Abstract
The aim was to investigate transgenerational effects of feeding genetically modified (GM) maize expressing a truncated form of Bacillus thuringiensis Cry1Ab protein (Bt maize) to sows and their offspring on maternal and offspring intestinal microbiota. Sows were assigned to either non-GM or GM maize dietary treatments during gestation and lactation. At weaning, offspring were assigned within sow treatment to non-GM or GM maize diets for 115 days, as follows: (i) non-GM maize-fed sow/non-GM maize-fed offspring (non-GM/non-GM), (ii) non-GM maize-fed sow/GM maize-fed offspring (non-GM/GM), (iii) GM maize-fed sow/non-GM maize-fed offspring (GM/non-GM), and (iv) GM maize-fed sow/GM maize-fed offspring (GM/GM). Offspring of GM maize-fed sows had higher counts of fecal total anaerobes and Enterobacteriaceae at days 70 and 100 postweaning, respectively. At day 115 postweaning, GM/non-GM offspring had lower ileal Enterobacteriaceae counts than non-GM/non-GM or GM/GM offspring and lower ileal total anaerobes than pigs on the other treatments. GM maize-fed offspring also had higher ileal total anaerobe counts than non-GM maize-fed offspring, and cecal total anaerobes were lower in non-GM/GM and GM/non-GM offspring than in those from the non-GM/non-GM treatment. The only differences observed for major bacterial phyla using 16S rRNA gene sequencing were that fecal Proteobacteria were less abundant in GM maize-fed sows prior to farrowing and in offspring at weaning, with fecal Firmicutes more abundant in offspring. While other differences occurred, they were not observed consistently in offspring, were mostly encountered for low-abundance, low-frequency bacterial taxa, and were not associated with pathology. Therefore, their biological relevance is questionable. This confirms the lack of adverse effects of GM maize on the intestinal microbiota of pigs, even following transgenerational consumption.
INTRODUCTION
Genetically modified (GM) maize is one of the most widely grown GM crops worldwide. A large proportion of this is maize that expresses a truncated form of the Cry1Ab protein from Bacillus thuringiensis (Bt maize), which confers resistance to certain maize pests (1, 2). While to date adverse effects of Bt maize consumption have not been definitively documented, the safety of GM food and feed is an intensely debated subject (3).
Numerous studies have investigated the effects of Bt maize consumption on production and health characteristics in different animal species (3–10); however, apart from those conducted by our group, few studies have been performed in pigs. Furthermore, while a number of studies have investigated the effects of Bt maize on the intestinal microbiota of ruminants (6, 11, 12), our group was the first to examine its impact on the porcine intestinal microbiota (13, 14). Moreover, we were the first to employ high-throughput 16S rRNA gene sequencing to determine if consumption of GM food/feed influences intestinal microbial communities. Such studies are warranted, considering in vitro observations that the Cry1Ab protein is antimicrobial against intestinal bacteria, such as Clostridium, both in its intact and fragmented forms, at concentrations of 25 to 63 μg/ml (15). Furthermore, the Cry1Ab protein is not completely degraded during intestinal transit when administered in feed at concentrations of 0.17 to 0.52 μg/g feed and persists in the intestine at concentrations of 0.003 to 0.03 μg/ml (8, 9, 16). In addition, the finding that indigenous gut bacteria contribute to B. thuringiensis susceptibility in the larvae of some insect species (17) suggests that Bt toxins in some way impact the intestinal microbiota. Furthermore, Finamore et al. revealed changes in local and systemic immune responses in mice associated with Bt MON810 maize feeding that were especially evident in young and old mice, i.e., at times of major shifts in the intestinal microbiota (18). As no clear hypothesis is provided to explain the outcomes of the study, it is possible that the differences observed by Finamore et al. may be due to indirect effects of the MON810 maize on the intestinal microbiota.
To date, the impact of GM feed consumption on the intestinal microbiota of pregnant females and/or their offspring has not been examined. Due to the physiological changes that occur during pregnancy and the extra demands of the developing fetuses, pregnant females may respond differently to the consumption of Bt maize. Furthermore, any dietary-induced perturbations at the level of the intestinal microbiota could potentially trigger an immune response in the pregnant female that could affect in utero development of the offspring. Any disturbance of the intestinal microbiota of pregnant females may also have consequences for establishment of the piglets' intestinal microbiota and indirectly for maturation of the piglet immune system. This is because the newborn's first microbial contact is with the microbiota of the mother at birth (19), and the maternal microbiota is the inoculum for colonization of the neonatal digestive tract, which is important for maturation of the neonatal immune system (20). Furthermore, as newborn pigs are in constant contact with the mother's feces until weaning, it is likely that establishment of their postnatal preweaning intestinal microbiota depends largely on that of the sow (20).
Therefore, the aim of this study was to investigate the transgenerational effects of feeding GM maize to sows during gestation and lactation and to their offspring from weaning for 115 days on sow fecal and offspring fecal, ileal, and cecal bacterial communities using culture-dependent and -independent approaches.
MATERIALS AND METHODS
Pig feeding study.
The pig study complied with European Union Council Directives 91/630/EEC (outlines minimum standards for the protection of pigs) and 98/58/EC (concerns the protection of animals kept for farming purposes). The study was approved by and a license was obtained from the Irish Department of Health and Children (license number B100/4147). Ethical approval was obtained from the Teagasc and Waterford Institute of Technology ethics committees.
The duration of the feeding study was 36 weeks. At insemination (day 0), 24 Large White × Landrace nulliparous sows (∼165 kg) were blocked by body weight and date of insemination before being randomly assigned to one of two dietary treatments: (i) near-isogenic parent line maize (Pioneer PR34N43; non-GM) or (ii) Bt maize (Pioneer PR34N44 event MON810; GM). Sows were fed diets from insemination throughout pregnancy and lactation until weaning at ∼day 28 postparturition. At weaning, offspring (n = 20/sow treatment) were selected and blocked by sow treatment, sex, and body weight and randomly assigned within sow treatment to either a non-GM or GM maize-based diet for 115 days, giving rise to four dietary treatments: (i) non-GM maize-fed sow/non-GM maize-fed offspring (non-GM/non-GM), (ii) non-GM maize-fed sow/GM maize-fed offspring (non-GM/GM), (iii) GM maize-fed sow/non-GM maize-fed offspring (GM/non-GM), and (iv) GM maize-fed sow/GM maize-fed offspring (GM/GM). Sow and offspring housing and management have previously been described by Walsh et al. (21) and Buzoianu et al. (22), respectively.
Maize and diets.
Seeds derived from the Bt MON810 and non-GM parent line maize (PR34N44 and PR34N43, respectively; Pioneer Hi-Bred, Sevilla, Spain) were grown simultaneously side by side in Valtierra, Navarra, Spain, under similar management conditions by independent tillage farmers in 2007. This was done to avoid, insofar as possible, compositional differences between the non-GM and the GM maize due to differences in environmental exposure, soil composition, and management practices. The GM and non-GM maize were purchased by the authors from the tillage farmers for use in this animal study. Samples from the non-GM and GM maize were tested for the presence of the cry1Ab gene, pesticide contaminants, mycotoxins, and carbohydrate composition as previously described by Walsh et al. (10). All diets were manufactured, and proximate and amino acid analysis was performed as previously described by Walsh et al. (10). All diets were formulated to meet or exceed the National Research Council requirements for pigs at each production stage (23). Details of animal feeding have previously been described by Walsh et al. (21) and Buzoianu et al. (22).
Sample collection and analysis.
Fecal samples were collected by rectal stimulation from 12 sows per treatment group at insemination (day 0), day 110 of gestation (∼1 week prior to parturition), and day 28 of lactation. Samples were collected into sterile plastic containers and stored at 4°C in anaerobic jars containing Anaerocult A gas packs (Merck, Darmstadt, Germany) until analysis (within 12 h). Fecal samples were collected in a similar manner from offspring at weaning and at days 30, 70, and 100 postweaning. At day 115 postweaning, i.e., when pigs had reached the normal slaughter weight (∼105 kg), the offspring were harvested in a commercial abattoir using electrical stunning followed by exsanguination. Digesta samples were removed aseptically from the terminal tip of the cecum and from the ileum (15 cm proximal to the ileocecal junction), placed in sterile plastic containers, and stored anaerobically, as described for the fecal samples. The last meal was provided 3 h before euthanasia.
Enumeration of Lactobacillus (indicator of beneficial bacteria [24]) and Enterobacteriaceae (indicator of pathogenic bacteria [24]) from individual fecal samples and ileal and cecal digesta was performed as described by Gardiner et al. (56). To inhibit growth of yeasts and molds, nystatin (Sigma-Aldrich Ireland Ltd., Wicklow, Ireland) was added to Lactobacillus selective agar (Becton, Dickinson, Cockeysville, MD) at a concentration of 50 units/ml. Total anaerobic bacteria from individual fecal samples were enumerated as previously described by Rea et al. (25). To maintain anaerobiosis, manipulation of all samples was performed in a Whitley A85 anaerobic workstation (DW Scientific, Shipley, United Kingdom), and the plates were also incubated anaerobically within the workstation.
Fecal samples from sows at day 110 of gestation and from offspring at weaning and day 100 postweaning and cecal samples from day 115 postweaning were frozen at −20°C for subsequent 16S rRNA gene sequencing.
DNA extraction and PCR.
DNA was extracted from fecal samples of sows at day 110 of gestation and from fecal samples of offspring at weaning and day 100 postweaning and from offspring cecal samples at day 115 postweaning as described by Buzoianu et al. (13). PCR amplification of the 239-bp V4 region of the 16S rRNA gene, gel electrophoresis, amplicon quantification, and purification were performed as previously outlined by Buzoianu et al. (13).
16S rRNA gene sequencing and bioinformatics analysis.
Sequencing was performed on a 454 Genome Sequencer FLX platform (Roche Diagnostics Ltd., Burgess Hill, West Sussex, United Kingdom), according to the manufacturer's protocols. Resulting raw sequences were quality trimmed as previously described (26). A BLAST search of trimmed FASTA sequences was then performed against a locally installed version of the SILVA 16S rRNA database, with the top 50 hits against the database selected. The taxonomic distribution of reads was determined using MEGAN, with modified accession lookup tables for mapping the SILVA assignments to NCBI taxonomy. MEGAN assigns reads to NCBI taxonomies by employing the lowest common ancestor algorithm. Bit scores were used from within MEGAN for filtering the results before tree construction and summarization. A bit score of 86 was selected as previously used for 16S ribosomal sequence data (27). Phylum, family, and counts for each subject were extracted from MEGAN. MOTHUR software was used to compute alpha diversity indices. Sequence reads were clustered into operational taxonomical units (OTUs) using the QIIME suite of software tools. OTUs were aligned, and chimeric OTUs were removed using the ChimeraSlayer program. A phylogenetic tree was generated using the FastTreeMP tool. Subsequently, beta diversities of the samples were calculated. Principal coordinate analysis (PCoA) and hierarchical clustering of samples were implemented. PCoA plots were visualized with the KiNG viewer. The number of reads assigned to each taxonomic rank was divided by the number of reads assigned to the highest rank (phylum) to obtain percent relative abundance values.
Statistical analysis.
Statistical analysis of data was performed using SAS/STAT, version 9.2 (2010) (SAS Institute, Inc., Cary, NC). Data were checked for normality using the Shapiro-Wilk test within PROC UNIVARIATE in SAS. In an attempt to ensure normality, bacterial counts were log transformed to the base 10, and nonnormal relative abundance data were transformed using the Box-Cox transformation (28). Data that were initially normally distributed or were normalized with the Box-Cox transformation were analyzed using PROC MIXED while nonnormal data were analyzed using PROC NPAR1WAY. Data analyzed using PROC MIXED are presented as least-squares means and 5th to 95th confidence limits of raw data (for data that were initially normal) or of the back-transformed values (for data normalized using the Box-Cox transformation), while bacterial counts are presented as log values with their corresponding standard errors of the means. Data analyzed using nonparametric tests are presented as medians and 5th to 95th percentiles (the 5th percentile is larger than 5% of the values and the 95th percentile is larger than 95% of the values). Significance is reported at a P value of ≤0.05. For all variables, the individual pig was the experimental unit.
For sows, relative abundance data that were normally distributed and log-transformed microbial counts were analyzed using PROC MIXED in SAS with treatment as a fixed effect and block as a random effect in the statistical model. For microbial counts, day 0 values were included as covariates in the statistical model, and day was included as a repeated variable. The slice option was used to test for simple effects at individual time points. Nonnormal data were analyzed using the Kolmogorov-Smirnov test within PROC NPAR1WAY.
For offspring, data that were normally distributed or that were normalized were analyzed as a 2-by-2 factorial split-plot design with sow treatment regarded as the main plot and offspring treatment as the subplot. Sow and offspring treatment and their interaction were included in the statistical model as fixed effects, and block, sow block, and the sow block-sow treatment interaction were included as random effects. Data that were normal or that were normalized for both day 0 and day 100 were analyzed as repeated measures, with day included in the statistical model as a repeated variable. The slice option was used to test for simple effects at individual time points. P values were adjusted for multiple comparisons using the Tukey-Kramer adjustment. For bacterial counts, day 0 values were included as a covariate in the statistical model. For nonnormal data, a nonparametric Kolmogorov-Smirnov test was used for sow treatment and offspring treatment effects, and a nonparametric Kruskal-Wallis test was used to analyze differences among the four combinations of treatments.
RESULTS
One sow from the non-GM treatment group received antibiotic treatment on days 105 to 107 of gestation. Likewise, two of the offspring (one from each of the non-GM/GM and the non-GM/non-GM treatment groups) were treated with antibiotics for 3 days between day 70 and day 100. As a result, data from any of these treated animals were not included for analysis at subsequent sampling points.
Maize and diets.
No major compositional differences were observed between the GM and non-GM maize (10) or between the non-GM and GM diets (see Tables S1 and S2 in the supplemental material). The Bt maize was found to have a 2.63 percentage units (as a percentage of dry matter) lower enzyme-resistant starch content and a 3.08 percentage units (as a percentage of dry matter) higher overall starch content than the non-GM maize. However, the values remained within the natural variability for maize varieties cited in the literature (13).
Effects of feeding GM maize-based diets to sows during gestation and lactation and to offspring for 115 days postweaning on selected culturable fecal and intestinal microbiota.
Bacterial counts from sow fecal samples are presented in Table S3 in the supplemental material. There was no effect of feeding GM maize-based diets to sows on counts of culturable fecal Enterobacteriaceae, Lactobacillus bacteria, or total anaerobes on day 110 of gestation or day 28 of lactation. While Lactobacillus counts increased from day 28 of gestation to day 28 of lactation (P < 0.05) (data not shown), Enterobacteriaceae and total anaerobe counts decreased between these two sampling points (P < 0.05) (data not shown).
Bacterial counts from offspring fecal and intestinal digesta samples are presented in Table 1. There was no sow treatment-offspring treatment interaction or effect of offspring treatment on fecal Enterobacteriaceae counts at any time during the study (P > 0.05). At day 100 postweaning, offspring from GM maize-fed sows had higher fecal counts of Enterobacteriaceae than offspring from non-GM maize-fed sows (P ≤ 0.05) (Table 1). No sow treatment-offspring treatment interaction was observed for Lactobacillus counts in the feces of offspring, nor were any sow or offspring treatment effects seen. A sow treatment effect was observed for fecal total anaerobes at day 70 postweaning when offspring of GM maize-fed sows had higher counts than offspring of non-GM maize-fed sows (P < 0.05) (Table 1). Ileal Enterobacteriaceae counts were lower at day 115 postweaning in offspring on the GM/non-GM treatment diet than in offspring on the non-GM/non-GM and GM/GM treatment diet (P < 0.05). No treatment effect was observed for offspring ileal or cecal Lactobacillus counts at day 115 postweaning (P > 0.05). However, at day 115 postweaning ileal total anaerobe counts were lower in pigs on the GM/non-GM treatment than in pigs on all other treatments (P < 0.05), and this led to an offspring treatment effect, with GM maize-fed offspring having higher counts than non-GM maize-fed offspring (P < 0.05; Table 1). A sow treatment-offspring treatment interaction was also observed for cecal total anaerobes at day 115 postweaning, with offspring on the non-GM/GM and GM/non-GM treatments having lower counts than offspring on the non-GM/non-GM treatment (P < 0.05).
Table 1.
Bacterial counts in offspring of sows fed a GM or non-GM maize diet
| Sample source, bacteria, and sampling daya | Counts in offspring of sows fedb: |
P value |
||||||
|---|---|---|---|---|---|---|---|---|
| Non-GM diet |
GM diet |
SEM | ||||||
| Non-GMc | GMc | Non-GM | GM | Sow treatment | Offspring treatment | Treatment interactiond | ||
| Fecal samples | ||||||||
| Enterobacteriaceae | ||||||||
| Day 30 | 5.61 | 5.94 | 5.80 | 6.24 | 0.196 | 0.51 | 0.29 | 0.64 |
| Day 70 | 6.72 | 6.73 | 6.61 | 6.78 | 0.154 | 0.91 | 0.73 | 0.97 |
| Day 100 | 6.78 | 6.50 | 7.15 | 7.18 | 0.139 | 0.05 | 0.61 | 0.16 |
| Lactobacillus | ||||||||
| Day 30 | 8.92 | 9.03 | 8.94 | 8.98 | 0.090 | 0.94 | 0.68 | 0.97 |
| Day 70 | 8.48 | 8.98 | 8.71 | 8.42 | 0.096 | 0.41 | 0.58 | 0.18 |
| Day 100 | 8.69 | 8.90 | 8.76 | 8.17 | 0.171 | 0.34 | 0.59 | 0.41 |
| Total anaerobes | ||||||||
| Day 30 | 9.41 | 9.20 | 9.60 | 9.27 | 0.120 | 0.41 | 0.09 | 0.29 |
| Day 70 | 9.20 | 9.51 | 9.70 | 9.66 | 0.116 | 0.03 | 0.35 | 0.09 |
| Day 100 | 8.99 | 9.72 | 9.53 | 9.15 | 0.224 | 0.96 | 0.68 | 0.59 |
| Digesta samples | ||||||||
| Enterobacteriaceae (day 115) | ||||||||
| Ileum | 7.91 A | 7.34 AB | 6.31 B | 8.30 A | 0.432 | 0.37 | 0.06 | 0.01 |
| Cecum | 7.85 | 7.18 | 7.05 | 7.50 | 0.302 | 0.41 | 0.69 | 0.06 |
| Lactobacillus (day 115) | ||||||||
| Ileum | 7.01 | 7.25 | 7.02 | 7.19 | 0.383 | 0.95 | 0.52 | 0.91 |
| Cecum | 8.34 | 7.78 | 7.90 | 7.92 | 0.320 | 0.62 | 0.38 | 0.34 |
| Total anaerobes (day 115) | ||||||||
| Ileum | 8.70 A | 8.63 A | 7.96 B | 8.87 A | 0.192 | 0.14 | 0.02 | 0.01 |
| Cecum | 9.54 A | 8.97 B | 9.15 B | 9.19 AB | 0.148 | 0.55 | 0.07 | 0.03 |
Day postweaning.
Sows were fed either a non-GM or a GM maize-based diet during gestation and lactation. Data are presented as treatment least-squares means (log10 CFU g−1). Variability present at weaning has been accounted for by including weaning counts as covariates in the statistical model. Within a row, the same letter is placed next to values whose means are not significantly different (P ≤ 0.05). Means separation was performed using a Tukey-Kramer adjustment for multiple comparisons.
Diet of offspring. Within sow treatment, offspring were fed either a non-GM or a GM maize-based diet from weaning for 115 days.
Sow treatment-offspring treatment interaction.
Effects of feeding GM maize-based diets to sows during gestation and lactation and to offspring for 115 days postweaning on microbial population indices.
A total of 65,890 reads (average of 2,864 [range, 1,068 to 4,176] reads/sample) of the V4 region of the 16S rRNA gene were generated from high-throughput sequencing of fecal samples from sows. In offspring at weaning, 188,583 (average of 4,715 reads [range, 3,048 to 6,827]/sample) reads were generated, while at day 100 postweaning, 175,133 reads were generated (average of 4,609 [range, 2,509 to 6,881] reads/sample). High-throughput sequencing of cecal samples at day 115 postweaning yielded 100,601 reads (average of 2,719 [range, 1,400 to 4,682] reads/sample).
Population indices were similar among treatments (Table 2). Unweighted beta diversity plots did not show clustering specific to any treatment group (see Fig. S1, S2, S3, and S4 in the supplemental material).
Table 2.
Bacterial diversity within samples from offspring fed non-GM or GM maize-based dietsa
| Microbiota source and diversity measure | Value for offspring of sows fed:b |
|||
|---|---|---|---|---|
| Non-GM dietc |
GM dietd |
|||
| Non-GMe | GMe | Non-GM | GM | |
| Offspring feces at weaning | ||||
| Chao 1 richness estimation | 806 | 812 | 953 | 820 |
| Shannon diversity index | 5.06 | 5.02 | 5.30 | 5.16 |
| Good's coverage | 0.95 | 0.95 | 0.94 | 0.95 |
| Offspring feces at day 100 postweaning | ||||
| Chao 1 richness estimation | 1,106 | 1,098 | 1,125 | 1,156 |
| Shannon diversity index | 5.68 | 5.61 | 5.54 | 5.58 |
| Good's coverage | 0.93 | 0.93 | 0.93 | 0.93 |
| Offspring cecal digesta at day 115 postweaning | ||||
| Chao 1 richness estimation | 846 | 938 | 784 | 869 |
| Shannon diversity index | 5.40 | 5.53 | 5.46 | 5.44 |
| Good's coverage | 0.91 | 0.90 | 0.90 | 0.91 |
Estimates of diversity were computed using MOTHUR software and are presented as treatment means.
Sows were fed either a non-GM or a GM maize-based diet during gestation and lactation.
Diversity values for sows on a non-GM diet were 1,267 (Chao 1 richness estimation), 5.92 (Shannon diversity index), and 0.87 (Good's coverage).
Diversity values for sows on a GM diet were 1,242 (Chao 1 richness estimation), 5.86 (Shannon diversity index), and 0.88 (Good's coverage).
Diet fed to offspring. Within sow treatment, offspring were fed either a non-GM or a GM maize-based diet from weaning at ∼28 days for 115 days.
Effect of feeding a GM maize-based diet to sows on the relative abundance of sow fecal microbiota at day 110 of gestation.
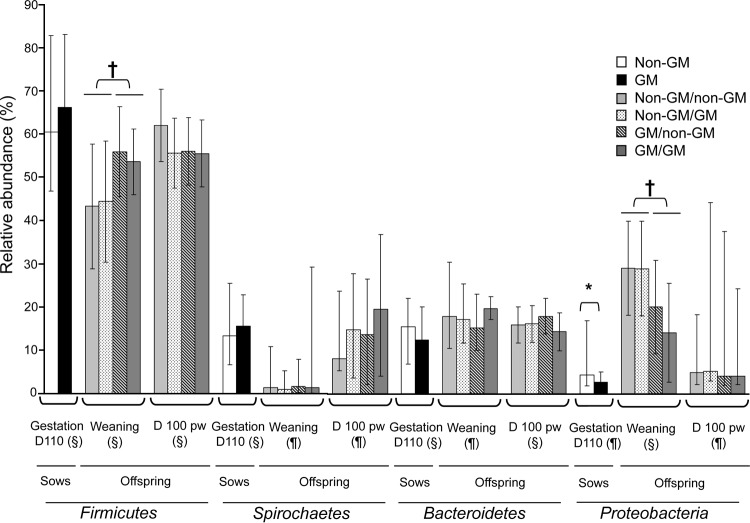
Results for taxa with abundances that differed significantly among sow treatments are presented in Table 3 and Fig. 1 (only main phyla are shown in the figure). A full outline of all taxa detected in fecal samples of sows can be found in Table S4 in the supplemental material. At the phylum level, the relative abundance of Proteobacteria was lower in the feces of GM maize-fed sows (P < 0.05) (Fig. 1). Verrucomicrobia were detected in only four sows on the non-GM treatment and in none on the GM treatment, resulting in a lower relative abundance of this phylum in GM maize-fed sows (P < 0.05) (Table 3).
Table 3.
Relative abundance (%) of sow fecal bacteria at day 110 of gestationa
| Taxon | Abundance (%) in feces of sows fed:b |
P value | No. of animalsc |
||
|---|---|---|---|---|---|
| Non-GM diet | GM diet | Non-GM diet | GM diet | ||
| Phylum | |||||
| Verrucomicrobia# | 0 (0–0.4) | 0 (0) | 0.04 | 4 | 0 |
| Family | |||||
| Rikenellaceae† | 5.9 (1.7–11.9) | 3.6 (1.9–6.5) | 0.05 | 11 | 12 |
| Prevotellaceae† | 2.4 (1.3–4.4) | 1.5 (0.7–2.9) | 0.01 | 11 | 12 |
| Succinivibrionaceae# | 1.9 (0–14.8) | 0.5 (0–2.3) | 0.01 | 10 | 10 |
| Rikenellaceae-related bacteria† | 1.6 (0–4.7) | 0.6 (0.2–1.2) | 0.04 | 10 | 12 |
| Lactobacillaceae# | 0.2 (0–1.0) | 0 (0–0.4) | 0.03 | 9 | 3 |
| Genus | |||||
| Prevotella† | 2.4 (1.3–4.4) | 1.5 (0.7–2.9) | 0.01 | 11 | 12 |
| Lachnospiraceae incertae sedis# | 1.3 (0.5–2.8) | 0.8 (0.6–1.5) | 0.01 | 11 | 12 |
| Anaerobiospirillum# | 1.8 (0–14.3) | 0.5 (0–2.3) | 0.01 | 10 | 10 |
| Lactobacillus# | 0.2 (0–1.0) | 0 (0–0.4) | 0.03 | 9 | 3 |
Data are presented as treatment means (95% confidence intervals are given in parentheses) for data analyzed using parametric tests (†) or medians (with 5th to 95th percentiles) for data analyzed using nonparametric tests (#). The main phyla are shown in Fig. 1, and a full outline of the relative abundance of all bacterial taxa detected in sow feces is available in Table S4 in the supplemental material.
Sows were fed a non-GM or a GM maize-based diet during gestation and lactation.
Number of animals in which the bacterial taxon was present (total of 11 and 12 sows on the non-GM and GM treatments, respectively).
Fig 1.
Main phyla detected in feces of sows and their offspring. Data are presented as treatment means or medians, with whiskers corresponding to the 95% confidence interval or 5th to 95th percentiles, respectively. Sows were fed either a non-GM or a GM maize-based diet during gestation and lactation. Within sow treatment, offspring were fed either a non-GM or a GM maize-based diet from weaning for 115 days, giving rise to four dietary treatments: non-GM maize-fed sow/non-GM maize-fed offspring (non-GM/non-GM), non-GM maize-fed sow/GM maize-fed offspring (non-GM/GM), GM maize-fed sow/non-GM maize-fed offspring (GM/non-GM), and GM maize-fed sow/GM maize-fed offspring (GM/GM). *, P < 0.05 for the treatment effect; †, P < 0.05 for the maternal treatment effect. The numbers of animals per treatment group were as follows: 11 sows on the non-GM treatment, 12 sows on the GM treatment, 10 offspring per treatment at weaning, 9 offspring per treatment on the non-GM/non-GM and non-GM/GM treatments, and 10 offspring per treatment on the GM/non-GM and GM/GM treatments at day 100 postweaning (pw). A full outline of all taxa detected can be found in Tables S4, S5, and S6 in the supplemental material.
A total of 40 bacterial families were identified in the feces of sows at day 110 of gestation (see Table S4 in the supplemental material). The Rikenellaceae differed in their relative abundances among treatments, with GM maize-fed sows having lower abundance than the non-GM maize-fed sows (P < 0.05). Counts of the Prevotellaceae, Succinivibrionaceae, Rikenellaceae-related bacteria, and Lactobacillaceae were also lower in the feces of GM maize-fed sows than in feces of non-GM maize-fed sows (P < 0.05).
Forty genera were detected in the feces of sows at day 110 of gestation. The relative abundances of the genus Prevotella were significantly different between non-GM maize- and GM maize-fed sows, with a lower relative abundance found in the latter (P < 0.05) (Table 3). The relative abundances of Lachnospiraceae incertae sedis, Anaerobiospirillum, and Lactobacillus were also lower in GM maize-fed sows (P < 0.05); with respect to Lactobacillus this may have resulted from the low frequency of detection in this treatment group.
Effect of feeding a GM maize-based diet to sows on the relative abundance of fecal microbiota of offspring at weaning.
Results for taxa with abundances that differed significantly among treatments are presented in Table 4 and Fig. 1 (showing only the main phyla) and in Tables S8 and S9 in the supplemental material. A full outline of all taxa detected in offspring fecal samples at weaning can be found in Table S5. At the phylum level, the relative abundance of Firmicutes was higher for offspring of GM maize-fed sows than for offspring of non-GM maize-fed sows (P ≤ 0.05) (Fig. 1). A sow treatment effect was also observed for fecal Proteobacteria, with lower relative abundance in offspring from GM maize-fed sows than in offspring of non-GM maize-fed sows (P < 0.05) (Fig. 1).
Table 4.
Relative abundance of fecal bacteria in offspring at weaning and 100 days postweaning and of cecal bacteria at day 115 postweaninga
| Sample source, sampling date, and taxon | Abundance (%) in sows fedb: |
P value (treatment interaction)d | No. of animals for the combinatione |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Non-GM diet |
GM diet |
||||||||
| Non-GMc | GMc | Non-GM | GM | Non-GM/non-GM | Non-GM/GM | GM/non-GM | GM/GM | ||
| Feces at weaning (∼28 days of age) | |||||||||
| Family | |||||||||
| Clostridiaceae# | 0.3 (0–5.5) AB | 0 (0–0.4) B | 0.9 (0–2.4) A | 0.3 (0–1.3) AB | 0.01 | 7 | 3 | 9 | 9 |
| Genus | |||||||||
| Clostridium# | 0.1 (0–0.6) AB | 0 (0–0.2) B | 0.4 (0–2.3) A | 0.1 (0–0.4) AB | 0.03 | 5 | 2 | 8 | 5 |
| Butyricimonas# | 0 (0) B | 0 (0) B | 0 (0) B | 0 (0–0.2) A | 0.02 | 0 | 0 | 0 | 3 |
| Roseburia# | 0 (0) B | 0.1 (0–1.4) A | 0 (0–0.1) B | 0 (0–0.2) B | 0.02 | 0 | 5 | 2 | 1 |
| Feces at day 100 postweaning | |||||||||
| Phylum | |||||||||
| Fibrobacteres# | 0.4 (0–0.8) A | 0 (0–1.1) B | 0 (0–0.4) B | 0.2 (0–1.9) AB | 0.02 | 8 | 3 | 4 | 7 |
| Family | |||||||||
| Fibrobacteraceae# | 0.4 (0–0.8) A | 0 (0–1.1) B | 0 (0–0.4) B | 0.2 (0–1.9) AB | 0.02 | 8 | 3 | 4 | 7 |
| Helicobacteraceae# | 0 (0–0.1) B | 0 (0–0.2) A | 0 (0) B | 0 (0) B | 0.01 | 1 | 4 | 0 | 0 |
| Genus | |||||||||
| Ruminococcus# | 0.8 (0.3–4.1) AB | 1.0 (0.6–3.3) A | 0.7 (0.2–1.4) AB | 0.5 (0.3–2.2) B | 0.05 | 9 | 9 | 10 | 10 |
| Fibrobacter# | 0.4 (0–0.8) A | 0 (0–1.1) B | 0 (0–0.4) B | 0.2 (0–1.9) AB | 0.02 | 7 | 3 | 4 | 7 |
| Helicobacter# | 0 (0–0.1) B | 0 (0–0.2) A | 0 (0) B | 0 (0) B | 0.01 | 1 | 4 | 0 | 0 |
| Solobacterium# | 0 (0–0.2) A | 0 (0–0.1) AB | 0 (0) B | 0 (0) B | 0.02 | 4 | 2 | 0 | 0 |
| Cecal digesta at day 115 postweaning | |||||||||
| Family | |||||||||
| Clostridiaceae† | 1.2 (0.3–2.1) B | 2.3 (1.4–3.3) A | 2.1 (1.2–3.0) AB | 1.5 (0.6–2.4) AB | 0.04 | 8 | 8 | 10 | 9 |
| Genus | |||||||||
| Phascolarctobacterium† | 5.5 (3.9–7.1) A | 3.6 (2.0–5.2) B | 4.0 (2.5–5.5) AB | 5.3 (3.9–6.8) A | 0.03 | 9 | 8 | 10 | 10 |
Data are presented as treatment means (95% confidence intervals are given in parentheses) for data analyzed using parametric tests (†) or medians (with 5th to 95th percentiles) for data analyzed using nonparametric tests (#). The main phyla are shown in Fig. 1, and a full outline of the relative abundance of all bacterial taxa detected in the feces of offspring at weaning and at day 100 postweaning is available in Tables S5 and S6 in the supplemental material. A full outline of the relative abundance of all bacterial taxa detected in the cecum of offspring at day 115 postweaning is available in Table S7 in the supplemental material. Within a row, the same letter is placed next to values whose means are not significantly different (P ≤ 0.05). Means separation was performed using Tukey-Kramer adjustment for multiple comparisons.
Sows were fed either a non-GM or a GM maize-based diet during gestation and lactation.
Diet of offspring. Within sow treatment, offspring were fed either a non-GM or a GM maize-based diet for 115 days from weaning.
Sow treatment-offspring treatment interaction. A true sow treatment-offspring treatment interaction could be modeled only for data that underwent parametric statistical analysis. For nonparametric analysis, the P value from the comparison among the four combinations (non-GM/non-GM, non-GM/GM, GM/non-GM, and GM/GM) of treatments is presented.
Number of animals in which the bacterial taxon was present (n = 10 per group at weaning; n = 9 per treatment for the non-GM/non-GM and non-GM/GM treatments; n =10 per treatment for the GM/non-GM and GM/GM treatments at days 100 and 115 postweaning).
Analysis of fecal bacterial communities in weanling pigs revealed 43 families (see Table S5 in the supplemental material). A higher relative abundance was observed in offspring from GM maize-fed sows than in offspring from non-GM maize-fed sows for the families Ruminococcaceae (P < 0.05) (see Table S8), Lachnospiraceae (P < 0.05) (see Table S8), and Clostridiaceae (P < 0.05) (see Table S8). Fecal Clostridiaceae were also present at a higher relative abundance in the feces of pigs on the GM/non-GM treatment than in pigs on the non-GM/GM treatment (P < 0.05) (Table 4). There was also an overall offspring treatment effect for the Rikenellaceae family, with GM maize-fed offspring having higher relative abundance than non-GM maize-fed offspring (P < 0.05) (see Table S9). Offspring of GM maize-fed sows also had higher relative abundance of Victivallaceae than offspring of non-GM maize-fed sows (P < 0.05) (see Table S8).
A total of 73 genera were detected in the feces of weanling pigs, with offspring of GM maize-fed sows having a higher relative abundance of Subdoligranulum than offspring of non-GM maize-fed sows (P < 0.05) (see Table S8 in the supplemental material). Relative abundance of fecal Clostridium was higher in pigs on the GM/non-GM treatment than in pigs on the non-GM/GM treatment (P < 0.05) (Table 4). Fecal Butyricimonas bacteria were present at a higher relative abundance in pigs on the GM/GM treatment than in pigs on any of the other treatments (P < 0.05) (Table 4). Likewise, Roseburia was more abundant in feces of pigs on the non-GM/GM treatment than in pigs on all other treatments (P < 0.05) (Table 4).
Effect of feeding a GM maize-based diet to sows and their offspring on the relative abundance of fecal microbiota in offspring at day 100 postweaning.
As noted above, results for taxa with abundances that differed significantly among treatments are presented in Table 4 and in Tables S8 and S9 in the supplemental material. A full outline of all taxa detected in offspring fecal samples at day 100 postweaning can be found in Table S6. At the phylum level, only the Fibrobacteres were affected by treatment, with lower counts in pigs on the non-GM/GM and the GM/non-GM treatments than in pigs on the non-GM/non-GM treatment (P < 0.05) (Table 4).
Thirty-five bacterial families were detected in the feces of pigs at 100 days postweaning. The relative abundance of fecal Fibrobacteraceae was lower in pigs on the non-GM/GM and GM/non-GM treatments than in pigs on the non-GM/non-GM treatment (P < 0.05) (Table 4). Fecal Helicobacteraceae counts were higher in pigs on the non-GM/GM treatment than pigs on all other treatments (P < 0.05) (Table 4).
Fecal Ruminococcus was less abundant in pigs on the GM/GM treatment than in pigs on the non-GM/GM treatment (P < 0.05) (Table 4), leading to a lower relative abundance of this genus in offspring of GM maize-fed sows than in offspring of non-GM maize-fed sows (P < 0.05) (see Table S8 in the supplemental material). Oscillospira and Faecalibacterium were less abundant in feces of GM maize-fed offspring than in non-GM maize-fed offspring (P < 0.05) (see Table S9). Fibrobacter was less abundant in offspring on the non-GM/GM and GM/non-GM treatments than in offspring on the non-GM/non-GM treatment (P < 0.05) (Table 4). GM maize-fed offspring had lower fecal Thalassospira counts than non-GM maize-fed offspring (P < 0.05) (see Table S9). Helicobacter was present at a higher relative abundance in pigs on the non-GM/GM treatment than in pigs on any of the other treatments (P < 0.05) (Table 4). Solobacterium was less abundant in pigs on the GM/non-GM and GM/GM treatments than in pigs on the non-GM/non-GM treatment (P < 0.05) (Table 4).
Effect of feeding a GM maize-based diet to sows and their offspring on the relative abundance of cecal microbiota of offspring at day 115 postweaning.
Results for taxa with abundances that differed significantly among treatments are presented in Table 4 and in Tables S8 and S9 in the supplemental material, as noted above, and a full outline of all taxa detected in offspring cecal samples can be found in Table S7. A total of 13 phyla were detected at 115 days postweaning. However, no phylum was affected by treatment (P > 0.05).
At the family level, a sow treatment-offspring treatment interaction was observed for Clostridiaceae, with a higher relative abundance observed in the ceca of pigs on the non-GM/GM treatment than in pigs on the non-GM/non-GM treatment (P < 0.05) (Table 4). The offspring of GM maize-fed sows had lower cecal Enterobacteriaceae counts than the offspring of non-GM maize-fed sows, irrespective of offspring treatment (P < 0.05) (see Table S8 in the supplemental material).
At the genus level, a sow treatment-offspring treatment interaction was observed for cecal Phascolarctobacterium, with pigs on the non-GM/GM treatment having lower relative abundance than those on the non-GM/non-GM and the GM/GM treatments (P < 0.05) (Table 4). The relative abundance of Anaerotruncus was also higher in GM maize-fed pigs than in non-GM maize-fed pigs (P < 0.05) (see Table S9 in the supplemental material).
DISCUSSION
It is well recognized that the intestinal microbiota has a major influence on host health (19, 29). As the maternal microbiota provides the inoculum for colonization of the offspring digestive tract (19), any effect that GM maize consumption may have on the intestinal microbiota of pregnant sows could affect the health and lifelong performance of their offspring. A number of studies have investigated the impact of Bt maize consumption on the intestinal microbiota of ruminants (11, 12, 30), and our group has evaluated its influence on the porcine microbiota (13, 14). However, results from transgenerational studies are absent from the literature, and, to our knowledge, this is the first study to investigate the effects of GM maize on the intestinal microbiota of pregnant sows and their offspring.
While some differences were observed in selected groups of culturable bacteria in the offspring feces, these are not likely to have originated from the mother, as no differences were observed for these groups within the sow feces when analyzed at the end of gestation. Furthermore, the differences in Enterobacteriaceae observed using culturing were not evident when the unculturable component of this family was accounted for using 16S rRNA gene sequencing analysis. Moreover, these differences, as well as the treatment differences observed for total anaerobes in the ileal and cecal digesta, were not associated with intestinal dysfunction or organ pathology (22) and are, therefore, not believed to be of biological relevance. This is in agreement with findings from our previous studies that showed a lack of adverse effects on culturable intestinal microbiota in pigs fed GM maize for 31 or 110 days (14, 22).
Sows.
If GM maize were to affect the intestinal microbiota, it would be most evident in sows as they had the highest dietary inclusion of maize. However, GM maize consumption had no significant effects on selected culturable microbiota or on the dominant bacterial phyla, as assessed by high-throughput sequencing. Proteobacteria were less abundant in GM maize-fed sows, most likely as a result of lower relative abundance of members of this phylum (i.e., the family Succinivibrionaceae and its member genus Anaerobiospirillum). A lower relative abundance of Proteobacteria was also observed at weaning for offspring of GM maize-fed sows, and this may have been due to the influence of the maternal microbiota; however, it was not associated with treatment differences in any member taxa. These reductions can be considered beneficial as increased Proteobacteria have been linked with intestinal inflammation (31), and this phylum encompasses bacteria known to cause intestinal pathology in humans and animals (32, 33). Interestingly, this reduction in Proteobacteria was associated with beneficial immune effects in the same sows (34).
The lower relative abundance of Prevotellaceae and its member genus Prevotella in GM maize-fed sows could be a result of the lower enzyme-resistant starch content of the GM maize used in the present study (10) as these bacteria are known to be involved in intestinal fiber fermentation (35). However, this was not reflected in the offspring microbiota at weaning, 100 days later, or at slaughter age. The lack of effect at weaning may be explained by the fact that pigs were still suckling up to this point and had therefore not consumed any solid feed. Furthermore, fiber-degrading populations in the gastrointestinal tract (GIT) do not stabilize before 6 weeks postweaning (36). The fact that offspring abundances of Prevotellaceae and Prevotella were not impacted by maternal dietary treatment later in life may be due to the fact that sows are known to have a fully developed hindgut fiber-fermenting microbiota, while hindgut fermentation is not at its peak in younger pigs (37). These data also provide evidence that, while the maternal microbiota is important for initial colonization of the digestive tract, other factors, such as diet composition and rearing environment, have a major influence on lifelong structure of offspring microbiota, which is in agreement with the findings of Schmidt et al. (38) and Mulder et al. (39).
The lower abundance of the family Lactobacillaceae in the feces of GM maize-fed sows resulted from a lower abundance of its member genus Lactobacillus. Although no difference was observed in fecal Lactobacillus counts using traditional culturing methods, this can be attributed to the inability of culture-based approaches to account for unculturable lactobacilli (40). Lower intestinal Lactobacillus counts would generally be perceived as negative as this genus is considered to have a beneficial role in the porcine, as well as the human, intestine (41, 42); however, no health abnormalities were observed in the sows (21, 34).
Offspring at weaning.
Fecal Firmicutes were more abundant in the offspring of GM maize-fed sows at weaning as a result of differences in member taxa, such as the Ruminococcaceae, Lachnospiraceae, and Clostridiaceae families and their member genera, Subdoligranulum and Clostridium. This increase in Firmicutes, although not found in the sow feces, was accompanied by a reduction in Proteobacteria, as outlined above. A higher abundance of Firmicutes has been associated with a higher energy intake and increased deposition of body mass (43). This is in agreement with our data as offspring of GM maize-fed sows consumed more feed and grew faster than offspring from non-GM maize-fed sows (22) even though there were no major nutritional differences between the non-GM and GM diets. These animals had improved lifelong performance even though the increase in Firmicutes did not persist to day 100 postweaning. This study was not designed to investigate sow reproductive performance, and there was insufficient replication to allow statistical analysis of the litter size of sows. However, a higher number of pigs were born to GM maize-fed sows (21). This may have led to marginal in utero growth retardation in piglets, which is known to induce leptin resistance and increased appetite in offspring (44, 45). The latter was in fact observed postweaning in the offspring of GM maize-fed sows (22). Interestingly, the intestinal microbiota of leptin-deficient mice has also been shown to be higher in Firmicutes and lower in Proteobacteria (43).
Offspring at day 100 postweaning.
At day 100 postweaning, the genus Helicobacter and its corresponding family, the Helicobacteraceae, were also altered in the feces of pigs on the non-GM/GM treatment compared to all other treatments, in that they were found at higher relative abundances. However, while these bacteria are normally associated with gastrointestinal pathology, in this instance no health abnormalities or disruptions in intestinal architecture were observed in these pigs (22).
While the pattern of relative abundance for Ruminococcus resembles the pattern of feed intake for these pigs (22), there are no data in the literature to correlate this fiber-fermenting genus with feed intake. Faecalibacterium and Oscillospira are detected with low abundance in the human and ruminant digestive tracts and seem to be associated with a high-fiber diet (46, 47). Therefore, the lower relative abundances of these genera observed in the feces of GM maize-fed pigs at day 100 postweaning may be a result of the slightly lower enzyme-resistant starch (which is considered dietary fiber) in the GM maize used in this study (13).
Offspring at day 115 postweaning.
The large intestine is the main microbial fermentation site in pigs, with the highest bacterial load (37). However, no major effects of GM maize feeding were observed within the cecal microbiota at slaughter at day 115 postweaning; for example, no differences were uncovered for the major bacterial phyla or families. Relative abundance of the family Clostridiaceae, however, was increased in pigs on the non-GM/GM treatment, and Enterobacteriaceae counts were lower in offspring of GM maize-fed sows. However, while members of both of these families are known to cause intestinal pathology in humans and animals (48–50), the differences were numerically small, and no changes in intestinal architecture or any adverse health effects were observed in these treatment groups (22).
Overall observations.
Overall, of the total of 436 taxa (phyla, families, and genera) detected in this study in sows and their offspring, differences among treatments were observed for only 36 taxa, and most of the differences occurred for minor taxa with low frequencies of detection whose role in the intestine is not fully established. Some of the differences observed in the offspring microbiota might be attributable to the unexpected and most likely treatment-independent differences in the in utero environment experienced by pigs on each sow treatment; however, a much more likely explanation is that they were due to changes in the maternal microbiota in response to feeding GM maize. This is because the sow plays a major role in microbial colonization of the digestive tract of preweaning pigs (20). However, a clear effect of feeding GM maize to sows on the intestinal microbiota of their newly weaned offspring was not observed, and the treatment differences observed at weaning did not persist. Furthermore, no adverse effects were observed on the immune system or organ health in either sows or their offspring (21, 22).
The absence of adverse effects of GM maize consumption supports our previous conclusions that cecal bacteria are not majorly affected by either short- or long-term GM maize consumption (13, 14). Similar results have been obtained in cows, where short-term consumption of GM maize did not affect rumen microbiota as assessed by 16S rRNA gene sequencing (11), quantitative PCR (12), or ribosomal intergenic spacer analysis (30). Culturable amylolytic and cellulolytic bacteria were also unaffected by 3 years of feeding GM maize to sheep (6).
Apart from investigating the impact of GM maize consumption, the present study also provides, for the first time, an insight into the intestinal microbiota of sows at the end of pregnancy and its impact on offspring microbiota using 16S rRNA gene sequencing. The fecal microbiota of sows and their offspring and the offspring cecal microbiota were dominated for the most part by Firmicutes and Bacteroidetes, similar to previous findings for the cecal and fecal microbiota of weanling and finisher pigs (13, 14, 51, 52) and in agreement with data from humans (47, 53). Some similarities are evident between the fecal microbiota of sows and their offspring, providing support for the assumption that maternal flora influences the microbiota of progeny (19). For example, Lachnospiraceae and Ruminococcaceae were the dominant families in sows prior to delivery and in offspring at weaning and thereafter. However, differences were also observed, perhaps the most obvious being the relatively low abundance of the Spirochaetaceae family in offspring at weaning and the fact that Bacteroidetes were replaced by Proteobacteria as the second most dominant phylum in these animals while the Spirochaetes were the second most dominant phylum in sows. At the genus level, Prevotella bacteria are among the most abundant genus within both the sow and offspring microbiota. However, in accordance with its corresponding family, Spirochaetaceae, and phylum, Spirochaetes, Treponema are detected at low relative abundance in offspring at weaning in comparison to sows and offspring at later time points. Another difference is that the fecal microbiota of pigs at weaning is low in Ruminococcus-related bacteria, while they were a major genus in growing pigs and sows. Overall, these data demonstrate perturbations in the intestinal microbiota at weaning, which is most likely due to maternal separation as well as social and environmental changes and is in agreement with the findings of other investigators (54, 55). Furthermore, weaning is a time of flux for the intestinal microbiota, and while the maternal microbiota provides the seed inoculum, adult-type bacterial communities take time to establish. However, the data also indicate that most of the main phyla and families were already established and resembled adult patterns as early as day 28 of life, providing further evidence of the influence of early life environment on gut microbiota composition in adult pigs (39).
While long-term GM maize consumption by sows and their offspring impacted the intestinal microbiota, the effects were limited and were not associated with any health abnormalities in either sows or their progeny. Furthermore, differences observed in GM maize-fed sows did not transfer to offspring, and effects in offspring were not consistently detected across sampling points. This helps to confirm the lack of adverse effects of GM maize consumption even following long-term exposure in immunodeficient animals, i.e., pregnant females and newly weaned pigs.
Supplementary Material
ACKNOWLEDGMENTS
The research leading to these results received funding from the European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement number 211820 and the Teagasc Walsh Fellowship Programme.
None of the authors had a financial or personal conflict of interest regarding the present study and the work was conducted independently of any commercial input, financial or otherwise.
We acknowledge Fiona Crispie for assistance with 16S rRNA gene sequencing, Lidia Teixeira, Julie Ledoux, Marie Laetitia Gilliot, and Julia Chaumier for assistance with sampling, and Laurie O'Sullivan, Gemma McCarthy, and Maria Luz Prieto for technical assistance with sample processing at harvest.
Author contributions were as follows: P. G. Lawlor and R. P. Ross secured the funding for the research; P. G. Lawlor and G. E. Gardiner designed the study; S. G. Buzoianu, M. C. Walsh, P. G. Lawlor, G. E. Gardiner, and M. C. Rea conducted the study; S. G. Buzoianu, M. C. Walsh, G. E. Gardiner, and M. C. Rea conducted the laboratory analysis; S. G. Buzoianu, O. O'Sullivan, and L. Quigley analyzed the data; S. G. Buzoianu wrote the manuscript; S. G. Buzoianu, G. E. Gardiner, P. G. Lawlor, P. D. Cotter, and M. C. Rea revised the manuscript. All authors read and approved the final manuscript.
Footnotes
Published ahead of print 4 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02937-13.
REFERENCES
- 1.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.James C. 2010. Global status of commercialized biotech/GM crops: 2010. ISAAA brief no. 42 ISAAA, Ithaca, NY [Google Scholar]
- 3.Domingo JL, Giné Bordonaba J. 2011. A literature review on the safety assessment of genetically modified plants. Environ. Int. 37:734–742 [DOI] [PubMed] [Google Scholar]
- 4.Chowdhury EH, Shimada N, Murata H, Mikami O, Sultana P, Miyazaki S, Yoshioka M, Yamanaka N, Hirai N, Nakajima Y. 2003. Detection of Cry1Ab protein in gastrointestinal contents but not visceral organs of genetically modified Bt11-fed calves. Vet. Hum. Toxicol. 45:72–74 [PubMed] [Google Scholar]
- 5.Adel-Patient K, Guimaraes VD, Paris A, Drumare M-F, Ah-Leung S, Lamourette P, Nevers M-C, Canlet C, Molina J, Bernard H, Créminon C, Wal J-M. 2011. Immunological and metabolomic impacts of administration of Cry1Ab protein and MON 810 maize in mouse. PLoS One 6:e16346. 10.1371/journal.pone.0016346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trabalza-Marinucci M, Brandi G, Rondini C, Avellini L, Giammarini C, Costarelli S, Acuti G, Orlandi C, Filippini G, Chiaradia E, Malatesta M, Crotti S, Antonini C, Amagliani G, Manuali E, Mastrogiacomo AR, Moscati L, Naceur Haouet M, Gaiti A, Magnani M. 2008. A three-year longitudinal study on the effects of a diet containing genetically modified Bt176 maize on the health status and performance of sheep. Livest. Sci. 113:178–190 [Google Scholar]
- 7.Buzoianu SG, Walsh MC, Rea MC, Cassidy JP, Ross RP, Gardiner GE, Lawlor PG. 2012. Effect of feeding genetically modified Bt MON810 maize to ∼40 day old pigs for 110 days on growth and health indicators. Animal 6:1609–1619 [DOI] [PubMed] [Google Scholar]
- 8.Walsh MC, Buzoianu SG, Gardiner GE, Rea MC, Gelencsér E, Jánosi Epstein AMM, Ross RP, Lawlor PG. 2011. Fate of transgenic DNA from orally administered Bt MON810 maize and effects on immune response and growth in pigs. PLoS One 6:27177. 10.1371/journal.pone.0027177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walsh MC, Buzoianu SG, Rea MC, O'Donovan O, Gelencsér E, Ujhelyi G, Ross RP, Gardiner GE, Lawlor PG. 2012. Effects of feeding Bt MON810 maize to pigs for 110 days on peripheral immune response and digestive fate of the cry1Ab gene and truncated Bt toxin. PLoS One 7:e36141. 10.1371/journal.pone.0036141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh MC, Buzoianu SG, Gardiner GE, Cassidy JP, Rea MC, Ross RP, Lawlor PG. 2012. Effects of short-term feeding of Bt MON810 maize on growth performance, organ morphology and function in pigs. Br. J. Nutr. 107:364–371 [DOI] [PubMed] [Google Scholar]
- 11.Einspanier R, Lutz B, Rief S, Berezina O, Zverlov V, Schwarz W, Mayer J. 2004. Tracing residual recombinant feed molecules during digestion and rumen bacterial diversity in cattle fed transgene maize. Eur. Food Res. Technol. 218:269–273 [Google Scholar]
- 12.Wiedemann S, Gurtler P, Albrecht C. 2007. Effect of feeding cows genetically modified maize on the bacterial community in the bovine rumen. Appl. Environ. Microbiol. 73:8012–8017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buzoianu SG, Walsh MC, Rea MC, O'Sullivan O, Cotter PD, Ross RP, Gardiner GE, Lawlor PG. 2012. High-throughput sequence-based analysis of the intestinal microbiota of weanling pigs fed genetically modified MON810 maize expressing Bacillus thuringiensis Cry1Ab (Bt maize) for 31 days. Appl. Environ. Microbiol. 78:4217–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buzoianu SG, Walsh MC, Rea MC, O'Sullivan O, Crispie F, Cotter PD, Ross RP, Gardiner GE, Lawlor PG. 2012. The effect of feeding Bt MON810 maize to pigs for 110 days on intestinal microbiota. PLoS One 7:e33668. 10.1371/journal.pone.0033668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yudina TG, Brioukhanov AL, Zalunin IA, Revina LP, Shestakov AI, Voyushina NE, Chestukhina GG, Netrusov AI. 2007. Antimicrobial activity of different proteins and their fragments from Bacillus thuringiensis parasporal crystals against clostridia and archaea. Anaerobe 13:6–13 [DOI] [PubMed] [Google Scholar]
- 16.Chowdhury EH, Kuribara H, Hino A, Sultana P, Mikami O, Shimada N, Guruge KS, Saito M, Nakajima Y. 2003. Detection of corn intrinsic and recombinant DNA fragments and Cry1Ab protein in the gastrointestinal contents of pigs fed genetically modified corn Bt11. J. Anim. Sci. 81:2546–2551 [DOI] [PubMed] [Google Scholar]
- 17.Broderick N, Robinson C, McMahon M, Holt J, Handelsman J, Raffa K. 2009. Contributions of gut bacteria to Bacillus thuringiensis-induced mortality vary across a range of Lepidoptera. BMC Biol. 7:11. 10.1186/1741-7007-7-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finamore A, Roselli M, Britti S, Monastra G, Ambra R, Turrini A, Mengheri E. 2008. Intestinal and peripheral immune response to MON810 maize ingestion in weaning and old mice. J. Agric. Food Chem. 56:11533–11539 [DOI] [PubMed] [Google Scholar]
- 19.Kelly D, King T, Aminov R. 2007. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat. Res. 622:58–69 [DOI] [PubMed] [Google Scholar]
- 20.Thompson CL, Wang B, Holmes AJ. 2008. The immediate environment during postnatal development has long-term impact on gut community structure in pigs. ISME J. 2:739–748 [DOI] [PubMed] [Google Scholar]
- 21.Walsh MC, Buzoianu SG, Gardiner GE, Rea MC, O'Donovan O, Ross RP, Lawlor PG. 2013. Effects of feeding Bt MON810 maize to sows during first gestation and lactation on maternal and offspring health indicators. Br. J. Nutr. 109:873–881 [DOI] [PubMed] [Google Scholar]
- 22.Buzoianu SG, Walsh MC, Rea MC, Cassidy JP, Ryan TP, Ross RP, Gardiner GE, Lawlor PG. 2013. Trans-generational effects of feeding genetically modified maize to nulliparous sows and offspring on offspring growth and health. J. Anim. Sci. 91:318–330 [DOI] [PubMed] [Google Scholar]
- 23.National Research Council 1998. Nutrient requirements of swine, 10th ed. National Academy of Sciences, Washington, DC [Google Scholar]
- 24.Castillo M, Martín-Orúe SM, Manzanilla EG, Badiola I, Martín M, Gasa J. 2006. Quantification of total bacteria, enterobacteria and lactobacilli populations in pig digesta by real-time PCR. Vet. Microbiol. 114:165–170 [DOI] [PubMed] [Google Scholar]
- 25.Rea MC, Clayton E, O'Connor PM, Shanahan F, Kiely B, Ross RP, Hill C. 2007. Antimicrobial activity of lacticin 3147 against clinical Clostridium difficile strains. J. Med. Microbiol. 56:940–946 [DOI] [PubMed] [Google Scholar]
- 26.Claesson MJ, O'Sullivan O, Wang Q, Nikkilä J, Marchesi JR, Smidt H, de Vos WM, Ross RP, O'Toole PW. 2009. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. 10.1371/journal.pone.0006669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urich T, Lanzen A, Qi J, Huson DH, Schleper C, Schuster SC. 2008. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527. 10.1371/journal.pone.0002527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osborne JW. 2010. Improving your data transformations: applying the Box-Cox transformation. Pract. Assess. Res. Eval. 15:1–9 [Google Scholar]
- 29.Sekirov I, Russell SL, Antunes LC, Finlay BB. 2010. Gut microbiota in health and disease. Physiol. Rev. 90:859–904 [DOI] [PubMed] [Google Scholar]
- 30.Brusetti L, Crotti E, Tamburini A, Cittaro D, Garavaglia V, Rolli E, Sorlini C, Daffonchio D, Borin S. 2011. Influence of transgenic Bt176 and non-transgenic corn silage on the structure of rumen bacterial communities. Ann. Microbiol. 61:925–930 [Google Scholar]
- 31.Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. 2012. IBD: what role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 9:219–230 [DOI] [PubMed] [Google Scholar]
- 32.Salyers AA, Whitt DD. 2002. Bacterial pathogenesis: a molecular approach. ASM Press, Washington, DC [Google Scholar]
- 33.De Cock HEV, Marks SL, Stacy BA, Zabka TS, Burkitt J, Lu G, Steffen DJ, Duhamel GE. 2004. Ileocolitis associated with Anaerobiospirillum in cats. J. Clin. Microbiol. 42:2752–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buzoianu SG, Walsh MC, Rea MC, O'Donovan O, Gelencsér E, Ujhelyi G, Szabó E, Nagy A, Ross RP, Gardiner GE, Lawlor PG. 2012. Effects of feeding Bt maize to sows during gestation and lactation on maternal and offspring immunity and fate of transgenic material. PLoS One 7:e47851. 10.1371/journal.pone.0047851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6:121–131 [DOI] [PubMed] [Google Scholar]
- 36.Castillo M, Martín-Orúe SM, Anguita M, Pérez JF, Gasa J. 2007. Adaptation of gut microbiota to corn physical structure and different types of dietary fibre. Livest. Sci. 109:149–152 [Google Scholar]
- 37.Lewis AJ, Southern LL. 2001. Swine nutrition, 2nd ed. CRC Press, Boca Raton, FL [Google Scholar]
- 38.Schmidt B, Mulder IE, Musk CC, Aminov RI, Lewis M, Stokes CR, Bailey M, Prosser JI, Gill BP, Pluske JR, Kelly D. 2011. Establishment of normal gut microbiota is compromised under excessive hygiene conditions. PLoS One 6:e28284. 10.1371/journal.pone.0028284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mulder I, Schmidt B, Stokes C, Lewis M, Bailey M, Aminov R, Prosser J, Gill B, Pluske J, Mayer C-D, Musk C, Kelly D. 2009. Environmentally acquired bacteria influence microbial diversity and natural innate immune responses at gut surfaces. BMC Biol. 7:79. 10.1186/1741-7007-7-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clarridge JE., III 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reuter G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr. Issues Intest. Microbiol. 2:43–53 [PubMed] [Google Scholar]
- 42.Walter J. 2008. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl. Environ. Microbiol. 74:4985–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy EF, Cotter PD, Healy S, Marques TM, O'Sullivan O, Fouhy F, Clarke SF, O'Toole PW, Quigley EM, Stanton C, Ross PR, O'Doherty RM, Shanahan F. 2010. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut 59:1635–1642 [DOI] [PubMed] [Google Scholar]
- 44.Attig L, Djiane J, Gertler A, Rampin O, Larcher T, Boukthir S, Anton PM, Madec JY, Gourdou I, Abdennebi-Najar L. 2008. Study of hypothalamic leptin receptor expression in low-birth-weight piglets and effects of leptin supplementation on neonatal growth and development. Am. J. Physiol. Endocrinol. Metab. 295:E1117–E1125 [DOI] [PubMed] [Google Scholar]
- 45.Ross MG, Beall MH. 2008. Adult sequelae of intrauterine growth restriction. Semin. Perinatol. 32:213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mackie RI, Aminov RI, Hu W, Klieve AV, Ouwerkerk D, Sundset MA, Kamagata Y. 2003. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl. Environ. Microbiol. 69:6808–6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flint HJ, Duncan SH, Scott KP, Louis P. 2007. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ. Microbiol. 9:1101–1111 [DOI] [PubMed] [Google Scholar]
- 48.Leman AD, Straw BE, Mengeling WL, D'Allaire S, Taylor DJ. 1992. Diseases of swine. Wolfe Publishing, London, United Kingdom [Google Scholar]
- 49.Schierack P, Walk N, Reiter K, Weyrauch KD, Wieler LH. 2007. Composition of intestinal Enterobacteriaceae populations of healthy domestic pigs. Microbiology 153:3830–3837 [DOI] [PubMed] [Google Scholar]
- 50.Walk ST, Young VB. 2008. Emerging insights into antibiotic-associated diarrhea and Clostridium difficile infection through the lens of microbial ecology. Interdiscip. Perspect. Infect. Dis. 2008:125081. 10.1155/2008/125081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poroyko V, White JR, Wang M, Donovan S, Alverdy J, Liu DC, Morowitz MJ. 2010. Gut microbial gene expression in mother-fed and formula-fed piglets. PLoS One 5:e12459. 10.1371/journal.pone.0012459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim HB, Borewicz K, White BA, Singer RS, Sreevatsan S, Tu ZJ, Isaacson RE. 2011. Longitudinal investigation of the age-related bacterial diversity in the feces of commercial pigs. Vet. Microbiol. 153:124–133 [DOI] [PubMed] [Google Scholar]
- 53.Larsen N, Vogensen FK, van den Berg FWJ, Nielsen DS, Andreasen AS, Pedersen BK, Al-Soud WA, Sørensen SJ, Hansen LH, Jakobsen M. 2010. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One 5:e9085. 10.1371/journal.pone.0009085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Konstantinov SR, Awati AA, Williams BA, Miller BG, Jones P, Stokes CR, Akkermans ADL, Smidt H, De Vos WM. 2006. Post-natal development of the porcine microbiota composition and activities. Environ. Microbiol. 8:1191–1199 [DOI] [PubMed] [Google Scholar]
- 55.Konstantinov SR, Favier CF, Zhu WY, Williams BA, Kluß J, Souffrant W-B, de Vos WM, Akkermans ADL, Smidt H. 2004. Microbial diversity studies of the porcine gastrointestinal ecosystem during weaning transition. Anim. Res. 53:317–324 [Google Scholar]
- 56.Gardiner GE, Casey PG, Casey G, Lynch PB, Lawlor PG, Hill C, Fitzgerald GF, Stanton C, Ross RP. 2004. Relative ability of orally administered Lactobacillus murinus to predominate and persist in the porcine gastrointestinal tract. Appl. Environ. Microbiol. 70:1895–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.