Abstract
Type II DNA methyltransferases (MTases) are enzymes found ubiquitously in the prokaryotic world, where they play important roles in several cellular processes, such as host protection and epigenetic regulation. Three classes of type II MTases have been identified thus far in bacteria which function in transferring a methyl group from S-adenosyl-l-methionine (SAM) to a target nucleotide base, forming N-6-methyladenine (class I), N-4-methylcytosine (class II), or C-5-methylcytosine (class III). Often, these MTases are associated with a cognate restriction endonuclease (REase) to form a restriction-modification (R-M) system protecting bacterial cells from invasion by foreign DNA. When MTases exist alone, which are then termed orphan MTases, they are believed to be mainly involved in regulatory activities in the bacterial cell. Genomes of various lytic and lysogenic phages have been shown to encode multi- and mono-specific orphan MTases that have the ability to confer protection from restriction endonucleases of their bacterial host(s). The ability of a phage to overcome R-M and other phage-targeting resistance systems can be detrimental to particular biotechnological processes such as dairy fermentations. Conversely, as phages may also be beneficial in certain areas such as phage therapy, phages with additional resistance to host defenses may prolong the effectiveness of the therapy. This minireview will focus on bacteriophage-encoded MTases, their prevalence and diversity, as well as their potential origin and function.
RESTRICTION-MODIFICATION SYSTEMS
Genes encoding restriction-modification (R-M) systems are present on approximately 90% of currently available bacterial and archaeal genome sequences (1). These systems can be encoded by genes on plasmids or chromosomes, and their general role is to recognize and target invading foreign DNA with restriction enzymes, while simultaneously protecting the host's DNA by methyltransferase (MTase) activity. Bacterial R-M systems have been comprehensively reviewed (2–4), and therefore, only the main characteristics of these R-M systems will be summarized here. Four types of R-M systems (I, II, III, and IV) are currently recognized (5), differing in the functional arrangement of the restriction endonuclease (REase) and methyltransferase activities, as well as the requirement for specificity subunits or additional cofactors (6). Typical type I R-M systems consist of three subunits, the S (specificity subunit), M (methyltransferase), and R (restriction endonuclease) subunits, where the S subunit determines the target recognition specificity of the system, while the M and R subunits are required for methylation activity and DNA restriction, respectively (7). Type II R-M systems are the most prevalent type and generally function as two individual proteins (8), where the REase cleaves the target DNA at defined positions within or close to their recognition site, while the MTase protects host DNA by methylation. Type III R-M systems are composed of the products of at least two genes, res and mod, where Mod binds to and methylates DNA, while Res functions in DNA restriction. Mod can function independently of Res; however, Mod is required for Res activity (9, 10). The type IV restriction systems differ from the other types in that the methyltransferase and endonuclease activities are combined in a single enzyme (11, 12), which exclusively cleaves modified DNA (methylated, glucosyl-hydroxymethylated, and hydroxymethylated) (5).
As this minireview focuses on orphan MTases, the majority of which belong to type II MTases, it is necessary to first define this type in more detail. Type II DNA MTases are enzymes found ubiquitously in the prokaryotic world and play important additional roles (other than in host protection from invading DNA) in several cellular processes, such as replication, transcription, and population evolution (13). Type II methyltransferases function by transferring a methyl group from S-adenosyl-l-methionine (SAM) to a target sequence, and these MTases can be divided into three functional classes. Two target the exocyclic nitrogen atoms in certain bases of double-stranded DNA at position 4 in cytosine or position 6 in adenine to generate N-4-methylcytosine (m4c) or N-6-methyladenine (m6a), respectively. The third class is the MTase targeting the carbon 5 position of cytosine to generate C-5-methylcytosine (m5c) (14). MTases are further subclassified based on the presence of conserved amino acid motifs, which represent the DNA binding domain, the target recognition domain (TRD) and the catalytic domain. The C-5-MTases are found to contain 10 conserved domains (designated I through to X) (15), whereas the exocyclic N-targeting MTases harbor nine conserved domains and can be subdivided into 6 groups, α, β, γ, ζ, δ, and ε based on the SAM binding site, active site, and TRD (16, 17). At the time of writing, REBASE listed 9,789 type II REases, and 13,787 putative type II MTases (http://rebase.neb.com/rebase/rebase.html). The large number of MTases relative to that of REases may be the result of the toxicity of the latter on bacterial cells, and as a result, the MTases may have been retained more freely. It should be noted that REases may not be identified as such by bioinformatics due to limited overall sequence similarity (18), and as mentioned above, MTases have additional roles other than R-M systems, such as cell cycle regulation (19). While R-M systems typically consist of a combination of REase and MTase, both have been found to exist independently as orphan genes in bacterial genomes, and the remainder of this minireview will focus on the orphan MTases.
BACTERIAL ORPHAN MTases
While this minireview is focused on highlighting orphan MTases in bacteriophage genomes, it is first important to understand the role that they play in bacterial cells. Mobile genetic elements, including plasmids, prophages, insertion sequence elements (ISs), and transposons, harbor and may mobilize methyltransferase-encoding genes (sometimes accompanied by a cognate restriction endonuclease-encoding gene) and as a result can facilitate their spread among bacterial genomes via plasmid uptake and exchange or through integration of lysogenic phages (20, 21). It has been demonstrated that the genetic material encoding R-M systems can become integrated into host chromosomes where it replicates along with the host genome and may block other “parasitic” DNA attempting to integrate/enter into the host (22, 23). The genes encoding R-M systems in bacteria may be challenged by newly introduced DNA (e.g., “incompatible” plasmids or a transducing fragment of homologous DNA), which may attempt to displace the genes encoding R-M systems (20). In such cases, R-M systems may behave “selfishly” if the incoming parasitic DNA gains access to the host cell. Displacement of the R-M systems in members of the populations may cause the cells to die, a process known as postsegregational killing, a similar mechanism of plasmid maintenance that occurs with toxin-antitoxin systems (24–26). The daughter cells are no longer protected due to a reduction in methylation activity, and their genetic material is subjected to cleavage by the still present REase (27). An example of this phenomenon is the chromosomally encoded BamHI R-M system of Bacillus amyloliquefaciens, which was shown to resist replacement by homologous recombination (28). Recent studies have demonstrated that the presence of an orphan MTase targeting the same DNA sequence as a resident R-M system may protect the host in the event of the displacement of such an R-M system (18). For example, when the genes encoding the R and M subunits of the type II EcoRII system are expressed from a plasmid in the absence of selective pressure, the resulting segregational loss of the plasmid was shown to lead to incomplete methylation of host DNA and to cell death due to the persistent activity of the EcoRII REase. These detrimental effects can be counteracted by expressing an orphan MTase with the same recognition site as the EcoRII REase. These findings suggest that such events occur naturally in bacterial strains and that it may drive the acquisition and maintenance of orphan MTases. It has been shown that the acquisition of R-M systems can occur by means of horizontal gene transfer (HGT) (29), for example by being present on insertion sequence (IS) elements (30), and as a large number of annotated genes on bacterial genomes encode orphan MTases, it has been proposed that these genes have also been acquired by HGT or are due to genetic decay of R-M systems in the host cell (31).
While MTases are most often described in the context of R-M systems, MTases can exist as orphan MTases without a cognate REase partner and as such have been shown to be involved in cell regulation, replication, DNA repair, and population evolution (19, 32, 33). Understanding the roles of orphan MTases in bacterial cells may reveal the function they play in phage genomes. For the DAM (DNA adenine MTase) enzyme of Escherichia coli, these roles include DNA mismatch repair, a process requiring discrimination between the parental DNA strand and newly synthesized DNA behind the replication fork (34). Due to the DAM methyltransferase, the parental DNA is already methylated prior to replication, while the newly replicated strand is not, allowing the mismatch repair protein, MutH, to distinguish between the (presumed) correct sequence of the parental strand and noncomplementary bases on the newly synthesized strand. The mismatch repair protein can then utilize the parental strand as a template to fix such replication errors (35). The frequency by which the DAM recognition site (GATC) occurs in the origin of replication of E. coli allows tight regulation of the cell cycle and consequently chromosomal replication. The hemimethylated DNA prevents the replication initiation protein DnaA from acting more than once on the replication origin, oriC, in a given cell cycle, a process known as sequestration (36). Several examples of the role of DAM in bacterial virulence have also been reviewed (37).
Initially identified in Caulobacter crescentus, the cell cycle regulator methyltransferase (CcrM) targets the recognition sequence GANTC (38, 39) in a nonprocessive manner (40), in contrast to earlier findings (39). The CcrM methylase is essential for the proper control of the life cycle of Caulobacter, as methylation of the ori directs initiation by DnaA (41). Following cell division, which produces two morphologically distinct cell types, the stalk cell (DNA replication allowed) and the swarmer cell (flagellum, no DNA replication allowed), the CcrM methylase is degraded, as methylation of the ori is not required until late in the cycle (42). In the swarmer cell, CtrA (cell cycle regulator) prevents chromosome replication; however, when the swarmer cell changes to a stalk cell, this regulatory protein becomes subject to degradation. Transcription of the gene specifying the CtrA regulator protein is dependent on two promoter sequences, one of which contains a GANTC site (43). When this site is methylated, transcription of ctrA is repressed, allowing timed and controlled synthesis of CcrM, as CtrA accumulation in the cell is required to transcribe ccrM. As DNA replication proceeds, the replication forks pass the CtrA locus whereby it becomes hemimethylated and is subsequently expressed under the control of the GcrA protein (41, 42).
BACTERIOPHAGE MTases, POSSIBLE FUNCTION, AND PREVALENCE
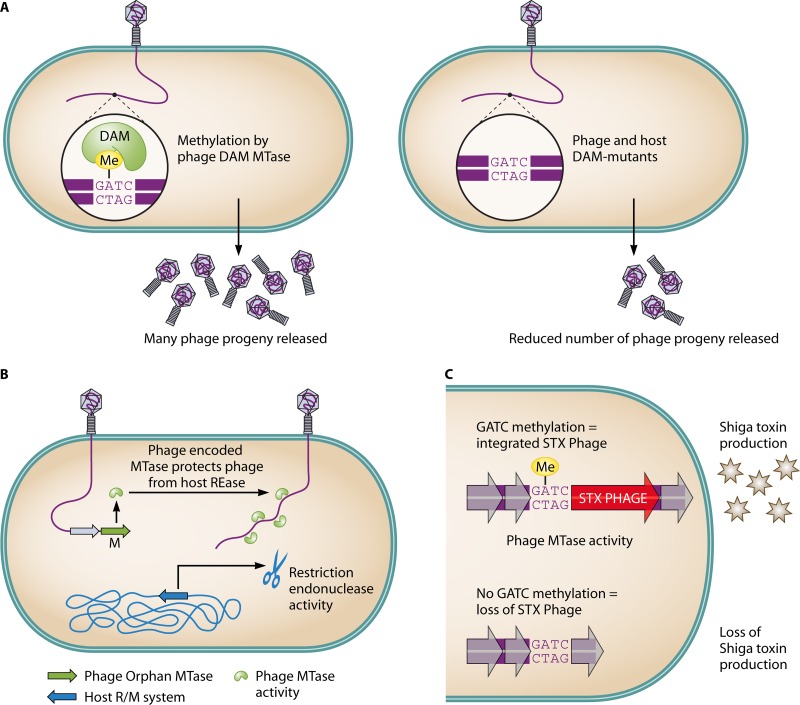
Currently, little is understood as to the function of orphan MTases when genes encoding them are present in the genomes of bacteriophages, but previous studies indicate that they may have functions that are similar to the functions of MTases found in bacterial cells. Well-studied phage-derived methyltransferases are those encoded by the Escherichia coli-infecting T-even bacteriophages. Previous in vitro studies have shown that phage T4 encodes a DAM MTase, which was found to target GATC sites (44) and protects phage DNA from restriction endonuclease recognizing this sequence (45). DAM activity is essential in the regulation of E. coli cellular functions (46), and while it does not appear to play a role in the lytic cycle of the T2 and T4 phage (47), it was found to play a role other than blocking host endonucleases in the E. coli temperate phage P1. P1 specifies a functional DAM enzyme (designated Dmt) of 754 amino acids (48, 49), which has been observed to be active only during the lytic stage of the phage cycle (50), and packaging of phage P1 DNA is dependent on methylated GATC sequences within the 162-bp pac site (51). Furthermore, in the presence of a dam mutant host and dmt− (phage DAM knockout) phage mutant, a significant reduction in phage progeny was observed in comparison to phage propagations produced in the presence of either the host or phage MTase (51) (Fig. 1 A). Additionally, the promoters for the site-specific recombinase-encoding gene, cre, were found to be regulated by DAM methylation (52), and it is suggested that several other areas of the P1 genome are under transcriptional control by DAM methylation (48).
Fig 1.
Potential advantageous effects of phage-encoded orphan MTases (see text for further details). (A) Methylation of the GATC sites within the pac region of the phage P1 genome by a self-encoded DAM MTase facilitates the efficient release of progeny phage. Generation of both dam mutant hosts and dmt− phage (phage DAM knockout) prevents methylation of the phage DNA during packaging, leading to a decreased level of phage progeny released. (B) Protection of phage genomes from host-encoded restriction endonucleases through the protection afforded by the phage-encoded orphan MTase. (C) DAM methylation is essential for lysogeny of the Shiga toxin-encoding phage 933W and release of Shiga toxin by EHEC. Loss of GATC methylation results in release of integrated phage and loss of Shiga toxin production by EHEC. Me, methyl group.
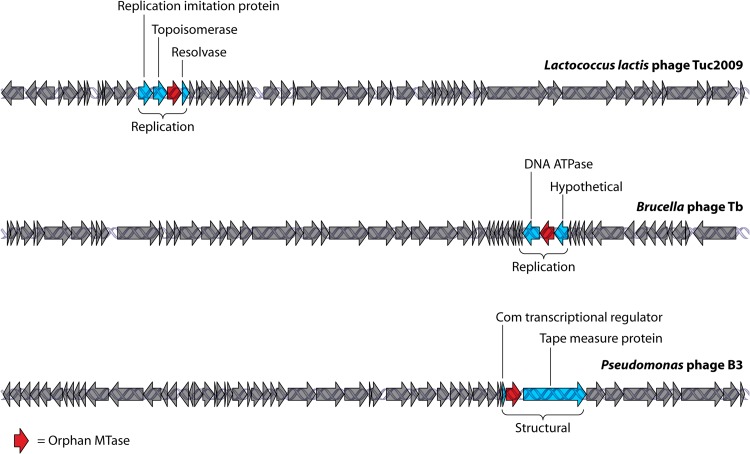
A number of orphan MTases have also been identified in Bacillus phages. Interestingly, these phages carry genes encoding MTases that can recognize more than one target sequence due to the presence of multiple TRDs (53). Such multispecific MTases have been identified in the temperate Bacillus subtilis phages ø3T, p11B, and SPβ (54), which each recognize two target sequences, while phage SPR carries a gene that encodes a type II MTase specific for three different recognition sites (55). Under conditions where the Bacillus prophage SPβ is induced, an orphan MTase becomes active during the lytic phage cycle and provides the phage with protection against digestion by various restriction endonucleases (56, 57) (Fig. 1B). B. amyloliquefaciens phage H2 carries a gene that encodes a multispecific m5c methyltransferase that was found to share high sequence homology with its counterparts on the genomes of ø3T, SPR, and p11s (58). Furthermore, the ability of these MTases to recognize more than a single target sequence may provide a particular phage with immunity to several R-M systems. DAM methylation plays a significant role in bacterial pathogenicity (59), and DAM-mediated methylation is also important for the maintenance of lysogeny of the 933W phage with a gene encoding Shiga toxin (60). As a result, the phage is kept in a temperate state allowing enterohemorrhagic E. coli (EHEC) to actively produce the virulent toxin (Fig. 1C). Although many of these proteins contain the conserved domains common among these MTases (61), the presence of such motifs is not essential for a functional enzyme. For example, the lactococcal phage ø50 was found to be resistant to the plasmid gene-encoded resistance systems (pTR2030) present in the parent strain, one being an R-M system (62). It was subsequently shown that both pTR2030 and ø50 shared sequence homology within a type II MTase-encoding gene, and furthermore, it was demonstrated that a single motif was sufficient to confer the phage with protection against the R gene of the R-M system (62). The Myxococcus xanthus temperate phage Mx8 was also found to contain an MTase-encoding gene, designated mox. This gene was found to be nonessential for lysogeny and presented no effect on the lytic phage cycle or superinfection immunity (lysogenic phage blocking subsequent infection of superinfecting phages) (63). Phage genomes are significantly smaller than their bacterial counterparts, and while the functions of many proteins in phage genomes are unknown, it is assumed that genes are retained only if they confer some benefit for continued survival (64–66). Therefore, the mox gene may play a role in phage genome protection. The locations of these MTase-encoding genes within phage genomes may provide an insight into their function. For example, the genome of lactococcal phage Tuc2009 harbors a putative MTase-encoding gene flanked by genes specifying a DNA replication protein, a putative topoisomerase, and a resolvase (67) (Fig. 2). A similar genetic arrangement is observed for Brucella phage Tb (68). The MTase encoded by the Tuc2009 phage gene may actively methylate DNA during replication to prevent digestion and protect progeny, but the MTase may also play a role in regulating DNA replication in terms of the timing of replication or potentially functioning in mismatch repair, which as mentioned above, is a trait also observed for certain bacterial MTases. Pseudomonas phage B3 possesses an orphan DAM-encoding gene within the structural module of the phage genome flanked by genes specifying a transcriptional regulator and the tape measure protein (TMP). It is imaginable that DAM methylation acts in a regulatory capacity to control the production of phage proteins such as the TMP, among others (69) (Fig. 2). Additionally, it was hypothesized that an adenine-specific MTase plays a role in regulating the cell cycle interactions of Drosophila and the intracellular symbiont Wolbachia pipientis, as prophages have been found only in strains that are in a symbiotic relationship (70), further highlighting that phage orphan MTases have roles similar to the roles of MTases found in bacterial cells.
Fig 2.
Genomic organization of lactococcal phage Tuc2009, Brucella phage Tb, and Pseudomonas phage B3 highlighting examples of orphan MTases and flanking genes to show that orphan MTases can be found in different locations in a given phage genome (regions encoding predicted structural and replication proteins).
The ability of phages to integrate the genes encoding orphan MTases into their genome may further contribute to the emergence of phages with broad host ranges, as the acquisition of an MTase-specifying gene would allow progeny phages to infect additional hosts by overcoming R-M systems. Such a notion is consistent with observations for Salmonella phage PVP-SE1 (71) and Yersinia phage PY100 (72), both of which possess an MTase and display a broad host range. While this minireview is primarily focused on orphan phage MTases, it is interesting that the Staphylococcus aureus quadruple-converting phage π42 was shown to harbor an R-M system, Sau42I, that confers resistance to the host against lysis by exogenous phages (73) and in doing so provides a selective advantage to the lysogenic phage. Burkholderia cepacia phage KL3 carries a gene that encodes an MTase that appears to be part of a functional defense module. Gene 47 encodes an EcoRII MTase associated with not only a cognate endonuclease but also a Vsr endonuclease (74). It has been proposed that the MTase functions in phage protection and that the endonuclease functions in bacterial DNA digestion, while the Vsr protein is thought to be responsible for postmethylation mismatch repair, the latter of which has been shown to be controlled by DAM-mediated methylation in bacterial cells as mentioned above (74). These studies indicate that phages have developed the ability to circumvent the R-M mechanisms of their hosts by the integration of orphan MTase-encoding genes and whole R-M systems, highlighting the ever-adapting nature of phages to become more advanced to outwit their bacterial hosts. While a number of studies on bacteriophage MTases have been performed, the abundance of these enzymes in phage genomes is relatively unknown.
To estimate the number of genes encoding putative orphan MTases among bacteriophage genomes, a manual search of the PubMed protein database (http://www.ncbi.nlm.nih.gov/pubmed/) was performed using the term “phage methyltransferase” which yielded 819 putative orphan phage MTases associated with a wide variety of bacterial species. Using the REBASE website (http://rebase.neb.com/rebase/rebase.html), a database was constructed to blast these 819 retrieved proteins to determine whether these putative MTases belong to one of the three subtypes of type II MTases (m6a, m4c, and m5c) (for examples from each species, see Table S1 in the supplemental material). It should be noted that some of these MTases may not possess all the conserved motifs found within their bacterial counterparts; however, as mentioned above, it has been shown that a single motif can be enough to provide phage protection against R-M systems (62). Orphan MTases found in the currently available, fully or partially annotated phage genomes are associated with a diverse range of hosts and environments. These hosts include members of the family Enterobacteriaceae, the pulmonary pathogens Burkholderia and Mycobacterium, and the marine bacterium Synechococcus (Table S1). Additionally, phages infecting industrial strains such as the dairy starter culture Lactococcus lactis were also found to carry genes that encode MTases. Acquisition of (apparently) orphan MTases by bacteriophages seems to occur in diverse phage-host ecosystems with high cell densities, causing a competitive, high-number propagation race between phages and bacteria: bacterial counts in the gastrointestinal tract range from 104 CFU ml−1 in the stomach to 1012 CFU ml−1 in the colon (75), and Synechococcus on occasions can form blooms with 106 CFU ml−1 (76). Likewise, the relatively closed environment of a fermentation facility creates very high cell densities, mostly represented by a small number of strains/species. It was found that despite being present in such a large variety of phages, with the exception of some outliers, methyltransferases that were predicted to belong to a particular methylation type (m5c, m4c, or m6a), for the most part, were shown to group together (see Fig. S1 in the supplemental material). Several of the m6a and m4c methyltransferases were shown to form a phylogenetic group (Fig. S1), which is consistent with their classification.
SOURCE OF BACTERIOPHAGE MTases
Understanding the role of these orphan MTases also involves trying to find the origins of the MTase-encoding genes. Bacterial hosts containing genes that encode an R-M system and are infected by phages may, at low frequency, produce progeny phages that have become methylated and thus are resistant to such an R-M system (77). The resulting “modified” phages are thus due to methylation of the phage genome by the host-encoded R-M system and did not arise through the acquisition of genomic material. From the data and studies presented in this minireview, it is apparent that phages have gained the advantageous ability to permanently overcome such an R-M hurdle via the integration of a cognate orphan MTase. Exchange of genetic material between bacteria occurs naturally via HGT processes such as transformation, transduction, and conjugation (78). Bacteriophages play an important role in the lateral exchange of genetic material between bacterial hosts, and while this occurs in lytic phage often by recognition of pseudo-pac sites on host chromosomes, it is more common among the lysogenic phages, as during lytic phage infection, the host DNA can be substantially degraded (79, 80). Temperate phages can pick up bacterial genes that flank the integration sites as a result of an excision error and transfer these genes to a new host, known as specialized transduction. Specialized transduction has been demonstrated in several studies including Brachyspira intermedia (81) and Bacillus phages (82). Temperate phages likely attained their orphan MTase-encoding genes in this way, as no homology was found between Bacillus temperate phages and their hosts (57). It has been demonstrated that E. coli phage P2 is capable of transferring the EcoT38I R-M system into chromosomal DNA by means of HGT (83): approximately 30% of the genome of phage P2 and sequences homologous to the phage P2 attachment site were found flanking the EcoT38I R-M system genes. Evidence of the uptake and transfer of the EcoO109I R-M by HGT was also shown for the E. coli-infecting bacteriophage P4 (21). This provides strong evidence that temperate phages transfer genetic information encoding DNA modification enzymes, and therefore, we hypothesize that in certain cases temperate phages may retain the MTase gene due to a conferred advantage such as the ability to overcome host R-M systems or improved genome replication and/or regulation.
Errors in the DNA packaging process during lytic phage replication may also lead to HGT whereby DNA from the bacterial host (genomic and/or plasmid) can become incorporated into the phage capsid and subsequently introduced into a bacterium, a process referred to as generalized transduction (84). Furthermore, nonhomologous recombination is known to play a role in the mosaic nature of many phage genomes, allowing the exchange of genes as well as complete functional modules (85), and may be behind the emergence of these orphan phage MTases (86). Previous studies have shown that phages can exchange genetic material with host bacteria through homologous recombination events, for example lactococcal phage ul36 was shown to be subject to two genetic exchanges with prophage-like DNA located within the host chromosome (87). Orphan MTases may thus be acquired by temperate phages which consequently become incorporated in the bacterial genome through lysogeny. Subsequent to infection by a lytic phage, the prophage-encoded MTase may be transferred to the lytic phage during the phage cycle, giving rise to R-M-resistant progeny. This may be the case for phage 4268 where comparative genomic analysis of its lactococcal host revealed high sequence similarity to the chromosomally encoded MTase (88). As mentioned above, the functional MTase, LlaI, was shown to be acquired by a phage genome from a conjugative plasmid. While the exact mechanism of transfer is unknown, it is likely that the acquisition of the gene by the phage was due to HGT, e.g., utilizing nonhomologous recombination (62). This highlights the apparent regular occurrence of such exchanges during phage-host interactions and the role HGT plays in the spread of orphan MTases. While bacteria continue to fight back against phage infection, it is clear that phages are driven to evolve mechanisms that will eventually allow them to overcome these resistance systems, thus reaching a stable coexistence with their hosts. The acquisition of orphan MTase-encoding genes is one such mechanism.
FUTURE PERSPECTIVES
Exploitation of phage MTases for use in phage therapy.
With the emergence of highly antibiotic-resistant bacteria, in particular among pathogens that are easily transmitted (Enterococcus, Streptococcus, and Mycobacterium tuberculosis) (89–91), renewed interest in phage therapy has emerged. A number of key elements have to be known before a phage can be applied as a therapeutic agent. These include using a well-characterized lytic phage with a broad host range, while avoiding the use of temperate phages. The determination of complete genome sequences of the phages is necessary to reveal the presence of any genes that may encode toxic or allergic compounds. Several recent reviews highlight the methods and challenges that accompany the use of phages as antimicrobial agents (92, 93). The success of phage therapy may be limited by the inherent or acquired phage defense mechanisms of a targeted bacterial species. Studies have demonstrated that phage cocktails can be effective in phage therapy to compensate for phage resistance, although eventually bacteria and phages achieve environmental coexistence, thus rendering the phage therapy ineffective (94). An approach to aid therapeutic phages in circumventing bacterial resistance mechanisms may be to include phage isolates harboring orphan MTases. By including such phages with orphan MTases to the selection criteria for phage therapy, it could aid in the rapid removal of infectious bacteria by delaying the ability of host R-M systems to attack the incoming phage, thus prolonging the treatment's effectiveness and improving the overall benefits of phage therapy.
SMRT DNA sequencing and functional analysis of phage MTases.
Characterization or identification/cloning of MTases using traditional molecular biology approaches can involve PCR amplification, restriction digestion, and overnight ligation of vector and target DNA, followed by transformation and selection via antibiotic plates. Furthermore, subsequent colonies have to be screened by PCR, plasmid profiling, and in most cases sequencing of the vector's multiple cloning site region to determine and verify the presence and sequence integrity of an insert (95). This approach, while successful, is rather laborious and time-consuming. A particular type of new generation sequencing technology has provided an alternative, fast, and attractive approach. Pacific Biosciences SR DNA sequencing technology is a recent high-throughput sequencing platform that can generate average read lengths of over 2,500 bp of whole phage genomes (96). Despite initial high error rates, on-going improvements of the technology and software have greatly increased read accuracy (97, 98). Each sequencing single-molecule real-time (SMRT) cell contains 150,000 zero-mode waveguides (ZMVs) (99), nanophotonic compartments containing a single DNA polymerase and a single strand of template DNA (Pacific Biosciences, Menlo Park, CA). The ZMVs have a tiny aperture that allows light to penetrate, creating a chamber for visualizing the activity of the DNA polymerase. Each nucleotide is linked to a unique fluorophore label attached to the phosphate of the base. As a nucleotide is incorporated, the fluorescent molecule is released and detected, thus allowing nucleotide sequence determination in real time. This sequencing technology can be applied to de novo sequencing and to base modification analysis (Pacific Biosciences, Menlo Park, CA). Recent studies have utilized this SMRT technology to characterize DNA methylation patterns by monitoring the time taken to incorporate each nucleotide by the DNA polymerase, the kinetic variation (KV) (100). Measuring the KV allows direct detection of modified nucleotides in the DNA template, including N-6-methyladenine and C-5-methylcytosine, as each has a unique kinetic signature (101). SMRT sequencing has enabled researchers to determine the identity and position of methylated bases, and from this information, the target sequence of the MTases encoded by genes on the genome can be derived (102). This technology can be easily applied to phage genomes, as they are relatively small, and a large quantity of data can be generated using a 5- to 10-kb insert library, ultimately to determine whether phage gene-encoded orphan MTases are functionally active and also to potentially establish the role they play.
CONCLUSION
The occurrence of genes that specify orphan MTases is relatively high at approximately 20% of the currently annotated phage genomes. While a number of studies have been carried out on MTases encoded by genes on temperate phages, little is known about their recognition sequences, source, or the precise role these genes play in enhancing the infectivity and survival of phage populations. It is apparent from Table S1 in the supplemental material that phages isolated from a diverse range of ecosystems possess integrated orphan MTase-encoding genes. However, it is unclear whether this is driven by the vast array of phage-phage and/or phage-host interactions or the necessity for survival or if certain phage species are better equipped for such genome modifications. More than likely, in the majority of cases, the incorporation of MTase-encoding genes into phage genomes may provide protection against host R-M systems. The continued isolation and whole-genome sequencing of phages as well as the use of next-generation sequencing may provide greater insight into the source of these MTases, their target specificities, and mechanisms of genome incorporation.
Supplementary Material
ACKNOWLEDGMENTS
D. van Sinderen is the recipient of a Science Foundation Ireland (SFI) principal investigator award (award 08/IN.1/B1909). J. Murphy is the recipient of an Irish Research Council Enterprise Partnership Scheme postgraduate scholarship.
Biographies

James Murphy received his B.Sc. in Microbiology from University College Cork, Ireland. In 2009 he completed a taught master's degree in Molecular Medicine at Trinity College Dublin with a research component focusing on Helicobacter pylori outer membrane vesicles. He returned to University College Cork to complete a research master's degree studying the biodiversity of bacteriophage in an Irish dairy plant under the supervision of Professor Douwe van Sinderen. He has remained a member of this research group and is currently an Irish Research Postgraduate Scholar via the Enterprise Partnership scheme, studying the 936-type lactococcal bacteriophages. The main focus of his current research is on phage biodiversity, whole-genome analysis, genomic adaptations, and evolution.

Jennifer Mahony obtained her B.Sc. in Applied Biosciences with Food Science & Technology from Cork Institute of Technology in 2003 and during her undergraduate studies worked in the phage research laboratory of Professor Douwe van Sinderen as a research assistant. Upon completion of her B.Sc. degree, with the financial support of an Irish Research Council scholarship, she completed a Ph.D. at University College Cork under the supervision of Professor Douwe van Sinderen on the subject of phage resistance systems encoded by dairy lactococcal prophages. Currently, as a postdoctoral researcher, she continues to work on lactococcal phages, focusing on phage-host interactions.

Stuart Ainsworth received his B.Sc. in Biotechnology from the University of Reading, United Kingdom, and is currently studying for a Ph.D. in microbiology at University College Cork, Ireland. His research interests focus on bacteriophage-host interactions, including the host response to bacteriophage infection and identification of bacteriophage-host receptors.

Arjen Nauta received a cum laude Ph.D. degree from the State University of Groningen in 1997 with his work on bacteriophages. After a postdoctoral fellowship, he joined FrieslandCampina in 1998, where he held various Research and Development positions. At present, he is the Senior Scientist of Nutritional Sciences at the FrieslandCampina Innovation Centre and is responsible for research activities and external collaborations in the field of dairy fermentations and microbiology of the gastrointestinal tract.

Douwe van Sinderen received his B.Sc./M.Sc. in Biochemistry from the University of Groningen, The Netherlands. He obtained a Ph.D. degree from the same university in 1994, after which he moved to Ireland to work as a postdoctoral scientist at the National Food Biotechnology Centre at University College Cork (UCC). He was awarded an EMBO fellowship to work in the Department of Microbiology at UCC, after which he joined the academic staff in this department (now School of Microbiology), where he currently works as an Associate Professor. He has worked on bacteriophages (infecting lactic acid bacteria) for 20 years and became interested in this research area not only because of their relevance to the food industry but also and especially out of scientific curiosity due to their prevalence, diversity, genomic plasticity, and intriguing functionality as parasitic, DNA-injecting, self-assembling nanomachines.
Footnotes
Published ahead of print 11 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02229-13.
REFERENCES
- 1.Roberts RJ, Vincze T, Posfai J, Macelis D. 2010. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38:D234–D236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327 [DOI] [PubMed] [Google Scholar]
- 3.Pingoud A, Fuxreiter M, Pingoud V, Wende W. 2005. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 62:685–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Youell J, Firman K. 2012. Mechanistic insight into type I restriction endonucleases. Front. Biosci. 17:2122–2139 [DOI] [PubMed] [Google Scholar]
- 5.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, Firman K, Gromova ES, Gumport RI, Halford SE, Hattman S, Heitman J, Hornby DP, Janulaitis A, Jeltsch A, Josephsen J, Kiss A, Klaenhammer TR, Kobayashi I, Kong H, Kruger DH, Lacks S, Marinus MG, Miyahara M, Morgan RD, Murray NE, Nagaraja V, Piekarowicz A, Pingoud A, Raleigh E, Rao DN, Reich N, Repin VE, Selker EU, Shaw PC, Stein DC, Stoddard BL, Szybalski W, Trautner TA, Van Etten JL, Vitor JM, Wilson GG, Xu SY. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31:1805–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson GG, Murray NE. 1991. Restriction and modification systems. Annu. Rev. Genet. 25:585–627 [DOI] [PubMed] [Google Scholar]
- 7.Murray NE. 2000. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev. 64:412–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sistla S, Rao DN. 2004. S-Adenosyl-l-methionine-dependent restriction enzymes. Crit. Rev. Biochem. Mol. Biol. 39:1–19 [DOI] [PubMed] [Google Scholar]
- 9.Bickle TA, Kruger DH. 1993. Biology of DNA restriction. Microbiol. Rev. 57:434–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su P, Im H, Hsieh H, Kang AS, Dunn NW. 1999. LlaFI, a type III restriction and modification system in Lactococcus lactis. Appl. Environ. Microbiol. 65:686–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lepikhov K, Tchernov A, Zheleznaja L, Matvienko N, Walter J, Trautner TA. 2001. Characterization of the type IV restriction modification system BspLU11III from Bacillus sp LU11. Nucleic Acids Res. 29:4691–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janulaitis A, Petrusyte M, Maneliene Z, Klimasauskas S, Butkus V. 1992. Purification and properties of the Eco57I restriction endonuclease and methylase–prototypes of a new class (type IV). Nucleic Acids Res. 20:6043–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Driscoll J, Heiter DF, Wilson GG, Fitzgerald GF, Roberts R, van Sinderen D. 2006. A genetic dissection of the LlaJI restriction cassette reveals insights on a novel bacteriophage resistance system. BMC Microbiol. 6:40. 10.1186/1471-2180-6-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson GG. 1991. Organization of restriction-modification systems. Nucleic Acids Res. 19:2539–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Cheng X, Klimasauskas S, Mi S, Posfai J, Roberts RJ, Wilson GG. 1994. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 22:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malone T, Blumenthal RM, Cheng X. 1995. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 253:618–632 [DOI] [PubMed] [Google Scholar]
- 17.Bujnicki JM. 2002. Sequence permutations in the molecular evolution of DNA methyltransferases. BMC Evol. Biol. 2:3. 10.1186/1471-2148-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi N, Naito Y, Handa N, Kobayashi I. 2002. A DNA methyltransferase can protect the genome from postdisturbance attack by a restriction-modification gene complex. J. Bacteriol. 184:6100–6108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boye E, Lobner-Olesen A. 1990. The role of dam methyltransferase in the control of DNA replication in E. coli. Cell 62:981–989 [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kita K, Tsuda J, Kato T, Okamoto K, Yanase H, Tanaka M. 1999. Evidence of horizontal transfer of the EcoO109I restriction-modification gene to Escherichia coli chromosomal DNA. J. Bacteriol. 181:6822–6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sekizaki T, Otani Y, Osaki M, Takamatsu D, Shimoji Y. 2001. Evidence for horizontal transfer of SsuDAT1I restriction-modification genes to the Streptococcus suis genome. J. Bacteriol. 183:500–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Claus H, Friedrich A, Frosch M, Vogel U. 2000. Differential distribution of novel restriction-modification systems in clonal lineages of Neisseria meningitidis. J. Bacteriol. 182:1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakayama D, Kobayashi I. 1998. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl. Acad. Sci. U. S. A. 95:6442–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, Storz G. 2010. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 38:3743–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mruk I, Kobayashi I. 13 August 2013. To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. [Epub ahead of print.] 10.1093/nar/gkt711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fukuda E, Kaminska KH, Bujnicki JM, Kobayashi I. 2008. Cell death upon epigenetic genome methylation: a novel function of methyl-specific deoxyribonucleases. Genome Biol. 9:R163. 10.1186/gb-2008-9-11-r163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sadykov M, Asami Y, Niki H, Handa N, Itaya M, Tanokura M, Kobayashi I. 2003. Multiplication of a restriction-modification gene complex. Mol. Microbiol. 48:417–427 [DOI] [PubMed] [Google Scholar]
- 29.Jeltsch A, Pingoud A. 1996. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J. Mol. Evol. 42:91–96 [DOI] [PubMed] [Google Scholar]
- 30.Takahashi N, Ohashi S, Sadykov MR, Mizutani-Ui Y, Kobayashi I. 2011. IS-linked movement of a restriction-modification system. PLoS One 6:e16554. 10.1371/journal.pone.0016554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seshasayee AS, Singh P, Krishna S. 2012. Context-dependent conservation of DNA methyltransferases in bacteria. Nucleic Acids Res. 40:7066–7073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer BR, Marinus MG. 1994. The dam and dcm strains of Escherichia coli–a review. Gene 143:1–12 [DOI] [PubMed] [Google Scholar]
- 33.Schlagman SL, Hattman S, Marinus MG. 1986. Direct role of the Escherichia coli Dam DNA methyltransferase in methylation-directed mismatch repair. J. Bacteriol. 165:896–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marinus MG, Morris NR. 1973. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J. Bacteriol. 114:1143–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pukkila PJ, Peterson J, Herman G, Modrich P, Meselson M. 1983. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics 104:571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boye E, Lobner-Olesen A, Skarstad K. 2000. Limiting DNA replication to once and only once. EMBO Rep. 1:479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marinus MG, Casadesus J. 2009. Roles of DNA adenine methylation in host-pathogen interactions: mismatch repair, transcriptional regulation, and more. FEMS Microbiol. Rev. 33:488–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zweiger G, Marczynski G, Shapiro L. 1994. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J. Mol. Biol. 235:472–485 [DOI] [PubMed] [Google Scholar]
- 39.Berdis AJ, Lee I, Coward JK, Stephens C, Wright R, Shapiro L, Benkovic SJ. 1998. A cell cycle-regulated adenine DNA methyltransferase from Caulobacter crescentus processively methylates GANTC sites on hemimethylated DNA. Proc. Natl. Acad. Sci. U. S. A. 95:2874–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Albu RF, Jurkowski TP, Jeltsch A. 2012. The Caulobacter crescentus DNA-(adenine-N6)-methyltransferase CcrM methylates DNA in a distributive manner. Nucleic Acids Res. 40:1708–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reisenauer A, Kahng LS, McCollum S, Shapiro L. 1999. Bacterial DNA methylation: a cell cycle regulator? J. Bacteriol. 181:5135–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marczynski GT, Shapiro L. 2002. Control of chromosome replication in Caulobacter crescentus. Annu. Rev. Microbiol. 56:625–656 [DOI] [PubMed] [Google Scholar]
- 43.Reisenauer A, Shapiro L. 2002. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 21:4969–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller ES, Kutter E, Mosig G, Arisaka F, Kunisawa T, Ruger W. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kossykh VG, Schlagman SL, Hattman S. 1995. Phage T4 DNA [N6-adenine]methyltransferase. Overexpression, purification, and characterization. J. Biol. Chem. 270:14389–14393 [DOI] [PubMed] [Google Scholar]
- 46.Messer W, Noyer-Weidner M. 1988. Timing and targeting: the biological functions of Dam methylation in E. coli. Cell 54:735–737 [DOI] [PubMed] [Google Scholar]
- 47.Hattman S. 1970. DNA methylation of T-even bacteriophages and of their non-glucosylated mutants: its role in P1-directed restriction. Virology 42:359–367 [DOI] [PubMed] [Google Scholar]
- 48.Lobocka MB, Rose DJ, Plunkett G, III, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR. 2004. Genome of bacteriophage P1. J. Bacteriol. 186:7032–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coulby JN, Sternberg NL. 1988. Characterization of the phage P1 dam gene. Gene 74:191. [DOI] [PubMed] [Google Scholar]
- 50.Citron M, Velleman M, Schuster H. 1989. Three additional operators, Op21, Op68, and Op88, of bacteriophage P1. Evidence for control of the P1 dam methylase by Op68. J. Biol. Chem. 264:3611–3617 [PubMed] [Google Scholar]
- 51.Sternberg N, Coulby J. 1990. Cleavage of the bacteriophage P1 packaging site (pac) is regulated by adenine methylation. Proc. Natl. Acad. Sci. U. S. A. 87:8070–8074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sternberg N, Sauer B, Hoess R, Abremski K. 1986. Bacteriophage P1 cre gene and its regulatory region. Evidence for multiple promoters and for regulation by DNA methylation. J. Mol. Biol. 187:197–212 [DOI] [PubMed] [Google Scholar]
- 53.Wilke K, Rauhut E, Noyer-Weidner M, Lauster R, Pawlek B, Behrens B, Trautner TA. 1988. Sequential order of target-recognizing domains in multispecific DNA-methyltransferases. EMBO J. 7:2601–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noyer-Weidner M, Pawlek B, Jentsch S, Gunthert U, Trautner TA. 1981. Restriction and modification in Bacillus subtilis: gene coding for a BsuR-specific modification methyltransferase in the temperate bacteriophage phi 3T. J. Virol. 38:1077–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gunthert U, Reiners L. 1987. Bacillus subtilis phage SPR codes for a DNA methyltransferase with triple sequence specificity. Nucleic Acids Res. 15:3689–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trautner TA, Pawlek B, Gunthert U, Canosi U, Jentsch S, Freund M. 1980. Restriction and modification in Bacillus subtilis: identification of a gene in the temperate phage SP beta coding for a BsuR specific modification methyltransferase. Mol. Gen. Genet. 180:361–367 [DOI] [PubMed] [Google Scholar]
- 57.Gunthert U, Trautner TA. 1984. DNA methyltransferases of Bacillus subtilis and its bacteriophages. Curr. Top. Microbiol. Immunol. 108:11–22 [DOI] [PubMed] [Google Scholar]
- 58.Lange C, Noyer-Weidner M, Trautner TA, Weiner M, Zahler SA. 1991. M.H2I, a multispecific 5C-DNA methyltransferase encoded by Bacillus amyloliquefaciens phage H2. Gene 100:213–218 [DOI] [PubMed] [Google Scholar]
- 59.Balbontin R, Rowley G, Pucciarelli MG, Lopez-Garrido J, Wormstone Y, Lucchini S, Garcia-Del Portillo F, Hinton JC, Casadesus J. 2006. DNA adenine methylation regulates virulence gene expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:8160–8168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murphy KC, Ritchie JM, Waldor MK, Lobner-Olesen A, Marinus MG. 2008. Dam methyltransferase is required for stable lysogeny of the Shiga toxin (Stx2)-encoding bacteriophage 933W of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 190:438–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lauster R, Trautner TA, Noyer-Weidner M. 1989. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J. Mol. Biol. 206:305–312 [DOI] [PubMed] [Google Scholar]
- 62.Hill C, Miller LA, Klaenhammer TR. 1991. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J. Bacteriol. 173:4363–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Magrini V, Salmi D, Thomas D, Herbert SK, Hartzell PL, Youderian P. 1997. Temperate Myxococcus xanthus phage Mx8 encodes a DNA adenine methylase, Mox. J. Bacteriol. 179:4254–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scholl D, Merril C. 2005. The genome of bacteriophage K1F, a T7-like phage that has acquired the ability to replicate on K1 strains of Escherichia coli. J. Bacteriol. 187:8499–8503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandmeier H. 1994. Acquisition and rearrangement of sequence motifs in the evolution of bacteriophage tail fibres. Mol. Microbiol. 12:343–350 [DOI] [PubMed] [Google Scholar]
- 66.Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, Chisholm SW. 2011. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. U. S. A. 108:E757–E764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van de Guchte M, Daly C, Fitzgerald GF, Arendt EK. 1994. Identification of int and attP on the genome of lactococcal bacteriophage Tuc2009 and their use for site-specific plasmid integration in the chromosome of Tuc2009-resistant Lactococcus lactis MG1363. Appl. Environ. Microbiol. 60:2324–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flores V, Lopez-Merino A, Mendoza-Hernandez G, Guarneros G. 2012. Comparative genomic analysis of two brucellaphages of distant origins. Genomics 99:233–240 [DOI] [PubMed] [Google Scholar]
- 69.Braid MD, Silhavy JL, Kitts CL, Cano RJ, Howe MM. 2004. Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J. Bacteriol. 186:6560–6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saridaki A, Sapountzis P, Harris HL, Batista PD, Biliske JA, Pavlikaki H, Oehler S, Savakis C, Braig HR, Bourtzis K. 2011. Wolbachia prophage DNA adenine methyltransferase genes in different Drosophila-Wolbachia associations. PLoS One 6:e19708. 10.1371/journal.pone.0019708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Santos SB, Kropinski AM, Ceyssens PJ, Ackermann HW, Villegas A, Lavigne R, Krylov VN, Carvalho CM, Ferreira EC, Azeredo J. 2011. Genomic and proteomic characterization of the broad-host-range Salmonella phage PVP-SE1: creation of a new phage genus. J. Virol. 85:11265–11273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwudke D, Ergin A, Michael K, Volkmar S, Appel B, Knabner D, Konietzny A, Strauch E. 2008. Broad-host-range Yersinia phage PY100: genome sequence, proteome analysis of virions, and DNA packaging strategy. J. Bacteriol. 190:332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dempsey RM, Carroll D, Kong H, Higgins L, Keane CT, Coleman DC. 2005. Sau42I, a BcgI-like restriction-modification system encoded by the Staphylococcus aureus quadruple-converting phage Phi42. Microbiology 151:1301–1311 [DOI] [PubMed] [Google Scholar]
- 74.Lynch KH, Stothard P, Dennis JJ. 2010. Genomic analysis and relatedness of P2-like phages of the Burkholderia cepacia complex. BMC Genomics 11:599. 10.1186/1471-2164-11-599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blaut M, Clavel T. 2007. Metabolic diversity of the intestinal microbiota: implications for health and disease. J. Nutr. 137:751S–755S [DOI] [PubMed] [Google Scholar]
- 76.Wang K, Wommack KE, Chen F. 2011. Abundance and distribution of Synechococcus spp. and cyanophages in the Chesapeake Bay. Appl. Environ. Microbiol. 77:7459–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Enikeeva FN, Severinov KV, Gelfand MS. 2010. Restriction-modification systems and bacteriophage invasion: who wins? J. Theor. Biol. 266:550–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bushman F. 2002. Lateral DNA transfer: mechanisms and consequences. CSH Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 79.Thiyagarajan S, Chrisolite B, Alavandi SV, Poornima M, Kalaimani N, Santiago TC. 2011. Characterization of four lytic transducing bacteriophages of luminescent Vibrio harveyi isolated from shrimp (Penaeus monodon) hatcheries. FEMS Microbiol. Lett. 325:85–91 [DOI] [PubMed] [Google Scholar]
- 80.Waddell TE, Franklin K, Mazzocco A, Kropinski AM, Johnson RP. 2009. Generalized transduction by lytic bacteriophages. Methods Mol. Biol. 501:293–303 [DOI] [PubMed] [Google Scholar]
- 81.Hafstrom T, Jansson DS, Segerman B. 2011. Complete genome sequence of Brachyspira intermedia reveals unique genomic features in Brachyspira species and phage-mediated horizontal gene transfer. BMC Genomics 12:395. 10.1186/1471-2164-12-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Noyer-Weidner M, Jentsch S, Kupsch J, Bergbauer M, Trautner TA. 1985. DNA methyltransferase genes of Bacillus subtilis phages: structural relatedness and gene expression. Gene 35:143–150 [DOI] [PubMed] [Google Scholar]
- 83.Kita K, Kawakami H, Tanaka H. 2003. Evidence for horizontal transfer of the EcoT38I restriction-modification gene to chromosomal DNA by the P2 phage and diversity of defective P2 prophages in Escherichia coli TH38 strains. J. Bacteriol. 185:2296–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fineran PC, Petty NK, Salmond GPC. 2009. Transduction: host DNA transfer by bacteriophages, 3rd ed. Academic Press, Cambridge, United Kingdom [Google Scholar]
- 85.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. U. S. A. 96:2192–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hendrix RW. 2002. Bacteriophages: evolution of the majority. Theor. Popul. Biol. 61:471–480 [DOI] [PubMed] [Google Scholar]
- 87.Bouchard JD, Moineau S. 2000. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology 270:65–75 [DOI] [PubMed] [Google Scholar]
- 88.Trotter M, McAuliffe O, Callanan M, Edwards R, Fitzgerald GF, Coffey A, Ross RP. 2006. Genome analysis of the obligately lytic bacteriophage 4268 of Lactococcus lactis provides insight into its adaptable nature. Gene 366:189–199 [DOI] [PubMed] [Google Scholar]
- 89.Byers KE, Anglim AM, Anneski CJ, Germanson TP, Gold HS, Durbin LJ, Simonton BM, Farr BM. 2001. A hospital epidemic of vancomycin-resistant Enterococcus: risk factors and control. Infect. Control Hosp. Epidemiol. 22:140–147 [DOI] [PubMed] [Google Scholar]
- 90.Knudsen JD, Odenholt I, Erlendsdottir H, Gottfredsson M, Cars O, Frimodt-Moller N, Espersen F, Kristinsson KG, Gudmundsson S. 2003. Selection of resistant Streptococcus pneumoniae during penicillin treatment in vitro and in three animal models. Antimicrob. Agents Chemother. 47:2499–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Amaral L, Kristiansen JE, Viveiros M, Atouguia J. 2001. Activity of phenothiazines against antibiotic-resistant Mycobacterium tuberculosis: a review supporting further studies that may elucidate the potential use of thioridazine as anti-tuberculosis therapy. J. Antimicrob. Chemother. 47:505–511 [DOI] [PubMed] [Google Scholar]
- 92.Keen EC. 2012. Phage therapy: concept to cure. Front. Microbiol. 3:238. 10.3389/fmicb.2012.00238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kutateladze M, Adamia R. 2010. Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 28:591–595 [DOI] [PubMed] [Google Scholar]
- 94.Kunisaki H, Tanji Y. 2010. Intercrossing of phage genomes in a phage cocktail and stable coexistence with Escherichia coli O157:H7 in anaerobic continuous culture. Appl. Microbiol. Biotechnol. 85:1533–1540 [DOI] [PubMed] [Google Scholar]
- 95.Sambrook J, Fritsch E, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 96.Mason CE, Elemento O. 2012. Faster sequencers, larger datasets, new challenges. Genome Biol. 13:314. 10.1186/gb-2012-13-3-314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Au KF, Underwood JG, Lee L, Wong WH. 2012. Improving PacBio long read accuracy by short read alignment. PLoS One 7:e46679. 10.1371/journal.pone.0046679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Klumpp J, Fouts DE, Sozhamannan S. 2012. Next generation sequencing technologies and the changing landscape of phage genomics. Bacteriophage 2:190–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Korlach J, Marks PJ, Cicero RL, Gray JJ, Murphy DL, Roitman DB, Pham TT, Otto GA, Foquet M, Turner SW. 2008. Selective aluminum passivation for targeted immobilization of single DNA polymerase molecules in zero-mode waveguide nanostructures. Proc. Natl. Acad. Sci. U. S. A. 105:1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Feng Z, Losic B, Mahajan MC, Jabado OJ, Deikus G, Clark TA, Luong K, Murray IA, Davis BM, Keren-Paz A, Chess A, Roberts RJ, Korlach J, Turner SW, Kumar V, Waldor MK, Schadt EE. 2012. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat. Biotechnol. 30:1232–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Clark TA, Murray IA, Morgan RD, Kislyuk AO, Spittle KE, Boitano M, Fomenkov A, Roberts RJ, Korlach J. 2012. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 40:e29. 10.1093/nar/gkr1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murray IA, Clark TA, Morgan RD, Boitano M, Anton BP, Luong K, Fomenkov A, Turner SW, Korlach J, Roberts RJ. 2012. The methylomes of six bacteria. Nucleic Acids Res. 40:11450–11462 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.