Abstract
Bacillus thuringiensis is an important source of insect resistance traits in commercial crops. In an effort to prolong B. thuringiensis trait durability, insect resistance management programs often include combinations of insecticidal proteins that are not cross resistant or have demonstrable differences in their site of action as a means to mitigate the development of resistant insect populations. In this report, we describe the activity spectrum of a novel B. thuringiensis Cry protein, Cry1Bh1, against several lepidopteran pests, including laboratory-selected B. thuringiensis-resistant strains of Ostrinia nubilalis and Heliothis virescens and progeny of field-evolved B. thuringiensis-resistant strains of Plutella xylostella and Spodoptera frugiperda. Cry1Bh1 is active against susceptible and B. thuringiensis-resistant colonies of O. nubilalis, P. xylostella, and H. virescens in laboratory diet-based assays, implying a lack of cross-resistance in these insects. However, Cry1Bh1 is not active against susceptible or Cry1F-resistant S. frugiperda. Further, Cry1Bh1 does not compete with Cry1Fa or Cry1Ab for O. nubilalis midgut brush border membrane binding sites. Cry1Bh1-expressing corn, while not completely resistant to insect damage, provided significantly better leaf protection against Cry1Fa-resistant O. nubilalis than did Cry1Fa-expressing hybrid corn. The lack of cross-resistance with Cry1Ab and Cry1Fa along with independent membrane binding sites in O. nubilalis makes Cry1Bh1 a candidate to further optimize for in-plant resistance to this pest.
INTRODUCTION
Insect-resistant Bacillus thuringiensis-treated corn (Bt corn) is a commercial success due to the efficacy, convenience, and safety of these products. Today, Bt corn is grown on more than 65% of U.S. corn acreage (1). Ostrinia nubilalis is historically one of the most damaging lepidopteran insect pests of corn throughout the United States and Canada. Annual revenue loss due to O. nubilalis resulting from reduced crop yield and expenditures for insect management was previously estimated at U.S. $1 billion (2). At present, economic losses due to O. nubilalis have been greatly reduced due to widespread, successful use of Bt corn (3, 4).
Cry1Ab was the first B. thuringiensis trait commercialized for control of O. nubilalis and continues to be used in a number of transgenic corn events approved by the U.S. Environmental Protection Agency (EPA) (5). B. thuringiensis Cry1Fa protein in maize event TC1507 is another important tool for transgenic control of lepidopteran pests due its broad spectrum of activity, which includes O. nubilalis and Spodoptera frugiperda (6–9). Other B. thuringiensis proteins used for transgenic control of O. nubilalis in the United States include Cry1A.105 and Cry2Ab (5).
Despite the success of Bt corn, the long-term reliance on a limited number of Cry proteins can lead to development of resistant insect populations in the field. For example, Cry1Fa-resistant populations of S. frugiperda were isolated from areas of Puerto Rico where transgenic Cry1Fa maize was extensively cultivated (8, 10). Ostrinia nubilalis resistance to Cry1Ab has also been detected among individuals selected from field populations (11).
Combining two or more B. thuringiensis insecticidal proteins that are not cross resistant and have independent sites of action is one strategy to delay the development of resistant insect populations (12, 13). Previous studies demonstrated that Cry1B-type insecticidal toxins are active on a broad spectrum of lepidopteran pests (for an overview, see reference 9). Cry1Ba1 (14) is the best-studied Cry1B-type protein. Several reports indicated that Cry1Ba is toxic to Cry1A-resistant colonies of Plutella xylostella (15–20) and Heliothis virescens (21). Cry1Ba is also active against Ostrinia nubilalis (22, 23) and does not share O. nubilalis midgut binding sites with Cry1A proteins (22). Cry1Ba has been successfully expressed in transgenic maize to provide protection against Diatraea saccharalis, Diatraea grandiosella, and Spodoptera frugiperda (24). In this report, we describe the characterization of a new B. thuringiensis insecticidal protein (25). The primary aim of this work was to assess the spectrum, potency, and potential resistance management utility for Cry1Bh1.
MATERIALS AND METHODS
Preparation of B. thuringiensis genomic DNA.
B. thuringiensis strain PS46L was cultured in 4 ml of LB (Luria-Bertani) medium overnight at 28°C with shaking at 150 rpm and then centrifuged at 2,000 rpm for 20 min. The supernatant was removed and the cell pellet was resuspended in the residual volume. Next, 150 μl of the cell suspension was transferred to a 96-well plate. Total DNA was isolated by using the Qiagen blood and tissue kit (catalog number 69582) according to the manufacturer's protocol.
PCR and genome walking.
An internal fragment of a putative cry1B allele was amplified by PCR with cry1B degenerate primers (forward, GAGGYASCCTTMTCTATACTATAGGG, and reverse, GGACTCGGTACCTGGAACATCC) using standard PCR conditions and the TaKaRa LA taq kit (catalog number RR013B). Codes for degenerate base pairs were as follows: m = A or C, y = C or T, and s = C or G. The resulting 1.2-kb amplified DNA fragment was cloned into plasmid vector pCR2.1 (Invitrogen catalog number K4500-01) according to the manufacturer's instructions. The amplification product was determined to contain a portion of the cry1Bh1 gene. The complete Cry1Bh gene sequence was elucidated using the Clontech GenomeWalker kit (catalog number 638904).
Cry1Bh1 protein expression and purification.
Standard methods were used to construct recombinant Pseudomonas fluorescens expression plasmids containing a synthetic cry1Bh1 gene codon optimized for plant expression (26). The expression plasmid was transformed by electroporation into DC454 (a near-wild-type P. fluorescens strain having mutations ΔpyrF and lsc::lacIQI), recovered in SOC-Soy hydrolysate medium, and plated on M9 glucose agar lacking uracil (27). Cry1Bh1 protein inclusion bodies from recombinant P. fluorescens were prepared essentially as described by Gao et al. (28).
Cry1Bh1 inclusion body preparations in 10 mM CAPS (N-cyclohexyl-3-aminopropanesulfonic acid, pH 10.0; Sigma-Aldrich Corporation) were diluted to desired concentrations in the same buffer. Protein concentrations were estimated by gel electrophoresis using bovine serum albumin (BSA; Sigma-Aldrich Corporation) to create a standard curve for gel densitometry with a Bio-Rad imaging system (Fluor-S MultiImager with Quantity One software, version 4.5.2).
Diet-based bioassay of Cry1Bh1.
B. thuringiensis-resistant insect colonies used in this study are described in Table 1. Cry1A-resistant P. xylostella (16) eggs were obtained from Benzon Research, Inc. The B. thuringiensis-resistant colonies of H. virescens and O. nubilalis were provided by North Carolina State University and University of Nebraska—Lincoln, respectively. The H. virescens YHD2 strain had mutations in both cadherin (29) and the ABC transporter (30) (F. Gould, personal communication).
Table 1.
Cry toxin resistance levels of P. xylostella, S. frugiperda, O. nubilalis, and H. virescens
| Insect species | Strain | Cry toxin resistance | Reference |
|---|---|---|---|
| P. xylostella | NO-QAGE | Cry1Aa (>20,000-fold), Cry1Ab (>10,000-fold), Cry1Ac (>40,000-fold), Cry1Fa (>10,000-fold), Cry1Ja (>2,000-fold); low resistance to Cry1Bb, Cry1Ca, Cry1Da, Cry2Aa, and Cry9Ca | 16 |
| S. frugiperda | Cry1Fa-resistant SI | Cry1Fa (>100-fold for growth inhibition rate) | 8 |
| O. nubilalis | Cry1Fa resistant | Cry1Fa (>3,000-fold) | 42 |
| H. virescens | CXC | Cry1Ac (50-fold), Cry1Ab (12-fold), Cry2Aa (52-fold), Cry1Aa (low level), Cry1B (low level), Cry1C (low level) | 44 |
| H. virescens | YHD2 | Cry1Ac (≅10,000-fold); cross resistant to Cry1Aa (>2,000-fold) and Cry1Ab and Cry1Fa (>3,500-fold); slightly cross resistant to Cry2A, Cry1B, and Cry1C | 21 |
Larval neonates of O. nubilalis, S. frugiperda, Cry1Fa-resistant S. frugiperda (8), Plutella xylostella (Linnaeus), B. thuringiensis HD-1-selected P. xylostella (16), Helicoverpa zea (Boddie), Heliothis virescens (Fabricius), and Diabrotica virgifera virgifera (LeConte) were tested with purified Cry1Bh1 for insecticidal activity in diet overlay bioassays at a single dose of 9,000 ng/cm2 (considered a high dose), except for O. nubilalis and H. virescens, which were tested at 3,000 ng/cm2. Cry1Fa-resistant S. frugiperda and O. nubilalis, as well as CXC and YHD2 strains of H. virescens, were maintained at the Dow AgroSciences LLC insectary; eggs for all other lepidopteran species were obtained from Benzon Research Inc. (Carlisle, PA). Nondiapausing D. virgifera virgifera worms were obtained from Crop Characteristics, Inc. (Farmington, MN).
Cry-resistant strains of H. virescens, CXC and YHD2, and Cry1Fa-resistant O. nubilalis were tested with Cry1Bh1 at 1,000 and 3,000 ng/cm2 in diet overlay bioassays, along with susceptible H. virescens and O. nubilalis. Cry1Ac, Cry1Fa, and Cry2Aa were used as positive controls, while water and 10 mM CAPS (pH 10.0) buffer were used as negative controls. Susceptible and Cry1Fa-resistant O. nubilalis were tested in a series of Cry toxin amounts (0, 4.1, 12, 37, 111, 333, 1,000, and 3,000 ng/cm2) to generate dose-response data used to calculate specific activity values for mortality, growth inhibition, and resistance ratio.
Bioassays were conducted in 128-well bioassay trays (C-D International, Pitman, NJ). A 40-μl aliquot of protein sample was delivered onto the surface of multispecies lepidopteran diet (Southland Products, Lake Village, AR) in each well. The treated trays were air dried, and one individual larva (24 to 48 h after eclosion) was deposited on the treated diet surface. The infested wells were then sealed with adhesive sheets of clear plastic vented to allow gas exchange (C-D International, Pitman, NJ). Bioassay trays were held under controlled environmental conditions (28°C, ∼40% relative humidity, 16:8 h light/dark) for 5 days. For D. virgifera virgifera, methods similar to the lepidopteran insect bioassays were followed, except that a Dow AgroSciences LLC proprietary rootworm diet was used and 80 to 100 μl of aliquot solution was used to treat the diet surface.
The total number of insects exposed to each protein sample, the number of dead insects, and the weight of surviving insects were recorded in all insect bioassays. Larvae which weighed <0.1 mg were considered moribund insects and were included in the percent mortality computation for the dose-response study with the susceptible and Cry1Fa-resistant strains of O. nubilalis as well as the single-dose bioassays with the B. thuringiensis-resistant H. virescens strains (CXC and YHD2) and Cry1Fa-resistant O. nubilalis tested with Cry1Bh1 at 1,000 and 3,000 ng/cm2. Percent mortality and percent growth inhibition were calculated for each treatment. Growth inhibition (GI) was calculated as follows: GI = 1 − (TWIT/TNIT)/(TWIBC/TNIBC) × 100, where TWIT is the total weight of insects in the treatment, TNIT is the total number of insects in the treatment, TWIBC is the total weight of insects in the buffer control, and TNIBC is the total number of insects in the buffer control. Control mortality did not exceed 10%. Bioassays were replicated at least 4 times, with 8 lepidopteran larvae or 17 D. virgifera virgifera larvae per replicate.
Initial insect screening at a single high dose, 3,000 or 9,000 ng/cm2, of Cry1Bh1 and subsequent exposures of three resistant insect species to various B. thuringiensis toxins were analyzed with a one-way analysis of variance (ANOVA) and means separation by using the Tukey-Kramer HSD test. With both susceptible and Cry1Fa-resistant strains of O. nubilalis, the slope and growth inhibition concentration-response curves were determined by using a nonlinear logistic 3-parameter model, and the effective concentrations required to cause 50% growth inhibition (GI50) and 90% growth inhibition (GI90) were estimated. These analyses were performed by using JMP Pro, version 9.0.3, software (SAS Institute Inc., Cary, NC). Probit analyses (31) of the pooled mortality data were conducted using POLO-PC (LeOra Software) to estimate the 50% lethal concentration (LC50), 90% lethal concentration (LC90), and slope of the concentration-response curves. For O. nubilalis, a resistance ratio was estimated by dividing the LC50 for the resistant strain by that for the susceptible strain when their dose-response slopes were overlapping based on the likelihood ratio test of parallelism tests (32).
Cry1Bh1 brush border membrane vesicle (BBMV) binding site interactions.
Purified trypsin-activated Cry1Ab toxin was iodinated using Iodo-Beads (Pierce). Three Iodo-Beads were washed twice with 500 μl of phosphate-buffered saline (PBS; 20 mM sodium phosphate, 0.15 M NaCl [pH 7.5]) and placed into a 1.5-ml centrifuge tube behind lead shielding. To this were added 100 μl of PBS and 1 mCi of Na125I (17.4 Ci/mg; Amersham). The components were allowed to react for 5 min at room temperature, and then 2 μg of highly pure trypsin-activated Cry1Ab protein in PBS was added to the solution and allowed to react for an additional 5 min. The reaction was terminated by removing the solution from the Iodo-Beads and applying it to a PD-10 desalting column containing 20 mM CAPS, pH 10.5. Radiolabeled protein was collected by taking 0.2-ml fractions. The radiopurity of the iodinated Cry proteins was determined by SDS-PAGE and phosphorimaging the gel. Specific activity was determined to be 3.1 μCi/μg of protein.
Standard radioiodination methods inactivate Cry1Fa, so we instead radiolabeled fluorescein-5-maleimide with 125I and allowed this molecule to attach to Cry1Fa, which labels the single cysteine residue (C205) located in the toxin core. Cry1Fa labeled by this protocol is biologically active (33). Briefly, fluorescein-5-maleimide was dissolved to 10 mM (4.27 mg/ml) in dimethyl sulfoxide (DMSO) and then diluted to 1 mM in PBS as determined by its molar extinction coefficient of 68,000 M−1cm−1. A total of 0.5 mCi of Na125I was added behind lead shielding to a 70-μl solution of PBS containing two Iodo-Beads (Pierce). The solution was allowed to mix at room temperature for 5 min, and then 10 μl of the 1 mM fluorescein-5-maleimide was added. The reactants were allowed to react for 10 min and then removed from the Iodo-Beads. To the reacted solution was added 2 μg of highly purified trypsin-activated Cry1Fa core toxin in PBS. The protein was incubated with the iodinated fluorescein-5-maleimide solution for 48 h at 4°C, and the reaction was stopped by adding 2-mercaptoethanol to 14 mM. The reaction mixture was then added to a Zebra spin column (Invitrogen) equilibrated in 20 mM CAPS–150 mM KCl (pH 9) and centrifuged at 1,500 × g for 2 min to separate nonreacted iodinated dye and 2-mercaptoethanol from the protein. The 125I-radiolabeled fluorescein-Cry1Fa was counted in a gamma counter to determine its specific activity. Typically, specific activities of 1.1 μCi/μg of Cry1Fa protein were obtained. The protein was also characterized by SDS-PAGE and visualized by phosphorimaging to ensure that the radioactivity measured was covalently associated with the Cry1Fa protein, which migrated at a distance associated with 65-kDa proteins.
Reciprocal competition binding studies with labeled Cry1Bh1 were not done.
BBMV preparation.
Last-instar O. nubilalis larvae were starved overnight and then dissected in the morning after chilling on ice for 15 min. The midgut tissue was removed from the body cavity, leaving behind the hindgut attached to the integument. The midgut was placed in 9× volume of ice-cold homogenization buffer (300 mM mannitol, 5 mM EGTA, 17 mM Tris base [pH 7.5]) supplemented with protease inhibitor cocktail (Sigma P-2714) diluted as recommended by the supplier. The tissue was homogenized with 15 strokes of a glass tissue homogenizer. BBMVs were prepared by the MgCl2 precipitation method of Wolfersberger (34). Briefly, an equal volume of a 24 mM MgCl2 solution in 300 mM mannitol was mixed with the midgut homogenate, stirred for 5 min, and allowed to stand on ice for 15 min. The solution was centrifuged at 2,500 × g for 15 min at 4°C. The supernatant was saved and the pellet suspended in the original volume of 2-fold-diluted homogenization buffer and centrifuged again. The two supernatants were combined and centrifuged at 27,000 × g for 30 min at 4°C to form the BBMV fraction. The pellet was suspended into BBMV storage buffer (10 mM HEPES, 130 mM KCl, 10% glycerol [pH 7.4]) to a concentration of about 3 mg/ml of protein. Protein concentration was determined by using the Bradford method (35) with BSA as the standard. Alkaline phosphatase (ALP) determination was made prior to freezing the samples using the Sigma assay by following the manufacturer's instructions. The specific activity of this marker enzyme in the BBMV fraction typically increased 7-fold compared to that found in the midgut homogenate fraction. The BBMVs were aliquoted into 250-μl samples, flash frozen in liquid N2, and stored at −80°C.
The binding of 125I-radiolabeled Cry1Ab or Cry1Fa to BBMVs prepared from O. nubilalis was measured in triplicate by incubating 0.5 nM 125I-Cry1Ab or 2 nM 125I-Cry1Fa with 0.15 mg/ml of BBMV protein in a total volume of 300 μl in binding buffer (PBS plus 0.1% BSA) for 1 h at 28°C. The samples were centrifuged at full speed in a microcentrifuge (16,000 × g) for 8 min, and the supernatant was removed. The protein pellet was washed twice with ice-cold binding buffer, and the bottom of the microcentrifuge tube containing the pellet was cut out, placed in a glass test tube, and counted in a gamma counter. Total radioactivity measured in the pellet in the absence of any competitor minus the amount of radioactivity measured in the presence of 1,000 nM unlabeled Cry1Ab due to nonspecific binding was considered 100% specific binding. The amount of total binding was measured in triplicate in the presence of various concentrations of nonlabeled competing proteins and plotted. Nonspecific binding represented 20.3 and 23.2% of total binding for 125I Cry1Ab and 125I Cry1Fa, respectively.
Plant expression vector construction.
Agrobacterium tumefaciens standard binary vectors harboring expression cassettes encoding the activated form of Cry1Bh1 (amino acid residues 1 to 652) were engineered using Gateway Technology (Invitrogen, Carlsbad, CA) and used in Agrobacterium-mediated plant transformation. Standard cloning methods were used in the construction of entry vectors containing cry1Bh1 expression cassettes. Restriction endonucleases were obtained from New England BioLabs (NEB; Ipswich, MA), and T4 DNA ligase (Invitrogen) was used for DNA ligation. PCR amplification was performed using Phusion high-fidelity DNA polymerase (NEB) and primers synthesized by Integrated DNA Technologies Inc. (IDT; Coralville, IA). Gateway reactions were performed using Gateway LR Clonase enzyme mix (Invitrogen) for assembling entry and destination vectors. Plasmid preparations were performed using either the NucleoSpin plasmid kit (Macherey-Nagel Inc., Bethlehem, PA) or the Plasmid Midi Kit (Qiagen) by following the instructions of the suppliers. DNA fragments were isolated using a QIAquick gel extraction kit (Qiagen) after agarose Tris-acetate gel electrophoresis.
Briefly, a synthetic cry1Bh1 gene optimized for maize codon usage was obtained from DNA 2.0 (Menlo Park, CA) in plasmid DASDNA362. Plant transformation vector construction was initiated with PCR amplification of the maize-optimized cry1Bh1v3 gene from DASDNA362 with primers DASDNA362 F2 and DASDNA362 R2 to generate the 1,959-bp cry1Bh1v5 gene fragment encoding the insecticidal core protein. Primer sequences were as follows: DASDNA362F2, CATTTTGACGGAGCTCTAGGTGATTAAGCTAACTATC, and DASDNA362R2, TGACGGAGCTCTAGGTGATTAAGCTAACTATCACCTTTCGAGGTCGTACTTCG.
The amplified fragment containing cry1Bh1v5 was digested with BbsI and SacI and inserted into pDAB107604 digested with NcoI and SacI to create entry vector pDAB109887 containing the ZmUbi1v2::cry1Bh1v5::ZmLipv1 expression cassette flanked by attL1 and attL2 Gateway recombination sites. The promoter in this expression cassette was the maize ubiquitin promoter (36), and the 3′ untranslated region was from the Zea mays Viviparous-1 (Vp1) gene (GenBank accession number L35913). pDAB109887 was recombined using Gateway technology with destination vector pDAB109805, which contained a proprietary selectable marker cassette for haloxyfop resistance to create final binary vector pDAB109890, coding for the approximate 65-kDa Cry1Bh1 insecticidal core protein fragment. Constructs were transformed into maize via Agrobacterium-mediated transformation of immature embryos isolated from the maize inbred line B104. The method used was similar to those published by Ishida et al. and Frame et al. (37, 38).
Insect bioassay of Cry1Bh1-expressing corn lines.
Five individual T1 generation corn plants per event, each hemizygous for the cry1Bh1 gene, were evaluated at the V5 stage of development for resistance to feeding damage by susceptible and Cry1Fa-resistant O. nubilalis. Events expressing yellow fluorescent protein (YFP) and a nontransformed isogenic corn line (B104) were used as negative controls in the assay. A commercially available Cry1Fa-expressing hybrid (event TC1507, Herculex I; HX1) was used as a positive control in the assay. Cry1Bh1 protein expression was performed via enzyme-linked immunosorbent assay (ELISA) (EnviroLogix, Portland, ME). Cry1Fa average protein expression in leaves of V3 to V5 plants grown in the greenhouse was used for comparison.
Thirty-two-well trays (C-D International, Pitman, NJ) were partially filled with a 2% agar solution. Leaf sections from the 3rd- to 4th-collar leaves, approximately 1 square inch in size, were taken from each plant, and one was placed into each well of the 32-well trays. Three leaf pieces were tested per event and per insect strain. Ten neonatal larvae (24 to 48 h old) were added into each well using a paintbrush. Trays were sealed with perforated sticky lids (C-D International), which allowed ventilation during the test. Trays were placed in growth chambers at 28°C with 60% relative humidity and 16:8 h of light/dark for 3 days. After the test, a visual percent damage score was taken for each leaf piece. Percent leaf damage for each event was averaged, and the leaf protection between the transgenic constructs was analyzed with ANOVA and mean separation using the Tukey-Kramer honestly significant difference (HSD) test. Statistical analyses were performed via JMP Pro 9.0.3 (SAS Institute Inc., NC).
Quantification of Cry1Bh1 protein expression levels in transgenic maize.
The amount of Cry1Bh1 in leaf material was determined by quantitative liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) peptide analysis (39). Protein samples extracted from maize leaves in 25 mM ammonium bicarbonate plus 0.05% Tween 20 were analyzed on an Agilent 1200 binary pump liquid chromatography system and a QTRAP 5500 (model number 1024945-AA) using an Acquity ultraperformance liquid chromatography (UPLC) BEH130 C18, 1.7-μm, 2.1- by 50-mm column. Two peptides were tracked (T37 [435.3 m/z] and T24 [682.9 m/z]) for each sample. T37 was the most sensitive peptide; thus, quantitation was based on the peptide transition 435.3/655.4.
Nucleotide sequence accession number.
The sequence obtained for Cry1Bh1 has been deposited in GenBank under accession number ADT78693.
RESULTS
Sequence identity of Cry1Bh1.
Cry1Bh1 shares 87% sequence identity to its nearest neighbor, Cry1Bd2, and 72% sequence identity to Cry1Ba2 (25). Protein domain comparison to Cry1Bd2 revealed sequence identity scores of 98.7%, 75.5%, and 98.6% for domains 1, 2, and 3, respectively. Domain 2 is associated with Cry protein receptor binding (40, 41), and this domain contains most of the sequence differences between Cry1Bh1 and Cry1Bd2. Further, while Cry1Bh1 domain 3 is approximately 99% identical to Cry1Bd2, it is interesting that the next nearest sequence identity of approximately 81% is with domain 3 from Cry1Ac (data not shown).
Insecticidal activity of Cry1Bh1 against susceptible and B. thuringiensis-resistant insects.
Cry1Bh1 insecticidal activity was initially assayed at a dose of 9,000 or 3,000 ng/cm2 on several lepidopteran pests. Cry1Bh1 caused 100% mortality on both susceptible and B. thuringiensis HD-1-resistant strains of P. xylostella, as well as H. virescens (Table 2). Treatment of diet with 3,000 ng/cm2 of Cry1Bh1 resulted in 69% larval mortality of O. nubilalis. Cry1Bh1 caused insignificant larval mortality of H. zea and was not active on susceptible or Cry1Fa-resistant colonies of S. frugiperda. Cry1Bh1 also lacked activity on an important coleopteran pest of corn, D. virgifera virgifera.
Table 2.
Insecticidal activity spectrum of Cry1Bh1 protein tested at 9,000 ng/cm2 on diet overlay bioassays
| Insect strain | % mortalityb |
|---|---|
| Plutella xylostella | 100* |
| B. thuringiensis HD-1-resistant P. xylostella | 100* |
| Helicoverpa zea | 3ns |
| Ostrinia nubilalisza | 69* |
| Heliothis virescensa | 100* |
| Spodoptera frugiperda | 0ns |
| Cry1Fa resistant S. frugiperda | 0ns |
| Diabrotica virgifera virgiferaa | 7ns |
O. nubilalis and H. virescens were tested at 3,000 ng/cm2, and D. virgifera virgifera was tested at 400 μg/cm2.
Values with asterisks are significantly different from the buffer control, while a superscript “ns” indicates a comparable response according to Fisher protected ANOVA (P < 0.05).
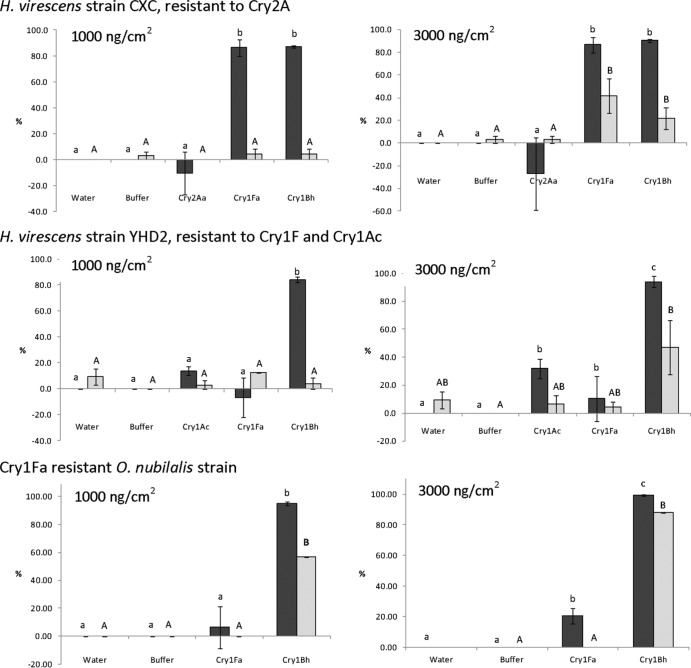
Additional bioassays were conducted at 1,000 ng/cm2 and 3,000 ng/cm2 on Cry1Fa-resistant O. nubilalis, Cry1Ac/Cry1Fa-resistant H. virescens strain YHD2, and H. virescens strain CXC, which is resistant to both Cry1Ac and Cry2A. These Cry-resistant insect colonies were all susceptible to Cry1Bh1, as indicated by high larval growth inhibition (>80%) at both toxin doses (Fig. 1). However, Cry1Bh1 caused higher mortality of Cry1Fa-resistant O. nubilalis than of the two Cry-resistant H. virescens strains. At 3,000 ng/cm2, Cry1Bh1 caused growth inhibition and mortality of the CXC strain comparable to those caused by Cry1Fa. Further, at 3,000 ng/cm2 the YHD2 strain was insensitive to either Cry1Ac or Cry1Fa, whereas Cry1Bh1 exhibited significant toxicity against this strain. Resistance of the H. virescens strains to the Cry proteins used for selection, i.e., Cry1Ac or Cry2Aa, was confirmed in these bioassays, as indicated by negligible larval mortality or growth inhibition for these proteins. In fact, the CXC-resistant H. virescens showed an increased growth rate when presented with Cry2Aa, indicated by the negative growth inhibition values.
Fig 1.
Mean percent growth inhibition (dark gray bars) and mortality (light gray bars) of the B. thuringiensis-resistant H. virescens strains CXC and YHD2 and Cry1Fa-resistant O. nubilalis when fed with Cry1Fa, Cry2Aa, Cry1Ac, and Cry1Bh1 toxins at 1,000 and 3,000 ng/cm2. Bars followed by the same letters are not significantly different according to Fisher protected ANOVA and the Tukey-Kramer HSD test (P > 0.05). Lowercase a, b, and c indicate mean percent growth inhibition, while A and B indicate mean percent mortality (4 replications, n = 8).
Next, the potency (LC50) of Cry1Bh1 against O. nubilalis and Cry1Fa-resistant O. nubilalis was determined (Table 3). Cry1Bh1 protoxin was approximately 6.6-fold less potent than Cry1Fa protoxin on susceptible O. nubilalis. Overlapping confidence intervals of both lethal concentration and growth inhibition parameters in the dose-response study indicated that Cry1Bh1 has similar potencies against both the susceptible and Cry1Fa-resistant O. nubilalis strains. Note that the Cry1Fa-resistant O. nubilalis strain is more than 3,000-fold resistant to Cry1Fa (42).
Table 3.
Susceptibilities of Ostrinia nubilalis and Cry1Fa-resistant O. nubilalis neonates exposed to the Bacillus thuringiensis Cry1Bh1 and Cry1Fa protoxins measured by mortality and growth inhibition
| Insect | Protein | Dose response | 50% estimation (95% FL)a | 90% estimation (95% FL)a | Nb | Slope (SE) | χ2c | r2d | df |
|---|---|---|---|---|---|---|---|---|---|
| O. nubilalis | Cry1Fa | Lethality (LCe) | 253.2 (185.7–335.4) | 1,004.9 (707.6–1,683.6) | 216 | 2.1 (0.3) | 2.1 | 3 | |
| Growth inhibition (ECf) | 2.5 (1.3–4.9) | 33.8 (6.3–182.5) | 336 | 0.9 (0.4) | 0.6193 | 21 | |||
| Cry1Bh | Lethality (LC) | 1,679.7 (1,239.0–2,493.6) | 6,552.1 (3,936.4–17,158.0) | 160 | 2.2 (0.4) | 0.4 | 2 | ||
| Growth inhibition (EC) | 293.7 (215.3–400.6) | 1,658.0 (1,009.5–2,723.0) | 224 | 1.0 (0.2) | 0.9788 | 25 | |||
| Cry1Fa-resistant O. nubilalis | Cry1Bh | Lethality (LC) | 1,008.1 (766.2–1,355.2) | 3,397.6 (2,316.2–6,299.2) | 192 | 2.4 (0.4) | 2.7 | 3 | |
| Growth inhibition (EC) | 216.3 (170.4–274.7) | 1,136.2 (718.2–1,797.7) | 159 | 1.2 (0.2) | 0.9629 | 14 |
ng of Cry toxin/cm2 of treated artificial diet surface, with 95% fiducial limits (FL) in parentheses. Mortality was calculated by probit analysis, and growth inhibition relative to untreated control was calculated by nonlinear regression fitted to a logistic 3-parameter model.
Total number of larvae tested in bioassay.
χ2 values from the goodness-of-fit test indicate a significant (P < 0.05) fit of the probit model for lethal-concentration statistics.
r2 of ≅1 from the nonlinear regression tests indicate significant fit to a logistic 3-parameter model for growth inhibition statistics.
LC, lethal concentration.
EC, effective concentration.
Toxin binding studies.
Radiolabeled Cry1Ab bound to BBMV proteins from O. nubilalis larvae in a specific manner, with nonspecific binding representing 20.3% of total binding. Total binding was competed in a dose-dependent manner by the addition of unlabeled Cry1Ab (Fig. 2). Total binding was reduced by 50% at 0.5 nM concentration of unlabeled Cry1Ab, the same concentration of radiolabeled Cry1Ab as used in the binding assay. Cry1Bh1, on the other hand, had no effect on the total binding of Cry1Ab to the BBMV proteins when added at concentrations as high as 100 nM (1,000 times the concentration of the labeled ligand) and reduced the binding of 125I-Cry1Ab by only about 25% at 300 nM. With 125I-Cry1Fa used as the radioligand, unlabeled Cry1Fa competed for total binding in a concentration-dependent manner; however, Cry1Bh1 did not compete with the binding of 125I-Cry1Fa even at 1,000 nM (Fig. 2), which is 500-fold higher than the concentration of the radiolabeled substrate used in the assay. We also used an alternative binding assay, where O. nubilalis BBMV protein bound with 125I-Cry1Fa was extensively washed, solubilized, and separated by SDS-PAGE. In this assay format we detected the presence of 125I-Cry1Fa bound to the BBMV by phosphorimaging the gel. In the presence of unlabeled Cry1Fa at 1,000 nM, the amount of radiolabeled Cry1Fa isolated bound to the BBMV pellet was reduced to the level of nonspecific binding but the presence of 1,000 nM Cry1Bh1 did not compete with the binding of the 125I-Cry1Fa to the BBV protein (data not shown).
Fig 2.
Competition binding of 125I-Cry1Ab and 125I Cry1Fa with Cry1Ab or Cry1Bh to BBMVs prepared from Ostrinia nubilalis. Circles, Cry1Ab competed against 0.1 nM 125I Cry1Ab; squares, Cry1Bh competed against 0.1 nM Cry1Ab; triangles, Cry1Fa competed against 2 nM 125I Cry1Fa; diamonds, Cry1Bh competed against 2 nM 125I Cry1Fa.
Protection of Cry1Bh1-containing corn to damage by O. nubilalis.
Transgenic corn lines expressing Cry1Bh1 demonstrated a range of protection to leaf damage by susceptible (Fig. 3A) and Cry1Fa-resistant (Fig. 3B) O. nubilalis. For susceptible O. nubilalis, the activated form of Cry1Bh1 expressed in transgenic maize events provided significant leaf protection compared to negative controls. Against Cry1Fa-resistant O. nubilalis, leaf protection was also significantly higher in maize expressing Cry1Bh than in maize expressing Cry1Fa. Leaf damage to the experimental Cry1Bh1 events by susceptible O. nubilalis was greater than the commercial standard (Herculex I).
Fig 3.
Mean percent leaf damage and protein expression for the active form of Cry1Bh1 T1 transgenic events (5 plants per event) infested with O. nubilalis (A) and Cry1Fa-resistant O. nubilalis (B). Controls were B104, a nontransformed negative-control maize line; YFP, a negative transgenic control expressing yellow fluorescent protein; and HX1, a commercial Cry1F hybrid (Herculex I). The center vertical error line denotes the standard error of the average of mean percent leaf damage or protein expression among the events.
Protein expression across Cry1Bh1 events ranged from approximately 10 to 30 ng/cm2 of protein except for event 2, which provided elevated protein expression (∼75 ng/cm2). However, event 2 did not exhibit significantly less leaf damage than did the rest of the Cry1Bh1 events. Across events other than event 2, Cry1Bh1 protein expression levels were approximately 25 to 75% of the level of Cry1Fa (∼ 40 ng/cm2) in Herculex I. Cry1Bh1 events provided less leaf protection (approximately 30% insect feeding damage) against O. nubilalis than for Herculex 1 (<5% insect feeding damage).
DISCUSSION
As a class, Cry1B-type B. thuringiensis insecticidal proteins are active against a broad spectrum of lepidopteran pests. Cry1B proteins show little to no cross-resistance with other Cry1 proteins when tested on Cry1A-resistant colonies of some important insect pests, e.g., P. xylostella and H. virescens (15–21, 43). Further, the most studied toxin of this family, Cry1Ba, was shown to recognize different midgut binding sites than Cry1A toxins in P. xylostella (15) and O. nubilalis (22) as well as D. saccharalis, D. grandiosella, and S. frugiperda (24).
The primary objective of the work presented here was to determine whether a new Cry1B family member, Cry1Bh1, has cross-resistance and competitive binding properties compatible with other Cry1 proteins for insect resistance management (IRM) in economically important lepidopteran pests. First, in laboratory bioassays we examined the spectrum of Cry1Bh1 lepidopteran insecticidal activity, including four insect colonies resistant to Cry1 proteins. Results demonstrated that Cry1Bh1 potencies were statistically similar for both Cry1Fa-susceptible and Cry1Fa-resistant O. nubilalis, with a resistance ratio of approximately 0.6 to 0.7. In comparison, the Cry1Fa-resistant O. nubilalis strain was 3,000-fold resistant to Cry1Fa (42). This suggests that Cry1Bh1 and Cry1Fa are not cross resistant in O. nubilalis. We also demonstrated that Cry1Bh1 has activity on Cry1-resistant P. xylostella (selected on B. thuringiensis strain HD-1), H. virescens strain YDH2 (Cry1Ac resistant), and H. virescens strain CXC (Cry1Ac resistant and Cry2A resistant). The absence, or very low levels, of cross-resistance for Cry1Bh1 on these insect colonies is consistent with the results for other Cry1B-type proteins tested previously (16, 21, 44).
The susceptibility of the B. thuringiensis-resistant insect strains to Cry1Bh1 suggests a different site or mechanism of action compared to Cry1Ac in P. xylostella NO-QAGE and to both Cry1Ac or Cry2Aa proteins in H. virescens YHD2 and CXC colonies. Mutations in membrane-bound protein receptors are implicated as Cry1A resistance determinants in these colonies. An ABC transporter was reported to be involved in Cry1Ac resistance in P. xylostella NO-QAGE (45). Both cadherin and ABC transporters have been implicated in resistance to Cry1Ac in H. virescens YHD2 (29, 30). It follows that the Cry1Bh1 membrane interactions are distinct from these characterized resistance determinants. In addition, reduced membrane-bound alkaline phosphatase (ALP) protein is correlated with resistance phenotypes for both YHD2 (46) and CXC strains of H. virescens (47), but reduction in ALP levels is apparently independent of the resistance mechanism in these colonies.
To further assess whether Cry1Bh1 is compatible in combination with Cry1Ab or Cry1Fa for resistance management of O. nubilalis, we studied the binding site interactions of this protein on midgut BBMVs. Our results indicated that Cry1Bh1 does not compete with the binding of either Cry1Ab or Cry1Fa in O. nubilalis BBMVs. This suggests differences in binding site interactions for Cry1Bh1 compared to Cry1Ab or Cry1Fa in O. nubilalis that could be useful for mitigation of receptor-mediated mechanisms of resistance. That said, the resistance mechanism for the Cry1Ab- or Cry1Fa-resistant colonies tested in this study is unknown, and as yet, there is no evidence for binding site-mediated resistance in either colony (48, 49).
Finally, we demonstrated that transgenic corn events expressing the Cry1Bh1 toxic core fragment had much less foliar damage caused by Cry1Fa-susceptible or Cry1Fa-resistant O. nubilalis than with nontransgenic isolines. The Cry1Bh1 level of leaf protection against susceptible O. nubilalis was, however, less than that of the commercial standard, Herculex I. The less than acceptable levels of insect control, compared to the commercialized corn hybrid, could be the result of lower expression levels combined with lower inherent potency of Cry1Bh1 than for Cry1Fa.
In conclusion, the results presented here suggest that Cry1Bh1 has biological properties that make it interesting to consider as a candidate for gene pyramids when combined with Cry1 proteins. However, further work is needed to optimize Cry1Bh1 insect resistance trait efficacy to control against Cry1Ab or Cry1Fa resistance in O. nubilalis.
ACKNOWLEDGMENTS
We thank Tom Meade and Nick Storer, Dow AgroSciences, for their support and helpful suggestions to improve the manuscript. We thank Fred Gould for H. virescens colonies YHD2 and CXC.
Footnotes
Published ahead of print 27 September 2013
REFERENCES
- 1.USDA-ERS 2012. 5 July 2012, posting date Adoption of genetically engineered crops in the U.S.: recent trends in GE adoption. USDA, Washington, DC: http://www.ers.usda.gov/data-products/adoption-of-genetically-engineered-crops-in-the-us/recent-trends-in-ge-adoption.aspx [Google Scholar]
- 2.Mason CE, Rice ME, Calvin DD, Van Duyn JW, Showers WB, Hutchison WD, Witkowski JF, Higgins RA, Onstad DW, Dively GP. 1996. European corn borer ecology and management. North Central Regional Extension Publication no. 327. Iowa State University, Ames, IA. [Google Scholar]
- 3.Hutchison WD, Burkness EC, Mitchell PD, Moon RD, Leslie TW, Fleischer SJ, Abrahamson M, Hamilton KL, Steffey KL, Gray ME, Hellmich RL, Kaster LV, Hunt TE, Wright RJ, Pecinovsky K, Rabaey TL, Flood BR, Raun ES. 2010. Areawide suppression of European corn borer with Bt maize reaps savings to non-Bt maize growers. Science 330:222–225 [DOI] [PubMed] [Google Scholar]
- 4.Siegfried BD, Hellmich RL. 2012. Understanding successful resistance management: the European corn borer and Bt corn in the United States. GM Crops Food 3:184–193 [DOI] [PubMed] [Google Scholar]
- 5.US Environmental Protection Agency 30 November 2011, posting date Current and previously registered section 3 PIP registrations. US Environmental Protection Agency, Washington, DC: http://www.epa.gov/pesticides/biopesticides/pips/pip_list.htm [Google Scholar]
- 6.Siebert MW, Babock JM, Nolting S, Santos AC, Adamczyk JJ, Neese PA, King JE, Jenkins JN, McCarty J, Lorenz GM, Fromme DD, Lassiter RB. 2008. Efficacy of Cry1F insecticidal protein in maize and cotton for control of fall armyworm (Lepidoptera: Noctuidae). Fla. Entomol. 91:555–565 [Google Scholar]
- 7.Siebert MW, Tindal KV, Leonard BR, Van Duyn JW, Babcock JM. 2008. Evaluation of corn hybrids expressing Cry1F (Herculex (R) I insect protection) against fall armyworm (Lepidoptera: Noctuidae) in the southern United States. J. Entomol. Sci. 43:41–51 [Google Scholar]
- 8.Storer NP, Babcock JM, Schlenz M, Meade T, Thompson GD, Bing JW, Huckaba RM. 2010. Discovery and characterization of field resistance to Bt maize: Spodoptera frugiperda (Lepidoptera: Noctuidae) in Puerto Rico. J. Econ. Entomol. 103:1031–1038 [DOI] [PubMed] [Google Scholar]
- 9.van Frankenhuyzen K. 2009. Insecticidal activity of Bacillus thuringiensis crystal proteins. J. Invertebr. Pathol. 101:1–16 [DOI] [PubMed] [Google Scholar]
- 10.Storer NP, Kubiszak ME, King JE, Thompson GD, Santos AC. 2012. Status of resistance to Bt maize in Spodoptera frugiperda: lessons from Puerto Rico. J. Invertebr. Pathol. 110:294–300 [DOI] [PubMed] [Google Scholar]
- 11.Siegfried BD, Spencer T, Crespo AL, Storer NP, Head GP, Owens ED, Guyer D. 2007. Ten years of Bt resistance monitoring in the European corn borer: what we know, what we don't know, and what we can do better. Am. Entomol. 53:208–214 [Google Scholar]
- 12.Roush RT. 1998. Two-toxin strategies for management of insecticidal transgenic crops: can pyramiding succeed where pesticide mixtures have not? Philos. Trans. R. Soc. Lond. B Biol. Sci. 353:1777–1786 [Google Scholar]
- 13.Head GP, Greenplate J. 2012. The design and implementation of insect resistance management programs for Bt crops. GM Crops Food 3:144–153 [DOI] [PubMed] [Google Scholar]
- 14.Brizzard BL, Whiteley HR. 1988. Nucleotide sequence of an additional crystal protein gene cloned from Bacillus thuringiensis subsp. thuringiensis. Nucleic Acids Res. 16:2723–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferré J, Real MD, Van Rie J, Jansens S, Peferoen M. 1991. Resistance to the Bacillus thuringiensis bioinsecticide in a field population of Plutella xylostella is due to a change in a midgut membrane receptor. Proc. Natl. Acad. Sci. U. S. A. 88:5119–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabashnik BE, Johnson KW, Engleman JT, Baum JA. 2000. Cross-resistance to Bacillus thuringiensis toxin Cry1Ja in a strain of diamondback moth adapted to artificial diet. J. Invertebr. Pathol. 76:81–83 [DOI] [PubMed] [Google Scholar]
- 17.Tabashnik BE, Malvar T, Liu YB, Finson N, Borthakur D, Shin BS, Park SH, Masson L, de Maagd RA, Bosch D. 1996. Cross-resistance of the diamondback moth indicates altered interactions with domain II of Bacillus thuringiensis toxins. Appl. Environ. Microbiol. 62:2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang JD, Shelton AM, Van Rie J, De Roeck S, Moar WJ, Roush RT, Peferoen M. 1996. Toxicity of Bacillus thuringiensis spore and crystal protein to resistant diamondback moth (Plutella xylostella). Appl. Environ. Microbiol. 62:564–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu YB, Tabashnik BE, Meyer SK, Crickmore N. 2001. Cross-resistance and stability of resistance to Bacillus thuringiensis toxin Cry1C in diamondback moth. Appl. Environ. Microbiol. 67:3216–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong Y, Wang C, Yang Y, Wu S, Wu Y. 2010. Characterization of resistance to Bacillus thuringiensis toxin Cry1Ac in Plutella xylostella from China. J. Invertebr. Pathol. 104:90–96 [DOI] [PubMed] [Google Scholar]
- 21.Gould F, Anderson A, Reynolds A, Bumgarner L, Moar W. 1995. Selection and genetic analysis of a Heliothis virescens (Lepidoptera: Noctuidae) strain with high levels of resistance to Bacillus thuringiensis toxins. J. Econ. Entomol. 88:1545–1559 [Google Scholar]
- 22.Denolf P, Jansens S, Peferoen M, Degheele D, Van Rie J. 1993. Two Different Bacillus thuringiensis delta-endotoxin receptors in the midgut brush border membrane of the European corn borer, Ostrinia nubilalis (Hubner) (Lepidoptera: Pyralidae). Appl. Environ. Microbiol. 59:1828–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan SY, Cayabyab BF, Alcantara EP, Ibrahim YB, Huang F, Blankenship EE, Siegfried BD. 2011. Comparative susceptibility of Ostrinia furnacalis, Ostrinia nubilalis and Diatraea saccharalis (Lepidoptera: Crambidae) to Bacillus thuringiensis Cry1 toxins. Crop Prot. 30:1184–1189 [Google Scholar]
- 24.Rang C, Bergvingson D, Bohorova N, Hoisington D, Frutos R. 2004. Competition of Bacillus thuringiensis Cry1 toxins for midgut binding sites: a basis for the development and management of transgenic tropical maize resistant to several stemborers. Curr. Microbiol. 49:22–27 [DOI] [PubMed] [Google Scholar]
- 25.Crickmore N, Zeigler D, Schnepf E, Van Rie J, Lereclus D, Baum J, Bravo A, Dean DH. 2012, posting date Bacillus thuringiensis toxin nomenclature. http://www.btnomenclature.info/
- 26.Squires CH, Retallack DM, Chew LC, Ramseier TM, Schneider JC, Talbot HW. 2004. Heterologous protein production in P. fluorescens. BioProcesss Int. 2004:54–58 [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed, vol 1 to 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28.Gao Y, Schafer BW, Collins RA, Herman RA, Xu XP, Gilbert JR, Ni WT, Langer VL, Tagliani LA. 2004. Characterization of Cry34Ab1 and Cry35Ab1 insecticidal crystal proteins expressed in transgenic corn plants and Pseudomonas fluorescens. J. Agric. Food Chem. 52:8057–8065 [DOI] [PubMed] [Google Scholar]
- 29.Gahan LJ, Gould F, Heckel DG. 2001. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293:857–860 [DOI] [PubMed] [Google Scholar]
- 30.Gahan LJ, Pauchet Y, Vogel H, Heckel DG. 2010. An ABC transporter mutation is correlated with insect resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 6:e1001248. 10.1371/journal.pgen.1001248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finney DJ. 1971. Probit analysis, 3rd ed. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 32.Robertson JL, Russell RM, Preisler HK, Savin NE. 2007. Bioassays with arthropods, 2nd ed. CRC Press, Boca Raton, FL [Google Scholar]
- 33.Evans SL, Li J, Sheets JJ. June 2012. Biologically-active radiolabeled Cry1Fa and receptor binding assay methods. US patent US20120156803A1
- 34.Wolfersberger MG. 1993. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the gypsy moth (Lymantria dispar). Arch. Insect Biochem. Physiol. 24:139–147 [DOI] [PubMed] [Google Scholar]
- 35.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 36.Christensen AH, Sharrock RA, Quail PH. 1992. Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 18:675–689 [DOI] [PubMed] [Google Scholar]
- 37.Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T. 1996. High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nat. Biotechnol. 14:745–750 [DOI] [PubMed] [Google Scholar]
- 38.Frame BR, McMurray JM, Fonger TM, Main ML, Taylor KW, Torney FJ, Paz MM, Wang K. 2006. Improved Agrobacterium-mediated transformation of three maize inbred lines using MS salts. Plant Cell Rep. 25:1024–1034 [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Filiou MD, Reckow S, Gormanns P, Maccarrone G, Kessler MS, Frank E, Hambsch B, Holsboer F, Landgraf R, Turck CW. 2011. Proteomic and metabolomic profiling of a trait anxiety mouse model implicate affected pathways. Mol. Cell. Proteomics 10(12):M111.008110. 10.1074/mcp.M111.008110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dean DH, Rajamohan F, Lee MK, Wu SJ, Chen XJ, Alcantara E, Hussain SR. 1996. Probing the mechanism of action of Bacillus thuringiensis insecticidal proteins by site-directed mutagenesis—a minireview. Gene 179:111–117 [DOI] [PubMed] [Google Scholar]
- 41.Bravo A, Gill SS, Soberon M. 2007. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 49:423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira EJG, Lang BA, Storer NP, Siegfried BD. 2008. Selection for Cry1F resistance in the European corn borer and cross-resistance to other Cry toxins. Entomol. Exp. Appl. 126:115–121 [Google Scholar]
- 43.Tabashnik BE, Finson N, Groeters FR, Moar WJ, Johnson MW, Luo K, Adang MJ. 1994. Reversal of resistance to Bacillus thuringiensis in Plutella xylostella. Proc. Natl. Acad. Sci. U. S. A. 91:4120–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gould F, Martinez-Ramirez A, Anderson A, Ferre J, Silva FJ, Moar WJ. 1992. Broad-spectrum resistance to Bacillus thuringiensis toxins in Heliothis virescens. Proc. Natl. Acad. Sci. U. S. A. 89:7986–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baxter SW, Badenes-Perez FR, Morrison A, Vogel H, Crickmore N, Kain W, Wang P, Heckel DG, Jiggins CD. 2011. Parallel evolution of Bacillus thuringiensis toxin resistance in lepidoptera. Genetics 189:675–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jurat-Fuentes JL, Adang MJ. 2004. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur. J. Biochem. 271:3127–3135 [DOI] [PubMed] [Google Scholar]
- 47.Jurat-Fuentes JL, Karumbaiah L, Jakka SR, Ning C, Liu C, Wu K, Jackson J, Gould F, Blanco C, Portilla M, Perera O, Adang M. 2011. Reduced levels of membrane-bound alkaline phosphatase are common to lepidopteran strains resistant to Cry toxins from Bacillus thuringiensis. PLoS One 6:e17606. 10.1371/journal.pone.0017606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pereira EJ, Siqueira HA, Zhuang M, Storer NP, Siegfried BD. 2010. Measurements of Cry1F binding and activity of luminal gut proteases in susceptible and Cry1F resistant Ostrinia nubilalis larvae (Lepidoptera: Crambidae). J. Invertebr. Pathol. 103:1–7 [DOI] [PubMed] [Google Scholar]
- 49.Coates BS, Sumerford DV, Lopez MD, Wang H, Fraser LM, Kroemer JA, Spencer T, Kim KS, Abel CA, Hellmich RL, Siegfried BD. 2011. A single major QTL controls expression of larval Cry1F resistance trait in Ostrinia nubilalis (Lepidoptera: Crambidae) and is independent of midgut receptor genes. Genetica 139:961–972 [DOI] [PubMed] [Google Scholar]