Abstract
Feed supplementation with the probiotic Enterococcus faecium for piglets has been found to reduce pathogenic gut microorganisms. Since Escherichia coli is among the most important pathogens in pig production, we performed comprehensive analyses to gain further insight into the influence of E. faecium NCIMB 10415 on porcine intestinal E. coli. A total of 1,436 E. coli strains were isolated from three intestinal habitats (mucosa, digesta, and feces) of probiotic-supplemented and nonsupplemented (control) piglets. E. coli bacteria were characterized via pulsed-field gel electrophoresis (PFGE) for clonal analysis. The high diversity of E. coli was reflected by 168 clones. Multilocus sequence typing (MLST) was used to determine the phylogenetic backgrounds, revealing 79 sequence types (STs). Pathotypes of E. coli were further defined using multiplex PCR for virulence-associated genes. While these analyses discerned only a few significant differences in the E. coli population between the feeding groups, analyses distinguishing clones that were uniquely isolated in either the probiotic group only, the control group only, or both groups (shared group) revealed clear effects at the habitat level. Interestingly, extraintestinal pathogenic E. coli (ExPEC)-typical clones adhering to the mucosa were significantly reduced in the probiotic group. Our data show a minor influence of E. faecium on the overall population of E. coli in healthy piglets. In contrast, this probiotic has a profound effect on mucosa-adherent E. coli. This finding further substantiates a specific effect of E. faecium strain NCIMB 10415 in piglets against pathogenic E. coli in the intestine. In addition, these data question the relevance of data based on sampling fecal E. coli only.
INTRODUCTION
In-feed antibiotics have been used in livestock for decades to decrease the risk of infectious diseases and promote growth performance (1, 2). However, in 2006, the use of antibiotics as growth promoters was prohibited in the European Union (3); instead, feed supplements, such as prebiotics, probiotics, or cations, like zinc, are used. In pigs, probiotics, such as Enterococcus faecium or Bacillus cereus variant Toyoi, are commonly used, based on prior reports of positive effects against microbial infections (4–6, 8).
In previous studies, we investigated the probiotic E. faecium strain NCIMB 10415 as a feed supplement in piglets. We observed that E. faecium did not change the general swine intestinal microbiota (9) but showed specific effects reducing natural infections by Chlamydia spp. and pathogenic intestinal Escherichia coli serotypes (10, 11).
E. coli is a member of the gastrointestinal autochthonous microbiota of pigs and contributes to the maintenance of the microbial gut balance (12). However, in addition to commensal strains, pathogenic strains causing intestinal or extraintestinal diseases are a great health concern for both humans and animals (13, 14). Intestinal pathogenic (InPEC) and extraintestinal pathogenic E. coli (ExPEC) strains are classified into certain pathotypes according to possession of virulence-associated genes (VAGs). Well-known InPEC pathotypes are enterotoxigenic (ETEC), enteropathogenic (EPEC), or Shiga-toxin producing (STEC), while typical ExPEC ones are uropathogenic (UPEC), newborn meningitis-causing (NMEC), septicemia-associated (SePEC), and avian-pathogenic (APEC) E. coli (13–15).
Porcine intestinal E. coli populations have been described as being highly individual and dynamic (9, 16) and are influenced by diet, climate, age, and particularly weaning, which initiates a massive change in the intestinal microbiota (18, 19). Schierack et al. (21) showed that the swine gut acts as a reservoir for ExPEC and suggested that high numbers of ExPEC-typical VAGs correlate with intestinal colonization. This finding was possible only because of detailed analysis, which had not been considered previously for the analysis of intestinal microbiota of conventionally raised swine (9, 20, 21).
To gain further insight into the effects between E. faecium and E. coli, we investigated the E. coli population of young piglets in more detail by testing the influence of this probiotic at three different age periods and by sampling three different habitats of the gut: mucosa and digesta of the colon ascendens and feces. By quantifying defined E. coli clones and linking their phylogenetic background with possession of 69 VAGs, we aimed to identify possible shifts in the occurrence of certain clones between these three habitats. We hypothesized that E. faecium has an influence on E. coli colonization in the porcine intestine. While our data did not show changes in the overall diversity of E. coli, E. faecium feeding caused a specific reduction in clones displaying ExPEC-typical virulence-associated factors. Since ExPEC-typical VAGs are known to promote colonization, this finding occurred particularly with clones adhering to the mucosa. Our results suggest a specific prophylactic effect of E. faecium against E. coli, with VAG profiles similar to those of ExPEC at the gut epithelia.
MATERIALS AND METHODS
Animal housing.
Sixteen pregnant purebred landrace sows were divided into two groups: a control group (n = 8) and a probiotic group (n = 8). Sows of the probiotic group were fed a diet containing 4.2 × 106 to 4.3 × 106 CFU/g E. faecium NCIMB 10415 (Cylactin, Cerbios-Pharma SA, Lugano, Switzerland) from 28 days ante partum (a.p.) onwards as described previously (22). All animals were kept under similar conditions but in different stables to prevent the transmission of E. faecium via feces from the probiotic group to the control group. After birth, piglets were kept with their dams until weaning at the age of 26 ± 1 days. After weaning, when sows were separated from their litters, the piglets were kept in commercial flat deck pens in two different buildings with two animals per pen. This is a customary procedure in animal feeding trials to reduce cage effects. From the age of 12 days onward, piglets had access to a prestarter diet. Postweaning, the piglets were fed a starter diet. The starter diets of the probiotic supplemented group contained 5.1 × 106 CFU/g (prestarter) and 3.6 × 106 CFU/g (starter) of E. faecium NCIMB 10415. No antibiotics, either for therapeutic or prophylactic purposes, were applied to any of the animals used in the study.
Of all piglets from 16 sows (8 sows per feeding group), 24 piglets were randomly chosen and assigned to the probiotic and control groups. The influence of E. faecium was examined at three different ages (12, 26, and 34 days) and in two different samples from intestine and feces. Samples taken at the ages of 12 ± 1 (n = 4/feeding group), 26 ± 1 (n = 4/feeding group), and 34 ± 1 (n = 4/feeding group) days were used to obtain intestinal digesta and mucosal samples. Euthanasia and sampling were performed as described previously (22). In brief, following a midline abdominal incision, the small intestine was dissected from the large intestine at the ileocecal junction and both segments were dissected from the mesentery. Digesta and mucosal scrapings were taken from the colon ascendens. Fecal samples were obtained from the ampulla recti prior to euthanasia.
The study was approved by the local state office of occupational health and technical safety, Landesamt für Gesundheit und Soziales Berlin (LaGeSo no. 0347/09).
Isolation of E. coli.
Isolation of E. coli has been described previously (23, 24). Briefly, intestinal contents from colon ascendens and feces were suspended in phosphate-buffered saline (PBS) buffer (0.2 g in 6 ml PBS), and serial dilutions were plated to different solid media to identify as many phenotypically diverse E. coli isolates as possible. To achieve this, sheep blood and Gassner agar plates (Sifin, Berlin, Germany), as well as CHROMagar orientation plates (CHROMagar, Paris, France[25]), were chosen. Additionally, CHROMagar orientation plates containing five different antibiotics (one per plate) were used: ampicillin (≥32 mg/ml), streptomycin (≥64 mg/ml), chloramphenicol (≥32 mg/ml), gentamicin (≥16 mg/ml), and tetracycline (≥16 mg/ml). The breakpoint concentrations were estimated based on previously published data (26). Thus, a total of eight different media were used to isolate E. coli bacteria from the three different samples from a total of 24 piglets. Colonies showing a typical pink color on CHROMagar orientation and/or a blue color on Gassner agar plates after incubation at 37°C for 24 h were assumed to be E. coli isolates. Approximately 20 pink or blue colonies per specimen (each representing a single isolate) were randomly picked from the plates (3 to 4 colonies per plate) for subcultivation onto CHROMagar orientation and sheep blood agar plates and incubated at 37°C for 24 h.
For mucosal samples, an approximately 2- by 5-cm section was washed twice in 1× PBS to remove visible fecal material. These short washing steps are not expected to affect mucosa-attached bacteria, since other studies have reported high numbers of mucosa-attached or epithelial cell-attached E. coli bacteria after up to four to six washing steps with physiological saline (27, 28). Approximately 0.5 g of each mucosal sample was removed from connective tissue by scraping with a glass microscope slide. Mucosal samples were transferred to a Dounce homogenizer and homogenized in 5 ml 1× PBS, and serial dilutions of the homogenates were plated to agar plates as described above.
A total of 20 to 24 E. coli isolates (each) were collected from the mucosa and intestinal content of the colon ascendens as well as the feces of each piglet.
Assignment of E. coli isolates to clones.
Macrorestriction analysis by pulsed-field gel electrophoresis (PFGE) (23) using the restriction endonuclease XbaI was initially used to define clones to exclude analysis of duplicates. PFGE profiles were compared using BioNumerics software, version 6.6 (Applied Maths, Belgium), with the unweighted-pair group using average linkages method. Dice similarity indices (complete linkage; optimization, 1%; position tolerance, 1.5%) were also calculated.
Each E. coli colony was regarded as an individual isolate. A clone was defined as an E. coli group of isolates with a specific macrorestriction pattern, whereas two clones differed by more than one band (23). The diversity of the E. coli population was determined using Simpson's index of diversity (Di). This method enables comparison of the diversities of populations with different numbers of isolates and has been described previously (29, 30).
A major clone was defined as a clone which represented ≥50% of typed isolates in one sample, and a minor clone was defined as a clone which represented ≤10% of typed isolates in one sample (31). One representative isolate for each clone was randomly chosen for further analysis via multilocus sequence typing (MLST) and multiplex PCR.
MLST.
MLST of one representative of each clone was performed to analyze the phylogenetic background. MLST was carried out as described previously (32). Gene amplification and sequencing were performed using primers available on the E. coli MLST website (http://mlst.ucc.ie/mlst/mlst/dbs/Ecoli). Sequences were analyzed using the software package SeqSphere 0.9.19 (http://www3.ridom.de/seqsphere). Sequence types (STs) were computed automatically. The phylogenetic group of the E. coli isolates was determined using the software program Structure 2.3.4 based on the sequences of the seven housekeeping genes used for MLST (http://pritch.bsd.uchicago.edu/structure.html). A spanning tree was constructed using BioNumerics (version 6.6; Applied Maths, Belgium).
Virulence-associated gene determination using PCR.
The presence of a total of 69 VAGs was tested by multiplex PCR as previously described (33, 34). In addition, the presence of stx2e, faeG, fanA, fasA, fedA, fimF41a, est-Ib, est-II, elt-Ia (typical for ETEC and edema disease E. coli [EDEC]), aggR, and the virulence plasmid pAA (typical for enteroaggregative E. coli [EAEC]) was assayed using primers and conditions described previously (35, 36). A representative isolate subjected to MLST was tested for VAGs, and results were considered to hold true for all isolates of the respective clone.
Statistical analysis.
Statistical analyses were carried out using the software program SPSS 19.0 (IBM SPSS Statistics) and the R software environment, version 2.15.2 (http://www.r-project.org). The prevalence of 69 VAGs in the control and probiotic groups was determined and compared for both groups using a permutation test. In a second approach, we categorized all clones into three groups. The first group consisted of clones occurring in the control group only. The second group consisted of clones that were found only in samples from piglets of the probiotic group, and for a third group, only clones that occurred in piglets of both study groups (shared clones) were assigned (see Fig. 1). The prevalences of the genes considered for each VAG in these three groups were determined and also compared by a permutation test. We permuted the allocation of the piglets to the feeding groups 10,000 times and calculated chi-square statistics for each of the permuted samples. The proportion of the chi-square values that were greater than the chi-square statistic of the original sample was determined. A P value of <0.05 was considered significant. We also used the permutation test approach to compare the three habitats with respect to the occurrence of clonal STs. We used the permutation test approach since the analyses were carried out at the level of clones. Since clones belonging to the same piglet are not independent statistical units, we could not use the classical tests. Phylogenetic analysis was carried out using the software program STRUCTURE (37, 38). This software applies Bayesian methods to predict distinct groupings of the E. coli population. Minimum spanning trees were created by the program BioNumerics (version 6.6; Applied Maths, Belgium). The level of significance was α = 0.05. Since the statistical analysis is exploratory, we did not perform a Bonferroni adjustment of the level of significance.
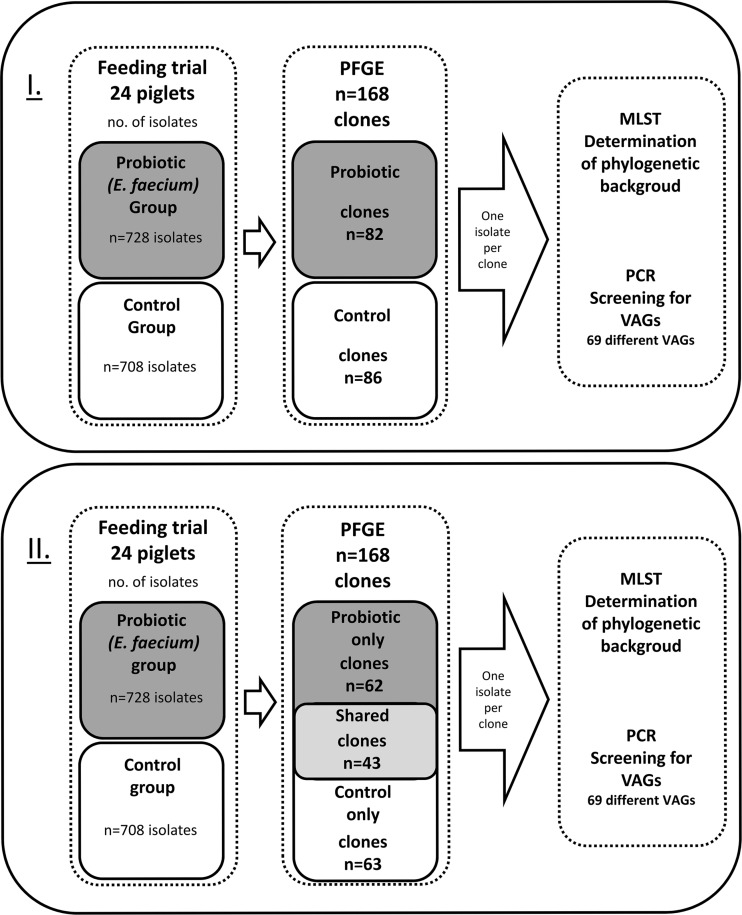
Fig 1.
Schematic work flow of clones and isolates included and methods used in this work. Two different approaches were used: I, comparing clones from the two feeding groups, the probiotic and control groups; II, comparing clones present in either the control or probiotic group or shared by both groups.
RESULTS
E. coli isolates from the probiotic group versus those from the control group. (i) Isolation of E. coli.
A total of 1,436 E. coli isolates were obtained from digesta, mucosa, and fecal samples from 24 clinically healthy piglets, 12 each from the control and the E. faecium (probiotic) groups. From each piglet, between 60 and 70 (20 to 24 per sample) E. coli isolates were analyzed, with approximately equal numbers of isolates from each feeding group (708 from the control versus 728 from the probiotic group) (see Fig. 1). The proportion of isolates originating with digesta was 36.1%; 35.1% were from mucosa, and 28.8% were from feces. As outlined in Table 1, the habitats of the isolates were also equally distributed between the feeding groups.
Table 1.
Overview of distribution of all isolates, clones, and STs for the three different ages and habitats of the control and probiotic groups
| Sample location | Age of piglets (days)a | No. of pigletsb |
Total no. of: |
||||||
|---|---|---|---|---|---|---|---|---|---|
|
E. coli isolatesc |
Clones identifiedd |
STs identifiede |
|||||||
| Control | Probiotic | Control | Probiotic | Control | Probiotic | Control | Probiotic | ||
| Colon (mucosa) | 12 ± 1 | 4 | 4 | 93 | 83 | 26 | 30 | 21 | 24 |
| 26 ± 1 | 4 | 4 | 88 | 83 | 35 | 31 | 26 | 22 | |
| 33 ± 1 | 4 | 4 | 78 | 81 | 28 | 24 | 21 | 18 | |
| Total (mucosa) | 12 | 12 | 259 | 247 | 89 | 85 | 68 | 64 | |
| Colon (digesta) | 12 ± 1 | 4 | 4 | 83 | 83 | 24 | 24 | 20 | 18 |
| 26 ± 1 | 4 | 4 | 89 | 90 | 34 | 37 | 24 | 25 | |
| 33 ± 1 | 4 | 4 | 83 | 89 | 26 | 23 | 21 | 17 | |
| Total (digesta) | 12 | 12 | 255 | 262 | 84 | 84 | 65 | 60 | |
| Feces | 12 ± 1 | 4 | 4 | 43 | 42 | 16 | 14 | 12 | 11 |
| 26 ± 1 | 4 | 86 | 88 | 37 | 35 | 29 | 25 | ||
| 33 ± 1 | 4 | 4 | 65 | 89 | 25 | 28 | 18 | 23 | |
| Total (feces) | 12 | 12 | 194 | 219 | 78 | 77 | 59 | 59 | |
| Total (control/probiotic) | 12 | 12 | 708 | 728 | 251 | 246 | 192 | 183 | |
Mean ± SD.
Samples were obtained from a total of 24 piglets.
A total of 1,436 E. coli isolates were analyzed.
Clones are defined as an E. coli group of isolates with a specific macrorestriction pattern, wherein two clones differed by more than one band. A total of 499 clones were found, with 168 different clones identified.
STs, sequence types, based on seven housekeeping genes. A total of 375 STs were found, with 79 different STs identified.
(ii) Clonal analysis of E. coli.
Macrorestriction analysis of the 1,436 isolates identified 168 clones, with numbers of isolates per clone ranging between 1 and 181. Both feeding groups displayed a high clonal diversity, and their diversity indices (Di) were nearly equal (for E. coli populations from both feeding groups, Di = 0.967; for E. coli populations from the control group, Di = 0.954; for E. coli populations from the probiotic group, Di = 0.962). The diversity indices of the E. coli populations from the three different habitats also showed no major differences (for E. coli populations from digesta of the control group, Di = 0.947; for E. coli populations from mucosa of the control group, Di = 0.946; for E. coli populations from feces of the control group, Di = 0.969; for E. coli populations from digesta of the probiotic group, Di = 0.961; for E. coli populations from mucosa of the probiotic group, Di = 0.965; for E. coli populations from feces of the probiotic group, Di = 0.958). Of the 168 E. coli clones, 63 were isolated only from the control and 62 only from the probiotic group. Forty-three clones appeared in both groups, leading to a total of 86 clones in the control group and 82 clones in the probiotic group (see Fig. 1).
When each clone was counted once per group, a total of 106 clones were identified in the control and 105 in the probiotic group. In general, some clones occurred only in one animal and one sample, while other clones were present in up to 20 animals and 48 samples. On average, 2.4 clones per animal and 1.3 clones per sample were found. Two different major clones and 165 minor clones were detected, using the previous definition of minor and major clones (31). One representative isolate of each clone was randomly chosen for subsequent analyses, including multilocus sequence typing (MLST) and PCR for detection of virulence-associated genes (VAG-PCR).
(iii) Phylogenetic analysis.
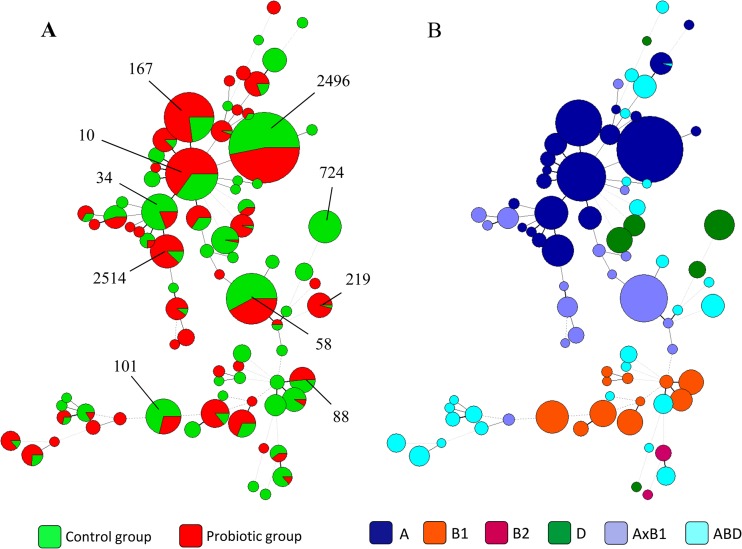
Macrorestriction analysis has a higher discriminatory power than MLST, conferring the ability to subdivide sequence types (STs) into clones. With the stringent definition of the clone, on which this study is based, we performed MLST (32) analysis to assign an ST to each of the 168 clones. This resulted in the identification of 79 distinct STs. Eighteen of these showed allele combinations which had not been reported as of 2 March 2013 on the MLST website (http://mlst.ucc.ie/mlst/mlst/dbs/Ecoli). For relatedness of all STs, see Fig. 2A. The minimum spanning tree (MSTree) comprises all 1,436 isolates, belonging to the 168 clones, which are assigned to 79 STs. Forty-four different STs occurred in the probiotic group and 50 different STs in the control group, with 30 STs occurring in both feeding groups. STs were nonrandomly distributed, since some STs were overrepresented, e.g., ST10 (n = 18 clones; n = 125 isolates), ST58 (n = 10 clones; n = 118 isolates), ST167 (n = 3 clones; n = 114 isolates), and ST2496 (n = 4 clones; n = 233 isolates). The number of isolates per ST varied from a maximum of 233 (ST2496) to a minimum of just 1. These singletons were found in a total of 17 STs; however, an association with a particular feeding groups was not defined.
Fig 2.
(A) Minimum spanning tree (MSTree) of STs from 168 clones defined by PFGE, assuming the results for one representative for the complete group. Green, control; red, probiotic group. (B) Minimum spanning tree of phylogenetic groups, assuming the results for one representative for the complete group. Both MSTrees were calculated using Bionumerics 6.6.
Evidence suggests that specific E. coli phylotypes are associated with certain ecological adaptations (39). To define phylotypes, we analyzed the recombination events of E. coli by comparing the polymorphisms in the concatenated sequences of the seven gene fragments used for MLST using the STRUCTURE software program (37, 38). This software applies Bayesian methods to predict distinct groupings of the E. coli population. Using the linkage model of STRUCTURE, we were able to assign the isolates to ECOR groups A (59 clones, 710 isolates), B1 (27 clones, 197 isolates), B2 (4 clones, 12 isolates), and D (8 clones, 116 isolates). In addition, 31 clones (203 isolates) were assigned to the hybrid group AxB1 and 39 clones (198 isolates) to hybrid group ABD. Thus, we defined a remarkably low number of isolates belonging to ECOR B2 and D. Both phylogroups are known to harbor particularly virulent E. coli strains (15, 32, 40, 41). Most importantly, no differences were found between the two feeding groups, since all phylogenetic groups were distributed equally between the probiotic group and the control group (for data, see Fig. 2A and B).
We further analyzed the distribution of STs in association with the habitat the E. coli bacteria were isolated from, namely, mucosa, digesta, or feces. However, no differences were seen in the distribution (see Fig. S1 in the supplemental material). Also, we found no age-related association of the piglets with the appearance of certain STs (data not shown).
(iv) VAG determination.
As outlined in Fig. 1, one representative isolate of each of the 168 E. coli clones was further tested by PCR for the presence of 69 virulence-associated genes (VAGs) to assign InPEC and ExPEC pathotypes. Using PCR-based detection of est-Ia, est-II, eltB, fedA, fasA, stx1, stx2, or eae and bfp, none of the isolates could be assigned to the highly pathogenic edema disease E. coli (EDEC) or the enteropathogenic E. coli (EPEC), while 1.2% of the clones and 0.2% of the isolates were identified as Shiga toxin-producing E. coli (STEC) (detection of stx1 and stx2), 6.5% clones and 1.5% isolates as atypical EPEC (aEPEC) (eae-positive and bfp-negative), and 4.8% clones and 1.5% isolates as enterotoxigenic E. coli (ETEC) (detection of est-II, est-Ia, eltB, fedA, or fasA). Isolates with VAGs described as associated with ExPEC pathotypes were more frequently detected. However, compared to findings of previous studies, the number of intestinal pathogenic E. coli identified in piglets in this study was low (20, 21).
In general, we detected only a limited number of VAGs in all isolates from the feeding groups, with a tendency toward lower numbers in isolates from the probiotic group. However, a more detailed analysis revealed differences in the appearance of six VAGs between all isolates of the probiotic group on the one hand and all isolates from the control group on the other hand: hlyF (P = 0.011), focG (P = 0.015), papC (P = 0.008), papGIII (P = 0.028), iroN (P = 0.04), and cvaC (P = 0.002) were significantly less frequent in the probiotic group (Table 2). These genes represent four different categories of VAGs (toxin, adhesion, iron acquisition, and bacteriocin), all of which are typical for ExPEC (33, 34). Some of these genes, like iroN, tsh, or colV, do not appear independently from each other since they are located on plasmids (15). Dividing the isolates of the two feeding groups according to the habitat they had been isolated from, namely, mucosa, digesta, and feces, a more distinct picture arises. Concentrating on the isolates from fecal samples, no significant differences appeared. In contrast, E. coli isolated from digesta showed significantly different distributions of the six VAGs mentioned above. The isolates from mucosa showed nearly equal significant associations, with only one exception: papGIII (P = 0.138) was not significantly different in occurrence (not shown), but another gene, sitA (P = 0.045), was reduced in these specific isolates (Table 2).
Table 2.
VAGs that are differently distributed between control and probiotic groupsa
| Origin | Gene (function/location) | % positive samples for group |
P valueb | |
|---|---|---|---|---|
| Probiotic | Control | |||
| All | hlyF (toxin) | 14.1 | 23.3 | 0.011 |
| focG (adhesin) | 1.9 | 6.8 | 0.015 | |
| papC (adhesin) | 0.4 | 4.1 | 0.008 | |
| papGIII (adhesin) | 0.3 | 2.8 | 0.028 | |
| iroN (iron acquisition/plasmid) | 23.4 | 33.3 | 0.040 | |
| cvaC (bacteriocin/plasmid) | 12.9 | 23.9 | 0.002 | |
| Mucosa | hlyF (toxin) | 15.0 | 23.6 | 0.046 |
| focG (adhesin) | 0.4 | 7.0 | 0.002 | |
| papC (adhesin) | 0.0 | 4.3 | 0.009 | |
| sitA (iron acquisition) | 42.7 | 57.4 | 0.045 | |
| cvaC (bacteriocin/plasmid) | 13.8 | 23.6 | 0.041 | |
| Digesta | hlyF (toxin) | 14.1 | 25.5 | 0.012 |
| focG (adhesin) | 2.7 | 8.6 | 0.037 | |
| papC (adhesin) | 0.4 | 3.5 | 0.027 | |
| papGIII (adhesin) | 0.4 | 2.7 | 0.023 | |
| iroN (iron acquisition/plasmid) | 22.9 | 34.5 | 0.011 | |
| cvaC (bacteriocin/plasmid) | 12.6 | 25.9 | 0.001 | |
Calculations based on all isolates of each group for VAGs that are differently distributed between mucosa and digesta from control and probiotic groups.
A total of 69 VAGs were screened; here, only significant results are listed (permutation test).
Further analyses were performed to detect possible differences between isolates in association with the age of the piglets (before, at the time of, and after weaning). Here, the total number of E. coli isolates was higher in weaned piglets, and they harbored more toxin and adhesin genes, with the toxin genes astA (P = 0.005) and est-II (P = 0.028) and adhesion genes mat (P = 0.040) and traT (P = 0.000) being significantly increased after weaning (see Table S1 in the supplemental material).
E. coli isolates unique to the probiotic group versus those unique to the control group versus the shared group.
In summary, we detected only minor differences when comparatively analyzing those E. coli isolated from the control group versus those isolated from the probiotic feeding group. To gain further insight into possible group specificities of the identified E. coli clones, we divided the 168 clones into three groups depending on their occurrence: those restricted to piglets from the probiotic (probiotic only; n = 62) or the control group (control only; n = 63) and those shared by both groups (shared; n = 43) (Fig. 1). The distribution of STs was as follows: n = 20, probiotic only; n = 29, control only; n = 30, shared group. Thus, despite this new assignment, the clones and STs were equally distributed between the three groups (Fig. 2).
Virulence-associated gene determination.
Based on assigning the clones into these three groups (probiotic only, control only, and shared), the occurrences of the 69 VAGs revealed only marginal differences. As outlined in Table S2 in the supplemental material, only seven of these VAGs, mainly genes coding for adhesins, showed significant differences. However, when looking at the distribution of the E. coli VAGs in association with the ecological-habitat origin of the samples, namely, mucosa, digesta, or feces, clear differences were found. A total of 11 VAGs (tsh [P = 0.017], mat [P = 0.001], focG [P = 0.002], papC [P = 0.037], colV ′[P = 0.048], ompT [P = 0.003], cvaC [P = 0.004], iroN [P = 0.000], etsB [P = 0.003], etsC [P = 0.003], and hlyF [P = 0.001]) were significantly reduced in clones adhering to the mucosa from the probiotic or shared groups compared to results for the control group (Table 3). As mentioned above, some of the named genes are located on plasmids and are nonrandomly distributed. Nevertheless, ExPEC-typical VAGs for adhesins, serum resistance, and iron acquisition appeared to be affected, indicating a reduction in the frequency of E. coli bacteria similar to ExPEC in the E. faecium-supplemented group of piglets.
Table 3.
Association between mucosa-associated E. coli isolates from the probiotic, control, or shared group and the occurrence of VAGsa
| Gene(s) (function/location) | Mucosa-associated E. coli isolated from: |
P valuee | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Probiotic-only groupb |
Control-only groupc |
Shared groupd |
||||||||
| No. neg. | No. pos. | % pos. | No. neg. | No. pos. | % pos. | No. neg. | No. pos. | % pos. | ||
| tsh (adhesin/plasmid) | 84 | 21 | 20.0 | 104 | 38 | 26.8 | 253 | 4 | 1.6 | 0.017 |
| mat (adhesin) | 22 | 83 | 79.0 | 6 | 136 | 95.8 | 102 | 155 | 60.3 | 0.001 |
| focG (adhesin) | 104 | 1 | 1.0 | 124 | 18 | 12.7 | 257 | 0 | 0.0 | 0.002 |
| papC (adhesin) | 105 | 0 | 0.0 | 131 | 11 | 7.7 | 257 | 0 | 0.0 | 0.037 |
| ColV genes: cvi, cva (plasmid) | 89 | 16 | 15.2 | 103 | 39 | 27.5 | 233 | 24 | 9.3 | 0.048 |
| ompT (outer membrane protein) | 80 | 25 | 23.8 | 91 | 51 | 35.9 | 233 | 24 | 9.3 | 0.003 |
| cvaC (bacteriocin/plasmid) | 92 | 13 | 12.4 | 90 | 52 | 36.6 | 227 | 30 | 11.7 | 0.004 |
| iroN (iron acquisition/plasmid) | 73 | 32 | 30.5 | 59 | 83 | 58.5 | 220 | 37 | 14.4 | 0.000 |
| etsB (ABC transporter) | 72 | 33 | 31.4 | 89 | 53 | 37.3 | 226 | 31 | 12.1 | 0.003 |
| etsC (ABC transporter) | 72 | 33 | 31.4 | 89 | 53 | 37.3 | 226 | 31 | 12.1 | 0.003 |
| hlyF (toxin) | 87 | 18 | 17.1 | 86 | 56 | 39.4 | 233 | 24 | 9.3 | 0.001 |
neg., negative; pos., positive.
Probiotic only; n = 105.
Control only; n = 142.
Shared group; n = 257.
A total of 69 VAGs were screened; here, only significant results are listed (permutation test).
DISCUSSION
The probiotic E. faecium NCIMB 10415 is widely used as a feed supplement for different animals and as a pharmaceutical in humans. Previous studies have reported favorable effects on both health and shedding of pathogenic microorganisms (8, 10, 11, 42–45). We had previously observed a reduction of E. coli serotype O141 during supplementation of E. faecium NCIMB 10415 in piglets (10). However, serotyping is not sufficient for defining the pathogenic potential or the phylogenetic background of the respective E. coli bacteria. We therefore investigated the influence of E. faecium supplementation on the porcine intestinal E. coli populations by both clonal and phylogenetic analysis of 1,436 E. coli isolates and PCR-based typing of virulence-associated genes (VAGs) typical for intestinal pathogenic E. coli (InPEC) and extraintestinal pathogenic E. coli (ExPEC). Our analysis focused on three main approaches, namely, comparison of the following E. coli populations: (i) those originating with the control group versus those from the probiotic group, (ii) those originating with feces versus digesta versus mucosa, and (iii) those present either in the control or the probiotic group or shared by both groups.
Only by this extensive clonal analysis of each of the 1,436 isolates were we able to show that E. faecium feeding is associated with reduced appearance of pathogenic E. coli bacteria, in particular those with features of ExPEC, on the mucosa and the digesta of the colon ascendens.
Macrorestriction analysis revealed an overall high clonal diversity of E. coli in the 24 tested piglets, confirming previous observations (9, 16, 46). Nevertheless, our results indicated that the supplementation of E. faecium NCIMB 10415 to healthy piglets did not influence the overall intestinal E. coli diversity, corroborating previous data (9, 10). Likewise, a detailed breakdown of the E. coli clones into three different groups (control only, probiotic only, and shared), did not indicate any significant changes in diversity.
Determination of 69 VAGs revealed an overall low number of VAGs regardless of the feeding regimen. The low occurrence of pathogenic E. coli harboring VAGs correlates with a low number of isolates belonging to ECOR B2 and D, two phylotypes which are known to harbor virulent E. coli types (15, 32, 41, 47). We consider the low number of VAGs identified to be at least partly due to the optimized hygienic conditions under which the animals were housed. Genes associated with InPEC were also very rarely detected. Nevertheless, ExPEC-typical genes were present in the E. coli isolates, and differences were evident in our three main approaches. Concerning the two feeding groups, significantly lower numbers of six different VAGs (Table 2) were identified in the probiotic feeding group. Focusing on the habitats (feces versus digesta versus mucosa), significant changes were detected for the genes mentioned above in the digesta. Isolates originating from the mucosa differed by one gene. Most of the clones with ExPEC-typical VAGs appeared in the mucosa as well as the digesta. We therefore speculate that there is a dynamic exchange between the adherent and nonadherent E. coli in the colon ascendens. This could be influenced by E. faecium NCIMB 10415.
Applying a third analytical approach of dividing the clones into the three groups (probiotic only, control only, and shared), we found the same and even more significant reductions only of VAGs for mucosa-associated isolates. The occurrence of 11 genes was reduced in both the probiotic and shared groups; in particular, genes for adhesion, serum resistance, and iron acquisition were significantly decreased (Table 3). This finding was perhaps not unexpected, since E. coli bacteria adhering to the mucosa should have adhesive properties. The reduction in genes associated with extraintestinal virulence suggests that feeding of E. faecium can reduce the number of E. coli bacteria harboring ExPEC-typical genes in the intestine, especially those adhering to the mucosa of the colon ascendens.
Although some of the genes examined are linked to each other via their plasmid-borne nature, most of these genes are not perfectly linked, resulting in different P values from those which are linked due to their colocalization on plasmids.
Several hypotheses regarding how the probiotic E. faecium NCIMB 10415 may influence the bacterial colonization of piglets have been suggested. One hypothesis is based on the assumption that enterococci can inhibit strains in the gut by production of organic acids and by lowering the pH (44). This explanation would appear to be unlikely, since we did not observe an overall reduction of ExPEC features; rather, the decrease of genes associated with extraintestinal virulence was restricted to the mucosa and digesta only. Were this explanation correct, we should have observed a reduction of E. coli with ExPEC-typical VAGs in the feces as well. One might hypothesize that a pH effect of the probiotic has a greater influence in the colon ascendens, an effect which decreases during transit through the intestine. Due to the short generation time of E. coli, the ExPEC-typical subpopulation in the feces may already have recovered, masking an effect of the probiotic strain on ExPEC in the feces.
Another explanation for the probiotic effect is the inhibition of attachment by the pathogen to the mucosa of the gastrointestinal tract (48). This has been described for members of the order Lactobacillales, including the lactobacilli (49, 50) and enterococci (51, 52). According to this hypothesis, probiotic bacteria compete with pathogens for binding sites on the host cell surfaces or bind to the pathogen itself, blocking adhesive surface structures or preventing binding of the pathogen to binding sites by sterical interference (51, 53). The competition leads to a reduction in the adhesive properties of the pathogen and thus to its decreased virulence. This assumption could explain our results, since a higher adhesion rate of E. faecium than of E. coli similar to ExPEC to gut epithelium or binding of E. faecium to E. coli would lower the number of E. coli bacteria with ExPEC-typical genes adhering to the mucosa. In a previous study, gnotobiotic pigs were infected with pathogenic E. coli. Pigs supplemented with E. faecium NCIMB 10415 showed a reduction in diarrhea caused by E. coli and a greater increase in body mass (43).
The finding that ExPEC-typical genes were reduced in piglets supplemented with E. faecium was possible only due to the fact that we sampled the mucosa and digesta of the piglets and did not limit the sampling to feces. Nevertheless, most studies focus on the collection and analysis of fecal samples (17, 30, 46, 54) or the intestinal content (46, 55). The practice of examining feces is common because it presents a rapid and easy method for conducting such studies, avoiding euthanasia of the tested animals. However, our data question the relevance of fecal sampling only, since our results clearly show that fecal samples do not appear to include the full population of E. coli. Since important subpopulations might not be sampled with this method, changes in their genotypic or phenotypic appearance would not be detected.
Weaning presents a stress factor for piglets and is known to influence the intestinal microbiota (8, 12, 17, 30, 56). Therefore, we expected higher numbers of pathogenic E. coli bacteria in the intestine of the weaned animals. A possible explanation for the observation of low pathogen numbers in the animals in this study might be effective stall management, resulting in improved health status. Beginning with weaning until the end of the study, the piglets were kept in pairs per pen. This situation is not comparable to the typical livestock breeding conditions. Whether the effect of the probiotic would be stronger or weaker when supplemented under typical farming conditions should be tested in further studies. Since E. coli harboring higher numbers of ExPEC-typical VAGs might have been expected, one might hypothesize that an effect on the mucosa-associated isolates would be stronger.
In conclusion, E. faecium feed supplementation caused no evident changes in the overall diversity of E. coli in healthy weaned piglets, corroborating previous data (9, 22, 57–59). However, more detailed analyses showed a reduction in isolates harboring ExPEC-typical virulence-associated factors, particularly for isolates adherent to the mucosa of the colon ascendens. Since ExPEC bacteria are known to have a significant colonization advantage (21), our results suggest a prophylactic effect of E. faecium NCIMB 10415 against potential pathogenic E. coli at the intestinal epithelial mucosa.
Supplementary Material
ACKNOWLEDGMENTS
The study was supported by the German Research Foundation (DFG) through grant SFB852/1. Torsten Semmler and Bianca Kinnemann were supported by the Federal Ministry of Education (BMBF) and Research Network Zoonosis (FBI-Zoo, grant 01KI1012A).
Footnotes
Published ahead of print 11 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03138-13.
REFERENCES
- 1.Gaskins HR, Collier CT, Anderson DB. 2002. Antibiotics as growth promotants: mode of action. Anim. Biotechnol. 13:29–42 [DOI] [PubMed] [Google Scholar]
- 2.Dibner JJ, Richards JD. 2005. Antibiotic growth promoters in agriculture: history and mode of action. Poult. Sci. 84:634–643 [DOI] [PubMed] [Google Scholar]
- 3.Casewell M, Friis C, Marco E, McMullin P, Phillips I. 2003. The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J. Antimicrob. Chemother. 52:159–161 [DOI] [PubMed] [Google Scholar]
- 4.Casey PG, Gardiner GE, Casey G, Bradshaw B, Lawlor PG, Lynch PB, Leonard FC, Stanton C, Ross RP, Fitzgerald GF, Hill C. 2007. A five-strain probiotic combination reduces pathogen shedding and alleviates disease signs in pigs challenged with Salmonella enterica serovar Typhimurium. Appl. Environ. Microbiol. 73:1858–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taras D, Vahjen W, Simon O. 2007. Probiotics in pigs—modulation of their intestinal distribution and of their impact on health and performance. Livestock Sci. 108:229–231 [Google Scholar]
- 6.Konstantinov SR, Smidt H, Akkermans AD, Casini L, Trevisi P, Mazzoni M, De Filippi S, Bosi P, de Vos WM. 2008. Feeding of Lactobacillus sobrius reduces Escherichia coli F4 levels in the gut and promotes growth of infected piglets. FEMS Microbiol. Ecol. 66:599–607 [DOI] [PubMed] [Google Scholar]
- 7. Reference deleted.
- 8.Zeyner A, Boldt E. 2006. Effects of a probiotic Enterococcus faecium strain supplemented from birth to weaning on diarrhoea patterns and performance of piglets. J. Anim. Physiol. Anim. Nutr. 90:25–31 [DOI] [PubMed] [Google Scholar]
- 9.Schierack P, Walk N, Reiter K, Weyrauch KD, Wieler LH. 2007. Composition of intestinal Enterobacteriaceae populations of healthy domestic pigs. Microbiology 153:3830–3837 [DOI] [PubMed] [Google Scholar]
- 10.Scharek L, Guth J, Reiter K, Weyrauch KD, Taras D, Schwerk P, Schierack P, Schmidt MF, Wieler LH, Tedin K. 2005. Influence of a probiotic Enterococcus faecium strain on development of the immune system of sows and piglets. Vet. Immunol. Immunopathol. 105:151–161 [DOI] [PubMed] [Google Scholar]
- 11.Pollmann M, Nordhoff M, Pospischil A, Tedin K, Wieler LH. 2005. Effects of a probiotic strain of Enterococcus faecium on the rate of natural chlamydia infection in swine. Infect. Immun. 73:4346–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon DM, Cowling A. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575–3586 [DOI] [PubMed] [Google Scholar]
- 13.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 14.Russo TA, Johnson JR. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753–1754 [DOI] [PubMed] [Google Scholar]
- 15.Ewers C, Li G, Wilking H, Kiessling S, Alt K, Antao EM, Laturnus C, Diehl I, Glodde S, Homeier T, Bohnke U, Steinruck H, Philipp HC, Wieler LH. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297:163–176 [DOI] [PubMed] [Google Scholar]
- 16.Katouli M, Lund A, Wallgren P, Kuhn I, Soderlind O, Mollby R. 1995. Phenotypic characterization of intestinal Escherichia coli of pigs during suckling, postweaning, and fattening periods. Appl. Environ. Microbiol. 61:778–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventre A, Elion J, Picard B, Denamur E. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147:1671–1676 [DOI] [PubMed] [Google Scholar]
- 18.Franklin MA, Mathew AG, Vickers JR, Clift RA. 2002. Characterization of microbial populations and volatile fatty acid concentrations in the jejunum, ileum, and cecum of pigs weaned at 17 vs 24 days of age. J. Anim. Sci. 80:2904–2910 [DOI] [PubMed] [Google Scholar]
- 19.Wu XY, Chapman T, Trott DJ, Bettelheim K, Do TN, Driesen S, Walker MJ, Chin J. 2007. Comparative analysis of virulence genes, genetic diversity, and phylogeny of commensal and enterotoxigenic Escherichia coli isolates from weaned pigs. Appl. Environ. Microbiol. 73:83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schierack P, Steinruck H, Kleta S, Vahjen W. 2006. Virulence factor gene profiles of Escherichia coli isolates from clinically healthy pigs. Appl. Environ. Microbiol. 72:6680–6686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schierack P, Walk N, Ewers C, Wilking H, Steinruck H, Filter M, Wieler LH. 2008. ExPEC-typical virulence-associated genes correlate with successful colonization by intestinal E. coli in a small piglet group. Environ. Microbiol. 10:1742–1751 [DOI] [PubMed] [Google Scholar]
- 22.Martin L, Pieper R, Kroger S, Boroojeni FG, Vahjen W, Neumann K, Van Kessel AG, Zentek J. 2012. Influence of age and Enterococcus faecium NCIMB 10415 on development of small intestinal digestive physiology in piglets. Anim. Feed Sci. Technol. 175:65–75 [Google Scholar]
- 23.Schierack P, Roemer A, Jores J, Kaspar H, Guenther S, Filter M, Eichberg J, Wieler LH. 2009. Isolation and characterization of intestinal Escherichia coli clones from wild boars in Germany. Appl. Environ. Microbiol. 75:695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bednorz C, Oelgeschlager K, Kinnemann B, Hartmann S, Neumann K, Pieper R, Bethe A, Semmler T, Tedin K, Schierack P, Wieler LH, Guenther S. 2013. The broader context of antibiotic resistance: zinc feed supplementation of piglets increases the proportion of multi-resistant Escherichia coli in vivo. Int. J. Med. Microbiol. 303:396–403 [DOI] [PubMed] [Google Scholar]
- 25.Merlino J, Siarakas S, Robertson GJ, Funnell GR, Gottlieb T, Bradbury R. 1996. Evaluation of CHROMagar Orientation for differentiation and presumptive identification of gram-negative bacilli and Enterococcus species. J. Clin. Microbiol. 34:1788–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guenther S, Bethe A, Fruth A, Semmler T, Ulrich RG, Wieler LH, Ewers C. 2012. Frequent combination of antimicrobial multiresistance and extraintestinal pathogenicity in Escherichia coli isolates from urban rats (Rattus norvegicus) in Berlin, Germany. PLoS One 7:e50331. 10.1371/journal.pone.0050331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartley CL, Neumann CS, Richmond MH. 1979. Adhesion of commensal bacteria to the large intestine wall in humans. Infect. Immun. 23:128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M, Lochs H. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44–54 [DOI] [PubMed] [Google Scholar]
- 29.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katouli M, Melin L, Jensen-Waern M, Wallgren P, Mollby R. 1999. The effect of zinc oxide supplementation on the stability of the intestinal flora with special reference to composition of coliforms in weaned pigs. J. Appl. Microbiol. 87:564–573 [DOI] [PubMed] [Google Scholar]
- 31.Schlager TA, Hendley JO, Bell AL, Whittam TS. 2002. Clonal diversity of Escherichia coli colonizing stools and urinary tracts of young girls. Infect. Immun. 70:1225–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ewers C, Antao EM, Diehl I, Philipp HC, Wieler LH. 2009. Intestine and environment of the chicken as reservoirs for extraintestinal pathogenic Escherichia coli strains with zoonotic potential. Appl. Environ. Microbiol. 75:184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roemer A, Wieler LH, Schierack P. 2012. Analyses of intestinal commensal Escherichia coli strains from wild boars suggest adaptation to conventional pig production conditions. Vet. Microbiol. 161:122–129 [DOI] [PubMed] [Google Scholar]
- 35.Moon HW, Hoffman LJ, Cornick NA, Booher SL, Bosworth BT. 1999. Prevalences of some virulence genes among Escherichia coli isolates from swine presented to a diagnostic laboratory in Iowa. J. Vet. Diagn. Invest. 11:557–560 [DOI] [PubMed] [Google Scholar]
- 36.Wieler LH, Semmler T, Eichhorn I, Antao EM, Kinnemann B, Geue L, Karch H, Guenther S, Bethe A. 2011. No evidence of the Shiga toxin-producing E. coli O104:H4 outbreak strain or enteroaggregative E. coli (EAEC) found in cattle faeces in northern Germany, the hotspot of the 2011 HUS outbreak area. Gut Pathog. 3:17. 10.1186/1757-4749-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Lin K. 2012. A phylogenomic analysis of Escherichia coli/Shigella group: implications of genomic features associated with pathogenicity and ecological adaptation. BMC Evol. Biol. 12:174. 10.1186/1471-2148-12-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8:207–217 [DOI] [PubMed] [Google Scholar]
- 41.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, Denamur E, Arlet G. 2008. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J. Antimicrob. Chemother. 61:1024–1028 [DOI] [PubMed] [Google Scholar]
- 42.Taras D, Vahjen W, Macha M, Simon O. 2006. Performance, diarrhea incidence, and occurrence of Escherichia coli virulence genes during long-term administration of a probiotic Enterococcus faecium strain to sows and piglets. J. Anim. Sci. 84:608–617 [DOI] [PubMed] [Google Scholar]
- 43.Underdahl NR, Torres-Medina A, Dosten AR. 1982. Effect of Streptococcus faecium C-68 in control of Escherichia coli-induced diarrhea in gnotobiotic pigs. Am. J. Vet. Res. 43:2227–2232 [PubMed] [Google Scholar]
- 44.Strompfova V, Marcinakova M, Simonova M, Gancarcikova S, Jonecova Z, Scirankova L, Koscova J, Buleca V, Cobanova K, Laukova A. 2006. Enterococcus faecium EK13—an enterocin A-producing strain with probiotic character and its effect in piglets. Anaerobe 12:242–248 [DOI] [PubMed] [Google Scholar]
- 45.Behnsen J, Deriu E, Sassone-Corsi M, Raffatellu M. 2013. Probiotics: properties, examples, and specific applications. Cold Spring Harb. Perspect. Med. 3:a010074. 10.1101/cshperspect.a010074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dixit SM, Gordon DM, Wu XY, Chapman T, Kailasapathy K, Chin JJ. 2004. Diversity analysis of commensal porcine Escherichia coli—associations between genotypes and habitat in the porcine gastrointestinal tract. Microbiology 150:1735–1740 [DOI] [PubMed] [Google Scholar]
- 47.Sun J, Liao X, Li L, Yang Y, Liu B, Wang Y, Zhu H, Zhu L, Tu C, Liu Y. 2011. Emergence of pandemic B2-O25b-ST131 Escherichia coli harbouring blaCTX-M-3, blaCTX-M-27 and qnrS1 genes. J. Med. Microbiol. 60:1711–1712 [DOI] [PubMed] [Google Scholar]
- 48.Candela M, Perna F, Carnevali P, Vitali B, Ciati R, Gionchetti P, Rizzello F, Campieri M, Brigidi P. 2008. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 125:286–292 [DOI] [PubMed] [Google Scholar]
- 49.Siggers RH, Siggers J, Boye M, Thymann T, Molbak L, Leser T, Jensen BB, Sangild PT. 2008. Early administration of probiotics alters bacterial colonization and limits diet-induced gut dysfunction and severity of necrotizing enterocolitis in preterm pigs. J. Nutr. 138:1437–1444 [DOI] [PubMed] [Google Scholar]
- 50.Collado MC, Grzeskowiak L, Salminen S. 2007. Probiotic strains and their combination inhibit in vitro adhesion of pathogens to pig intestinal mucosa. Curr. Microbiol. 55:260–265 [DOI] [PubMed] [Google Scholar]
- 51.Jin LZ, Marquardt RR, Zhao X. 2000. A strain of Enterococcus faecium (18C23) inhibits adhesion of enterotoxigenic Escherichia coli K88 to porcine small intestine mucus. Appl. Environ. Microbiol. 66:4200–4204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ditu LM, Chifiriuc MC, Bezirtzoglou E, Voltsi C, Bleotu C, Pelinescu D, Mihaescu G, Lazar V. 2011. Modulation of virulence and antibiotic susceptibility of enteropathogenic Escherichia coli strains by Enterococcus faecium probiotic strain culture fractions. Anaerobe 17:448–451 [DOI] [PubMed] [Google Scholar]
- 53.Ouwehand AC, Conway PL. 1996. Purification and characterization of a component produced by Lactobacillus fermentum that inhibits the adhesion of K88 expressing Escherichia coli to porcine ileal mucus. J. Appl. Bacteriol. 80:311–318 [DOI] [PubMed] [Google Scholar]
- 54.Pupo GM, Lan R, Reeves PR, Baverstock PR. 2000. Population genetics of Escherichia coli in a natural population of native Australian rats. Environ. Microbiol. 2:594–610 [DOI] [PubMed] [Google Scholar]
- 55.de la Fe Rodriguez PY, Martin LO, Munoz EC, Imberechts H, Butaye P, Goddeeris BM, Cox E. 2013. Several enteropathogens are circulating in suckling and newly weaned piglets suffering from diarrhea in the province of Villa Clara, Cuba. Trop. Anim. Health Prod. 45:435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wieler LH, Ilieff A, Herbst W, Bauer C, Vieler E, Bauerfeind R, Failing K, Klos H, Wengert D, Baljer G, Zahner H. 2001. Prevalence of enteropathogens in suckling and weaned piglets with diarrhoea in southern Germany. J. Vet. Med. B Infect. Dis. Vet. Public Health 48:151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szabo I, Wieler LH, Tedin K, Scharek-Tedin L, Taras D, Hensel A, Appel B, Nockler K. 2009. Influence of a probiotic strain of Enterococcus faecium on Salmonella enterica serovar Typhimurium DT104 infection in a porcine animal infection model. Appl. Environ. Microbiol. 75:2621–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guenther S, Filter M, Tedin K, Szabo I, Wieler LH, Nockler K, Walk N, Schierack P. 2010. Enterobacteriaceae populations during experimental Salmonella infection in pigs. Vet. Microbiol. 142:352–360 [DOI] [PubMed] [Google Scholar]
- 59.Kreuzer S, Janczyk P, Assmus J, Schmidt MF, Brockmann GA, Nockler K. 2012. No beneficial effects evident for Enterococcus faecium NCIMB 10415 in weaned pigs infected with Salmonella enterica serovar Typhimurium DT104. Appl. Environ. Microbiol. 78:4816–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.