Abstract
Lactobacillus panis PM1 belongs to the group III heterofermentative lactobacilli that use the 6-phosphogluconate/phosphoketolase (6-PG/PK) pathway as their central metabolic pathway and are reportedly unable to grow on fructose as a sole carbon source. We isolated a variant PM1 strain capable of sporadic growth on fructose medium and observed its distinctive characteristics of fructose metabolism. The end product pattern was different from what is expected in typical group III lactobacilli using the 6-PG/PK pathway (i.e., more lactate, less acetate, and no mannitol). In addition, in silico analysis revealed the presence of genes encoding most of critical enzymes in the Embden-Meyerhof (EM) pathway. These observations indicated that fructose was metabolized via two pathways. Fructose metabolism in the PM1 strain was influenced by the activities of two enzymes, triosephosphate isomerase (TPI) and glucose 6-phosphate isomerase (PGI). A lack of TPI resulted in the intracellular accumulation of dihydroxyacetone phosphate (DHAP) in PM1, the toxicity of which caused early growth cessation during fructose fermentation. The activity of PGI was enhanced by the presence of glyceraldehyde 3-phosphate (GAP), which allowed additional fructose to enter into the 6-PG/PK pathway to avoid toxicity by DHAP. Exogenous TPI gene expression shifted fructose metabolism from heterolactic to homolactic fermentation, indicating that TPI enabled the PM1 strain to mainly use the EM pathway for fructose fermentation. These findings clearly demonstrate that the balance in the accumulation of GAP and DHAP determines the fate of fructose metabolism and the activity of TPI plays a critical role during fructose fermentation via the EM pathway in L. panis PM1.
INTRODUCTION
Lactobacilli generate metabolic energy (i.e., ATP) mainly by substrate-level phosphorylation by relatively simple pathways: the Embden-Meyerhof (EM) pathway and/or the 6-phosphogluconate/phosphoketolase (6-PG/PK) pathway. The members of the genus Lactobacillus are subdivided into three groups according to their fermentative characteristics on the basis of the presence of these pathways (1, 2). Group I lactobacilli are obligatory homofermentative lactobacilli that exclusively ferment hexose sugars (e.g., glucose) to lactate via the EM pathway; however, they do not metabolize pentose sugars or gluconate. Group II lactobacilli are the facultative heterofermentative lactobacilli that ferment hexoses to lactate via the EM pathway. They can also ferment pentose sugars using an inducible phosphoketolase, producing lactate and acetate, and they can yield carbon dioxide from gluconate but not from glucose. Group III lactobacilli are the obligatory heterofermentative lactobacilli that ferment hexoses to lactate, ethanol and/or acetate, and carbon dioxide, which is a distinct feature of the group III lactobacilli. Group III lactobacilli exclusively use the 6-PG/PK pathway to achieve this metabolism.
Fructose, an abundant hexose sugar found in many plants, is one of the main monosaccharides for bacterial growth in most ecosystems associated with plants and in lactobacilli is typically fermented by the two metabolic pathways described above (3, 4). During homofermentative fructose metabolism in group I and II lactobacilli, fructose is converted by the phosphoenolpyruvate (PEP)-dependent sugar phosphotransferase system (PTS) and 6-phosphofructokinase (PFK) to fructose 1,6-diphosphate (FDP) and is then split by a fructose 1,6-diphosphate aldolase (FBA) into dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (GAP). Triosephosphate isomerase (TPI) isomerizes DHAP into GAP, which is then further metabolized into lactate with the production of ATP. These enzymes compose the EM pathway. In typical fructose fermentation by group III lactobacilli, fructose is taken up by fructose permease, and fructokinase (FK) and glucose 6-phosphate isomerase (PGI) convert the assimilated fructose to glucose 6-phosphate (G6P), which is further metabolized via the 6-PG/PK pathway, producing two intermediates: GAP and acetyl phosphate. GAP further follows the same metabolism as it does in the EM pathway and produces ATP, while acetyl phosphate is metabolized into either acetate or ethanol. Theoretically, fructose metabolism via the 6-PG/PK pathway has a low energy yield (1 ATP molecule per fructose molecule) compared to that of the EM pathway (2 ATP molecules per fructose molecule) (5). However, many group III lactobacilli can compensate for this disadvantage by using fructose not only as a carbon source but also as an electron acceptor, producing mannitol by a NAD+ mannitol dehydrogenase (6). This mannitol pathway allows NAD+ regeneration as an alternative to ethanol production. As a result, a part of the acetyl phosphate is redirected to acetate production, along with yielding one extra ATP.
Lactobacillus panis PM1 has been described as a group III lactobacillus that uses the 6-PG/PK and various external electron acceptor pathways (e.g., glycerol, citrate, and oxygen) for its central metabolism and NADH recycling system (7–9), and its inability to grow on fructose medium has been reported by our group (7). In this study, we isolated a variant PM1 strain able to grow on fructose as the sole carbon source. Growth on fructose revealed characteristics distinct from those of typical group III lactobacilli: sporadic growth, no mannitol but erythritol production, less acetate production, and more lactate production. Possible fructose metabolism and its regulation were studied using in silico and metabolic profile analyses, enzyme assays, and the introduction of an Escherichia coli TPI gene into L. panis PM1.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Lactobacillus panis PM1 strain was an isolate from bioethanol thin stillage (7) and has been deposited under accession number 180310-01 at the International Depository Authority of Canada, National Microbiology Laboratory, Public Health Agency of Canada (IDAC, NML-HCCC; Winnipeg, MB, Canada) under the provision of the Budapest Treaty. Lactobacillus panis PM1 was cultured at 37°C under microaerobic conditions until late log phase using commercial MRS medium (BD, Franklin Lakes, NJ), at which time 1% (vol/vol) of this preculture was transferred into fresh modified MRS (mMRS) medium. The mMRS medium consisted of 5 g yeast extract, 10 g peptone, 10 g meat extract, 2 g K2HPO4, 2 g ammonium citrate, 5 g sodium acetate, 100 mg MgSO4·7H2O, 50 mg MnSO4, and a defined concentration of fructose and/or glucose per liter. The cultures were incubated at 37°C under microaerobic conditions, unless otherwise stated. Airtight 15-ml tubes, filled to the two-thirds level, were incubated under static conditions to establish microaerobic conditions. Escherichia coli TOP 10 (Invitrogen, Carlsbad, CA) was used for cloning and preparation of the target plasmid and was cultured in LB medium at 37°C with vigorous shaking. The concentrations of erythromycin (Em) for selective plating were 10 μg/ml and 300 μg/ml for L. panis PM1 and E. coli, respectively.
Preparation of crude cell extracts.
Lactobacillus panis PM1 cells grown in mMRS medium containing different carbon sources (fructose and/or glucose) were disrupted by sonication as previously described (9). A crude cell extract was obtained by centrifugation for 10 min at 16,160 × g, and the protein concentration was determined using a protein assay kit (Bio-Rad, Hercules, CA) with bovine serum albumin (BSA) as a standard.
Enzyme assays.
All enzyme assays were spectrophotometrically evaluated at 37°C by measuring the changes in absorbance at 340 nm (εNADH = 6,220 M−1 cm−1). One unit of activity corresponds to the generation or consumption of 1 μmol of NADH per min, and specific activity was expressed as units per milligram of protein. For PGI (EC 5.3.1.9; fructose 6-phosphate [F6P] to G6P) activity, the reaction mixture was composed of 100 mM potassium phosphate, pH 7.0, 1.5 mM NAD+, 1.0 mM ATP, 1.0 unit/ml of hexokinase, 1.0 unit/ml of glucose 6-phosphate dehydrogenase, 2 mM d-fructose, 0 to 20 mM GAP (for tests of the inhibition or activation of PGI activity), and 0.05 to 0.1 mg of protein. Mannitol dehydrogenase (ManDH; EC 1.1.1.67) activity was measured in 90 mM Tris-HCl, pH 9.0, 3.0 mM NAD+, 1.5 mM d-mannitol, and 0.05 mg of protein. TPI (EC 5.3.1.1; GAP to DHAP) was assayed in 100 mM potassium phosphate (pH 7.4) containing 1.0 unit/ml of glycerol 3-phosphate dehydrogenase, 0.3 mM NADH, 0.4 mM GAP, and 0.05 to 0.1 mg of protein.
Determination of hexoses (glucose and fructose) and end products.
The culture optical density at 600 nm (OD600) was measured as an index of growth with a DU 800 spectrophotometer (Beckman Coulter, Mississauga, ON, Canada). After centrifugation, the supernatant was filtered through a 0.22-μm-pore-size filter and stored at −20°C for high-pressure liquid chromatography (HPLC) analysis. To quantify the concentrations of sugars (glucose and fructose), organic acids, and ethanol, samples were analyzed on an organic acid column (HPX-87H; Bio-Rad) using an HPLC system equipped with a refractive index detector (RID G1362A, 1100 series; Agilent Technologies, Palo Alto, CA). Operating conditions were determined by the method described in the column manual with minor modifications. Filtered culture medium (40 μl) was loaded on the column and eluted with 5 mM sulfuric acid at a flow rate of 0.6 ml/min at 55°C for 30 min. The mannitol concentration in the culture broth was measured in accordance with the instructions in the d-mannitol/l-arabitol kit (Megazymel Bray Co., Wicklow, Ireland).
Quantification of intracellular DHAP level.
For the quantification of intracellular DHAP levels, L. panis PM1 was cultured in mMRS medium containing either 60 mM fructose or glucose. After a 24-h culture, the cells were harvested and the crude extract was prepared as described above. A standard curve for the DHAP concentration was evaluated spectrophotometrically (at 340 nm, εNADH = 6,220 M−1 cm−1) in 100 mM potassium phosphate (pH 7.4) containing 1.0 unit/ml of glycerol 3-phosphate dehydrogenase, 0.2 mM NADH, and external DHAP standards (0, 10, 20, 40, and 80 μM). The DHAP concentrations in the crude extract were calculated from the standard curve. Intracellular DHAP concentrations were calculated on the basis of the dry cell mass conversion factor (0.255 g/liter/OD600 unit) and water content (3.53 μl/mg [dry weight]) of L. panis PM1.
General DNA techniques, plasmid construction, and bacterial transformation.
DNA work was carried out according to standard protocols described in Molecular Cloning (10). Restriction endonucleases were purchased from Fermentas (Waltham, MA), and digestions were performed as recommended by the manufacturer. A Bio-Basic gel extraction kit (Bio-Basic Inc., Markham, ON, Canada) was used to isolate specific restriction and PCR fragments from agarose gels. DNA ligations and subsequent transformations into competent E. coli TOP 10 cells (Invitrogen) were carried out according to standard protocols (10). Escherichia coli JM109 genomic DNA was used as a template for TPI gene amplification. For the PCR amplification, a 50-μl PCR mixture was prepared using 0.5 μM (each) primers fTPI and rTPI (Table 1), 1.25 U Pfu DNA polymerase (Fermentas), 1× Pfu buffer with MgCl2, and 0.2 mM deoxynucleotide triphosphates (dNTPs). The DNA template was denatured for 3 min at 95°C, and the PCR continued with 30 cycles of 30 s at 95°C, 30 s at 55°C, and 1 min at 72°C. After the final cycle, the mixtures were further incubated for 5 min at 72°C using a thermal cycler (FTGENE-5D; Techgene, Burlington, NJ). The amplified TPI gene was digested with NotI and SalI restriction endonucleases and cloned into the expression vector pCER-EGFP (Table 2). The resultant plasmids were transformed into L. panis PM1 by electroporation, as described previously (11).
Table 1.
Primers used in this study
| Primer | Tma (°C) | Nucleotide sequence (5′ → 3′)b | Target gene | Function |
|---|---|---|---|---|
| f16S | 58 | TGGCCCAACTGATATGAC | 16S rRNA | 16S rRNA |
| r16S | 58 | CTCTCATGCACGTTCTTCTT | ||
| fFBA | 60 | TAGTGGGGAACTGGCTTCAG | FBA gene | Fructose 1,6-diphosphate aldolase |
| rFBA | 60 | ATCCCTTGTGGTCCTTGTCA | ||
| fXFP | 60 | CATGGAACACAAGGACATGC | XFP gene | Xylulose 5-phosphate phosphoketolase |
| rXFP | 60 | AAGCCACGGAATGTATCTGG | ||
| fTPI | 69 | ATAAGAATGCGGCCGCATGCGACATCCTTTAGTGATGGGT | TPI gene | Triosephosphate isomerase |
| rTPI | 65 | ACGCGTCGACTTAAGCCTGTTTAGCCGCTTC |
Tm, melting temperature.
The underlined primer sequences represent the NotI and SalI restriction enzyme sites in fTPI and rTPI, respectively.
Table 2.
Plasmids and primers used in this study
| Plasmid | Relevant features | Source or reference |
|---|---|---|
| pEGFP-N1 | Source of EGFPa gene | Clontechb |
| pSIP411 | Source of PorfX promoter | 16 |
| pCER | E. coli-L. panis PM1 shuttle vector, Emr Cmr, 5.0 kb | 11 |
| pCER-EGFP | pCER derivative expressing the EGFP gene under the control of the PorfX promoter, Emr Cmr, 6.0 kb | This study |
| pCER-TPI | pCER-EGFP derivative in which the EGFP gene is replaced with the TPI gene, Emr Cmr, 6.0 kb | This study |
EGFP, enhanced green fluorescent protein.
Clontech (Mountain View, CA).
RNA extraction and reverse transcription.
RNA was extracted by the acid-hot phenol method as described previously (9). First-strand cDNA was synthesized using reverse primers of the selected genes (Table 1) as described by Kang et al. (9).
Quantitative PCR.
Real-time PCR amplification was performed in a CFX96 real-time detection system (Bio-Rad) using SsoFast EvaGreen Supermix (Bio-Rad). The total volume of the PCR master mixture was 20 μl, to which cDNA template equivalent to 25 ng RNA starting material and 0.5 μM each primer (Table 1) was added. PCR amplification was initiated at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 10 s. Amplification was followed by a melting curve analysis between 65°C and 95°C using a 0.5°C increment. All sample and primer combinations were assessed in three biological replicates with two technical replicates per biological replicate. A no-template control was used for the negative-control PCR, and PCR specificity and product detection were verified by examining the temperature-dependent melting curves of the PCR products as well as ethidium bromide staining on 1% agarose gels. For relative gene expression, the 2−ΔΔCT threshold cycle (CT) method, using the 16S rRNA gene for normalization, was performed as described by Livak and Schmittgen (12). The steps for calculating the expression ratio were as follows: ΔCT(test) = CT(FBA and XFP, test) − CT(16S rRNA, test), ΔCT(control) = CT(FBA and XFP, control) − CT(16S rRNA, control), and ΔΔCT = ΔCT(test) − ΔCT(control), where test and control refer to the test and control reactions for the indicated genes and XFP indicates xylulose 5-phosphate phosphoketolase. The normalized expression ratio of FBA(test) and XFP(test) was equal to 2−ΔΔCT. The real-time PCR data were processed using CFX Manager software (Bio-Rad).
Statistical analysis.
For determinations of end product concentrations and enzyme activities, data were presented as mean values ± standard errors of the means calculated from at least three independent experiments. Differences in end products, enzyme specific activities, and transcriptional expression were analyzed by two-way analysis of variance with Bonferroni posttests for three groups or the unpaired t test for two groups using GraphPad Prism, version 5.0, software (GraphPad Software, Inc., San Diego, CA). A P value of <0.05 was considered significant.
Nucleotide sequence accession numbers.
Newly determined sequence data were deposited in GenBank under accession numbers KF312453 to KF312456 and KF384501 to KF384508.
RESULTS
Sporadic growth on fructose medium.
We previously reported that L. panis PM1 is unable to grow solely on fructose (7). However, we unexpectedly observed that a culture of over 1 month revealed sporadic growth of a variant PM1 strain on fructose. In the subsequent growth tests, this variant mainly showed three types of fructose consumption patterns (Fig. 1): (i) full growth, consuming all available fructose (60 mM), (ii) arrested cell growth at the mid- to late-log growth phase, consuming one-third of the available fructose, and (iii) early cell growth cessation, leaving most of the available fructose unconsumed. The strain showing the first type of growth was designated a fructose-utilizing variant. Pedersen et al. (13) have reported three different strains of L. panis, DAF 1, DAG 76, and DAF 355, showing different growth patterns, growth, weak growth, and no growth, respectively, in a fructose medium; however, their 16S rRNA sequences were identical. In the present study, the sequences of 16S rRNA, a possible sugar permease, and genes necessary for the EM and 6-PG/PK pathways of the variant and the parent strain were compared in order to identify the variant, and the sequences of these genes were identical in both strains (data not shown). The lag phase of the variant strain observed when it was grown in fructose medium was longer than that observed when it was grown on glucose (24 h versus 12 h). Once the isolated variant strain fully grew on fructose medium, successive subcultures in fructose medium showed a growth pattern similar to that observed in glucose medium; however, once it became dormant, i.e., it was frozen for storage, the cell cultures usually reacquired the same inability of parent strain L. panis PM1 (Fig. 1). Therefore, it was apparent that the two strains were naturally occurring phenotypic variants and, thus, the same L. panis strain, PM1.
Fig 1.
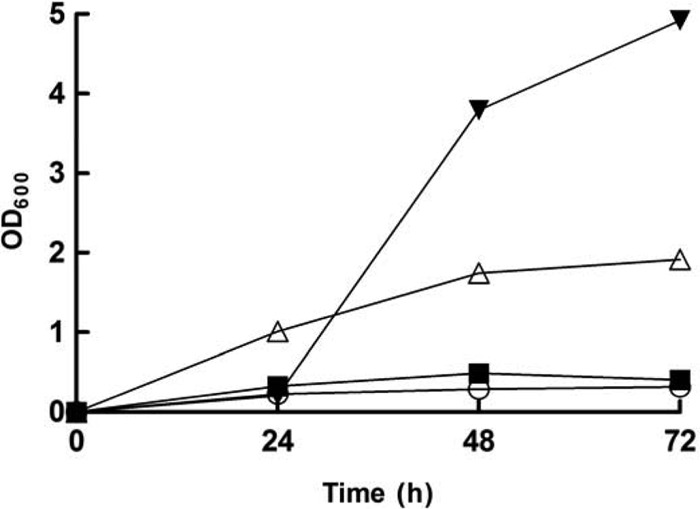
Growth pattern of L. panis PM1 on fructose medium. The PM1 strain was cultured in mMRS medium containing 60 mM fructose. Three types of patterns were observed: full growth (inverted triangles), arrested cell growth at mid- to late log phase (triangles), and early cell growth cessation (squares). mMRS medium without a carbon source was used as a negative control (circles).
Metabolic profiles of fructose fermentation.
When fructose-utilizing L. panis PM1 was cultured in mMRS medium with different carbon sources (fructose and/or glucose), each culture showed different end product production patterns. As shown in Fig. 2, fructose fermentation by L. panis PM1 significantly increased lactate production (25% in 60 mM glucose and fructose medium and 51% in 120 mM fructose medium), along with decreases in acetate (28% in 60 mM glucose and fructose medium and 50% in 120 mM fructose medium) (P < 0.01). In contrast, ethanol concentrations were comparable under all conditions (P > 0.05). Interestingly, mannitol, which is a typical main end product of fructose fermentation in group III lactobacilli, was not detected in any fermentation broths, and the activity of ManDH was not observed from cells grown on either glucose or fructose (data not shown), indicating that fructose is used only as a carbon source in L. panis PM1. Instead of mannitol, erythritol was detected in the glucose and fructose fermentation by the fructose-utilizing L. panis PM1, and the presence of fructose significantly increased the final yield of this end product (by 102% in 60 mM glucose and fructose medium and 210% in 120 mM fructose medium compared with that in 120 mM glucose medium) (P < 0.01). These changes in metabolites indicated that the variant used different pathways, in addition to the 6-PK/PG pathway, to metabolize fructose, suggesting the possibility of the simultaneous use of the 6-PG/PK and EM pathways for fructose metabolism.
Fig 2.
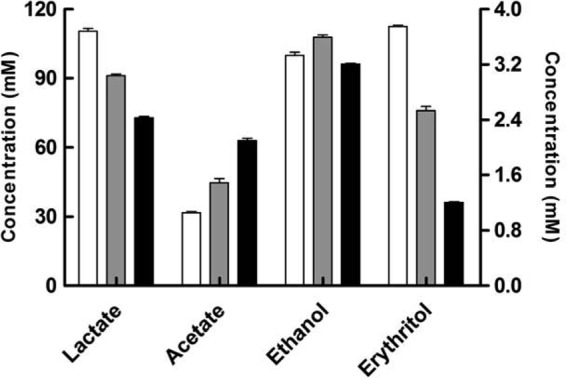
Main end products yielded from fructose and/or glucose fermentation by L. panis PM1. Lactobacillus panis PM1 was cultured in mMRS medium containing 120 mM fructose (white bars), 60 mM (each) fructose and glucose (gray bars), or 120 mM glucose (black bars) for 48 h. The concentrations of lactate, acetate, and ethanol are provided on the left-hand y axis, and the concentration of erythritol is provided on the right-hand y axis.
In silico analysis of the EM pathway and quantitative reverse transcription-PCR (qRT-PCR).
To identify genes encoding enzymes involved in the EM pathway, annotation data from the draft genome of L. panis PM1 (M. C. Haakensen, V. Pittet, D. A. S. Grahame, D. R. Korber, and T. Tanaka, unpublished data) were analyzed at the UniProt database (http://www.uniprot.org), identifying similarities with other proteins from related bacteria. This in silico analysis revealed the presence of the genes encoding the most critical enzymes of the EM pathway: FK (EC 2.7.1.4; GenBank accession no. KF312453), PGI (EC 5.3.1.9; GenBank accession no. KF312454), FBA (EC 4.1.2.13; GenBank accession no. KF312455), and TPI (EC 5.3.1.1; GenBank accession no. KF384506), but not PFK (EC 2.7.1.11). These genes were mainly clustered in two chromosomal loci. The PGI and FK genes were organized in a five-gene operon structure coding for glyoxalase (GenBank accession no. KF384502), sorbitol dehydrogenase (GenBank accession no. KF384503), and a small hypothetical protein. A gene annotated as a member of the major facilitator superfamily (GenBank accession no. KF384501) with a suggestive sugar transporter function was located in the upstream (1,446-bp) region of the PGI gene (14). Two other genes for the EM pathway were grouped in an operon structure coding for phosphoenolpyruvate synthase (GenBank accession no. KF384504), a hypothetical protein (GenBank accession no. KF384505), the TPI gene, the FBA gene, and phosphoglycerate mutase (GenBank accession no. KF384507). The sequences of the five key genes (those encoding FK, PFK, PGI, FBA, and TPI) were searched against and compared to those of 19 other lactobacillus strains released by the DOE Joint Genome Institute (http://www.jgi.doe.gov/) (see Table S1 in the supplemental material). Overall, the genes of the PM1 strain showed higher similarities with those of group III lactobacilli (64 to 88%) than those of group I and II lactobacilli (42 to 73%), and the sequence of the L. reuteri JCM 1112 strain showed the highest similarity (64, 83, and 88% in the TPI, FBA, and PGI genes, respectively) with that of the PM1 strain for all genes except the FK gene (28 and 30% to the two FK genes of the L. reuteri JCM 1112 strain). Our previous phylogenetic work showed higher identity (96%) in 16S rRNA sequences to the 16S rRNA sequence of L. reuteri (7). Combined with the present results, this indicates that L. panis PM1 and L. reuteri are close relatives. We could not find the PFK gene in the annotation data of L. panis PM1; however, this gene was found only in group I and II lactobacilli in our comparison. The PM1 strain possessed a gene annotated as a ribokinase (RBSK; EC 2.7.1.15; GenBank accession no. KF312456) showing 82% similarity to a pfkB orthologous gene (GenBank accession no. EF547651) in L. reuteri (15). The similarity of the FK gene of group I and II lactobacilli with that of strain PM1 was relatively low (61 to 29%). In addition, the TPI gene showed 39 to 64% similarity across all groups. In contrast, the PGI gene had two well-conserved sugar isomerase domains and showed higher similarity than other genes among groups I, II, and III (67 to 88%). Table S1 in the supplemental material shows that the FBA gene, containing a specific tetrameric class II aldolase, was found only in L. reuteri among the group III lactobacilli, and the FBA gene of strain PM1 had a higher similarity with the FBA gene of group I lactobacilli (66 to 72%) than with the FBA gene of group II lactobacilli (55 to 59%).
The activities of the EM and 6-PK/PG pathways were evaluated using qRT-PCR with RNA prepared from a variety of cultures. Two representative genes, the FBA gene and the xylulose 5-phosphate phosphoketolase gene (GenBank accession no. KF384508), were evaluated for their use of the EM and 6-PG/PK pathways, respectively, including analysis of (i) early- and mid-log-phase cells grown on fructose medium and (ii) mid-log-phase cells grown on different carbon sources (glucose and/or fructose) (Table 3). During growth on fructose medium, the expression of the XFP gene was 2.9-fold higher in the early log phase than in the mid-log phase, whereas the expression of the FBA gene was induced (4.1-fold) in the mid-log-phase (P < 0.01). The difference in carbon source upregulated the expression of both genes (1.5-fold in 30 mM fructose medium and 3.7-fold in 60 mM fructose medium relative to that in 60 mM glucose medium) (P < 0.01).
Table 3.
Growth phase- and carbon source-dependent expression of the FBA and XFP genes determined using qRT-PCR
| Growth phase or carbon sourcea | Relative gene expression level |
|
|---|---|---|
| FBA gene | XFP gene | |
| Early log phaseb (OD600, 0.65) | 1.00 ± 0.37 | 1.00 ± 0.31 |
| Mid-log phase (OD600, 1.86) | 4.09 ± 0.38 | 0.35 ± 0.04 |
| Glcb | 1.00 ± 0.16 | 1.00 ± 0.12 |
| Glc and Frc | 1.49 ± 0.31 | 1.52 ± 0.32 |
| Frc | 3.65 ± 0.33 | 3.63 ± 0.39 |
Strain PM1 was cultured in 60 mM glucose (Glc), 30 mM glucose and fructose (Glc/Frc), or 60 mM fructose (Frc) medium for 24 h.
Control condition for qRT-PCR.
Enzyme activities in the EM pathway.
The activity of PGI is essential to convert F6P to G6P for channeling of fructose fermentation to the 6-PG/PK pathway. PGI enzyme activity was monitored for 42 h in cultures grown on medium with different carbon sources (glucose and/or fructose). The activity of PGI was induced by the presence of glucose, and the activities observed in medium with 30 mM (each) fructose and glucose were 10 times higher than those observed in 60 mM fructose medium (P < 0.0001) (Fig. 3). Furthermore, the PGI activity on 60 mM glucose medium gradually increased from 0.76 to 2.25 units/mg protein until glucose depletion at the 32-h time point.
Fig 3.
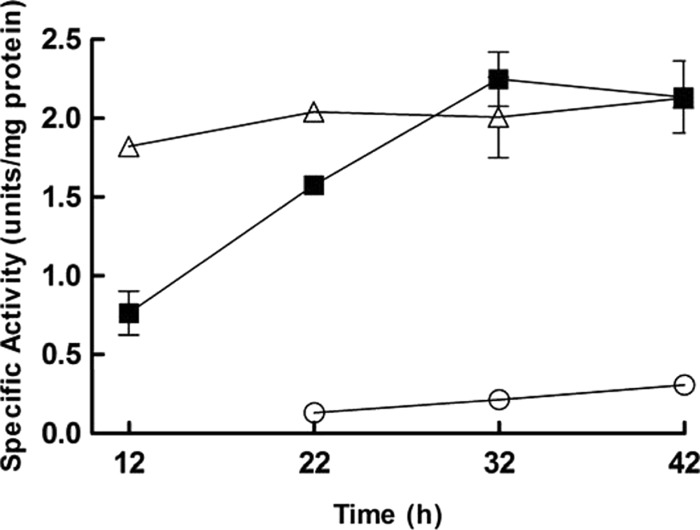
Changes in glucose 6-phosphate isomerase activity on different carbon sources. Lactobacillus panis PM1 was cultured in mMRS medium containing 60 mM fructose (circles), 60 mM glucose (squares), or 30 mM (each) fructose and glucose (triangles). At the 12-h time point, PGI activity could not be determined in 60 mM fructose medium due to sporadic growth; i.e., little growth was observed at 12 h.
In group I and II lactobacilli, TPI is a key enzyme necessary to metabolize fructose via the EM pathway without carbon loss. In the absence of TPI, one-half of the fructose mass is theoretically converted to a three-carbon intermediate (DHAP), reducing the yield of the end product (e.g., lactate). To detect this enzyme activity, the parent L. panis PM1 strain was cultured in MRS medium and was successively subcultured in mMRS medium containing either 60 mM glucose or fructose for 24 h. Cell growth on glucose medium followed a typical bacterial growth pattern, whereas cell growth on fructose as the sole carbon source ceased at the early log phase (OD600, 0.48). The specific activity of the TPI enzyme was not detected in the PM1 cells grown on glucose medium, whereas TPI activity was observed in cells grown on fructose medium. However, the specific activity was not very high and in fact was 65 times lower than that of the crude extracts of the E. coli culture (P < 0.0001), which was performed in LB medium for 12 h and served as the positive control. The specific activity of the TPI enzyme observed in L. panis PM1 cultured in 60 mM fructose for 24 h to a final cell density (OD600) of 0.48 was 0.91 ± 0.02 units/mg protein, where 1 unit of activity was defined as the amount of enzyme able to generate 1 μmol of DHAP per min. The specific activity for the E. coli culture was 59.17 ± 0.17 units/mg protein, and no detectable TPI enzyme activity was found for PM1 grown on 60 mM glucose for 24 h to a final cell density (OD600) of 1.58.
Repressor and inducer of fructose metabolism.
The enzyme assay data prompted us to focus on the two three-carbon intermediates, DHAP and GAP, which are substrates of TPI reactions. To investigate the roles of GAP on fructose metabolism, the PGI activity in crude extracts of PM1 cells cultured in glucose medium until mid-log phase was measured. PGI enzyme assays were conducted in the presence of 0, 5, 10, and 20 mM GAP. Figure 4 shows the increased activity of PGI in the presence of GAP. The enzyme reaction was accelerated by 34 times in the presence of 10 mM GAP. However, 20 mM GAP lost much of this stimulatory effect, with the activity of PGI being only 6 times that in the absence of GAP. This result clearly suggests a significant role of GAP on the activation of PGI, which can stimulate channeling of fructose to the 6-PK/PG pathway.
Fig 4.
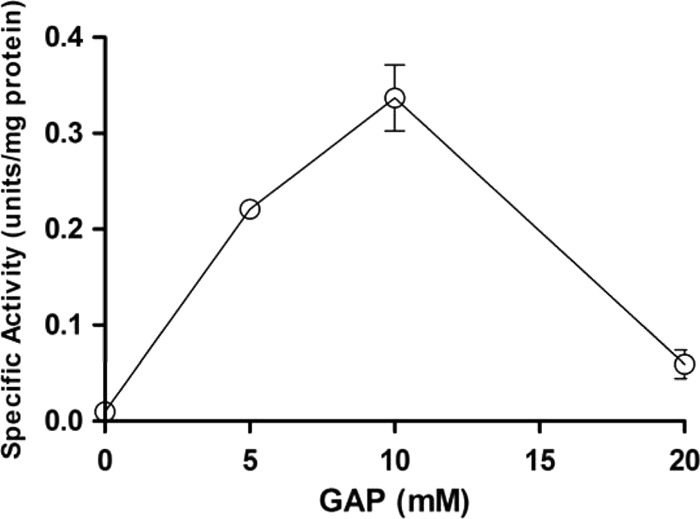
Relation of glucose 6-phosphate isomerase activity to the availability of glyceraldehyde 3-phosphate. The specific activities of glucose 6-phosphate isomerase were measured from crude extracts of L. panis PM1 cultured in the presence of glyceraldehyde 3-phosphate (GAP).
A significant level of intracellular DHAP (5.03 mM), along with the early cessation of growth, was detected in cells grown on fructose medium, whereas the presence of this chemical was not observed in cells grown on glucose medium, on which they exhibited a normal growth pattern. For cells grown on glucose and fructose, the OD600 (final cell density) values were 1.88 and 0.48, respectively, and the DHAP concentration was 5.03 ± 0.49 mM for cells grown on fructose, but it was not detectable for cells grown on glucose. The negative effect of dihydroxyacetone (DHA; a precursor and nonphosphorylated form of DHAP) on cell growth was observed in medium containing 0, 5.6, 11.2, 28, or 56 mM DHA. The growth of L. panis PM1 cells was completely inhibited by 28 mM DHA (Fig. 5). Generally, DHAP is formed by the EM or glycerol oxidative (glycerol-to-DHAP) pathway; however, L. panis PM1 does not possess the glycerol oxidative routes (7, 8). Thus, the accumulation of DHAP suggested the utilization of the EM pathway and provided a direct reason for growth cessation with fructose but not with glucose.
Fig 5.
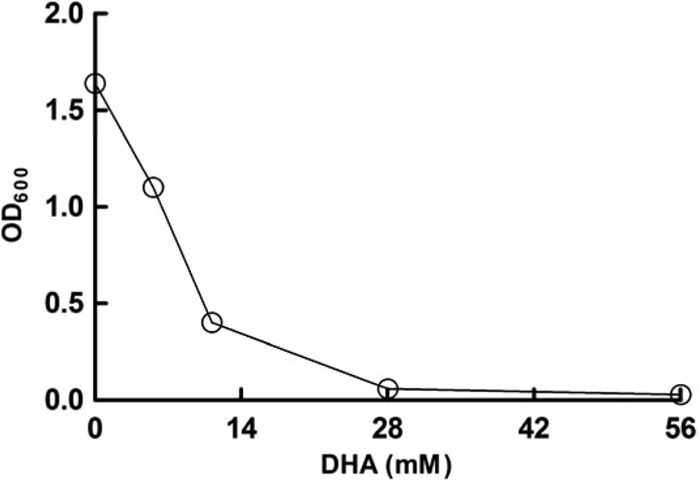
Growth inhibition by the presence of DHA in L. panis PM1 culture. Lactobacillus panis PM1 was cultured in MRS medium containing 0, 5.6, 11.2, 28, or 56 mM DHA, and the cell density was determined after 24 h of culture.
Role of TPI activity on fructose metabolism.
Although TPI genes were annotated from the genome data for L. panis PM1, functional activity was absent or very low in L. panis PM1, leading us to speculate that the accumulation of intracellular DHAP was occurring during fructose fermentation via the EM pathway. To circumvent DHAP accumulation, a TPI gene of E. coli was expressed in a wild-type L. panis PM1 strain. The TPI gene was cloned into the expression vector pCER-EGFP and was under the control of the PorfX promoter from plasmid pSIP144 (16) (Table 2), and the resulting plasmid, pCER-TPI, was introduced into a wild-type PM1 strain (i.e., a strain unable to readily utilize fructose). The resultant strain, PM1-TPI, was cultured in fructose medium, and cell growth and end product profiles were monitored. Interestingly, the introduction of the TPI gene enabled L. panis PM1 to grow on fructose medium without undergoing sporadic growth (Fig. 6) and was accompanied by a shift in the fructose fermentation pattern. Accordingly, at the 24-h culture time point, the recombinant strain mainly produced lactate (51.89 mM) and ethanol (10.82 mM) from the fructose consumed (30 mM) but did not produce acetate, representative of a typical homolactic fermentation pattern.
Fig 6.
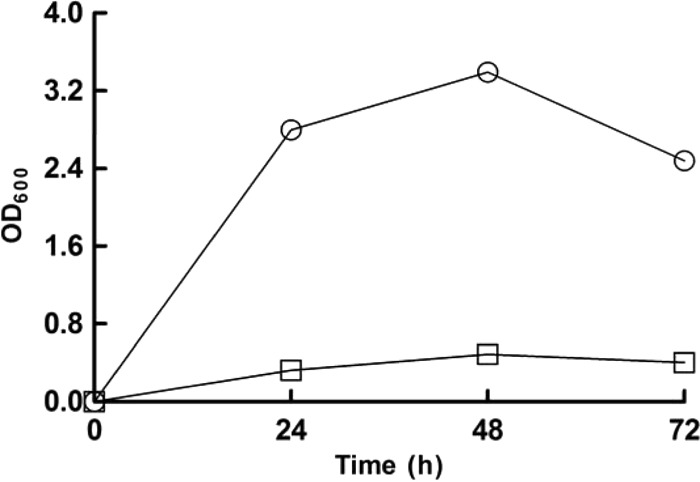
Effect of triosephosphate isomerase expression on L. panis PM1 growth on fructose-containing mMRS medium. PM1(pCER) (squares) and PM1(pCER-TPI) (circles) strains were cultured in mMRS medium containing 60 mM fructose, and samples taken from a 24-h culture were used for end product analysis.
DISCUSSION
Group III lactobacilli generally tend to lack PTS activity, and the reaction involving PTS is the first reaction in routing glucose into the EM pathway in group I and II lactobacilli. As a result, group III lactobacilli do not use the EM pathway but, rather, use the 6-PK/PG pathway to utilize glucose. However, the presence of fructose-specific PTS has been reported in group III lactobacilli, L. brevis, and L. reuteri CRL 1098 (17, 18). In addition, the utilization of both the EM and 6-PK/PG pathways has been evidenced in L. brevis and L. reuteri ATCC 55730 during their fructose fermentation (15, 18). In the present study, we also observed evidence of the utilization of dual pathways for fructose metabolism in L. panis PM1 and that its fructose-metabolizing patterns were unique compared with those of known fructose-utilizing lactobacilli. Further study elucidated the detailed mechanism of fructose fermentation, the regulation of this mechanism, and the role of the TPI enzyme in the shift between homolactic and heterolactic fermentation patterns.
End product pattern of fructose metabolism.
Fructose fermentation by L. panis PM1 did not produce mannitol due to the absence of ManDH activity, indicating that fructose is used only as a carbon source. This characteristic of L. panis PM1 would offer fewer benefits in terms of energy metabolism since the PM1 strain cannot regenerate NAD+ using fructose as an electron acceptor. Thus, the amount of NADH produced during rapid growth on fructose could exceed the capacities of NADH-consuming enzymes (lactate dehydrogenase, acetaldehyde dehydrogenase, and alcohol dehydrogenase) under the circumstances described above without external electron acceptors (8). Instead of mannitol, L. panis PM1 produced erythritol, and its final yield was increased by the availability of fructose (Fig. 2). The activity of phosphoketolase cleaves fructose 6-phosphate into acetyl phosphate and erythrose 4-phosphate, which are used for the disposal of 1 mol (or 3 mol, if acetyl phosphate is converted to ethanol) of NADH per mol of glucose consumed in various heterofermentative lactobacilli (19–21). Thus, erythritol production by L. panis PM1 suggests the role of this pathway as an alternate route for NAD+ regeneration. However, the contribution of this pathway to NAD+ regeneration is limited when the low yield of erythritol (3.75 mM) relative to that of lactate (110.47 mM) is considered.
If fructose is solely fermented by the 6-PG/PK pathway, the end product pattern should be the same as that for glucose metabolism. However, fructose fermentation by strain PM1 did not follow this model and produced more lactate and less acetate than the amounts seen during glucose fermentation (Fig. 2). If utilization of the EM pathway is assumed, fructose would be processed only to lactate and would contribute to the increased lactate observed in Fig. 2. These observations strongly suggest the utilization of both the EM and 6-PK/PG pathways during fructose fermentation by L. panis PM1.
Possible EM pathway for fructose metabolism.
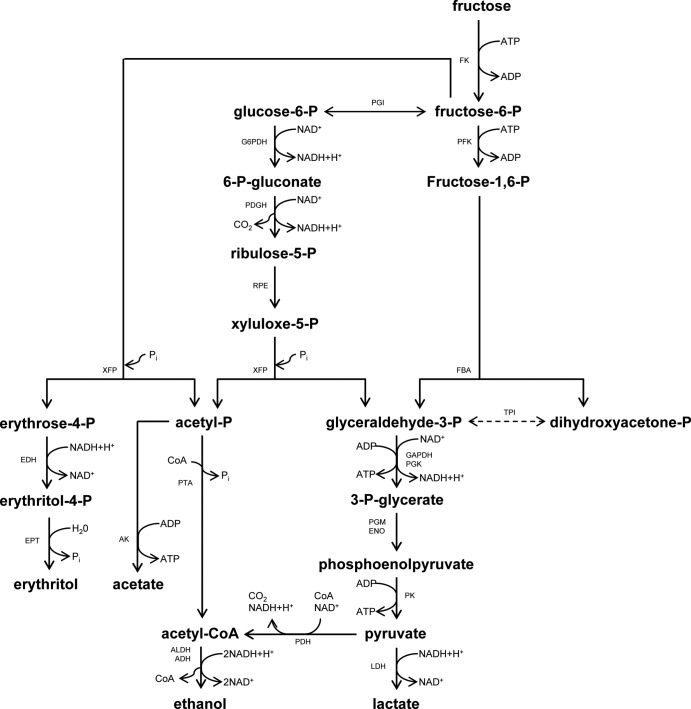
A possible fructose metabolic pathway was examined on the basis of in silico and metabolite analyses (Fig. 7). Draft genomic information of L. panis PM1 revealed the presence of the genes encoding most of the critical enzymes of the EM pathway (FK, PGI, FBA, and TPI); the exception was PFK. The absence of this gene has also been reported in the genomes of group III lactobacilli released in the GenBank database (15). However, Årsköld et al. (15) found that L. reuteri with orthologs to pfkB, which encodes a minor PFK in E. coli, possesses PFK activity. Since relatively high similarities of the key EM pathway genes were observed among L. panis PM1 and L. reuteri strains and the rbsK gene showed a similarly high degree of similarity to the pfkB gene, it is suggested that rbsK also functions as an ortholog of PFK. The lower similarity of the FK gene in groups I and II indicated the presence of a fructose-importing system different from that in group III; i.e., the PTS systems of group I and II strains take up fructose in the form of fructose 6-phosphate, followed by intracellular phosphorylation by PFK, and thus require little FK activity during the conversion of fructose to fructose 6-phosphate. The similarity of the TPI gene was relatively low among the three groups of lactobacilli, and the TPI gene of L. panis PM1 showed 33% similarity to the TPI gene of E. coli, explaining the low or absent TPI activity, as described above, whereas the higher similarities of the PGI and FBA genes among the three groups suggest that these annotated genes function in the upper EM pathway.
Fig 7.
Schematic diagram of the fructose metabolic pathway in L. panis PM1. Dashed line, the absence of enzyme activity in L. panis PM1. Abbreviations, ADH, alcohol dehydrogenase; AK, acetate kinase; ALDH, acetaldehyde dehydrogenase; CoA, coenzyme A; EDH, erythritol dehydrogenase; ENO, enolase; EPT, erythrose 4-phosphate phosphotransferase; FBA, fructose 1,6-diphosphate aldolase; FK, fructokinase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; G6PDH, glucose 6-phosphate dehydrogenase; LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; PFK, 6-phosphofructokinase; PGDH, 6-phosphogluconate dehydrogenase; PGI, glucose 6-phosphate isomerase; PGK, phosphoglycerate kinase; PGM, phosphoglyceromutase; PK, pyruvate kinase; PTA, phosphotransacetylase; RPE, ribulose 5-phosphate 3-epimerase; TPI, triosephosphate isomerase; and XFP, xylulose 5-phosphate phosphoketolase.
FBA and XFP were chosen as representative enzymes in the EM and 6-PK/PG pathways, respectively, due to their critical roles in the fate of carbon (i.e., lactate, acetate, or ethanol). The FBA and XFP genes were upregulated by the availability of fructose, and both genes were differently activated according to the growth phase on fructose medium (Table 3). This observation suggests two important aspects of fructose utilization: (i) the presence of dual pathways for fructose metabolism and (ii) the fructose-directed regulation of those pathways. However, a question still remains: why does L. panis PM1 sporadically grow on fructose medium, even though it possesses these two pathways?
Regulation of fructose metabolism.
To address this question, it was determined that the activities of two enzymes, PGI and TPI, are critical to the functioning of these two pathways. These enzymes are critical because PGI channels fructose to the 6-PK/PG pathway and TPI governs the fate of the two split products of FBA activity to prevent the overaccumulation of DHAP, i.e., the loss of carbon. The activity of PGI was induced by glucose, and 10 times less activity was observed in cells grown on medium containing fructose as the sole carbon source than on medium containing glucose as the sole carbon source (Fig. 3), suggesting that fructose fermentation through the 6-PK/PG pathway did not occur at a high rate and that glucose could accelerate the channeling of fructose to the 6-PG/PK pathway. In addition, the glucose concentration significantly affected the activity of this enzyme. At the 12-h time point, the PGI activity in cells grown on medium containing 30 mM glucose was two times higher than that in cells grown on 60 mM glucose. PGI enzyme activity on 60 mM glucose increased with glucose consumption and peaked at the time of glucose depletion (32 h). Therefore, these enzyme assay data indicate that intermediates from glucose metabolism indeed induce the activity of the PGI enzyme.
While PGI enables the utilization of fructose, cell growth on fructose medium halted at early log phase, suggesting that initial fructose fermentation can repress growth. If fructose is fully introduced into the 6-PK/PG pathway, there should not be any repression of growth, as it would essentially follow the same reaction flow that it does during glucose fermentation. This observation further suggested that the PM1 strain uses the EM pathway like group I and II lactobacilli do and that the cessation of growth on fructose medium could be related to the accumulation of intermediates through utilization of the EM pathway. The significant accumulation of DHAP was observed only from cells grown on fructose medium. In this case, the low TPI activity was insufficient to process the DHAP produced by FBA; thus, fructose utilization through the EM pathway resulted in DHAP accumulation. The toxicity of DHAP was confirmed by a growth inhibition test with the nonphosphorylated form of this chemical (DHA) (Fig. 5; see Results). Combining these observations, the accumulation of DHAP by the EM pathway caused early growth cessation during fructose metabolism. Interestingly, the other three-carbon metabolite, GAP, made from FDP by FBA, enhanced the activity of PGI at 5 and 10 mM; however, a higher GAP concentration (20 mM) inhibited the activity of this enzyme. These findings clearly demonstrate that the balance in the accumulation of the three-carbon intermediates (GAP and DHAP) functions to control fructose metabolism patterns in L. panis PM1, as follows. Initially, fructose is fermented via the EM pathway, producing GAP and DHAP. GAP increases the activity of PGI and is further metabolized to lactate. This enhanced PGI activity then allows F6P to be processed through the 6-PG/PK pathway for further fructose fermentation, producing lactate, ethanol, and/or acetate. It should be noted that once PGI is activated, the 6-PK/PG pathway supplies GAP, keeping fructose channeled to the 6-PK/PG pathway. When this scheme is compared with glucose fermentation solely through the 6-PK/PG pathway, the production of acetate is reduced by the amount of reactants via the EM pathway, where no acetate is produced. When the majority of carbon is fructose (as in the culture on 120 mM fructose medium, for which the results are shown in Fig. 2), the initial EM pathway fermentation provided a certain concentration of DHAP, and due to low or absent TPI activity, this DHAP accumulated, resulting in growth cessation at an early stage. Thus, this regulation nicely explains the sporadic growth and the concentrations of the different end products (more lactate and less acetate) observed during fructose fermentation (Fig. 2).
Role of the TPI gene on fermentation pattern.
Various NAD+ regeneration routes (e.g., mannitol production) enable group III lactobacilli to compensate for the reduced ATP yield that occurs via the 6-PG/PK pathway by shifting acetyl phosphate to ATP production, producing acetate instead of ethanol (3, 22–24). However, the absence of ManDH activity in L. panis PM1 made fructose metabolism less energy efficient than that in group III lactobacilli, which can produce mannitol. The absence of TPI activity was a key bottleneck which hampered the potential yield of energy from fructose metabolism via the EM pathway; i.e., DHAP accumulation keeps one-half of the carbon from generating ATP, resulting in a zero-sum ATP balance. To overcome this reduced efficiency and the sporadic growth on fructose medium, the TPI gene of E. coli was cloned into and expressed in L. panis PM1. The introduction of this gene eliminated the early growth cessation seen on fructose medium (Fig. 6) and altered fructose metabolism from heterolactic to homolactic fermentation, wherein most fructose was processed to lactate. Also, citrate (initial concentration, 16.03 mM) was not converted to succinate, which is an alternate NAD+ regeneration route for the 6-PG/PK pathway in the presence of citrate (11), suggesting no NADH regeneration provision from this pathway. Therefore, this change in fermentation pattern indicated that in this recombinant PM1 strain, fructose was mostly fermented via the EM pathway, generating 2 ATP molecules per fructose molecule.
Under certain conditions (e.g., low pH or glucose limitation), a shift from homolactic to heterolactic fermentation has been observed in homofermentative lactobacilli, producing end products such as acetate, ethanol, diacetyl, acetoin, and 2,3-butanediol (25, 26). The PM1-TPI strain also produced approximately 11 mM ethanol from fructose fermentation. This ethanol may have been produced via two possible routes: (i) the pyruvate-to-acetyl coenzyme A pathway from the activity of pyruvate dehydrogenase (11) or (ii) the 6-PG/PK pathway via the activity of PGI (Fig. 7). Considering the low level of PGI activity in L. panis PM1 grown on fructose medium (Fig. 3), part of the available pyruvate could have been directed to ethanol via the former route to meet the redox balance needs under conditions of rapid fructose metabolism.
Overall, our findings clearly indicate that L. panis PM1 can ferment fructose via both the EM and 6-PG/PK pathways and that the TPI enzyme plays a critical role in the shift between the 6-PK/PG and EM pathways, leading to an altered fermentation pattern. Despite the improved energy efficiencies of the EM pathway, the reason why L. panis PM1 loses its TPI activity requires further study.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Saskatchewan Agriculture Development Fund and the Agricultural Bioproducts Innovation Program of Agriculture and Agri-Food Canada.
The genome DNA sequence of L. panis PM1 is the result of a collaboration among the University of Saskatchewan, Contango Strategies (Saskatoon, SK, Canada), and Milligan Bio-Tech (Foam Lake, SK, Canada).
We acknowledge the contribution of D. A. S. Grahame, V. Pittet, and M. C. Haakensen to genome sequencing. We thank N. Low and X. Qiu of the University of Saskatchewan for providing HPLC and qRT-PCR machines, respectively. L. Axelsson of Nofima Mat, Norway, kindly provided the pSIP411 plasmid.
Footnotes
Published ahead of print 4 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02377-13.
REFERENCES
- 1.Stiles ME, Holzapfel WH. 1997. Lactic acid bacteria of foods and their current taxonomy. Int. J. Food Microbiol. 36:1–29 [DOI] [PubMed] [Google Scholar]
- 2.Kandler O. 1983. Carbohydrate metabolism in lactic acid bacteria. Antonie Van Leeuwenhoek 49:209–224 [DOI] [PubMed] [Google Scholar]
- 3.Wisselink HW, Weusthuis RA, Eggink G, Hugenholtz J, Grobben GJ. 2002. Mannitol production by lactic acid bacteria: a review. Int. Dairy J. 12:151–161 [Google Scholar]
- 4.Saha BC, Racine FM. 2011. Biotechnological production of mannitol and its applications. Appl. Microbiol. Biotechnol. 89:879–891 [DOI] [PubMed] [Google Scholar]
- 5.Heinrich R, Montero F, Klipp E, Waddell TG, Melendez-Hevia E. 1997. Theoretical approaches to the evolutionary optimization of glycolysis: thermodynamic and kinetic constraints. FEBS J. 243:191–201 [DOI] [PubMed] [Google Scholar]
- 6.Poolman B. 1993. Energy transduction in lactic acid bacteria. FEMS Microbiol. Rev. 12:125–147 [DOI] [PubMed] [Google Scholar]
- 7.Khan NH, Kang TS, Grahame DA, Haakensen MC, Ratanapariyanuch K, Reaney MJ, Korber DR, Tanaka T. 2013. Isolation and characterization of novel 1,3-propanediol-producing Lactobacillus panis PM1 from bioethanol thin stillage. Appl. Microbiol. Biotechnol. 97:417–428 [DOI] [PubMed] [Google Scholar]
- 8.Kang TS, Korber DR, Tanaka T. 2013. Glycerol and environmental factors: effects on 1,3-propanediol production and NAD+ regeneration in Lactobacillus panis PM1. J. Appl. Microbiol. 115:1003–1011 [DOI] [PubMed] [Google Scholar]
- 9.Kang TS, Korber DR, Tanaka T. 2013. Influence of oxygen on NADH recycling and oxidative stress resistance systems in Lactobacillus panis PM1. AMB Express 3:10. 10.1186/2191-0855-3-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 11.Kang TS, Korber DR, Tanaka T. 2013. Contributions of citrate in redox potential maintenance and ATP production: metabolic pathways and their regulation in Lactobacillus panis PM1. Appl. Microbiol. Biotechnol. 97:8693–8703 [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 13.Pedersen C, Jonsson H, Lindberg JE, Roos S. 2004. Microbiological characterization of wet wheat distillers' grain, with focus on isolation of lactobacilli with potential as probiotics. Appl. Environ. Microbiol. 70:1522–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saier MH., Jr 2000. Families of transmembrane sugar transport proteins. Mol. Microbiol. 35:699–710 [DOI] [PubMed] [Google Scholar]
- 15.Årsköld E, Lohmeier-Vogel E, Cao R, Roos S, Radstrom P, van Niel EW. 2008. Phosphoketolase pathway dominates in Lactobacillus reuteri ATCC 55730 containing dual pathways for glycolysis. J. Bacteriol. 190:206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sørvig E, Mathiesen G, Naterstad K, Eijsink VG, Axelsson L. 2005. High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors. Microbiology 151:2439–2449 [DOI] [PubMed] [Google Scholar]
- 17.Taranto MP, Font de Valdez G, Perez-Martinez G. 1999. Evidence of a glucose proton motive force-dependent permease and a fructose phosphoenolpyruvate:phosphotransferase transport system in Lactobacillus reuteri CRL 1098. FEMS Microbiol. Lett. 181:109–112 [DOI] [PubMed] [Google Scholar]
- 18.Saier MH, Ye J-J, Klinke S, Nino E. 1996. Identification of an anaerobically induced phosphoenolpyruvate-dependent fructose-specific phosphotransferase system and evidence for the Embden-Meyerhof glycolytic pathway in the heterofermentative bacterium Lactobacillus brevis. J. Bacteriol. 178:314–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veiga-da-Cunha M, Santos H, Van Schaftingen E. 1993. Pathway and regulation of erythritol formation in Leuconostoc oenos. J. Bacteriol. 175:3941–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stolz P, Vogel RF, Hammes WP. 1995. Utilization of electron acceptors by lactobacilli isolated from sourdough. Z. Lebensm. Unters. Forsch. 201:402–410 [Google Scholar]
- 21.Richter H, Vlad D, Unden G. 2001. Significance of pantothenate for glucose fermentation by Oenococcus oeni and for suppression of the erythritol and acetate production. Arch. Microbiol. 175:26–31 [DOI] [PubMed] [Google Scholar]
- 22.Zaunmuller T, Eichert M, Richter H, Unden G. 2006. Variations in the energy metabolism of biotechnologically relevant heterofermentative lactic acid bacteria during growth on sugars and organic acids. Appl. Microbiol. Biotechnol. 72:421–429 [DOI] [PubMed] [Google Scholar]
- 23.Condon S. 1987. Responses of lactic acid bacteria to oxygen. FEMS Microbiol. Rev. 46:269–280 [Google Scholar]
- 24.Hugenholtz J. 1993. Citrate metabolism in lactic acid bacteria. FEMS Microbiol. Rev. 12:165–178 [Google Scholar]
- 25.Borch E, Berg H, Holst O. 1991. Heterolactic fermentation by a homofermentative Lactobacillus sp. during glucose limitation in anaerobic continuous culture with complete cell recycle. J. Appl. Microbiol. 71:265–269 [Google Scholar]
- 26.Torino MI, Taranto MP, Sesma F, de Valdez GF. 2001. Heterofermentative pattern and exopolysaccharide production by Lactobacillus helveticus ATCC 15807 in response to environmental pH. J. Appl. Microbiol. 91:846–852 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.