Abstract
Cloacal swabs from carcasses of Dutch wild birds obtained in 2010 and 2011 were selectively cultured on media with cefotaxime to screen for the presence of extended-spectrum β-lactamase (ESBL)/AmpC-producing Escherichia coli. Subsequently, all cefotaxime-resistant E. coli isolates were tested by broth microdilution and microarray. The presence of ESBL/AmpC and coexisting plasmid-mediated quinolone resistance (PMQR) genes was confirmed by PCR and sequencing. To determine the size of plasmids and the location of ESBL and PMQR genes, S1 pulsed-field gel electrophoresis (PFGE) was performed on transformants, followed by Southern blot hybridization. The study included 414 cloacal swabs originating from 55 different bird species. Cefotaxime-resistant E. coli isolates were identified in 65 birds (15.7%) from 21 different species. In all, 65 cefotaxime-resistant E. coli ESBL/AmpC genes were detected, mainly comprising variants of blaCTX-M and blaCMY-2. Furthermore, PMQR genes [aac(6′)-lb-cr, qnrB1, and qnrS1] coincided in seven cefotaxime-resistant E. coli isolates. Overall, replicon typing of the ESBL/AmpC-carrying plasmids demonstrated the predominant presence of IncI1 (n = 31) and variants of IncF (n = 18). Our results indicate a wide dissemination of ESBL and AmpC genes in wild birds from The Netherlands, especially among aquatic-associated species (waterfowl, gulls, and waders). The identified genes and plasmids reflect the genes found predominantly in livestock animals as well as in humans.
INTRODUCTION
Contaminated surface water seems to be an important factor in the spread of antibiotic resistance, as bacteria from different origins are able to mix and exchange antibiotic resistance genes (1). Moreover, wild birds associated with aquatic environments are often considered indicators for this environmental pollution (2). In the last decade, antibiotic resistance, especially that conferred by extended-spectrum β-lactamases (ESBLs) and AmpC β-lactamases, has been studied extensively in Enterobacteriaceae from humans and livestock in The Netherlands (3–5). In contrast, information on antibiotic resistance in the environment is scarce. In one study, antibiotic-resistant bacteria, including ESBL-producing Escherichia coli, were reported from Dutch rivers, indicating a wide spread of resistance genes in surface water (6). However, information about ESBL and AmpC genes in wild birds from The Netherlands is lacking.
The Netherlands has a high population density (496 inhabitants per km2 in 2012 [http://statline.cbs.nl/]), together with a high livestock density (7). In addition, during the migration season, hundreds of thousands of birds (e.g., geese, ducks, and waders) forage in Dutch mud flats, wetlands, and meadows (8). During springtime and summer, these meadows are repeatedly fertilized with manure. As a consequence, soil and surface water can be contaminated by overspill. Due to antibiotic usage in animal husbandry, manure has become a reservoir of antibiotic compounds and resistant bacteria and has an unknown impact on the spread of antibiotic resistance genes in the environment (9). In more populated areas, surface water can also be contaminated by human sources such as untreated wastewater. Therefore, wild birds feeding near or in urban areas contaminated with resistant bacteria via surface water, soil, or manure have a risk of acquiring antibiotic-resistant bacteria. Subsequently, birds become vehicles for the spread of resistant bacteria over great geographic distances (10). In this study, we screened for the presence of ESBL/AmpC-producing E. coli obtained from Dutch wild birds. Subsequently, ESBL genes and plasmids were further characterized to reveal their genetic background.
MATERIALS AND METHODS
Carcasses of wild birds were sent to the Central Veterinary Institute (CVI), part of Wageningen UR, for different diagnostic purposes, including screening for avian influenza virus, botulism, or poisoning. From August 2011 until October 2012, cloacal swabs were collected and selectively cultured in Luria-Bertani (LB) broth (Becton, Dickinson, Germany) supplemented with 1 mg/liter cefotaxime (Sigma-Aldrich, Germany). After 16 to 20 h of incubation at 37°C, 1 μl broth was cultured on MacConkey agar plates (Becton, Dickinson, Germany) supplemented with 1 mg/liter cefotaxime and incubated for 16 to 20 h at 37°C. When growth appeared on the selective plate, one typical pink colony was pure cultured and confirmed to be E. coli by testing for tryptophan reduction. In addition, all presumed E. coli isolates were identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectroscopy (Biotyper MS; Bruker Daltonics GmbH, Germany) according to the instructions provided by the manufacturer. E. coli isolates were suspended in buffered peptone with 30% glycerol and stored at −80°C pending analysis. Susceptibility to 13 antimicrobials was tested by broth microdilution with a custom-made panel of dehydrated antibiotics (Sensititre; Trek Diagnostic Systems, United Kingdom) according to ISO standards (11), using epidemiological cutoff values recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (http://mic.eucast.org/Eucast2/) for interpretation. The panel included the following antibiotics: ampicillin, cefotaxime, ceftazidime, ciprofloxacin, chloramphenicol, florfenicol, gentamicin, kanamycin, nalidixic acid, streptomycin, sulfamethoxazole, trimethoprim, and tetracycline. Isolates were screened for carbapenemase production by disk diffusion tests with imipenem, meropenem, and ertapenem according to the EUCAST disk diffusion method (http://www.eucast.org/antimicrobial_susceptibility_testing/disk_diffusion_methodology/), using validated criteria (12). Next, all putative ESBL/AmpC-positive E. coli isolates were tested by microarray analysis (ATR0503; Alere Technologies, The Netherlands) to detect antibiotic resistance genes. The presence of ESBL genes and plasmid-mediated quinolone resistance (PMQR) genes was confirmed by PCR and sequencing using previously described primers (4). Plasmid DNA was purified with a modified miniprep method (13) and transferred into DH10B cells (Gibco Invitrogen, USA) by electroporation. Subsequently, transformants were cultured on LB agar with 1 mg/liter cefotaxime for selection of ESBL-positive transformants or cultured on LB agar with 0.03 mg/liter ciprofloxacin for selection of PMQR-positive transformants. In cases where transformation experiments were not successful, conjugation experiments with rifampin-resistant E. coli K-12 were performed by broth mating. Transconjugants were selected on LB agar supplemented with 1 mg/liter cefotaxime plus 100 mg/liter rifampin (Sigma-Aldrich) or selected on LB agar supplemented with 0.03 mg/liter ciprofloxacin plus 100 mg/liter rifampin. To identify the ESBL- or PMQR-carrying plasmids, PCR-based replicon typing (PBRT) (14) was performed by using a commercial kit (Diatheva, Italy). The size of nontypeable plasmids was estimated by S1 pulsed-field gel electrophoresis (PFGE). In cases where transformation and conjugation experiments were not successful, the location of the beta-lactamase gene was determined by PFGE of S1 and I-CeuI digests of total bacterial DNA followed by Southern blot hybridization. To determine the statistical difference in the isolation rates of ESBL/AmpC-positive E. coli between groups of birds, a Fisher exact test was used.
RESULTS
Origin of samples, bird species, and ESBL isolation rates.
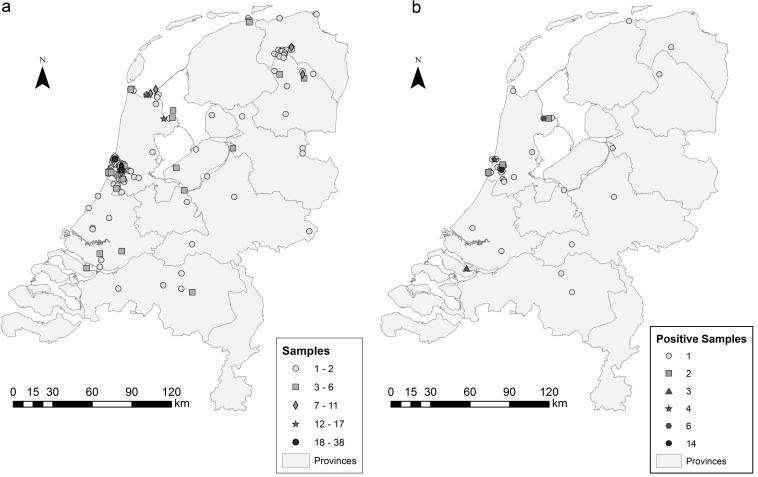
The study included 414 birds that belonged to 55 different species. During the study period, bird carcasses were sent in for screening for nonpathogenic avian influenza virus (84.1%), for diagnoses of botulism (8.2%), or for cases where an unnatural death was suspected (7.7%). Cloacal samples were predominately (90.1%) collected from birds associated with an aquatic environment (Table 1), which included gulls (n = 150), ducks (n = 67), sandpipers (n = 35), swans (n = 26), geese (n = 22), and rails (n = 15). Furthermore, samples were collected from grey heron (Ardea cinerea; n = 11), great cormorant (Phalacrocorax carbo; n = 7), common tern (Sterna hirundo; n = 4), great crested grebe (Podiceps cristatus; n = 2), ruff (Pilomachus pugnax; n = 5), northern lapwing (Vanellus vanellus; n = 2), and Eurasian oystercatcher (Haematopus ostralegus; n = 2). Also, typical seabirds were sampled, including auk (n = 19), northern gannet (Morus bassanus; n = 3), and northern fulmar (Fulmarus glacialis; n = 3). Birds not associated with an aquatic environment (Table 2) represented a relative small proportion (9.9%) in this study and were sent in mainly to examine their cause of death. This third category involved a large number of birds of prey (n = 24), including 8 species: common buzzard (Buteo buteo; n = 14), northern goshawk (Accipiter gentilis; n = 2), Eurasian sparrowhawk (Accipiter nisus; n = 1), western marsh harrier (Circus aeruginosus; n = 1), common kestrel (Falco tinnunculus; n = 1), barn owl (Tyto alba; n = 3), tawny owl (Strix aluco; n = 1), and Eurasian eagle-owl (Bubo bubo; n = 1). The latter group also involved feral pigeons (Columbia livia domesticus; n = 4), common wood pigeon (Columba palumbus; n = 1), stock dove (Columba oenas; n = 1), Eurasian woodcock (Scolopax rusticola; n = 8), common starling (Sturnus vulgaris; n = 1), and black crow (Corvus corone; n = 1). Although carcasses were collected from all over The Netherlands, most birds were sent in from the northwest part of the country (Fig. 1a). The majority of the birds were wildlife, but a small proportion (3.4%) were free-flying domesticated birds, including ducks (Anas platyrhynchos domesticus; n = 6), geese (Anser anser domesticus; n = 3), feral pigeons (Columbia livia domesticus; n = 4), and one captive black swan (Cygnus atratus).
Table 1.
Isolation rates of ESBL-positive E. coli in aquatic-associated birds from The Netherlands
| Bird (family or species) | No. of samples | No. of ESBL+ samples | ESBL ratio |
|---|---|---|---|
| Auk (Alcidae) | 19 | 3 | 0.16 |
| Duck (Anatidae) | 67 | 13 | 0.19 |
| Common tern (Sterna hirundo) | 4 | 0 | 0 |
| Eurasian oystercatcher (Hematopus ostralegus) | 2 | 0 | 0 |
| Goose (Anatidae/Anserini) | 22 | 3 | 0.14 |
| Great cormorant (Phalacrocorax carbo) | 7 | 0 | 0 |
| Great crested grebe (Podiceps cristatus) | 2 | 0 | 0 |
| Grey heron (Ardea cinerea) | 11 | 2 | 0.18 |
| Gull (Laridae) | 150 | 29 | 0.19 |
| Northern fulmar (Fulmarus glacialis) | 3 | 0 | 0 |
| Northern gannet (Morus bassanus) | 3 | 2 | 0.67 |
| Northern lapwing (Vanellus vanellus) | 2 | 1 | 0.50 |
| Rail (Rallidae) | 15 | 0 | 0 |
| Ruff (Philomachus pugnax) | 5 | 1 | 0.20 |
| Swan (Anatidae/Cygnini) | 26 | 5 | 0.19 |
| Sandpiper (Scolopacidae) | 35 | 5 | 0.14 |
| Total | 373 | 64 | 0.17 |
Table 2.
Isolation rates of ESBL-positive E. coli in non-aquatic-associated birds from The Netherlands
| Bird (species) | No. of samples | No. of ESBL+ samples | ESBL ratio |
|---|---|---|---|
| Birds of prey | 24 | 0 | 0 |
| Pigeons | 7 | 1 | 0.14 |
| Eurasian woodcock (Scolopax rusticola) | 8 | 0 | 0 |
| Black crow (Corvus corone) | 1 | 0 | 0 |
| Common starling (Sturnus vulgaris) | 1 | 0 | 0 |
| Total | 41 | 1 | 0.02 |
Fig 1.
(a) Location of bird carcasses collected in The Netherlands. (b) Location of bird carcasses with ESBL/AmpC-producing E. coli.
Cefotaxime-resistant E. coli isolates were identified in 65 samples (15.7%) from 21 different bird species from most areas included in the study (Fig. 1b). Except for one feral pigeon (Columbia livia domesticus), all ESBL/AmpC-positive birds were associated with a freshwater or saltwater environment. Moreover, the isolation rate of ESBL/AmpC-positive E. coli in aquatic-associated birds was significantly higher (P = 0.030) than that in non-aquatic-associated birds. The aquatic-associated birds comprised 20 different bird species, predominately gulls (n = 29) and ducks (n = 13). These gulls involved 5 different species: European herring gull (Larus argentatus; n = 14), black-headed gull (Chroicocephalus ridibundus; n = 6), lesser black-backed gull (Larus fuscus; n = 6), great black-backed gull (Larus marinus; n = 2), and common gull (Larus canus; n = 1). The ducks also included 5 species, predominately mallards (Anas platyrhynchos; n = 9) but also one common goldeneye (Bucephala clangula), one gadwall (Anas strepera), one tufted duck (Aythya fuligula), and one domesticated duck (Anas platyrhynchos domesticus). Furthermore, there were five swans, comprising mute swan (Cygnus olor; n = 4) and black swan (Cygnus atratus; n = 1), and three geese, comprising Egyptian goose (Alopochen aegyptiaca; n = 2) and barnacle goose (Branta leucopsis; n = 1). Other ESBL/AmpC-positive species were common redshank (Tringa totanus; n = 5), common guillemot (Uria aalge; n = 3), grey heron (Ardea cinerea; n = 2), northern gannet (Morus bassanus; n = 2), northern lapwing (Vanellus vanellus; n = 1), and ruff (Philomachus pugnax; n = 1).
Non-wild-type susceptibility and resistance genes of ESBL-producing E. coli.
All 65 isolates were resistant to cefotaxime and ampicillin (see Table S1 in the supplemental material), and almost all isolates were resistant to ceftazidime (96.9%). Moreover, a high proportion (70.8%) of the isolates were non-wild-type susceptible to three or more antibiotic classes, resulting in high levels of non-wild-type susceptibility to most antibiotics tested, including sulfamethoxazole (66.2%), tetracycline (61.5%), trimethoprim (58.5%), streptomycin (56.9%), ciprofloxacin (47.7%), nalidixic acid (44.6%), kanamycin (38.5%), chloramphenicol (33.8%), and gentamicin (21.5%). A relative low rate of the isolates was non-wild-type susceptible for florfenicol (7.7%). Finally, all isolates were susceptible to the carbapenem antibiotics tested (meropenem, imipenem, and ertapenem).
Microarray analysis (see Table S1 in the supplemental material) demonstrated the presence of ESBL (blaCTX-M and blaSHV) or AmpC (blaCMY) genes in all 65 isolates next to narrow-spectrum β-lactamase genes (blaTEM-1 and blaOXA-1). Besides integrons (intI1 and intI2), genes conferring resistance to tetracyclines [tet(A), tet(B), tet(D), and tet(E)], sulfonamides (sul1 and sul2), trimethoprim (dfrA1, dfrA7, dfrA12, dfrA14, and dfrA17), chloramphenicol (catA1 and cmlA1), florfenicol (floR), aminoglycosides [aadA1, aadA2, aadA4, aac(6′)-lb, strA, and strB], and quinolones (qnrB and qnrS) were detected.
ESBL/AmpC and PMQR gene detection and sequencing.
The presence of ESBL or AmpC genes was confirmed for all 65 cefotaxime-resistant E. coli isolates by PCR and sequencing (Table 3). Different variants of blaCTX-M (n = 46) were most abundant: blaCTX-M-1 (n = 14), blaCTX-M-15 (n = 13), blaCTX-M-3 (n = 8), blaCTX-M-14 (n = 8), and blaCTX-M-32 (n = 3). Furthermore, blaCMY-2 (n = 14) was also frequently identified. Also, blaSHV-12 (n = 3) and blaTEM-52c (n = 1) and a concurrence of blaSHV-12 with blaTEM-52c (n = 1) were found.
Table 3.
ESBL and AmpC genes and their location in wild birds from The Netherlands
| ESBL/AmpC gene(s) | Location(s) of ESBL gene(s) | Bird species (no. of isolates) | Total no. of isolates |
|---|---|---|---|
| blaCTX-M-1 | IncI1 | European herring gull (3), mallard (3), black-headed gull (1), Egyptian goose (1), feral pigeon (1), grey heron (1), mute swan (1), northern gannet (1), northern lapwing (1) | 13 |
| blaCTX-M-1 | IncF | Gadwall (1) | 1 |
| blaCTX-M-14 | Chromosomal | Common redshank (1) | 1 |
| blaCTX-M-14 | IncB/O | Common redshank (1) | 1 |
| blaCTX-M-14 | IncF | Lesser black-backed gull (2), mute swan (2), European herring gull (1), tufted duck (1) | 6 |
| blaCTX-M-15 | IncF | Black-headed gull (2), northern gannet (1) | 3 |
| blaCTX-M-15 | IncHI2 | Black-headed gull (1) | 1 |
| blaCTX-M-15 | IncHI2 + IncF | European herring gull (1) | 1 |
| blaCTX-M-15 | IncI1 | Common gull (1), European herring gull (1), lesser black-backed gull (1), mallard (1) | 4 |
| blaCTX-M-15 | IncI2 | Lesser black-backed gull (1) | 1 |
| blaCTX-M-15 | None typeable | Mallard (2), European herring gull (1) | 3 |
| blaCTX-M-3 | Chromosomal | Common goldeneye (1) | 1 |
| blaCTX-M-3 | IncF | Common guillemot (3), Egyptian Goose (1), European herring gull (1), mallard (1) | 6 |
| blaCTX-M-3 | IncI1 | Lesser black-backed gull (1) | 1 |
| blaCTX-M-32 | Chromosomal | Common redshank (1), mallard (1) | 2 |
| blaCTX-M-32 | IncF | Black swan (1) | 1 |
| blaSHV-12 | IncF | Black-headed gull (1) | 1 |
| blaSHV-12 | IncI1 | Grey heron (1) | 1 |
| blaSHV-12 | IncN | Barnacle goose (1) | 1 |
| blaSHV-12, blaTEM-52c | IncI1 | European herring gull (1) | 1 |
| blaTEM-52c | IncI1 | Ruff (1) | 1 |
| blaCMY-2 | Chromosomal | European herring gull (1) | 1 |
| blaCMY-2 | IncB/O | Common redshank (1) | 1 |
| blaCMY-2 | IncI1 | European herring gull (3), great black-backed gull (2), black-headed gull (1), domesticated duck (1), herring gull (1), lesser black-backed gull (1), mute swan (1) | 10 |
| blaCMY-2 | IncK | Mallard (1), common redshank (1) | 2 |
PMQR genes were detected in seven (10.8%) of the ESBL/AmpC-positive isolates. The altered aminoglycoside resistance gene aac(6′)-lb-cr (n = 6) was found exclusively in isolates harboring blaCTX-M-15, blaTEM-1b, and blaOXA-1. In two cases, aac(6′)-lb-cr genes coincided with qnr genes: in one isolate (WF 1.35) from a European herring gull (Larus argentatus) with qnrB1 only and in one isolate (WF 1.56) from a black-headed gull (Chroicocephalus ridibundus) with both qnrB1 and qnrS1. In addition, qnrS1 was detected in one isolate (WF 1.46) from a northern lapwing (Vanellus vanellus) with blaCTX-M-1 without the presence of aac(6′)-lb-cr.
Location of ESBL/AmpC and PMQR genes and typing of plasmids.
From 59 of the 65 isolates, ESBL/AmpC-carrying plasmids were successfully transferred into DH10B cells by electroporation. In five isolates, I-CeuI digestion and Southern blot hybridization revealed the chromosomal location of the ESBL/AmpC genes blaCTX-M-32 (n = 2), blaCTX-M-3 (n = 1), blaCTX-M-14 (n = 1), and blaCMY-2 (n = 1). In addition, in one strain (WF 1.56), a large IncHI2 plasmid (320 kb) was successfully transferred to E. coli K-12 by conjugation, which was confirmed by S1 digestion and microarray analyses. Hence, ESBL/AmpC genes were present on plasmids in 60 out of the 65 isolates studied (92.3%). Remarkably, in one isolate (WF 1.35), additional conjugation experiments revealed the coexistence of IncHI2 next to IncF, both harboring blaCTX-M-15. Overall, replicon typing of the ESBL/AmpC-carrying plasmids demonstrated the predominant presence of IncI1 (n = 31) and variants of IncF (n = 18). Less frequently detected were IncB/O (n = 3), IncHI2 (n = 2), IncK (n = 1), IncN (n = 1), and IncI2 (n = 1) plasmids. A small proportion of plasmids (n = 3) were nontypeable by PBRT.
Remarkably, six isolates harboring blaCTX-M-32 on IncF plasmids from four different bird species showed identical phenotypic profiles. Moreover, microarray analysis identified identical resistance genes in five of the six isolates: blaTEM, sul1, sul2, tet(B), aadA4, catA1, dfrA17, strA, and strB (see Table S1 in the supplemental material). In addition, XbaI PFGE revealed identical restriction patterns of all six isolates (data not shown).
The aac(6′)-lb-cr gene was identified solely on plasmids that harbored blaCTX-M-15, blaTEM-1b, and blaOXA-1. However, these plasmids revealed diverse replicon types: IncHI2 (n = 2), IncF (n = 2), and nontypeable (n = 2) plasmids. Additional array analysis of the transconjugants showed that both IncHI2 plasmids carried qnrB1. Remarkably, one E. coli isolate from a black-headed gull (WF 1.56) possessed qnrS1 on an IncY plasmid next to aac(6′)-lb-cr and qnrB1 on IncHI2. For one isolate from a northern lapwing (WF 1.46), Southern blot experiments were not successful, and as a consequence, the location of qnrS1 remained unidentified.
DISCUSSION
The study population was highly variable and consisted of 414 birds from 55 different species. Therefore, the birds were categorized into two main groups: (i) aquatic-associated birds and (ii) non-aquatic-associated birds (Tables 1 and 2). Despite the relatively large difference in the numbers of birds per group, we determined a significantly higher isolation rate of ESBL/AmpC-positive E. coli in aquatic-associated birds than in non-aquatic-associated birds (Tables 1 and 2). Besides, in the two largest bird families in this study, represented by gulls (n = 150) and ducks (n = 67), the isolation rates were relatively high (19.3% and 19.4%, respectively). Similarly high rates were described previously for gulls from Portugal (15), France (16), the Czech Republic (17), and Sweden (18). However, similar studies of mallard ducks in Poland (19) showed much lower isolation rates. It is notable that 58.5% of birds in the non-aquatic-associated group (Table 2) consisted of birds of prey, which all tested negative for ESBL- or AmpC-producing E. coli. This result is in contrast to results of a recent study from Portugal (21), where 32 out of 119 samples (26.9%) originating from birds of prey tested positive for ESBL-producing E. coli. Although the sample size in our study was small (n = 24), it may indicate a relatively low prevalence of ESBLs in birds of prey in The Netherlands. In conclusion, our results confirm the previously suggested association between the high prevalence of ESBL- or AmpC-producing E. coli from wild birds and their aquatic-associated habitat (2).
To the best of our knowledge, this is the first report of ESBL- and AmpC-producing E. coli in different species of waders and plovers, including common redshank (Tringa totanus), northern lapwing (Vanellus vanellus), and ruff (Philomachus pugnax). These birds feed in natural areas such as wetlands and mud flats during the migration season but often breed in rural areas such as meadows and acres. Consequently, birds migrate between natural and rural areas and vice versa. Hence, birds can be exposed to resistant bacteria originating from livestock animals or humans and potentially spread such bacteria over large distances to other regions.
Although ESBL- or AmpC-producing E. coli isolates have been found in mallard ducks (19), we report ESBL-producing E. coli from three other duck species: tufted duck (Aythya fuligula), gadwall (Anas strepera), and common goldeneye (Bucephala clangula). Moreover, we also identified ESBL-producing E. coli in two typical seabird species: common guillemot (Uria aalge) and northern gannet (Morus bassanus). These birds were collected at the North Sea coast and usually do not enter the rural or urban areas of The Netherlands. These results indicate a wide dissemination of ESBL and AmpC genes among aquatic-associated birds both in freshwater and saltwater environments.
Although our study focused on cefotaxime-resistant E. coli, we identified a remarkably high rate of non-wild-type susceptibility to almost all antibiotic classes tested, including tetracyclines, sulfonamides, trimethoprim, chloramphenicol, florfenicol, aminoglycosides, and quinolones. Moreover, microarray experiments confirmed the presence of resistance genes against these antibiotic groups. These results demonstrate the coexistence of multiple resistance genes in ESBL/AmpC-positive E. coli isolates in wild birds. Considering the fact that wild bird are not exposed to high dosages of antibiotics, these results are worrying. Apparently, E. coli isolates carrying resistance plasmids are able to survive in a relatively large proportion of the birds examined.
The results of plasmid and gene typing (Table 3) demonstrate the diversity of ESBL and AmpC genes in wild birds from The Netherlands. The presence of E. coli harboring blaCTX-M-1 or blaCMY-2 on IncI1 reflects genes and plasmids found predominantly in poultry (3, 22). However, E. coli isolates with blaCTX-M-14 or blaCTX-M-15 on IncF plasmids are frequently reported from human sources (23, 24). In six out of eight isolates (originating from 5 different bird species), blaCTX-M-14 was identified on IncF plasmids. These six isolates exhibited an identical microarray profile, including the presence of blaTEM-1b, aac(6′)-lb, sul1, tet(B), cmlA1, catA1, dfrA17, strA, and strB (see Table S1 in the supplemental material). Moreover, all six isolates revealed a very similar restriction pattern by XbaI PFGE (results not shown) and were considered closely related. Both the array analysis and the PFGE results indicate a clonal spread of multidrug-resistant E. coli harboring blaCTX-M-14 in different bird species. In contrast, blaCTX-M-15 was identified in 13 isolates on different plasmids showing variable microarray profiles. In five isolates, blaCTX-M-15 coincided with blaTEM-1, blaOXA-1, and aac(6′)-lb-cr. One other isolate also harbored blaOXA-1 and aac(6′)-lb-cr but lacked blaTEM-1. The co-incidence of blaCTX-M-15 with blaTEM-1, blaOXA-1, and aac(6′)-lb-cr on IncF plasmids is frequently reported (23). However, we identified this combination of genes on both IncHI2 plasmids (n = 2) and nontypeable plasmids (n = 2) next to IncF (n = 2). These results indicate the dissemination of this complex of genes to other plasmids.
We identified blaCTX-M-3 predominately on IncF plasmids in E. coli. These isolates showed similar resistance phenotypes, and almost all of these isolates revealed identical genotypes. Moreover, indistinguishable XbaI PFGE restriction patterns of all six isolates indicate a common source for these clonally related isolates. This ESBL gene was first reported in clinical isolates of E. coli and Citrobacter freundii from Poland on an IncL/M plasmid (25). Later, blaCTX-M-3 was disseminated to other parts of Poland in several other Enterobacteriaceae species. However, blaCTX-M-3 isolates have also been found on IncF plasmids in clinical E. coli isolates from Croatia and Australia (26).
Among the cefotaxime-resistant isolates, we identified seven isolates with both ESBL genes and PMQR genes, originating from five gulls, one mallard (Anas platyrhynchos), and one northern lapwing (Vanellus vanellus). In almost all cases, the ESBL and PMQR genes were located on the same plasmid, confirming the previously described association between ESBL and PMQR genes (27). However, the aquatic environment is considered to be the original source of qnr genes (20). For this reason, qnr genes could possibly occur at a higher rate in water-related bird species.
In summary, our results demonstrate wild birds as indicators for environmental contamination with antibiotic resistance genes from both humans and livestock animals.
Supplementary Material
ACKNOWLEDGMENTS
We thank Alessandra Carattoli (Istituto Superiore di Sanità, Rome, Italy) for providing positive-control strains for replicon typing of the plasmids. We also thank Gert-Jan Boender for constructing the figures included in this paper.
This work was supported by the Dutch Ministry of Economic Affairs (grant number WOT-01-002-03.02).
Footnotes
Published ahead of print 13 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01880-13.
REFERENCES
- 1.Baquero F, Martinez JL, Canton R. 2008. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19:260–265 [DOI] [PubMed] [Google Scholar]
- 2.Guenther S, Ewers C, Wieler LH. 2011. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front. Microbiol. 2:246. 10.3389/fmicb.2011.00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dierikx C, van der Goot J, Fabri T, van Essen-Zandbergen A, Smith H, Mevius D. 2013. Extended-spectrum-beta-lactamase- and AmpC-beta-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J. Antimicrob. Chemother. 68:60–67 [DOI] [PubMed] [Google Scholar]
- 4.Dierikx CM, van Duijkeren E, Schoormans AH, van Essen-Zandbergen A, Veldman K, Kant A, Huijsdens XW, van der Zwaluw K, Wagenaar JA, Mevius DJ. 2012. Occurrence and characteristics of extended-spectrum-beta-lactamase- and AmpC-producing clinical isolates derived from companion animals and horses. J. Antimicrob. Chemother. 67:1368–1374 [DOI] [PubMed] [Google Scholar]
- 5.Hasman H, Mevius D, Veldman K, Olesen I, Aarestrup FM. 2005. Beta-lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56:115–121 [DOI] [PubMed] [Google Scholar]
- 6.Blaak H. 2011. Prevalence of antibiotic resistant bacteria in the rivers Meuse, Rhine and New Meuse. RIVM report 703719071/2011 RIVM, Bilthoven, The Netherlands [Google Scholar]
- 7.EFSA 2012. Technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in Salmonella, Campylobacter and indicator Escherichia coli and Enterococcus spp. bacteria transmitted through food. EFSA J. 10:2742. 10.2903/j.efsa.2012.2742 [DOI] [Google Scholar]
- 8.Bijlsma RG. 2001. Avifauna of The Netherlands, vol 2 Common and scarce birds of The Netherlands. KNNV Publishing, Zeist, The Netherlands [Google Scholar]
- 9.Heuer H, Schmitt H, Smalla K. 2011. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14:236–243 [DOI] [PubMed] [Google Scholar]
- 10.Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. 2010. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8:251–259 [DOI] [PubMed] [Google Scholar]
- 11.International Organization for Standardization 2006. Clinical laboratory testing and in vitro diagnostic test systems—susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices. Part 1: reference method for testing the in vitro activity of antimicrobial agents against rapidly growing aerobic bacteria involved in infectious diseases (ISO standard 20776-1:2006). International Organization for Standardization, Geneva, Switzerland [Google Scholar]
- 12.Cohen Stuart J, Leverstein-Van Hall MA, Dutch Working Party on the Detection of Highly Resistant Microorganisms 2010. Guideline for phenotypic screening and confirmation of carbapenemases in Enterobacteriaceae. Int. J. Antimicrob. Agents 36:205–210 [DOI] [PubMed] [Google Scholar]
- 13.Hordijk J, Wagenaar JA, Kant A, van Essen-Zandbergen A, Dierikx C, Veldman K, Wit B, Mevius D. 2013. Cross-sectional study on prevalence and molecular characteristics of plasmid mediated ESBL/AmpC-producing Escherichia coli isolated from veal calves at slaughter. PLoS One 8:e65681. 10.1371/journal.pone.0065681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63:219–228 [DOI] [PubMed] [Google Scholar]
- 15.Poeta P, Radhouani H, Igrejas G, Goncalves A, Carvalho C, Rodrigues J, Vinue L, Somalo S, Torres C. 2008. Seagulls of the Berlengas Natural Reserve of Portugal as carriers of fecal Escherichia coli harboring CTX-M and TEM extended-spectrum beta-lactamases. Appl. Environ. Microbiol. 74:7439–7441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonnedahl J, Drobni M, Gauthier-Clerc M, Hernandez J, Granholm S, Kayser Y, Melhus A, Kahlmeter G, Waldenstrom J, Johansson A, Olsen B. 2009. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the South of France. PLoS One 4:e5958. 10.1371/journal.pone.0005958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dolejska M, Bierosova B, Kohoutova L, Literak I, Cizek A. 2009. Antibiotic-resistant Salmonella and Escherichia coli isolates with integrons and extended-spectrum beta-lactamases in surface water and sympatric black-headed gulls. J. Appl. Microbiol. 106:1941–1950 [DOI] [PubMed] [Google Scholar]
- 18.Bonnedahl J, Drobni P, Johansson A, Hernandez J, Melhus A, Stedt J, Olsen B, Drobni M. 2010. Characterization, and comparison, of human clinical and black-headed gull (Larus ridibundus) extended-spectrum beta-lactamase-producing bacterial isolates from Kalmar, on the southeast coast of Sweden. J. Antimicrob. Chemother. 65:1939–1944 [DOI] [PubMed] [Google Scholar]
- 19.Literak I, Dolejska M, Janoszowska D, Hrusakova J, Meissner W, Rzyska H, Bzoma S, Cizek A. 2010. Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum beta-lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Appl. Environ. Microbiol. 76:8126–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poirel L, Cattoir V, Nordmann P. 2012. Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front. Microbiol. 3:24. 10.3389/fmicb.2012.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto L, Radhouani H, Coelho C, Martins da Costa P, Simoes R, Brandao RM, Torres C, Igrejas G, Poeta P. 2010. Genetic detection of extended-spectrum beta-lactamase-containing Escherichia coli isolates from birds of prey from Serra da Estrela Natural Reserve in Portugal. Appl. Environ. Microbiol. 76:4118–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dierikx C, van Essen-Zandbergen A, Veldman K, Smith H, Mevius D. 2010. Increased detection of extended spectrum beta-lactamase producing Salmonella enterica and Escherichia coli isolates from poultry. Vet. Microbiol. 145:273–278 [DOI] [PubMed] [Google Scholar]
- 23.Doumith M, Dhanji H, Ellington MJ, Hawkey P, Woodford N. 2012. Characterization of plasmids encoding extended-spectrum beta-lactamases and their addiction systems circulating among Escherichia coli clinical isolates in the UK. J. Antimicrob. Chemother. 67:878–885 [DOI] [PubMed] [Google Scholar]
- 24.Marcade G, Deschamps C, Boyd A, Gautier V, Picard B, Branger C, Denamur E, Arlet G. 2009. Replicon typing of plasmids in Escherichia coli producing extended-spectrum beta-lactamases. J. Antimicrob. Chemother. 63:67–71 [DOI] [PubMed] [Google Scholar]
- 25.Golebiewski M, Kern-Zdanowicz I, Zienkiewicz M, Adamczyk M, Zylinska J, Baraniak A, Gniadkowski M, Bardowski J, Ceglowski P. 2007. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum beta-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 51:3789–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carattoli A. 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53:2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robicsek A, Jacoby GA, Hooper DC. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6:629–640 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.