5 |
| OL + IH | 25 | 0.5 | 1 | |
| OL + IL | 25 | 0.5 | 0.5 | |
| OH + IH | 25 | 1 | 1 |
Organic nitrogen sources consisted of bacterial protein, algal exudate, and humic matter, and nitrate was the source of inorganic nitrogen. Three concentrations of organic carbon, in the form of glucose, were used for each nitrogen treatment. OH, organic nitrogen, high concentration; OL, organic nitrogen, low concentration; IH, inorganic nitrogen, high concentration; IL, inorganic nitrogen, low concentration.
Soluble bacterial proteins were obtained from cultures of Bacillus subtilis, Pseudomonas aeruginosa, and Staphylococcus aureus incubated at 27°C for 24 h. Proteins were extracted using the Qproteome Bacterial Protein Prep Kit (Qiagen, MD) according to the manufacturer's protocol, and proteins from the three bacteria were pooled. Algal exudates were prepared by growing Chlamydomonas, Chlorella, and Synedra (Carolina Biological Supplies, Burlington, NC) in ASW with 20 mg/liter of NaNO3. Cultures were grown under constant light for 35 days and processed as described below. Humic matter was derived from senescent red oak (Quercus rubra), witch hazel (Hamamelis virginiana), and corn (Zea mays) leaves extracted overnight in the dark in 0.027% NaCl and was pooled. Algal exudates and leaf leachates were filtered through GF/F filters (Whatman, Maidstone, United Kingdom) and filter sterilized with 0.02-μm Anodisc filters (Whatman, Maidstone, United Kingdom). Nonionic DAX-8 resin (Supelco; Sigma-Aldrich, MO) was used to separate the humic fraction of the leaf leachate from the nonhumic fraction. The total DON and DOC concentrations of the bacterial protein, algal exudate, and humic matter were measured with the Shimadzu TNM-1 and TOC-5000 analyzers, respectively (Shimadzu Corporation, Columbia, MD).
After 5 or 10 days, ASW from the experimental units was filtered through 0.22-μm-pore-size polycarbonate filters (Poretics, Livermore, CA) before total dissolved organic carbon (TDOC) and total nitrogen (TN) concentrations were determined as described above. Nitrate (NO3−) concentrations were measured via ion chromatography (Dionex chromatography system; Thermo Fisher Scientific Inc., CA). pH was measured with a Delta 320 pH meter (Mettler-Toledo, OH).
At each of the two sampling points, 5 g of beads and 5 ml of ASW from a given microcosm were preserved in 8% paraformaldehyde for bacterial enumeration. Subsequently, samples were treated with 0.1% tetrasodium pyrophosphate and sonicated at 40 kHz for 5 min (ultrasonic cleaner, model 2210; Branson Ultrasonics Co., Danbury, CT) to detach bacterial cells (54). Samples were then filtered through 0.2-μm black polycarbonate filters (Livermore, CA) and stained with DAPI (4,6-diamidino-2-phenylindole) (55). Bacteria in 10 fields per filter were enumerated via epifluorescence microscopy and used to calculate the total number of bacteria in each flask.
For DNA extraction, 5 g of beads from each microcosm were sonicated as described above, and bacteria were concentrated by filtration (0.22-μm-pore-size polycarbonate filters; Poretics). The filters were frozen at −80°C until DNA was extracted using a Power-Soil DNA extraction kit (MoBio Laboratories, Carlsbad, CA) following the manufacturer's instructions with minor modifications as described by Feinstein et al. (56).
PCR of the 16S rRNA genes followed by T-RFLP was used to examine the bacterial community structure. An equimolar mixture of 5′-ACTCCTACGGGAGGCWGC-3′ (Eub338F-0-III) and 5′-ACACCTACGGGTGGCWGC-3′ (Eub338F-I-II), labeled with 6-carboxyfluorescein, as forward primers and 5′-ACGGGCGGTGTGTACA-3′ (1392R) (W = A or T) (57) as the reverse primer was used. Each 25-μl reaction mixture contained GoTaq Flexi DNA polymerase (2.5 U), buffer (1×), MgCl2 (0.5 mM), bovine serum albumin (0.64 mg ml−1), deoxynucleoside triphosphates (0.2 mM each), and forward and reverse primers (0.2 μM each), along with 2 μl of template DNA. Positive controls (P. aeruginosa genomic DNA) and negative controls (sterile deionized water) were run with each set of PCRs. Five PCRs per sample were carried out in a PTC 200 DNA Engine Cycler (Bio-Rad, Hercules, CA) with a thermal profile of 94°C for 3 min and 40 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 90 s, followed by a final extension of 72°C for 7 min. The success of the PCR amplification was verified by pooling the five reaction mixtures per sample and performing gel electrophoresis. PCR products were purified using the Qiaquick PCR purification kit (Qiagen, Valencia, CA) and digested with the endonuclease HaeIII (2 U) at 37°C for 18 to 24 h, followed by another round of clean up with the Qiaquick PCR purification kit (Qiagen, Valencia, CA). Analysis of PCR products was performed at the Ohio State Plant Microbe Genomics Facility using a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA). The results were analyzed with Gene Mapper 4.0 (Applied Biosystems, Foster City, CA).
To identify bacterial sequences in microcosms treated with algal exudate and amended with a high glucose concentration, PCR products of 16S rRNA genes were cloned and sequenced. PCR was carried out with the universal primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1552R (5′-AAGGAGGTGATCCARCCGCA-3′) (58, 59) in a PTC 200 DNA Engine Cycler (Bio-Rad, Hercules, CA) with a thermal profile of 94°C for 3 min and 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 90 s, followed by a final extension of 72°C for 5 min. The amplified PCR products were purified using a Qiaquick PCR purification kit (Qiagen, Valencia, CA) before it was ligated into the pGEM-T vector, followed by overnight incubation and transformation of competent Escherichia coli cells according to the manufacturer's protocol (Promega, Madison, WI). Plasmids were isolated from transformants as described by Ausubel et al. (60). Sequencing was performed at the Advanced Genetic Technologies Center at the University of Kentucky in Lexington, KY, using M13 primers [M13 forward sequencing primer, 5′-d(GTTTTCCCAGTCACGAC)-3′; M13 reverse sequencing primer, 5′-d(CAGGAAACAGCTATGAC)-3′] (Promega, Madison, WI). The plasmid sequence was removed, and amplicon sequences were trimmed for quality in Sequencher (Gene Codes Corporation, Ann Arbor, MI) using the default settings. Sequences were analyzed using BLAST at the National Center for Biotechnology Information website (61).
Statistical analysis.
Bacterial abundance, DOC, organic N, the nitrate concentration, and terminal restriction fragment (T-RF) numbers were compared among treatments via three-way analysis of variance (ANOVA) using JMP statistical software (version 10; SAS Institute Inc., Cary, NC), followed by Student's t test and Tukey's test for post hoc analysis; P values of ≤0.05 were considered significant. To determine the relationship between the growth rate and chemical variables (the DOC and inorganic and organic N used), multiple linear regression with stepwise forward and backward selection was used. The values to enter and leave the analyses were 0.10 and 0.05, respectively.
The contributions of N treatments, glucose concentrations, and the interaction of these factors to variation in T-RFLP profiles were determined using redundancy analysis (62). R statistical software (version 2.15.1 for Windows) was used to determine differences in T-RFLP profiles. Variation in relative peak heights (the Hellinger distance) and the absence and presence of peaks (Jaccard's distance) were considered for such analysis. Prior to analysis, the T-RFLP relative peak heights were square root transformed.
In addition, the composition of the bacterial community was examined by calculating the relative abundance of dominant T-RF peaks (63, 64). Smaller T-RF peaks (which contributed less than 2% to the total) in a pattern were lumped together as “other T-RFs.” Three-way ANOVA, followed by Tukey post hoc tests, was used to identify differences in T-RF patterns among treatments and times (5 and 10 days); percent relative abundances of T-RFs were normalized by log transformation.
RESULTS
Bacterial abundance.
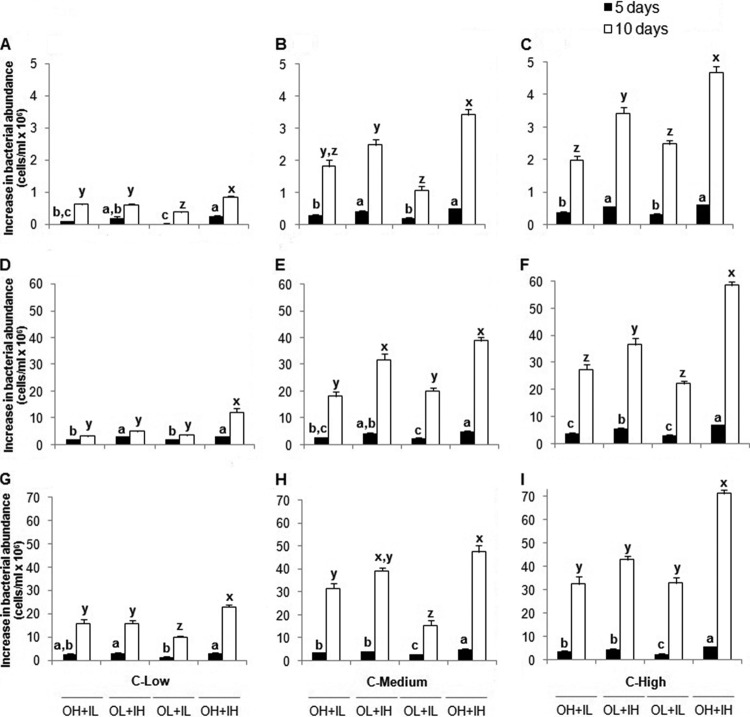
Growth of bacteria was determined based on changes in abundance of both the attached and planktonic cells. For each DON source separately, three-way ANOVAs were used to test for significant differences in bacterial abundance among treatments (Fig. 1). Growth of bacteria in microcosms treated with DON derived from humic matter was relatively modest, and at each of the three glucose concentrations, abundance at the end of the experiment (10 days) was highest in microcosms amended with the highest concentrations of both inorganic and organic N (Fig. 1A to C). Under high and medium glucose concentrations, bacterial growth in high-DIN and low-DON treatments was significantly lower than in treatments with both high DIN and high DON, revealing bacterial growth on DON. After 5 days of incubation under all three glucose concentrations, growth in treatments with high DIN and high DON was more similar to growth in high DIN and low DON, which changed after 10 days.
Fig 1.
Increase in bacterial abundance after 5 and 10 days of incubation. (A to I) Humic matter (A to C), bacterial protein (D to F), and algal exudate (G to I) as DON sources. C-Low, C-Medium, and C-High refer to glucose concentrations. O, organic N; I, inorganic N; H, high concentration; L, low concentration. The values are means and standard errors. Different letters above the bars indicate significant differences (P < 0.05).
When protein was the DON source, bacterial growth was 10-fold higher than in microcosms with humic matter. However, like humic matter, after 10 days of incubation, bacterial abundance was highest in the high organic and inorganic N treatment under high and low glucose concentrations (Fig. 1D to F). In contrast, under a medium glucose concentration, responses to this treatment were not significantly different from those under the high inorganic and low organic N treatment. Like humic matter, after 5 days of incubation, bacterial growth under high DIN and high DON was similar to growth in high DIN and low DON, except under a high glucose concentration.
Bacterial abundance in microcosms with algal exudate as the DON source was similar to that achieved by the protein amendment. Also, like protein, after 10 days of incubation, the greatest growth occurred in high-DIN and -DON treatments (Fig. 1G to I), except under a medium glucose concentration. After 5 days of incubation under high and medium glucose concentrations, growth was significantly higher in the high-DIN and -DON treatment than in the other treatments.
Controls were used to examine the effects of the glucose concentration alone on bacterial abundance. In these glucose-only controls, cell numbers were significantly lower (10-fold lower after 5 days and 1,000-fold lower after 10 days) than in glucose with N amendments (average abundance in glucose-only controls, 0.16 × 106 to 0.45 × 106 [5 days] to 0.07 × 106 to 0.36 × 106 cells/ml [10 days]). Negative controls were not amended with DOC or N, and cell abundances were low, ranging from 0.9 × 105 to 1.17 × 105 (5 days) to 0.15 × 105 to 0.3 × 105 cells/ml (10 days).
DOC utilization.
The average declines in DOC concentration were 67.9% and 83.7% after 5 and 10 days, respectively. Final DOC concentrations differed significantly among treatments. Regardless of the DON source, in positive controls with medium and high glucose amendments, the DOC concentration remaining was significantly higher than in the respective N treatments after both 5 and 10 days of incubation (Table 2). Maximum DOC loss was observed in treatments with low glucose concentrations and was lowest in treatments with high glucose, DON, and DIN, irrespective of the DON source and the duration of incubation. Also, microcosms with high and medium glucose had greater DOC loss (more bacterial uptake) in treatments with high DON and low DIN than in treatments with low DON and high DIN.
Table 2.
Final DOC concentrations in C- and N-amended microcosms after 5 and 10 days of incubation
| Carbon treatment | Nitrogen treatmenta | Final DOC concn (mg C/liter) |
|||||
|---|---|---|---|---|---|---|---|
| Humic |
Bacterial protein |
Algal exudate |
|||||
| 5 days | 10 days | 5 days | 10 days | 5 days | 10 days | ||
| C-low | OH + IL | 0.57 | 0.40 | 0.35 | 0.18 | 0.30 | 0.17 |
| OL + IH | 0.50 | 0.33 | 0.35 | 0.18 | 0.23 | 0.12 | |
| OL + IL | 0.41 | 0.14 | 0.27 | 0.16 | 0.25 | 0.10 | |
| OH + IH | 0.51 | 0.36 | 0.38 | 0.24 | 0.32 | 0.15 | |
| Positive control | 0.29 | 0.44 | 0.17 | 0.40 | 0.25 | 0.38 | |
| Negative control | 0.21 | 0.83 | 0.25 | 0.79 | 0.22 | 0.74 | |
| C-medium | OH + IL | 6.72 | 4.35 | 5.24 | 2.73 | 5.00 | 2.40 |
| OL + IH | 5.02 | 2.62 | 4.11 | 2.02 | 3.93 | 1.96 | |
| OL + IL | 8.13 | 6.01 | 5.13 | 2.96 | 5.95 | 2.59 | |
| OH + IH | 5.33 | 1.80 | 3.94 | 2.15 | 4.11 | 1.83 | |
| Positive control | 9.31 | 8.60 | 8.35 | 6.51 | 7.80 | 6.52 | |
| Negative control | 0.21 | 0.83 | 0.25 | 0.79 | 0.22 | 0.74 | |
| C-high | OH + IL | 12.42 | 9.42 | 10.33 | 4.79 | 10.35 | 4.22 |
| OL + IH | 10.54 | 5.87 | 9.75 | 3.63 | 8.83 | 3.02 | |
| OL + IL | 11.76 | 5.48 | 10.52 | 4.77 | 9.71 | 3.32 | |
| OH + IH | 9.36 | 3.31 | 8.92 | 3.32 | 7.95 | 3.18 | |
| Positive control | 22.18 | 19.07 | 19.33 | 17.16 | 18.22 | 6.52 | |
| Negative control | 0.21 | 0.83 | 0.25 | 0.79 | 0.22 | 0.74 | |
Three concentrations of organic carbon, in the form of glucose, were used for each nitrogen treatment. OH, organic nitrogen, high concentration; OL, organic nitrogen, low concentration; IH, inorganic nitrogen, high concentration; IL, inorganic nitrogen, low concentration.
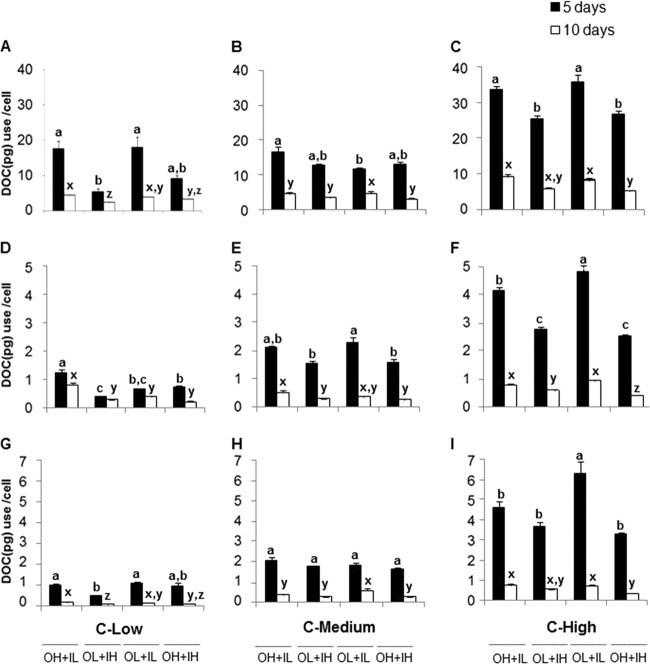
Because initial differences in DOC concentrations impacted the final concentrations, DOC loss was expressed as DOC utilization per bacterial cell (loss of DOC [i.e., the difference between initial and final DOC concentrations] divided by bacterial abundance) and differed significantly among treatments (Fig. 2). Generally, experimental microcosms with higher bacterial abundance had lower DOC use per cell than those with low bacterial abundance; bacterial DOC use per cell tracked with patterns of bacterial abundance, with some exceptions. At the end of the experiment, for all DON sources used, under low glucose concentrations, bacterial abundance in amendments with high DON and low DIN was similar to that with low DON and high DIN (Fig. 1A, D, and G). However, bacterial DOC use per cell was significantly lower in the low-DON and high-DIN treatment than in the high-DON and low-DIN treatment (Fig. 2A, D, and G).
Fig 2.
Utilization of DOC per bacterial cell after 5 and 10 days of incubation. (A to I) Humic matter (A to C), bacterial protein (D to F), and algal exudate (G to I) as DON sources. The values are means and standard errors. Different letters above the bars indicate significant differences (P < 0.05).
Since bacterial DOC use per cell was a function of abundance, in positive controls, DOC use per cell (average, 84.35 ± 9 pg/cell) was higher than DOC use in experimental microcosms. DOC was not added to the negative controls, and thus, DOC use per cell was below the detection limit.
N utilization.
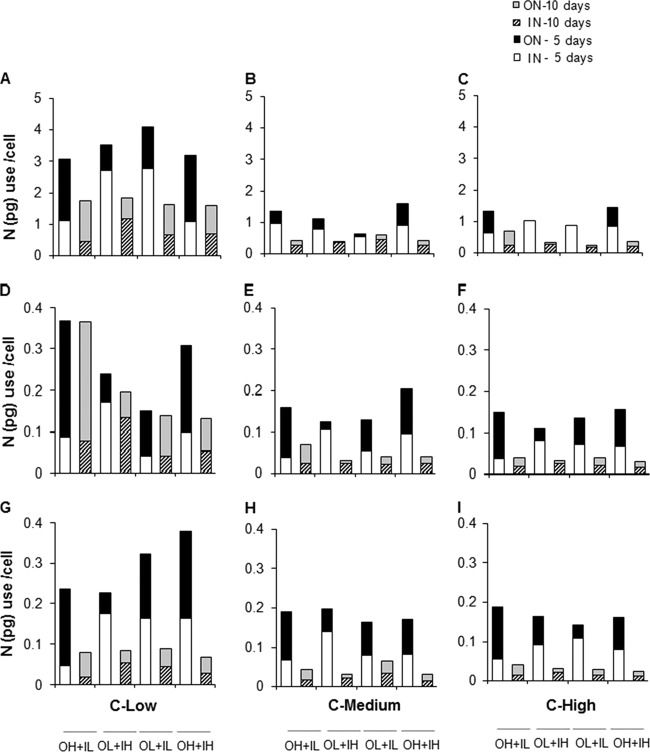
Bacterial utilization of organic and inorganic N (based on declines in DON and DIN concentrations in microcosms) differed significantly among treatments (Fig. 3). Irrespective of the DON source, under high DON, high DIN, and low glucose concentration conditions, bacterial use of organic N was significantly higher than use of inorganic N after both 5 and 10 days (Fig. 3A, D, and G). However, bacteria utilized organic and inorganic N equally well under medium and high glucose concentrations, when bacterial protein and algal exudate were the DON sources (Fig. 3E, F, H, and I). In microcosms treated with humic matter under both medium and high glucose concentrations, bacterial use of DON was lower than that of DIN in amendments with high DON and high DIN (Fig. 3B and C).
Fig 3.
Utilization of organic and inorganic nitrogen per bacterial cell after 5 and 10 days of incubation. (A to I) Humic matter (A to C), bacterial protein (D to F), and algal exudate (G to I) as DON sources.
To determine if changes in pH played a role in bacterial responses, pH was measured initially after 5 and 10 days. Initial pH values ranged from 6.50 to 7.45, and no significant differences in pH were observed; final values ranged from 6.25 to 7.20 (5 days) to 6.05 to 6.79 (10 days).
T-RFLP.
Based on redundancy analysis (RDA) of 16S rRNA gene T-RFLP, bacterial community structures differed significantly among treatments for all DON sources (P < 0.05). The largest percentage of variation in community structure was explained by the interaction between the N treatment and the glucose concentration (Table 3). Generally, the percentage of variation explained by this interaction was higher after 5 days (average, 83%) than after 10 days (average, 60%).
Table 3.
Percent variation in bacterial community structure after 5 and 10 days as explained by N treatments, glucose concentrations, and the interaction between thema
| DON source | Variation (%) explained by: |
|||||
|---|---|---|---|---|---|---|
| N treatment |
Glucose concn |
N treatment × glucose concn |
||||
| 5 days | 10 days | 5 days | 10 days | 5 days | 10 days | |
| Humic matter | 11 | 15 | 14 | 9 | 87 | 58 |
| Bacterial protein | 12 | 3.6 | 17 | 19 | 70 | 50 |
| Algal exudate | 13 | 8.1 | 20 | 54 | 91 | 72 |
Based on redundancy analysis of 16S rRNA gene T-RFLP profiles.
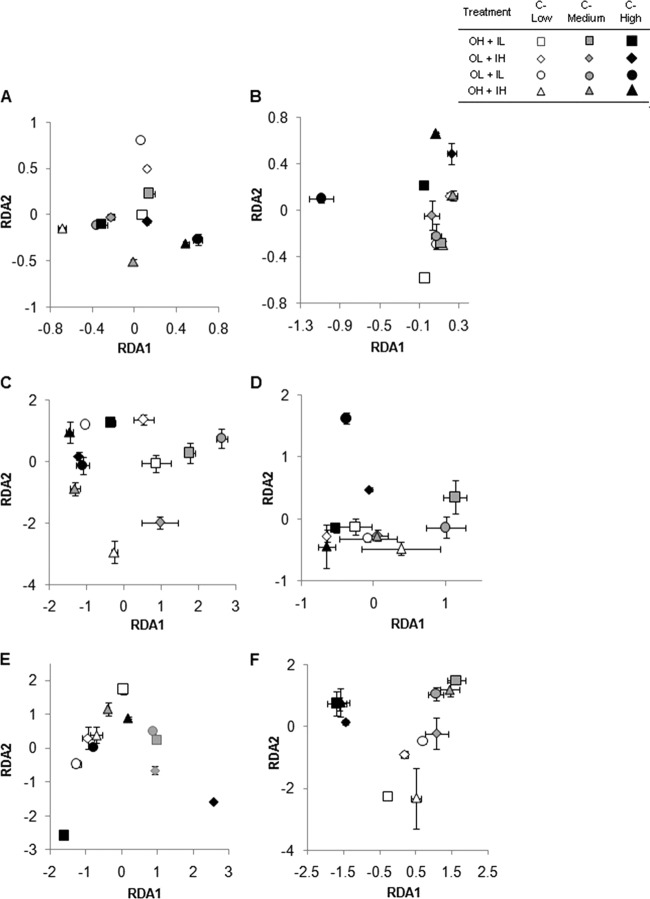
For bacterial communities in humic matter treatments, compositions varied among glucose concentrations and N treatments after 5 days (Fig. 4A). However, after 10 days, only bacterial communities under high glucose and low DIN and DON concentrations were spatially separated on RDA axis 1, whereas communities from other treatments were clustered relatively close together on this axis (Fig. 4B). Other treatments varied along RDA axis 2, but generally, those with the same glucose concentrations were the most similar.
Fig 4.
Ordination plots from redundancy analysis of 16S rRNA gene T-RFLP peak relative abundances after 5 (A, C, and E) and 10 (B, D, and F) days. (A and B) Humic matter. (C and D) Bacterial protein. (E and F) Algal exudate. The values are means and standard errors.
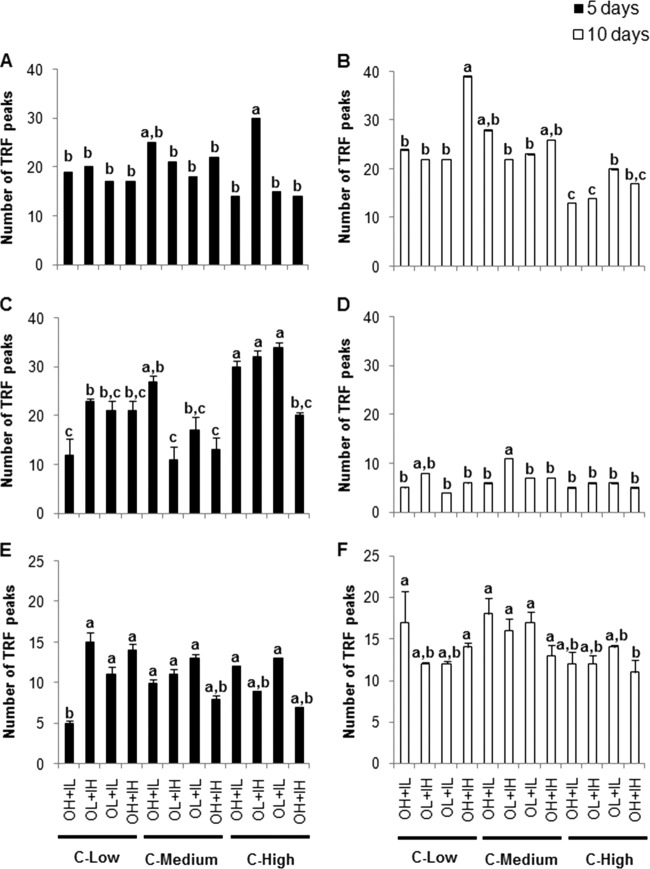
For the humic matter amendments, the number of T-RFs was highest in the treatment with low DON and high DIN under a high glucose concentration but did not differ significantly among the rest of the treatments after 5 days (Fig. 5A). However, after 10 days, there were significant differences in the numbers of T-RFs among treatments, with the highest numbers recorded in microcosms treated with high DON and DIN under low glucose concentrations (Fig. 5B).
Fig 5.
Numbers of peaks of 16S rRNA gene T-RFLP profiles after 5 and 10 days of incubation. (A and B) After 5 days (A) and after 10 days (B) with humic matter as the organic nitrogen source. (C and D) After 5 days (C) and after 10 days (D) with bacterial protein as the organic nitrogen source. (E and F) After 5 days (E) and after 10 days (F) with algal exudate as the organic nitrogen source. The values are means and standard errors. Different letters above the bars indicate significant differences (P < 0.05).
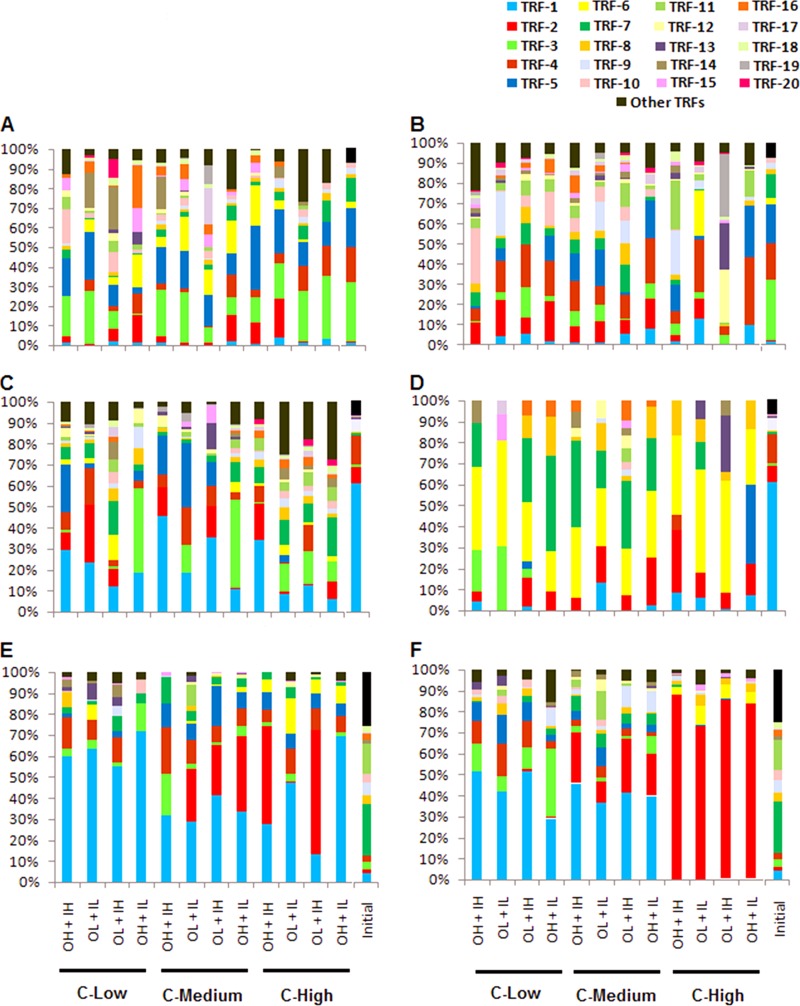
There were also significant differences in the relative abundances of T-RFs (Fig. 6A and B). For example, after 5 days under low and medium glucose concentrations, T-RF-2 and T-RF-4 had low relative abundances but were more dominant after 10 days. T-RF-3 had high relative abundance in all treatments except under high DON, low DIN, and low glucose. T-RF-12 and T-RF-19 were absent in all treatments after 5 days but together contributed 60% of the community in the treatment with low DON and high DIN under high glucose concentrations after 10 days.
Fig 6.
Percentages of dominant T-RFs after 5 and 10 days of incubation. (A and B) After 5 days (A) and after 10 days (B) with humic matter as the organic nitrogen source. (C and D) After 5 days (C) and after 10 days (D) with bacterial protein as the organic nitrogen source. (E and F) After 5 days (E) and after 10 days (F) with algal exudate as the organic nitrogen source.
When bacterial protein was the DON source, after 5 days, bacterial community structures differed among glucose and N treatments with no discernible pattern based on the treatment (Fig. 4C). However, after 10 days, communities became more similar, with varying degrees of overlap. Under high glucose concentrations, two N treatments (low DON plus high DIN and low DON plus low DIN) had distinct communities (Fig. 4D). The numbers of T-RFs differed significantly among treatments after 5 days, with maximum numbers under high glucose concentrations (Fig. 5C). However, after 10 days, the numbers of T-RFs decreased in all treatments, and there were few significant differences in the numbers of peaks (Fig. 5D).
Significant differences among treatments were also observed in relative abundances of T-RFs (Fig. 6C and D). Bacterial communities became more uniform in all the treatments, as they were dominated mostly by T-RF-2, -6, and -7 after 10 days.
The compositions of bacterial communities treated with algal exudate differed significantly among treatments after 5 days; a high glucose concentration with high-DON and low-DIN, as well as low-DON and high-DIN, conditions had the most distinct communities (Fig. 4E). However, after 10 days, communities in the high-glucose treatments clustered together regardless of the N treatment; this was also somewhat the case for the medium and low glucose concentrations (Fig. 4F). As observed in the humic matter amendments, the total number of T-RFs did not differ significantly between 5 and 10 days. Irrespective of the incubation period, no significant differences in numbers of peaks were observed among treatments, except for the low-glucose, high-DON, and low-DIN treatment after 5 days and the high-glucose, high-DON, and high-DIN treatment after 10 days, which had relatively low numbers of peaks (Fig. 5E and F). The relative abundances of specific T-RFs differed significantly among treatments (Fig. 6E and F). T-RF-1 was dominant in all treatments after 5 days. It continued to be the dominant T-RF at the end of 10 days in treatments with low and medium glucose amendments but was absent under high glucose concentrations. T-RF-2 was absent in all treatments with low-glucose amendments after 5 days. However, it showed a striking increase (accounting for 72% to 88% of the community) in treatments with high glucose after 10 days. Cloning and 16S rRNA sequencing followed by BLAST comparisons to GenBank performed on samples from this treatment revealed that 75% of the clones had a high degree of sequence similarity to Pseudomonas spp. while the other 25% had more similarity to members of the Enterobacteriaceae (Serratia spp., Klebsiella spp., and Proteus spp., as nearest neighbors) (Table 4). Out of 12 clones, 7 demonstrated maximum sequence similarity (95 to 97%) to strains of P. aeruginosa.
Table 4.
Best matches of the different sequences obtained from the microcosms treated with algal exudate and amended with high glucose, DON, and DIN
| Clone | Length of sequence (bp) | GenBank accession no. | Best match (BLASTN) | % identity/E value |
|---|---|---|---|---|
| CLAEhc1 | 1,200 | KC211291.1 | P. aeruginosa strain KLU02 | 97/0.0 |
| CLAEhc2 | 1,029 | JQ659882.1 | P. aeruginosa strain R7-521 | 96/0.0 |
| CLAEhc3 | 1,046 | JQ659882.1 | P. aeruginosa strain R7-521 | 97/0.0 |
| CLAEhc4 | 1,241 | JQ659882.1 | P. aeruginosa strain R7-521 | 960.0 |
| CLAEhc5 | 649 | JF708077.1 | P. aeruginosa strain mpc1 | 95/0.0 |
| CLAEhc6 | 641 | KC503912.1 | Pseudomonas sp. strain ZS-1 | 77/2e−10 |
| CLAEhc7 | 694 | JN622013.1 | Pseudomonas sp. strain WC-1 | 88/2e−23 |
| CLAEhc8 | 1,230 | JQ659882.1 | P. aeruginosa strain R7-521 | 97/0.0 |
| CLAEhc9 | 1,193 | KC211291.1 | P. aeruginosa strain KLU02 | 97/0.0 |
| CLAEhc10 | 378 | GU569122.1 | Uncultured Rahnella sp. clone JBXB28 | 80/9e−47 |
| CLAEhc11 | 1,292 | HQ018868.1 | Klebsiella sp. strain GX17 | 92/0.0 |
| CLAEhc12 | 1,300 | NR_041979.1 | Serratia ficaria strain DSM 4569 | 92/0.0 |
DISCUSSION
Carbon availability exerts strong controls on bacterial N dynamics (23, 65, 66). In aquatic ecosystems, heterotrophic bacteria meet their N demand via utilization of inorganic and organic N, but the interplay between DIN, DON, and labile DOC is not well understood. In this study, we manipulated the supply of labile C to the bacterial community and then followed loss of N and C and shifts in bacterial community abundance and composition. We found that the C supply impacted bacterial N utilization, and reliance on organic versus inorganic N was determined by changes in the labile-C concentration and the quality of organic N. Likewise, the availability of N can also affect C dynamics. Low N availability can result in bacterial sustenance by utilizing DOC for respiration and converting it to CO2, whereas increased N availability would allow bacteria to use DOC, not only for respiration, but also for growth.
In freshwater, more than 50% of total dissolved N is DON (25, 67, 68), which varies in composition and bioavailability (48). DON is an important N source for heterotrophic bacteria (69, 70). Since DON-derived N can be used either directly (71, 72) or indirectly via extracellular enzymes (73, 74), bacterial uptake depends on the nature of the DON pool. The stoichiometric ratio of C to N varies among different DON compounds and plays a critical role in nutrient mineralization by heterotrophic bacteria (75). While a high C/N ratio implies low N availability, as is found in recalcitrant compounds, like humic acids, a low C/N ratio implies increased availability of N, as is found in labile compounds, such as proteins. In our study, different DON sources elicited different bacterial responses. For example, bacteria preferentially used DIN over humic DON under medium and high glucose concentrations, since DON derived from humic matter is more resistant to biological degradation (52) and more energy is required (39). However, bacterial uptake of humic DON was higher than that of DIN under low glucose concentrations, suggesting that the humic compounds served as sources of both N and C (76) under carbon-limiting conditions. A high C/N ratio in humic matter coupled with the refractory nature of humic compounds accounts for the low bacterial abundance in microcosms treated with humic matter. Like humic DON, alga-derived DON and bacterial protein were used more than DIN under low glucose concentrations and were more labile (50), yielding greater bacterial abundances. In contrast, under medium and high glucose concentrations, DIN and DON were used equally, suggesting that even under conditions where C is less limited, these DON mixtures are useful as C and N sources.
In addition to impacts caused by differences in the type of DON, the effects of addition of labile DOC were also examined. DOC is an important driver of bacterial abundance (77), and as expected, bacterial growth was stimulated by the addition of labile DOC (78). In microcosms with high bacterial abundance, a rapid decline in DOC from the water column, along with low DOC use per cell, indicate strong competition for carbon within the bacterial community (79). In most treatments, it was also observed that DOC use per cell was lower after 10 days than after 5 days. This may result from a relatively slow response of bacterial biofilms to nutrient amendments compared to free-living bacteria in the water column. However, low bacterial abundance in treatments with low N availability and in positive controls without N amendment suggests that more of the DOC is utilized for bacterial respiration than for growth.
Heterotrophic bacteria differ in their utilization of C and N based on their metabolic capabilities, and thus, bacterial community composition plays a critical role in nutrient uptake (80). Bacterial community structures differed among DON sources and treatments. Previous research revealed that while differences in bacterial community composition can influence the metabolism of organic N (81, 82), in freshwater ecosystems, changes in resources (nutrient concentrations and quality) often trigger responses in community metabolism (83). Therefore, community composition is both a driver of differences in C and N utilization and an attribute that is influenced by C and N treatments.
Variation in community metabolism can result from physiological acclimation, changes in community composition, or a combination of the two (78, 84–86). In the experimental microcosms, differences in utilization of N under varying C concentrations is possibly due to changes in the abundance of bacteria with different nutritional needs, resulting in shifts in community composition. Community composition and functional response are highly correlated (87), and shifts in community composition can be accompanied by changes in the hydrolytic ectoenzyme activity (88).
We observed maximum differences in community structure among treatments after 5 days, which diminished after 10 days when bacterial protein and algal exudate were the DON sources. It is likely that, as resources were depleted, lower bioavailability of C and N led to slow-growing, stable communities that are dominated by fewer T-RFs, with similar capabilities to utilize the leftover resources (79). However, this was not observed in microcosms treated with humic matter. In this case, lower bioavailability of recalcitrant compounds possibly led to more diverse communities composed of different T-RFs, because enzymatic breakdown of complex compounds by some members allows the uptake of the products by others in the community (39, 89). This possibility is supported by the more balanced contributions of multiple T-RFs in the communities of humic treatments relative to other DON sources. Competition is another potential contributor to these differences in bacterial community composition among treatments as bacteria interact to obtain the C and N required for their growth (79).
The shift to dominance by a limited subset of T-RFs was most obvious in microcosms treated with high concentrations of glucose, with algal exudate as the DON source. Regardless of the N treatment, one T-RF accounted for approximately 80% of the community. Based on 16S rRNA gene sequencing, these dominant organisms were members of Pseudomonas, a genus that is widely distributed in aquatic ecosystems (90, 91). Being genetically and metabolically versatile, Pseudomonas spp. are ubiquitous and occupy multiple ecological niches (92). Several sequences had strong similarity to that of P. aeruginosa, which is an efficient competitor for resources; it produces antibiotics (79) and deploys toxins to attack the cell walls of other bacterial competitors in a community (93). In our experimental microcosms treated with algal exudate and amended with high glucose, Pseudomonas spp. possibly scavenged C and N, limiting their acquisition by other members of the bacterial community. Also, the bead method used may be selective or enriching for specific taxa, as in this laboratory study with the biofilm community in a closed system, which possibly contributed to lower bacterial diversity by selecting for a limited subset of taxa, such as Pseudomonas and members of the Enterobacteriaceae.
Overall, there was a strong linkage between C availability and bacterial utilization of DON over DIN. Bacterial utilization of DON was related not only to the availability of inorganic N, but also to the nature of the DON (labile versus recalcitrant). Differences in the use of organic N were strongly associated with differences in community composition. Most likely, C, DIN, and DON treatments drove changes in bacterial community composition that in turn affected the rates of DON and DIN utilization under various C concentrations.
ACKNOWLEDGMENTS
This work was supported by Kent State University via an Art and Margaret Herrick Research Grant.
We thank Christopher Blackwood, Moumita Moitra, Elizabeth Griffith, Cory Mahen, Erin Manis, Oscar Valverde, and Hasanti Widanagamage for their assistance.
Footnotes
Published ahead of print 13 September 2013
REFERENCES
- 1.Karlsson OM, Richardson JS, Kiffney PM. 2005. Modelling organic matter dynamics in headwater streams of south-western British Columbia, Canada. Ecol. Modell. 183:463–476 [Google Scholar]
- 2.Tank JL, Rosi-Marshall EJ, Griffiths NA, Entrekin SA, Stephen ML. 2010. A review of allochthonous organic matter dynamics and metabolism in streams. J. North Am. Benthol. Soc. 29:118–146 [Google Scholar]
- 3.del Giorgio PA, Cole JJ. 1998. Bacterial growth efficiency in natural aquatic systems. Annu. Rev. Ecol. Syst. 29:503–541 [Google Scholar]
- 4.Kritzberg ES, Cole JJ, Pace MM, Granéli W. 2005. Does autochthonous primary production drive variability in bacterial metabolism and growth efficiency in lakes dominated by terrestrial C inputs? Aquat. Microb. Ecol. 38:103–111 [Google Scholar]
- 5.Kritzberg ES, Cole JJ, Pace MM, Granéli W. 2006. Bacterial growth on allochthonous carbon in humic and nutrient-enriched lakes: results from whole-lake 13C addition experiments. Ecosystems 9:489–499 [Google Scholar]
- 6.Wallace JB, Eggert SL, Meyer JL, Webster JR. 1999. Effects of resource limitation on a detrital-based ecosystem. Ecol. Monogr. 69:409–442 [Google Scholar]
- 7.Hassan MA, Hogan DL, Bird SA, May CL, Gomi T, Campbell D. 2005. Spatial and temporal dynamics of wood in headwater streams of the Pacific Northwest. J. Am. Water Resour. Assoc. 41:899–919 [Google Scholar]
- 8.Sherr EB, Sherr BF. 1996. Temporal offset in oceanic production and respiration processes implied by seasonal changes in atmospheric oxygen: the role of heterotrophic microbes. Aquat. Microb. Ecol. 11:91–100 [Google Scholar]
- 9.Azam F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694–696 [Google Scholar]
- 10.Koetsier P, McArthur V, Leff LG. 1997. Spatial and temporal response of stream bacteria to sources of dissolved organic carbon in a blackwater stream system. Freshw. Biol. 37:79–89 [Google Scholar]
- 11.Leff LG. 2000. Longitudinal changes in microbial assemblages of the Ogeechee River. Freshw. Biol. 43:605–615 [Google Scholar]
- 12.Olapade OA, Leff LG. 2005. Seasonal response of stream biofilm communities to dissolved organic matter and nutrient enrichments. Appl. Environ. Microbiol. 71:2278–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turner RE. 2002. Element ratios and aquatic food webs. Estuaries 25:694–703 [Google Scholar]
- 14.Taylor PG, Townsend AR. 2010. Stoichiometric control of organic carbon-nitrate relationships from soils to the sea. Nature 464:1178–1181 [DOI] [PubMed] [Google Scholar]
- 15.Fisher SG, Likens GE. 1973. Energy flow in Bear Brook, New Hampshire: an integrative approach to stream ecosystem metabolism. Ecol. Monogr. 43:421–439 [Google Scholar]
- 16.McDowell WH, Fisher SG. 1976. Autumnal processing of dissolved organic matter in a small woodland stream ecosystem. Ecology 57:561–569 [Google Scholar]
- 17.Meyer JL, Tate CM. 1983. The effects of watershed disturbance on dissolved organic carbon dynamics of a stream. Ecology 64:33–44 [Google Scholar]
- 18.Grimm NB, Fisher SG, Minckley WL. 1981. Nitrogen and phosphorus dynamics in hot desert streams of Southwestern U. S. A. Hydrobiologia 83:303–312 [Google Scholar]
- 19.Triska FJ, Sedell JR, Cromack K, Jr, Gregory SV, McCorison FM. 1984. Nitrogen budget for a small coniferous forest stream. Ecol. Monogr. 54:119–140 [Google Scholar]
- 20.Mulholland PJ, Tank JL, Sanzone DM, Wollheim WM, Peterson BJ, Webster JR, Meyer JL. 2000. Nitrogen cycling in a forest stream determined by a 15N tracer addition. Ecol. Monogr. 70:471–493 [Google Scholar]
- 21.Bernhardt ES, Likens GE. 2002. Dissolved organic carbon enrichment alters nitrogen dynamics in a forest stream. Ecology 83:1689–1700 [Google Scholar]
- 22.Paerl HW. 1993. Interaction of nitrogen and carbon cycles in the marine environment, p 343–381 In Ford TE. (ed), Aquatic microbiology: an ecological approach. Blackwell Scientific, Oxford, United Kingdom [Google Scholar]
- 23.Seitzinger SP. 1994. Linkages between organic matter mineralization and denitrification in eight riparian wetlands. Biogeochemistry 25:19–39 [Google Scholar]
- 24.Wiegner TN, Seitzinger SP. 2004. Seasonal bioavailability of dissolved organic carbon and nitrogen from pristine and polluted freshwater wetlands. Limnol. Oceanogr. 49:1703–1712 [Google Scholar]
- 25.Wetzel RG. 2001. Limnology: lake and river ecosystems. Academic Press, San Diego, CA [Google Scholar]
- 26.Perakis SS, Hedin LO. 2002. Nitrogen losses from unpolluted South American forests mainly via dissolved organic compounds. Nature 415:416–419 [DOI] [PubMed] [Google Scholar]
- 27.Seitzinger SP, Sanders RW. 1997. Contribution of dissolved organic nitrogen from rivers to estuarine eutrophication. Mar. Ecol. Prog. Ser. 159:1–12 [Google Scholar]
- 28.Antia NJ, Harrison PJ, Oliveira L. 1991. The role of dissolved organic nitrogen in phytoplankton nutrition, cell biology and ecology. Phycologia 30:1–89 [Google Scholar]
- 29.Zehr JP, Ward BB. 2002. Nitrogen cycling in the ocean: new perspectives on processes and paradigms. Appl. Environ. Microbiol. 68:1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seitzinger SP, Sanders RW, Styles R. 2002. Bioavailability of DON from natural and anthropogenic sources to estuarine phytoplankton. Limnol. Oceanogr. 47:353–366 [Google Scholar]
- 31.Stepanauskas R, Laudon H, Jørgenson NOG. 2000. High DON bioavailability in boreal streams during a spring flood. Limnol. Oceanogr. 45:1298–1307 [Google Scholar]
- 32.Mulholland PJ. 2003. Large-scale patterns in dissolved organic carbon concentration, flux, and sources, p 139–159 In Findlay SEG, Sinsabaugh RL. (ed), Aquatic ecosystems: interactivity of dissolved organic matter. Academic Press, San Diego, CA [Google Scholar]
- 33.Kaplan LA, Bott TL. 1982. Diel fluctuations of DOC generated by algae in a piedmont stream. Limnol. Oceanogr. 27:1091–1100 [Google Scholar]
- 34.Mulholland PJ. 1992. Regulation of nutrient concentrations in a temperate forest stream: roles of upland, riparian, and instream processes. Limnol. Oceanogr. 37:1512–1526 [Google Scholar]
- 35.Wolter K. 1982. Bacterial incorporation of organic substances released by natural phytoplankton populations. Mar. Ecol. Prog. Ser. 7:287–295 [Google Scholar]
- 36.Bell WH. 1983. Bacterial utilization of algal extracellular products. 3. The specificity of algal-bacterial interaction. Limnol. Oceanogr. 28:1131–1143 [Google Scholar]
- 37.Sell AF, Overbeck J. 1992. Exudates: phytoplankton-bacterioplankton interactions in Pluβsee. J. Plankton Res. 14:1199–1215 [Google Scholar]
- 38.Brookshire ENJ, Valett HM, Thomas SA, Webster JR. 2005. Coupled cycling of dissolved organic nitrogen and carbon in a forest stream. Ecology 86:2487–2496 [Google Scholar]
- 39.Sinsabaugh RL, Findlay S, Franchini P, Fischer D. 1997. Enzymatic analysis of riverine bacterioplankton production. Limnol. Oceanogr. 42:29–38 [Google Scholar]
- 40.McCarthy JJ, Carpenter EJ. 1983. Nitrogen cycling in near-surface waters of the open ocean, p 487–512 In Carpenter EJ, Capone DG. (ed), Nitrogen in the marine environment. Academic Press, New York, NY [Google Scholar]
- 41.Wheeler PA, Kokkinakis SA. 1990. Ammonium recycling limits nitrate use in oceanic subarctic Pacific. Limnol. Oceanogr. 35:1267–1278 [Google Scholar]
- 42.Berman T, Bronk DA. 2003. Dissolved organic nitrogen: a dynamic participant in aquatic ecosystems. Aquat. Microb. Ecol. 31:279–305 [Google Scholar]
- 43.Kaushal SS, Lewis WM. 2005. Fate and transport of organic nitrogen in minimally disturbed montane streams of Colorado, U. S. A. Biogeochemistry 74:303–321 [Google Scholar]
- 44.Chrost RJ. 1991. Environmental control of the synthesis and activity of aquatic microbial ectoenzymes, p 29–59 In Chrost RJ. (ed), Microbial enzymes in aquatic environments. Springer-Verlag, New York, NY [Google Scholar]
- 45.Munster U, de Haan H. 1998. The role of microbial extracellular enzymes in the transformation of dissolved organic matter in humic waters. In Hessen DO, Tranvik LJ. (ed), Aquatic humic substances, Springer-Verlag, Berlin, Germany [Google Scholar]
- 46.Bott TL, Kaplan LA, Kuserk FT. 1984. Benthic bacterial biomass supported by streamwater dissolved organic matter. Microb. Ecol. 10:335–344 [DOI] [PubMed] [Google Scholar]
- 47.Liu WT, Marsh TL, Cheng H, Forney LJ. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Billen G. 1991. Protein degradation in aquatic environments, p 123–143 In Chrost RJ. (ed), Microbial enzymes in aquatic environments. Springer-Verlag, New York, NY [Google Scholar]
- 49.Hoppe HG. 1991. Microbial extracellular enzyme activity: a new key parameter in aquatic ecology, p 60–83 In Chrost RJ. (ed), Microbial enzymes in aquatic environments. Springer-Verlag, New York, NY [Google Scholar]
- 50.Bronk DA. 2002. Dynamics of organic nitrogen, p 153–247 In Hansell DA, Carlson CA. (ed), Biogeochemistry of marine dissolved organic matter. Academic Press, San Diego, CA [Google Scholar]
- 51.Pérez MT, Sommaruga R. 2006. Differential effect of algal- and soil-derived dissolved organic matter on alpine lake bacterial community composition and activity. Limnol. Oceanogr. 51:2527–2537 [Google Scholar]
- 52.Wetzel RG. 1992. Gradient-dominated ecosystems: sources and regulatory functions of dissolved organic matter in freshwater ecosystems. Hydrobiologia 229:181–198 [Google Scholar]
- 53.Santmire JA, Leff LG. 2007. The effect of sediment grain size on bacterial communities in streams. J. North Am. Benthol. Soc. 26:601–610 [Google Scholar]
- 54.McNamara CJ, Lemke MJ, Leff LG. 2002. Culturable and non-culturable fractions of bacterial populations in sediments of a South Carolina stream. Hydrobiologia 482:151–159 [Google Scholar]
- 55.Porter KG, Feig YS. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943–948 [Google Scholar]
- 56.Feinstein LM, Sul WJ, Blackwood CB. 2009. Assessment of bias associated with incomplete extraction of microbial DNA from soil. Appl. Environ. Microbiol. 75:5428–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blackwood CB, Oaks A, Buyer JS. 2005. Phylum- and class-specific PCR primers for general microbial community analysis. Appl. Environ. Microbiol. 71:6193–6198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acids techniques in bacterial systematics. Wiley, New York, NY [Google Scholar]
- 59.Johnson JL. 1994. Similarity analyses of rRNAs, p 683–700 In Gerhardt P, Murray RGE, Wood WA, Krieg NR. (ed), Methods for general and molecular bacteriology. ASM Press, Washington, DC [Google Scholar]
- 60.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. (ed). 1995. Short protocols in molecular biology. John Wiley & Sons Inc., New York, NY [Google Scholar]
- 61.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blackwood CB, Marsh T, Kim S-H, Paul EA. 2003. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69:926–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lueders T, Friedrich M. 2000. Archaeal population dynamics during sequential reduction processes in rice field soil. Appl. Environ. Microbiol. 66:2732–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kitts CL. 2001. Terminal restriction fragment patterns: a tool for comparing microbial communities and assessing community dynamics. Curr. Issues. Intest. Microbiol. 2:17–25 [PubMed] [Google Scholar]
- 65.Jones JB, Jr, Fisher SG, Grimm NB. 1995. Nitrification in the hyporheic zone of a desert stream ecosystem. J. North Am. Benthol. Soc. 14:249–258 [Google Scholar]
- 66.Currie WS. 1999. The responsive C and N biogeochemistry of the temperate forest floor. Trends Ecol. Evol. 14:316–320 [DOI] [PubMed] [Google Scholar]
- 67.Chapman PJ, Edwards AC, Reynolds B, Cresser MS, Neal C. 1998. The nitrogen content of rivers in upland Britain: the significance of organic nitrogen, p 443–450 In Kovar K, Tap-Peiner U, Peters NE, Craig RG. (ed), Hydrology, water resources and ecology in headwaters. Proceedings of the Head Water 98 Conference, Mereno, Italy, 20 to 23 April 1998 International Association of Hydrological Science, Wallingford, United Kingdom [Google Scholar]
- 68.Willett VB, Reynolds BA, Stevens PA, Ormerod SJ, Jones DL. 2004. Dissolved organic nitrogen regulation in freshwaters. J. Environ. Qual. 33:201–209 [DOI] [PubMed] [Google Scholar]
- 69.Middelboe M, Borch NH, Kirchman DL. 1995. Bacterial utilization of dissolved free amino acids, dissolved combined amino acids and ammonium in the Delaware Bay estuary: effects of carbon and nitrogen limitation. Mar. Ecol. Prog. Ser. 128:109–120 [Google Scholar]
- 70.Rosenstock B, Simon M. 2001. Sources and sinks of dissolved free amino acids and protein in a large and deep mesotrophic lake. Limnol. Oceanogr. 46:644–654 [Google Scholar]
- 71.Berg GM, Glibert PM, Lomas MW, Burford MA. 1997. Organic nitrogen uptake and growth by the chrysophyte Aureococcus anophagefferens during a brown tide event. Mar. Biol. 129:377–387 [Google Scholar]
- 72.Mulholland MR, Gobler CJ, Lee C. 2002. Peptide hydrolysis, amino acid oxidation, and nitrogen uptake in communities seasonally dominated by Aureococcus anophagefferens. Limnol. Oceanogr. 47:1094–1108 [Google Scholar]
- 73.Palenik B, Morel FMM. 1990. Amino-acid utilization by marine-phytoplankton—a novel mechanism. Limnol. Oceanogr. 35:260–269 [Google Scholar]
- 74.Mulholland MR, Lee C, Glibert PM. 2003. Extracellular enzyme activity and uptake of carbon and nitrogen along an estuarine salinity and nutrient gradient. Mar. Ecol. Prog. Ser. 258:3–17 [Google Scholar]
- 75.Manzoni S, Trofymow JA, Jackson RB, Porporato A. 2010. Stoichiometric controls on carbon, nitrogen, and phosphorus dynamics in decomposing litter. Ecol. Monogr. 80:89–106 [Google Scholar]
- 76.Carlsson P, Segatto AZ, Granéli E. 1993. Nitrogen bound to humic matter of terrestrial origin—a nitrogen pool for coastal phytoplankton? Mar. Ecol. Prog. Ser. 97:105–116 [Google Scholar]
- 77.Pomeroy LR, Wiebe WJ. 2001. Temperature and substrates as interactive limiting factor for marine heterotrophic bacteria. Aquat. Microb. Ecol. 23:187–204 [Google Scholar]
- 78.Eiler A, Langenheder S, Bertilsson S, Tranvik LJ. 2003. Heterotrophic bacterial growth efficiency and community structure at different natural organic carbon concentrations. Appl. Environ. Microbiol. 69:3701–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Reed HE, Martiny JBH. 2007. Testing the functional significance of microbial composition in natural communities. FEMS Microbiol. Ecol. 62:161–170 [DOI] [PubMed] [Google Scholar]
- 81.Guldberg LB, Finster K, Jorgenson NOG, Middelboe M, Lomstein BA. 2002. Utilization of marine sedimentary dissolved organic nitrogen by native anaerobic bacteria. Limnol. Oceanogr. 47:1712–1722 [Google Scholar]
- 82.Findlay S. 2003. Bacterial response to variation in dissolved organic matter, p 363–377 In Findlay SEG, Sinsabaugh RL. (ed), Aquatic ecosystems: interactivity of dissolved organic matter. Elsevier Science, New York, NY [Google Scholar]
- 83.Vrede K. 2005. Nutrient and temperature limitation of bacterioplankton growth in temperate lakes. Microb. Ecol. 49:245–256 [DOI] [PubMed] [Google Scholar]
- 84.Fisher MM, Klug JL, Lauster G, Newton M, Triplett MW. 2000. Effects of resources and trophic interactions on freshwater bacterioplankton diversity. Microb. Ecol. 40:125–138 [DOI] [PubMed] [Google Scholar]
- 85.del Giorgio PA, Gasol JM. 2008. Physiological structure and single-cell activity in marine bacterioplankton, p 243–298 In Kirchman DL. (ed), Microbial ecology of the oceans, 2nd ed. John Wiley & Sons Inc., Hoboken, NJ [Google Scholar]
- 86.Allison SD, Martiny JBH. 2008. Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U. S. A. 105:11512–11519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Comte J, del Giorgio PA. 2010. Linking the patterns of change in composition and functional capacities in bacterioplankton successions along environmental gradients. Ecology 91:1466–1476 [DOI] [PubMed] [Google Scholar]
- 88.Pinhassi J, Azam F, Hemphala J, Long RA, Martinez J, Zweifel UL, Hagstrom A. 1999. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat. Microb. Ecol. 17:13–26 [Google Scholar]
- 89.Morris CE, Rouse DI. 1985. Role of nutrients in regulating epiphytic bacterial populations, p 63–82 In Windels CE, Lindow SE. (ed), Biological control on the phylloplane. American Phytopathological Society, St. Paul, MN [Google Scholar]
- 90.Leclerc H. 2003. Relationships between common water bacteria and pathogens in drinking-water, p 80–118 In Bartram J, Cotruvo J, Exner M, Fricker C, Glasmacher A. (ed), Heterotrophic plate counts and drinking-water safety. IWA Publishing, London, United Kingdom [Google Scholar]
- 91.Tripathy S, Kumar N, Mohanty S, Samanta M, Mandal RN, Maiti NK. 2007. Characterisation of Pseudomonas aeruginosa isolated from freshwater culture systems. Microbiol. Res. 162:391–396 [DOI] [PubMed] [Google Scholar]
- 92.Bergan T. 1981. Human- and animal-pathogenic members of the genus Pseudomonas, p 666–700 In Starr MP, Stolp H, Tr̈uper HG, Balows A, Schlegel HG. (ed), The prokaryotes: a handbook on habitats, isolation, and identification of bacteria, vol 1 Springer-Verlag, Berlin, Germany [Google Scholar]
- 93.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]