Abstract
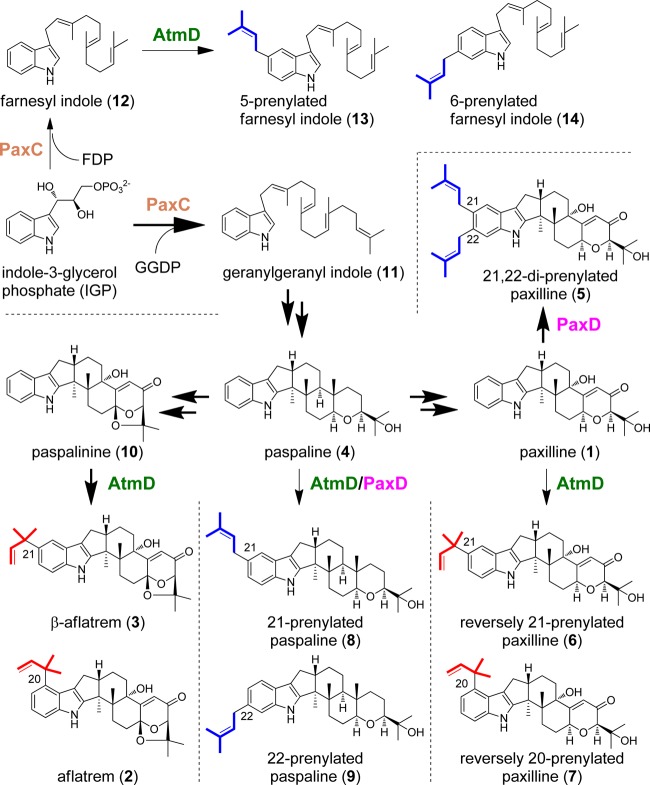
We recently reported the function of paxD, which is involved in the paxilline (compound 1) biosynthetic gene cluster in Penicillium paxilli. Recombinant PaxD catalyzed a stepwise regular-type diprenylation at the 21 and 22 positions of compound 1 with dimethylallyl diphosphate (DMAPP) as the prenyl donor. In this study, atmD, which is located in the aflatrem (compound 2) biosynthetic gene cluster in Aspergillus flavus and encodes an enzyme with 32% amino acid identity to PaxD, was characterized using recombinant enzyme. When compound 1 and DMAPP were used as substrates, two major products and a trace of minor product were formed. The structures of the two major products were determined to be reversely monoprenylated compound 1 at either the 20 or 21 position. Because compound 2 and β-aflatrem (compound 3), both of which are compound 1-related compounds produced by A. flavus, have the same prenyl moiety at the 20 and 21 position, respectively, AtmD should catalyze the prenylation in compound 2 and 3 biosynthesis. More importantly and surprisingly, AtmD accepted paspaline (compound 4), which is an intermediate of compound 1 biosynthesis that has a structure similar to that of compound 1, and catalyzed a regular monoprenylation of compound 4 at either the 21 or 22 position, though the reverse prenylation was observed with compound 1. This suggests that fungal indole diterpene prenyltransferases have the potential to alter their position and regular/reverse specificities for prenylation and could be applicable for the synthesis of industrially useful compounds.
INTRODUCTION
The isoprenoid compounds found in nature, with over 50,000 known examples, include industrially useful compounds like flavors, antibiotics, and plant hormones, among others (1–3). In some cases, isoprenoids are attached to other moieties, such as polyketide (4), indole/tryptophan (5), (iso)flavonoid (6), and phenazine moieties (7, 8). The isoprenoid moieties of these compounds are known to be important for their biological activities (9–11). For example, the presence of isoprenoid chains of various lengths and types is a major determinant of the bioactivity of prenylated flavonoids (12–14). The polyketide-isoprenoid hybrid compounds furaquinocin, naphterpin, marinone, and napyradiomycin have been reported to have antitumor (15), antioxidative (16), and anticancer (17) activities and to act as a nonsteroidal estrogen receptor antagonist (18), respectively. These molecules have similar polyketide moieties derived from 1,3,6,8-tetrahydroxynaphthalene (THN), showing that prenyl moieties play important roles in providing diverse biological activities. Therefore, prenyltransferases catalyzing the prenylation of various substrates at specific positions are very useful.
Recently, we characterized paxD (19), which is located next to paxQ in the paxilline (compound 1) biosynthetic gene cluster and has weak similarities to fungal prenyltransferase genes (20). Recombinant PaxD catalyzed the successive regular attachment of dimethylallyl diphosphate (DMAPP) to positions 21 and 22 of compound 1 to form compound 5 via a monoprenylated compound 1 intermediate (Fig. 1). A BLAST search showed that the enzyme most homologous to PaxD was the atmD product (32% amino acid identity) (19), which is located in the aflatrem (compound 2) biosynthetic gene cluster in Aspergillus flavus. However, compound 2 and β-aflatrem (compound 3), which are compound 1-related compounds, were reversely monoprenylated at the 20 and 21 position (21), respectively (Fig. 1). Therefore, we examined whether AtmD catalyzes the reverse prenylation to produce these compounds, or regular diprenylation, like PaxD. During the study, more importantly and surprisingly, we found that AtmD and PaxD accepted paspaline (compound 4), an intermediate of compound 1 biosynthesis that has a similar structure to compound 1, and that both enzymes unexpectedly showed different positions and regular/reverse specificities to those with compound 1. These results suggested that fungal indole diterpene prenyltransferases could be applicable for the synthesis of important compounds, including bioactive compounds.
Fig 1.
Summary of reactions catalyzed by AtmD and PaxD. The regular biosynthetic pathways are highlighted by bold arrows. Putative aflatrem and β-aflatrem biosynthetic pathways are also shown.
MATERIALS AND METHODS
General.
Sequence analysis of PCR fragments was performed by the dideoxy chain termination method with an automatic DNA sequencer (model 4000L; Li-Cor, Lincoln, NE). Cell disruption was performed with an ultrasonic disruptor (UD-200; Tomy, Tokyo, Japan). Analysis of the samples during protein purification was performed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the proteins were visualized by Coomassie brilliant blue staining. Protein concentrations were determined by the Bradford method (22) with bovine serum albumin as a standard. Plasmids from Escherichia coli were prepared using a Qiagen plasmid kit (Hilden, Germany). All restriction enzymes, T4 ligase, and calf intestinal alkaline phosphatase were obtained from Toyobo (Osaka, Japan) and used according to the manufacturer's instructions. Farnesyl indole (compound 12) and geranylgeranyl indole (compound 11) were synthesized according to previously reported methods (23).
Strain.
A. flavus NBRC 4295 was obtained from the Biological Resource Center, National Institute of Technology and Evaluation (NITE), Tokyo, Japan. This was used for the preparation of atmD cDNA because we could not obtain A. flavus NRRL6541, from which the compound 2 biosynthetic gene cluster was isolated by Nicholson et al. (21) The strain NBRC 4295 was suggested to produce compound 2 by liquid chromatography-electrospray ionization-mass spectrometry (LC-ESI-MS) analysis. The presence of the compound 2 biosynthetic gene cluster in the genome was confirmed by PCR with specific primers (see Table S1 and Fig. S1 in the supplemental material), which were designed based on the sequences of each atm gene in A. flavus NRRL6541. Cultivation and cDNA preparation were performed using the same method we used for Penicillium paxilli (19).
Cloning, overexpression, and purification of AtmD.
The cDNA carrying the atmD gene was amplified by PCR using the gene-specific primers 5′-TTGCATGCATGTCCACTCCCAAGTCGGATACATGC-3′ and 5′-ATCTGCAGCTACTTGGAAAGCCCCTTCACATCTGAC-3′. After subcloning and sequence confirmation, a 1.3-kb fragment obtained by SphI and PstI digestion was ligated into the same sites of the pQE30 plasmid (Qiagen) to construct pQE30-AtmD. E. coli M15 carrying pQE30 (a control) and pQE30-AtmD were grown separately in Luria-Bertani broth supplemented with 100 μg/ml ampicillin. Expression and purification procedures were the same as previously reported (19, 24). For molecular-mass and subunit structure determination of AtmD, the purified enzyme was loaded onto a HiLoad 26/60 Superdex 75 prep grade gel filtration column (Amersham Biosciences, Piscataway, NJ) and eluted with a buffer containing 50 mM Tris-HCl (pH 8.0) and 10 mM NaCl. The retention time of the eluted AtmD was compared with those of marker proteins. The markers applied to the column were aldolase (158 kDa), albumin (67 kDa), ovalbumin (43 kDa), and chymotrypsinogen A (25 kDa) (GE Healthcare, Little Chalfont, United Kingdom).
In vitro assay of prenyltransferases.
The standard assay mixture for AtmD and PaxD contained, in a final volume of 100 μl, 0.25 mM prenyl acceptors, 0.5 mM DMAPP, 50 mM Tris-HCl (pH 8.0), and 10 μM enzyme. This mixture was incubated at 30°C overnight, and the reaction was stopped by the addition of 100 μl methanol. The products were analyzed and purified by high-performance liquid chromatography (HPLC). The analytical conditions for the charts shown in panels A and B of Fig. 3, 4, and 5 were as follows: column, Merck Mightysil RP-18GP Aqua column (250 by 4.6 mm); mobile phase, acetonitrile in water (0 to 25 min, 70% acetonitrile; 25 to 40 min, 70 to 100%; 40 to 50 min, 100%); flow rate, 1.0 ml/min; detection, 230 nm. In the experiments whose results are shown in Fig. S4A, S5A, and S13 in the supplemental material, the conditions were as follows: column, Merck Mightysil RP-18GP Aqua column (250 by 4.6 mm); mobile phase, acetonitrile in water (0 to 35 min, 0 to 100%; 35 to 60 min, 100%); flow rate, 1.0 ml/min; detection, 278 nm. The analytical conditions for the Waters XBridge phenyl 5-μm column (250 by 4.6 mm) (see Fig. S6) were as follows: mobile phase, 65% (vol/vol) acetonitrile solution in water by isocratic flow; flow rate, 1.0 ml/min; detection, 230 nm.
Fig 3.
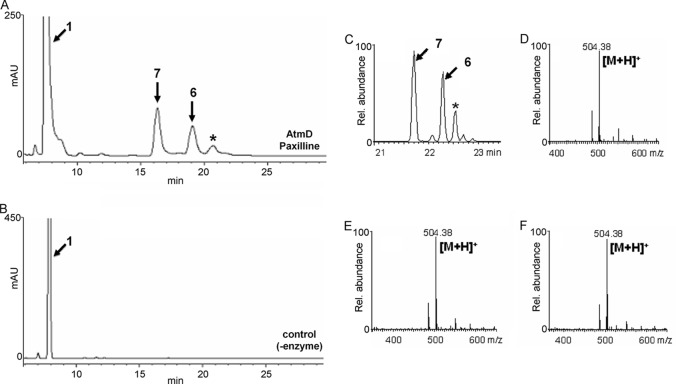
HPLC and LC-ESI-MS analysis of the reaction products formed from compound 1 and DMAPP by AtmD. The reaction products formed with (A, C) and without (B) AtmD were analyzed by HPLC (A, B) and LC-ESI-MS (C to F). Selected ion chromatograms (C) and spectra of the major products, compounds 7 (D) and 6 (E), and the minor product (F), indicated by the asterisk in panel C, are shown.
Fig 4.
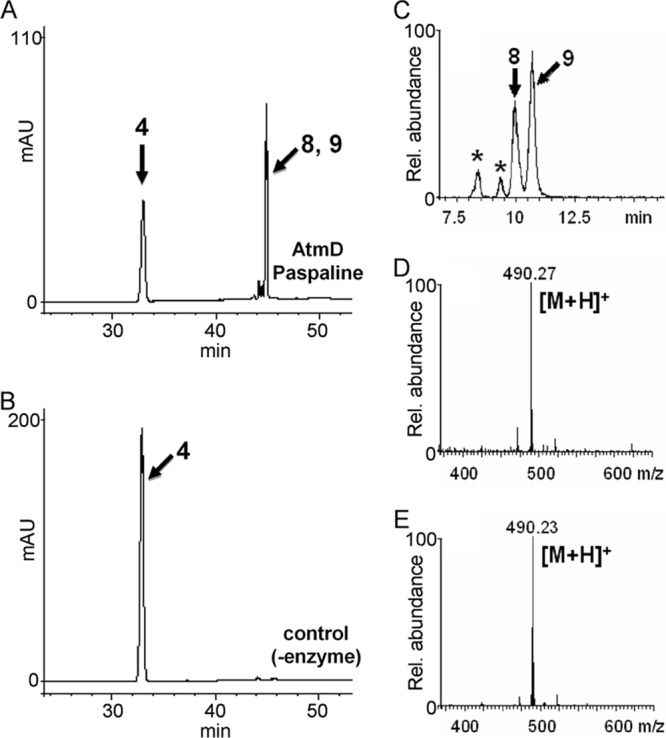
HPLC and LC-ESI-MS analysis of the reaction products formed from compound 4 and DMAPP by AtmD. The reaction products formed with (A, C) and without (B) AtmD were analyzed by HPLC (A, B) and LC-ESI-MS (C to E). Selected ion chromatograms (C) and spectra of compounds 8 (D) and 9 (E) are shown. Asterisks indicate unknown products.
Fig 5.
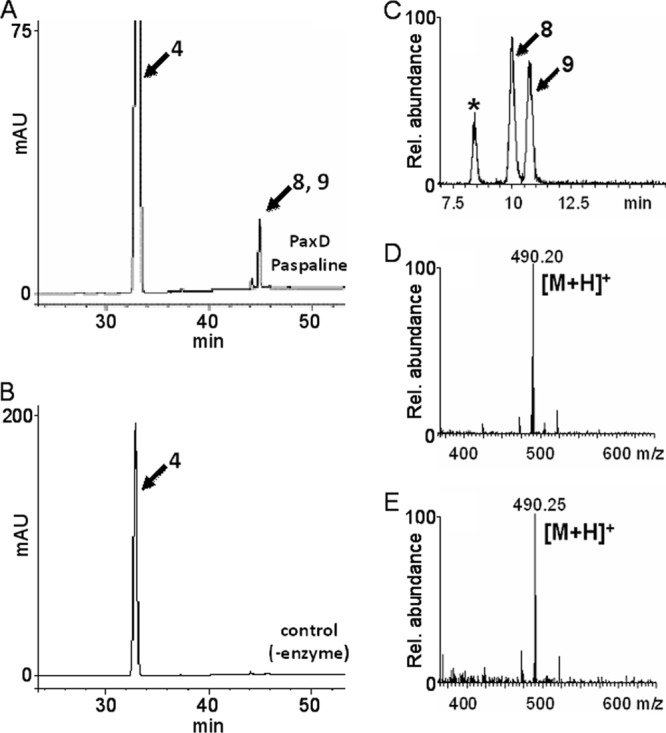
HPLC and LC-ESI-MS analysis of the reaction products formed from compound 4 and DMAPP by PaxD. The reaction products formed with (A, C) and without (B) PaxD were analyzed by HPLC (A, B) and LC-ESI-MS (C to E). Selected ion chromatograms (C) and spectra of compounds 8 (D) and 9 (E) are shown. The asterisk indicates an unknown product.
The following compounds were used to examine substrate specificity: compound 4 (purified from culture broth of Aspergillus oryzae carrying paxG, paxM, paxB, and paxC), cyclodipeptides (cyclo-l-Trp-l-Tyr, cyclo-l-Pro-l-Tyr, cyclo-l-His-l-Phe, cyclo-l-Phe-l-Pro, cyclo-l-Phe-l-Trp, and cyclo-l-Phe-l-Leu, all of which were kindly provided by H. Kanzaki of Okayama University, Japan), hydroxynaphthalenes (1-naphthol, 1,3-dihydroxynaphthalene, 2,6-dihydroxynaphthalene, 2,7-dihydroxynaphthalene, and 3,7-dihydroxy-2-naphtholic acid), indole, l-tryptophan, and l-tyrosine.
The steady-state kinetic parameters of AtmD and PaxD were determined by fitting to the Michaelis-Menten equation. The assay was linear with respect to protein concentration up to 5 μg for a 20-min incubation, and no substrate inhibition was observed with compound 1 or 4 or DMAPP up to 1.0 mM each substrate. The assay mixtures used to determine the kinetic parameters of AtmD with compound 1 as a substrate contained, in a final volume of 100 μl, 50 mM Tris-HCl (pH 8.0), 0.5 mM DMAPP, 0.5 μg of enzyme, and 0.5 μM to 0.1 mM compound 1. When the concentration of compound 1 was fixed at 0.25 mM, the concentration of DMAPP was varied from 0.02 μM to 10 μM. The mixtures were incubated at 30°C for 10 min. To determine the kinetic parameters with compound 4, 0.01 mM to 1 mM compound 4 with 0.5 mM DMAPP and 0.01 mM to 1.5 mM DMAPP with 0.25 mM compound 4 were used as substrates. The mixtures were incubated at 30°C for 20 min.
Metal dependency of AtmD.
Divalent metal ions (5 mM Mg2+, Ca2+, Fe2+, Cu2+, Zn2+, Mn2+, Ni2+, and Co2+) or 5 mM EDTA was added to the standard reaction mixture.
LC-ESI-MS analysis.
Products formed in the in vitro assays were analyzed by LC-ESI-MS (Waters Acquity UPLC equipped with an SQD2) with a Waters Acquity UPLC BEH C18 1.7-μm column (2.1 by 50 mm) (see Fig. 3C) or a Waters Acquity UPLC BEH phenyl 1.7-μm column (2.1 by 50 mm) (see Fig. 4C and 5C). The analytical conditions were as described previously (19).
Structural analysis of the reaction products formed from compounds 1, 4, and 12 with DMAPP.
The reaction products using compound 1, 4, or 12 with DMAPP were fractionated with HPLC. 1H- and 13C-NMR (nuclear magnetic resonance) spectra were recorded on a Bruker AMX-500 spectrometer as follows: reversely monoprenylated compound 1 at position 21 formed by AtmD (compound 6) (see Fig. S14 to S20 in the supplemental material), high-resolution (HR)-ESI-MS: [M+H]+ (calculated: 504.3108, observed: 504.3100); reversely monoprenylated compound 1 at position 20 formed by AtmD (compound 7) (see Fig. S14 and S21 to S26), HR-ESI-MS: [M+H]+ (calculated: 504.3108, observed: 504.3110); regularly monoprenylated compound 4 at position 21 formed by AtmD and PaxD (compound 8) (see Fig. S14 and S27 to S32), HR-ESI-MS: [M+H]+ (calculated: 490.3680, observed: 490.3675); regularly monoprenylated compound 4 at position 22 formed by AtmD and PaxD (compound 9) (see Fig. S14 and S33 to S38), HR-ESI-MS: [M+H]+ (calculated: 490.3680, observed: 490.3703); and regularly monoprenylated compound 12 at either the 5 (compound 13) or 6 position (compound 14) formed by AtmD (see Fig. S14 and S39 to S44), HR-ESI-MS: [M+H]+ (calculated: 390.3155, observed: 390.3121). Key signals of indole and the dimethylallyl chain of compounds 13 and 14 were assigned as follows (with the relevant compound indicated by suffix “-13” or “-14”): NMR δH (CDCl3, 500 MHz) (see Fig. S39) 1.60 (s), 1.68 (s), 1.75 (s), 1.76 (s), 1.90 to 2.20 (m), 3.44 (m, H1″ and H1′), 5.09 (m), 5.12 (m), 5.38 (m), 5.43 (m), 6.86 (brs, 1H, H2-14), 6.89 (brs, 1H, H2-13), 6.94 (brd, J = 8.1 Hz, 1H, H5-14), 7.00 (brd, J = 8.2 Hz, 1H, H6-13), 7.13 (s, 1H, H7-14), 7.24 (d, J = 8.2 Hz, 1H, H7-13), 7.36 (s, 1H, H4-13), 7.48 (d, J = 8.1 Hz, 1H, H4-14), 7.74 (s, 1H, NH-14), 7.77 (s, 1H, NH-13); NMR δC (CDCl3, 125 MHz) (see Fig. S40) 16.0, 16.1, 17.7, 17.8, 24.0, 24.1, 25.7, 25.8, 26.6, 26.7, 26.8, 34.5 (C1″-13), 34.6 (C1″-14), 39.7 (x2), 110.2 (C7-14), 110.8 (C7-13), 115.8 (C3-13), 116.0 (C3-14), 118.0 (C4-13), 118.9 (C4-14), 120.2 (C5-14), 120.6 (C2-14), 121.4 (C2-13), 122.8 (C6-13), 123.0 (x2), 124.1, 124.3, 124.4, 124.6, 125.7 (C3a-14), 127.8 (C3a-13), 131.3, 131.5, 131.9, 132.5 (C5-13), 135.0, 135.1 (C7a-13), 135.5 (x2), 135.8 (C6-14), 137.0 (C7a-14).
Nucleotide sequence accession number.
Newly determined sequence data have been deposited in GenBank under accession number AB778117.
RESULTS
Functional analysis of AtmD.
A cDNA of atmD was amplified based on the nucleotide sequence of the cDNA reported by Nicholson et al. (21) (GenBank accession number CAP53937). The predicted gene product consisted of 435 amino acids (GenBank accession number AB778117) and had 96% amino acid identity with the sequence with accession number CAP53937 (see Fig. S2 in the supplemental material). The AtmD cDNA was cloned into the pQE30 vector for protein expression in E. coli. His-tagged AtmD recombinant enzyme was successfully expressed as a soluble form. Purified enzymes were obtained by Ni2+ column chromatography and successive desalting with Amicon ultra devices. The recombinant AtmD obtained, with a calculated molecular mass of 46 kDa, was subjected to gel filtration and SDS-PAGE analyses. As shown by the data in Fig. 2, one major peak with a calculated molecular mass of 95 kDa and a band of approximately 46 kDa were detected by gel filtration and SDS-PAGE, respectively, suggesting that AtmD forms a homodimer, similar to PaxD.
Fig 2.
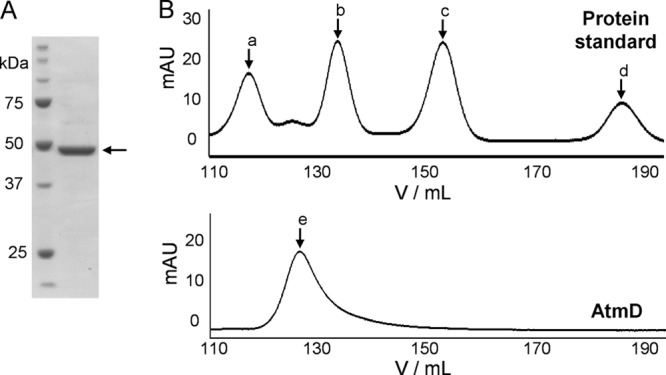
Purified AtmD was analyzed by SDS-PAGE and gel filtration chromatography. (A) Molecular mass markers (1st lane) and purified AtmD (2nd lane). (B) Elution profiles of the standard proteins (aldolase [a, 158 kDa], albumin [b, 67 kDa], ovalbumin [c, 43 kDa], and chymotrypsinogen A [d, 25 kDa]; top) and purified AtmD (bottom, e).
The recombinant AtmD was used for in vitro assay. We used as a prenyl acceptor commercially available compound 1, which has a structure similar to that of paspalinine (compound 10), a probable intrinsic substrate (Fig. 1). After the recombinant AtmD was incubated with compound 1 and DMAPP, the reaction products were analyzed by HPLC. Two major products and a trace of a minor product (Fig. 3A) were specifically detected. The total ion chromatograms obtained by LC-ESI-MS analysis showed three specific peaks with molecular masses corresponding to monoprenylated compound 1 (see Fig. S3 in the supplemental material). Moreover, selected ion chromatograms and their mass spectra strongly suggested that all products were monoprenylated compound 1 (Fig. 3C). Because the yield of the minor product was low, the exact structures of the two major products were analyzed. HR-ESI-MS of both products indicated the molecular formula C32H41NO4, supporting the idea that both products were monoprenylated compound 1. The 1H-NMR spectra of one major product (compound 6) showed new signals assigned to a reversely prenylated moiety at δ = 5.04 (dd, 1H), δ = 5.11 (dd, 1H), δ = 6.11 (dd, 1H), and 1.47 (s, 6H). Extensive NMR data analysis, including correlation spectroscopy (COSY), heteronuclear single-quantum correlation spectroscopy (HSQC), heteronuclear multiple-bond correlation spectroscopy (HMBC), and nuclear Overhauser effect spectroscopy (NOESY), proved that the structure was reversely monoprenylated compound 1 at position 21 (compound 6) (see Table S3 and Fig. S15 to S20). The 1H-NMR spectra of the other major product (compound 7) also showed new signals for a reversely prenylated moiety at δ = 4.88 (d, 1H), δ = 5.00 (d, 1H), δ = 6.26 (dd, 1H), 1.52 (s, 3H), and 1.53 (s, 3H). Subsequently, extensive NMR data analysis, including COSY, HSQC, HMBC, and NOESY, proved that the structure was reversely monoprenylated compound 1 at position 20 (compound 7) (see Table S4 and Fig. S21 to S26). Considering that the atmD gene is involved in the compound 2 biosynthetic gene cluster and that compounds 2 and 3 have a reversely attached prenyl moiety at the same positions as those formed with compound 1 (Fig. 1), AtmD should catalyze prenylation in compound 2 and 3 biosynthesis.
Biochemical characterization of AtmD.
The substrate specificity of the AtmD enzyme was investigated. For the prenyl acceptor, compounds 4, 11, and 12 (Fig. 1), related to indole diterpene biosynthesis, were examined with DMAPP as a prenyl donor. We also used several cyclodipeptides and hydroxynaphthalenes, because they were reported to be utilized by many fungal prenyltransferases (25–29). Of these compounds, compound 4 (Fig. 4), compound 11 (see Fig. S5 in the supplemental material), and compound 12 (see Fig. S4) were suggested by LC-ESI-MS analysis to be monoprenylated. Because the yield of prenylated compound 11 was low, the structures of prenylated compounds 4 and 12 were determined. HR-ESI-MS of the former and latter products indicated the molecular formulas C33H47NO2 and C28H39N, supporting the production of monoprenylated compounds 4 and 12. The exact structures of both products were elucidated by NMR analysis, but both samples were a mixture of two closely related compounds. We tried to separate each of the compounds using several different columns, and the XBridge phenyl column was found to be effective (see Fig. S6). In the case of monoprenylated compound 4, the yield was relatively high and each of the products was successfully separated and used for NMR analysis. Very interestingly and surprisingly, the 1H-NMR spectra of compound 8 showed new signals for a regularly prenylated moiety at δ = 3.40 (d, 2H), δ = 5.38 (m, 1H), δ = 1.73 (s, 3H), andδ = 1.75 (s, 3H) (see Table S5 and Fig. S27 to S32). Compound 9 also showed similar 1H-NMR spectra, with characteristic signals at δ = 3.41 (d, 2H), δ = 5.37 (m, 1H), δ = 1.73 (s, 3H), and δ = 1.73 (s, 3H) (see Table S6 and Fig. S33 to S38). Finally, one was determined to be regularly monoprenylated compound 4 at the 21 position (compound 8) and the other regularly monoprenylated compound 4 at the 22 position (compound 9). This was contrary to our expectations, because reverse prenylation at the 20 and 21 positions (compounds 7 and 6) occurred with compound 1 (Fig. 3). For monoprenylated compound 12, we conducted NMR analysis without separation of the two regioisomers (1:2.6 mixture) because the low yield prevented us from isolating a sufficient amount of each product. Typical signals for a regular dimethylallyl moiety were found at δ = 3.44 (m, 2H) and δ = 5.38 (m, 1H). Key HMBC, H-H COSY, and NOESY correlations were similar to those of compounds 8 and 9, suggesting that the prenylation takes place at the 5 and 6 positions on the indole moiety (see Fig. S39 to S44). Taking these results together, the products were determined to be regularly monoprenylated compound 12 at the 5 and 6 positions (compounds 13 and 14).
We next examined the substrate specificity of the prenyl donors. Aside from DMAPP, geranyl diphosphate, farnesyl diphosphate, and geranylgeranyl diphosphate were examined. However, no products were formed with compounds 1 and 4 as prenyl acceptors.
The biochemical properties of AtmD were investigated using compound 1 and DMAPP as substrates. Under the conditions described in Materials and Methods, product formation was optimal at 50°C and around pH 7.0 (see Fig. S7 and S8, respectively, in the supplemental material). The enzyme showed similar activity regardless of the presence of 5 mM EDTA, suggesting that it did not require Mg2+ for its activity. In contrast, Cu2+ and Zn2+ significantly inhibited its activity (see Fig. S9).
The kinetic parameters of AtmD were investigated. The enzyme reaction followed Michaelis-Menten kinetics. Using Hanes-Woolf plots (see Fig. S10 in the supplemental material), the Km values were calculated as 13.8 ± 0.9 μM (mean ± standard deviation) for compound 1 and 2.3 ± 0.1 μM for DMAPP. The kcat values were calculated as 0.38 ± 0.01/s. We also investigated the kinetic parameters with compound 4 as the substrate (see Fig. S11). The Km values were calculated as 131 ± 5 μM and 302 ± 11 μM for compound 4 and DMAPP, respectively. The kcat value was 0.09 ± 0.001/s, and the kcat/Km value was considerably lower than for compound 1. This low value was consistent with the fact that prenylated compound 4 has not been reported as a natural product.
PaxD also accepted compound 4.
Because AtmD accepted compounds 4, 11, and 12, we examined whether PaxD, which was previously shown to catalyze stepwise regular diprenylation at the 21 and 22 positions of compound 1 to form compound 5, was also able to use these compounds as prenyl acceptors. In this case, compounds 4 (Fig. 5) and 12 (see Fig. S4 in the supplemental material) were suggested by LC-ESI-MS analysis to be monoprenylated, and no diprenylated products were detected. The retention times and the observed mass spectra of both products were the same as those of the products formed by AtmD with compound 4. The products formed from compound 4 also contained two closely related compounds (see Fig. S6), and each of the products was purified and determined to be the same ones (compounds 8 and 9) formed by AtmD from compound 4 and DMAPP. Then, the kinetic parameters of PaxD were compared with those of AtmD. PaxD showed a similar Km value for compound 4 (124 ± 8 μM) and a very low value for DMAPP (7.9 ± 0.4 μM). The kcat value (0.07/s) was almost the same as that of AtmD (see Fig. S12).
DISCUSSION
In this study, we showed that AtmD and PaxD could accept the intermediate compounds of compound 1 biosynthesis. AtmD, whose intrinsic substrate is compound 10, utilized compounds 1, 4, 11, and 12. PaxD also accepted compounds 4 and 12 besides its real substrate, compound 1. These results suggested that the prenyltransferases responsible for indole diterpene biosynthesis possess broad substrate specificities. To examine this possibility, we investigated the substrate specificity of PaxC, which has been shown to catalyze the formation of geranylgeranyl indole from geranylgeranyl diphosphate and indole-3-glycerol phosphate (or indole) (30) and has no similarities to PaxD or AtmD. Although PaxC accepted none of the compounds used as prenyl acceptors for the same assay with AtmD and PaxD, the enzyme accepted farnesyl diphosphate as a prenyl donor to yield compound 12 (see Fig. S13 in the supplemental material) with a slightly lower kcat/Km value (16.6 s−1 mM−1) (see Table S2) than for geranyl geranyl diphosphate (278.1 s−1 mM−1) (30).
More importantly and surprisingly, AtmD catalyzed prenylation of compounds 1 and 4 at different positions and with regular/reverse specificities. AtmD catalyzed a reverse monoprenylation at either position 20 (compound 7) or 21 (compound 6) with compound 1 and DMAPP as substrates (Fig. 3). In contrast, regular monoprenylation at either position 21 or 22 (compounds 8 and 9) was observed with compound 4 as a substrate (Fig. 4). Moreover, PaxD, which had been shown to produce a regularly diprenylated product at the 21 and 22 positions (compound 5) from compound 1 (19), catalyzed the same reactions as those of AtmD with compound 4 (Fig. 5). We are unable to estimate the reaction mechanisms to explain why these enzymes altered their position specificity, the regular/reverse mode for prenylation, and the number of DMAPP introduced to structurally related compounds; additional experiments, such as molecular evolution engineering and site-directed mutagenesis based on X-ray structures of the enzymes, may give us an answer.
Besides the enzymes we studied, CdpNPT (31), AnaPT (32), and CdpC3PT (33), whose real substrates are probably cyclo-l-Trp-l-Tyr, (R)-benzodiazepinedione, and several cyclic dipeptides, respectively, were recently shown to accept hydroxynaphthalenes as substrates (28). FtmPT1 was also demonstrated to catalyze the prenylation of a nonaromatic carbon of an indole derivative to give α-prenylindolylbutenone (34). Considering these previous results and those of our current study together, some fungal prenyltransferases are suggested to have the potential to accept a variety of substrates with broad position and regular/reverse mode specificities. Such enzymes could therefore be applicable for the synthesis of industrially useful compounds.
Moreover, many cyclic dipeptide prenyltransferases have been reported to accept cyclic dipeptide/amino acid derivatives other than their intrinsic substrates (35–41). For example, FgaPT2, FtmPT1, and 7-DMATS have strict position specificities and essentially introduce DMAPP into the same positions as their intrinsic substrates (the C-4, C-2, and C-7 position, respectively, of the indole moiety). SirD also selectively introduces DMAPP into the C-4 benzene ring. Therefore, these enzymes could be applicable for position-specific prenylation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Grants-in-Aid for Scientific Research (grant 23108101 to T.D. and grant 22108002 to H.O.) from the Japan Society for the Promotion of Science (JSPS).
Footnotes
Published ahead of print 13 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02496-13.
REFERENCES
- 1.Christianson DW. 2007. Chemistry. Roots of biosynthetic diversity. Science 316:60–61 [DOI] [PubMed] [Google Scholar]
- 2.Connolly JD, Hill RA. 1992. Dictionary of terpenoids. Chapman and Hall, New York, NY [Google Scholar]
- 3.Dewick PM. 2002. The biosynthesis of C5-C25 terpenoid compounds. Nat. Prod. Rep. 19:181–222 [DOI] [PubMed] [Google Scholar]
- 4.Lo HC, Entwistle R, Guo CJ, Ahuja M, Szewczyk E, Hung JH, Chiang YM, Oakley BR, Wang CC. 2012. Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. J. Am. Chem. Soc. 134:4709–4720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steffan N, Grundmann A, Yin WB, Kremer A, Li SM. 2009. Indole prenyltransferases from fungi: a new enzyme group with high potential for the production of prenylated indole derivatives. Curr. Med. Chem. 16:218–231 [DOI] [PubMed] [Google Scholar]
- 6.Tahara S, Ibrahim RK. 1995. Prenylated isoflavonoids—an update. Phytochemistry 38:1073–1094 [Google Scholar]
- 7.Izumikawa M, Khan ST, Takagi M, Shin-ya K. 2010. Sponge-derived Streptomyces producing isoprenoids via the mevalonate pathway. J. Nat. Prod. 73:208–212 [DOI] [PubMed] [Google Scholar]
- 8.Saleh O, Haagen Y, Seeger K, Heide L. 2009. Prenyl transfer to aromatic substrates in the biosynthesis of aminocoumarins, meroterpenoids and phenazines: the ABBA prenyltransferase family. Phytochemistry 70:1728–1738 [DOI] [PubMed] [Google Scholar]
- 9.Dalziel JE, Finch SC, Dunlop J. 2005. The fungal neurotoxin lolitrem B inhibits the function of human large conductance calcium-activated potassium channels. Toxicol. Lett. 155:421–426 [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez MC, Lull C, Moya P, Ayala I, Primo J, Primo Yufera E. 2003. Insecticidal activity of penitrems, including penitrem G, a new member of the family isolated from Penicillium crustosum. J. Agric. Food Chem. 51:2156–2160 [DOI] [PubMed] [Google Scholar]
- 11.Singh SB, Ondeyka JG, Jayasuriya H, Zink DL, Ha SN, Dahl-Roshak A, Greene J, Kim JA, Smith MM, Shoop W, Tkacz JS. 2004. Nodulisporic acids D-F: structure, biological activities, and biogenetic relationships. J. Nat. Prod. 67:1496–1506 [DOI] [PubMed] [Google Scholar]
- 12.Wang BH, Ternai B, Polya G. 1997. Specific inhibition of cyclic AMP-dependent protein kinase by warangalone and robustic acid. Phytochemistry 44:787–796 [DOI] [PubMed] [Google Scholar]
- 13.Maitrejean M, Comte G, Barron D, El Kirat K, Conseil G, Di Pietro A. 2000. The flavanolignan silybin and its hemisynthetic derivatives, a novel series of potential modulators of P-glycoprotein. Bioorg. Med. Chem. Lett. 10:157–160 [DOI] [PubMed] [Google Scholar]
- 14.Murakami A, Gao G, Omura M, Yano M, Ito C, Furukawa H, Takahashi D, Koshimizu K, Ohigashi H. 2000. 1,1-Dimethylallylcoumarins potently suppress both lipopolysaccharide- and interferon-gamma-induced nitric oxide generation in mouse macrophage RAW 264.7 cells. Bioorg. Med. Chem. Lett. 10:59–62 [DOI] [PubMed] [Google Scholar]
- 15.Komiyama K, Funayama S, Anraku Y, Ishibashi M, Takahashi Y, Omura S. 1990. Novel antibiotics, furaquinocins A and B. Taxonomy, fermentation, isolation and physico-chemical and biological characteristics. J. Antibiot. 43:247–252 [DOI] [PubMed] [Google Scholar]
- 16.Shin-ya K, Imai Furihata S, Hayakawa K, Kato Y, Vanduyne Y, Clardy GD, Seto JH. 1990. Isolation and structural elucidation of an antioxidative agent, naphterpin. J. Antibiot. 43:444–447 [DOI] [PubMed] [Google Scholar]
- 17.Pathirana C, Jensen PR, Fenical W. 1992. Marinone and debromomarinone: Antibiotic sesquiterpenoid naphthoquinones of a new structure class from a marine bacterium. Tetrahedron Lett. 33:7663–7666 [Google Scholar]
- 18.Shiomi K, Nakamura H, Iinuma H, Naganawa H, Isshiki K, Takeuchi T, Umezawa H, Iitaka Y. 1986. Structures of new antibiotics napyradiomycins. J. Antibiot. 39:494–501 [DOI] [PubMed] [Google Scholar]
- 19.Liu C, Noike M, Minami A, Oikawa H, Dairi T. 24 March 2013. Functional analysis of a prenyltransferase gene (paxD) in the paxilline biosynthetic gene cluster. Appl. Microbiol. Biotechnol. 10.1007/s00253-013-4834-9 [DOI] [PubMed] [Google Scholar]
- 20.Young C, McMillan L, Telfer E, Scott B. 2001. Molecular cloning and genetic analysis of an indole-diterpene gene cluster from Penicillium paxilli. Mol. Microbiol. 39:754–764 [DOI] [PubMed] [Google Scholar]
- 21.Nicholson MJ, Koulman A, Monahan BJ, Pritchard BL, Payne GA, Scott B. 2009. Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl. Environ. Microbiol. 75:7469–7481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 23.Zhu X, Ganesan A. 2002. Regioselective synthesis of 3-alkylindoles mediated by zinc triflate. J. Org. Chem. 67:2705–2708 [DOI] [PubMed] [Google Scholar]
- 24.Noike M, Liu C, Ono Y, Hamano Y, Toyomasu T, Sassa T, Kato N, Dairi T. 2012. An enzyme catalyzing O-prenylation of the glucose moiety of fusicoccin A, a diterpene glucoside produced by the fungus Phomopsis amygdali. Chembiochem 13:566–573 [DOI] [PubMed] [Google Scholar]
- 25.Grundmann A, Li SM. 2005. Overproduction, purification and characterization of FtmPT1, a brevianamide F prenyltransferase from Aspergillus fumigatus. Microbiology 151:2199–2207 [DOI] [PubMed] [Google Scholar]
- 26.Haug-Schifferdecker E, Arican D, Bruckner R, Heide L. 2010. A new group of aromatic prenyltransferases in fungi, catalyzing a 2,7-dihydroxynaphthalene 3-dimethylallyl-transferase reaction. J. Biol. Chem. 285:16487–16494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin S, Yu X, Wang Q, Liu XQ, Li SM. 2013. Identification of a brevianamide F reverse prenyltransferase BrePT from Aspergillus versicolor with a broad substrate specificity towards tryptophan-containing cyclic dipeptides. Appl. Microbiol. Biotechnol. 97:1649–1660 [DOI] [PubMed] [Google Scholar]
- 28.Yu X, Xie X, Li SM. 2011. Substrate promiscuity of secondary metabolite enzymes: prenylation of hydroxynaphthalenes by fungal indole prenyltransferases. Appl. Microbiol. Biotechnol. 92:737–748 [DOI] [PubMed] [Google Scholar]
- 29.Zou HX, Xie XL, Linne U, Zheng XD, Li SM. 2010. Simultaneous C7- and N1-prenylation of cyclo-L-Trp-L-Trp catalyzed by a prenyltransferase from Aspergillus oryzae. Org. Biomol. Chem. 8:3037–3044 [DOI] [PubMed] [Google Scholar]
- 30.Tagami K, Liu C, Minami A, Noike M, Isaka T, Fueki S, Shichijo Y, Toshima H, Gomi K, Dairi T, Oikawa H. 2013. Reconstitution of biosynthetic machinery for indole-diterpene paxilline in Aspergillus oryzae. J. Am. Chem. Soc. 135:1260–1263 [DOI] [PubMed] [Google Scholar]
- 31.Yin WB, Ruan HL, Westrich L, Grundmann A, Li SM. 2007. CdpNPT, an N-prenyltransferase from Aspergillus fumigatus: overproduction, purification and biochemical characterisation. Chembiochem 8:1154–1161 [DOI] [PubMed] [Google Scholar]
- 32.Yin WB, Grundmann A, Cheng J, Li SM. 2009. Acetylaszonalenin biosynthesis in Neosartorya fischeri. Identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J. Biol. Chem. 284:100–109 [DOI] [PubMed] [Google Scholar]
- 33.Yin WB, Yu X, Xie XL, Li SM. 2010. Preparation of pyrrolo[2,3-b]indoles carrying a beta-configured reverse C3-dimethylallyl moiety by using a recombinant prenyltransferase CdpC3PT. Org. Biomol. Chem. 8:2430–2438 [DOI] [PubMed] [Google Scholar]
- 34.Chen J, Morita H, Wakimoto T, Mori T, Noguchi H, Abe I. 2012. Prenylation of a nonaromatic carbon of indolylbutenone by a fungal indole prenyltransferase. Org. Lett. 14:3080–3083 [DOI] [PubMed] [Google Scholar]
- 35.Unsold IA, Li SM. 2005. Overproduction, purification and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology 151:1499–1505 [DOI] [PubMed] [Google Scholar]
- 36.Steffan N, Unsold IA, Li SM. 2007. Chemoenzymatic synthesis of prenylated indole derivatives by using a 4-dimethylallyltryptophan synthase from Aspergillus fumigatus. Chembiochem 8:1298–1307 [DOI] [PubMed] [Google Scholar]
- 37.Wollinsky B, Ludwig L, Xie X, Li SM. 2012. Breaking the regioselectivity of indole prenyltransferases: identification of regular C3-prenylated hexahydropyrrolo[2,3-b]indoles as side products of the regular C2-prenyltransferase FtmPT1. Org. Biomol. Chem. 10:9262–9270 [DOI] [PubMed] [Google Scholar]
- 38.Kremer A, Westrich L, Li SM. 2007. A 7-dimethylallyltryptophan synthase from Aspergillus fumigatus: overproduction, purification and biochemical characterization. Microbiology 153:3409–3416 [DOI] [PubMed] [Google Scholar]
- 39.Kremer A, Li SM. 2008. Potential of a 7-dimethylallyltryptophan synthase as a tool for production of prenylated indole derivatives. Appl. Microbiol. Biotechnol. 79:951–961 [DOI] [PubMed] [Google Scholar]
- 40.Kremer A, Li SM. 2010. A tyrosine O-prenyltransferase catalyses the first pathway-specific step in the biosynthesis of sirodesmin PL. Microbiology 156:278–286 [DOI] [PubMed] [Google Scholar]
- 41.Zou HX, Xie X, Zheng XD, Li SM. 2011. The tyrosine O-prenyltransferase SirD catalyzes O-, N-, and C-prenylations. Appl. Microbiol. Biotechnol. 89:1443–1451 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.