Abstract
Broad-host-range self-transferable plasmids are known to facilitate bacterial adaptation by spreading genes between phylogenetically distinct hosts. These plasmids typically have a conserved backbone region and a variable accessory region that encodes host-beneficial traits. We do not know, however, how well plasmids that do not encode accessory functions can survive in nature. The goal of this study was to characterize the backbone and accessory gene content of plasmids that were captured from freshwater sources without selecting for a particular phenotype or cultivating their host. To do this, triparental matings were used such that the only required phenotype was the plasmid's ability to mobilize a nonconjugative plasmid. Based on complete genome sequences of 10 plasmids, only 5 carried identifiable accessory gene regions, and none carried antibiotic resistance genes. The plasmids belong to four known incompatibility groups (IncN, IncP-1, IncU, and IncW) and two potentially new groups. Eight of the plasmids were shown to have a broad host range, being able to transfer into alpha-, beta-, and gammaproteobacteria. Because of the absence of antibiotic resistance genes, we resampled one of the sites and compared the proportion of captured plasmids that conferred antibiotic resistance to their hosts with the proportion of such plasmids captured from the effluent of a local wastewater treatment plant. Few of the captured plasmids from either site encoded antibiotic resistance. A high diversity of plasmids that encode no or unknown accessory functions is thus readily found in freshwater habitats. The question remains how the plasmids persist in these microbial communities.
INTRODUCTION
Mobile genetic elements such as plasmids are thought to form communal gene pools that allow bacterial communities to rapidly adapt to changing environments (1). Plasmids that can transfer and replicate in phylogenetically diverse bacteria are called broad-host-range (BHR) plasmids (2, 3). BHR plasmids of Gram-negative bacteria with the widest host range belong to the incompatibility groups IncP (4), IncW (5), IncU (6), IncQ (7), and the recently defined PromA group (8). These plasmids are typically composed of two distinct regions: (i) the backbone region, which encodes genes responsible for plasmid replication, maintenance and gene regulation, as well as conjugative transfer, and (ii) the accessory region, which consists of genes that code for host-beneficial traits. BHR plasmids are thought to be particularly crucial in adaptation of bacterial communities to changing environments since they shuttle host-beneficial genes across taxonomic boundaries (9). Examples of exchanged traits are resistance to antibiotics or heavy metals and degradation of organic xenobiotics, like chlorinated aromatics.
A central question in plasmid ecology is whether or not plasmids can be maintained in bacterial communities without benefiting their hosts and thus be considered genetic parasites. Since in the absence of positive selection for plasmid-encoded traits plasmid carriage is assumed to impose a cost to the bacterial host, albeit sometimes small, plasmid-bearing bacteria are expected to be out-competed by their plasmid-free counterparts, which can arise spontaneously through incorrect plasmid segregation during cell division (10–14). Stewart and Levin (14) first argued that the conditions for plasmids to persist by infectious transfer alone, much like parasites, were very limited. However, later Lundquist and Levin (13) provided evidence through chemostat studies that naturally occurring plasmids may well be maintained by horizontal transfer due to high conjugation rates. We later showed that some BHR plasmids can invade bacterial populations grown on surfaces in the absence of any known selective pressure (15). The first description of completely sequenced self-transmissible plasmids that are devoid of identifiable accessory genes, e.g., IncP-1 plasmids pA1 (16) and pBP136 (17), supported the hypothesis that a parasitic existence through efficient horizontal transfer is possible for some BHR plasmids. However, while these plasmids are considered “cryptic” because they do not seem to confer any known benefits to their hosts, they still carry small open reading frames (ORFs) with unknown functions and represent a very small sample among the vast diversity of plasmids in various ecosystems. A better overview is needed of the natural occurrence of cryptic BHR plasmids.
The next logical question is in which environments such possibly cryptic plasmids are likely to be found. Most of our knowledge about accessory genes comes from plasmids that were isolated based on specific phenotypes that often match the environmental selection pressures. For example, plasmids isolated from bacteria found in soil contaminated with pesticides and heavy metals encode degradation of, or resistance to, these xenobiotics (18). The same is true for wastewaters known to contain pollutants such as antibiotics and organic and inorganic xenobiotics (19–22). Plasmids carrying genes conferring resistance to all major classes of antibiotics have been found in wastewater treatment plants (WWTP) (23, 24). In contrast, nonpolluted or moderately polluted environments may not impose strong selection for these traits and therefore may sustain bacteria with plasmids devoid of the corresponding genes. Understanding plasmid diversity in these environments is thus crucial to our understanding of plasmid parasitism.
Various methods exist for isolating plasmids from environmental bacteria. In traditional so-called endogenous methods, bacteria are isolated in pure culture, and this is followed by plasmid DNA extraction. Exogenous isolation methods capture plasmids from microbial communities without culturing the plasmid hosts and include biparental and triparental matings (for a review, see reference 25). Biparental matings typically capture plasmids that transfer genes encoding a specific phenotype to a recipient strain, such as mercury or antibiotic resistance. The method involves mixing the indigenous bacterial community with a plasmid-free recipient strain, allowing conjugation to occur, and selecting for recipients that acquired the plasmid-borne trait. Triparental matings include one additional parental population, i.e., a donor strain carrying a nonconjugative, mobilizable plasmid (Fig. 1). Here, plasmids are captured based solely on their ability to transfer this mobilizable plasmid and themselves into the recipient and not for specific accessory phenotypes they encode (26, 27). This method thus provides an assessment of plasmid diversity that is not biased for plasmid-borne traits. Using this method, BHR plasmids of the PromA group were obtained (pIPO2 and pMOL98) whose putative accessory genes currently have unknown functions (8, 28).
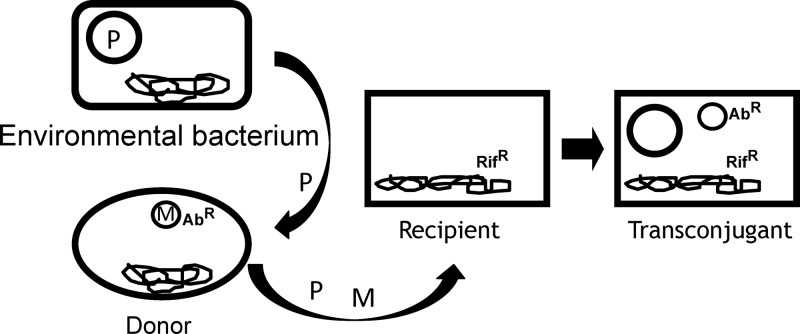
Fig 1.
Illustration of the triparental mating method. Environmental bacteria containing plasmids (P) are mixed with a donor bacterium, which carries a mobilizable plasmid (M) with a specific marker, such as antibiotic resistance (AbR), and a recipient bacterium, which has a second selectable marker, such as rifampin resistance (RifR). When plated on both antibiotics, only recipient strains that carry a mobilizable plasmid can grow, and since only the environmental plasmid (P) can move the mobilizable plasmid into the recipient strain, the environmental plasmid is sometimes present too. One pathway by which transconjugants can be formed is shown by the arrows.
In this study, we isolated and sequenced plasmids from three freshwater sites around Moscow, ID, using the triparental mating method. A diverse set of plasmids was found, and several plasmids appear to carry only genes involved in core plasmid functions, suggesting that they persist in these environments without conferring an accessory phenotype to their hosts. Given the low incidence of plasmids with drug resistance genes, we then compared antibiotic resistance profiles of plasmids newly captured from one of the creeks with those captured from the effluent of the downstream Moscow, ID, WWTP. Although we expected the proportion of resistance plasmids to be higher in the WWTP than in the creek, equally low proportions were found at both sites.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
A list of the strains and plasmids used in this study is provided in Table 1. Luria-Bertani (LB) broth was generally used to culture the strains. Solid medium was prepared by addition of 1.5% agar. Difco R2A agar (Becton, Dickinson, and Co., Franklin Lakes, NJ) was used for the triparental matings. Difco Mueller-Hinton agar (Becton, Dickinson, and Co., Franklin Lakes, NJ) was used for the antibiotics sensitivity test. Escherichia coli strains were incubated at 37°C, while other bacterial species and mating mixtures were incubated at 30°C. When necessary for selection, antibiotics were added to the medium at the following concentrations: chloramphenicol (Cm), 25 μg/ml; tetracycline (Tc), 10 μg/ml; streptomycin (Sm), 50 μg/ml; rifampin (Rif), 100 μg/ml; nalidixic acid (Nal), 20 μg/ml; ampicillin (Ap), 50 μg/ml; zeocin (Zeo), 100 μg/ml; kanamycin (Km), 50 μg/ml; gentamicin (Gm), 10 μg/ml. Mercury chloride was used at 5 μg/ml for selection of pQKH54. Cycloheximide (300 μg/ml) was added to LB agar (LBA) to prevent growth of fungus during screening of transconjugants from mating mixtures containing freshwater samples.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and relevant phenotypea | Reference or source |
|---|---|---|
| Strains (class) | ||
| E. coli (Gammaproteobacteria) | ||
| CV601gfp | Thr− Leu− Thi− chr::mini-Tn5-gfp-aphA; Rifr Kmr | 36 |
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1; Nalr | 35 |
| EC100/DH10B | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 deoR endA1 thi-1 recA1 gyrA96 relA1 hsdR17 supE44 Δ(ara-leu)7697 galU galK rpsL nupG; Smr | Epicentre, Madison, WI |
| HY841 | EC100 attTn7::mini-Tn7-zeo-PA1/04/03-eyfp; Smr Zeor | This study |
| HY842 | Rifr mutant of HY841; Smr Zeor Rifr | This study |
| JM109 | endA1 recA1 gyrA96 thi hsdR17 (rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB laqIqZΔM15] | 82 |
| K12Rif | Rifr mutant of MG1655 | 15 |
| K12Nal | Nalr mutant of MG1655 | 15 |
| Pseudomonas putida (Gammaproteobacteria) | ||
| UCW1gfp | UCW1 chr.::mini-Tn5-gfp-aphA; Rifr Kmr | 36 |
| Cupriavidus pinatubonensisb (Betaproteobacteria) | ||
| JMP228gfp | JMP228 chr.:: mini-Tn5-gfp-aphA; Rifr Kmr | 36 |
| Plasmids | ||
| pBBR1MCS | BHR mobilizable cloning vector; Cmr | 32 |
| pBBR1MCS-5 | BHR mobilizable cloning vector; Gmr | 34 |
| pMS0705 | p15A replicon carrying mini-Tn21OTcc; Cmr Tcr pMB1 oriV | M. Sota |
| pMT1247Tc | p15A replicon carrying mini-Tn21Tc; Cmr Tcr | M. Sota |
| pSU4814 | p15A replicon carrying CloDF13 oriT; Cmr | 30 |
| pQKH54 | IncP-1γ replicon; Hgr | 46 |
| pUC18T-mini-Tn7T-Zeo-eyfp | pUC replicon carrying mini-Tn7T-zeo-PA1/04/03-eyfp; Apr Zeor | 83 |
| pHY835 | R6K replicon carrying mini-Tn7 | 29 |
| pHY956 | pHY835::mini-Tn7T-zeo-PA1/04/03-eyfp; Apr Zeor | This study |
| pTNS2 | R6K replicon, Tn7 transposase expression vector | 84 |
Ap, ampicillin; Cm, chloramphenicol; Gm, gentamicin; Km, kanamycin; Nal, nalidixic acid; Rif, rifampin; Sm, streptomycin; Tc, tetracycline; Zeo, zeocin; Hgr, mercury resistance.
Previously named Cupriavidus necator.
Mini-Tn21OTc carries oriV from pBR322 (85).
A restriction system-free E. coli strain that expresses the enhanced yellow fluorescent protein (EYFP) was constructed to be used as a second recipient strain in the exogenous plasmid isolation experiments to compare creek water with WWTP effluent (see below). First, a mini-Tn7 region containing a zeocin resistance gene and eyfp was moved from pUC18T-mini-Tn7T-zeo-PA1/04/03-ecfp into an R6K replicon pHY835 (29). Then, the resulting plasmid, pHY956, and Tn7 transposase expression plasmid pTNS2 were simultaneously introduced into E. coli EC100 by electroporation. Subsequent screening for zeocin-resistant clones gave rise to HY841. Mini-Tn7 insertion into the specific site (attTn7) was confirmed by PCR using primers EcoliglmS (CATGCACATCATCGAGATGCC) and Tn7R109 (CAGCATAACTGGACTGATTTCAG). A spontaneous rifampin-resistant mutant of HY841 was designated HY842 and was used as the recipient strain.
Water samples.
Water was sampled from three sources in and near Moscow, ID. The first site was a pond in the University of Idaho's Arboretum and Botanical Garden. This pond receives its water supply from ground water, precipitation, and irrigation runoff from the University of Idaho. The water used by the university to irrigate its lawns is chlorinated effluent from the Moscow, ID, WWTP, which is chlorinated for a second time before irrigation. The second site was Paradise Creek in downtown Moscow, ID. The source of this stream is the Palouse Range from which the stream meanders down the mountain, collecting agricultural and urban runoff. Typical crops grown in this area include wheat, barley, peas, and lentils. The third site was Idler's Rest Creek in the Palouse Range. The site of water collection was a few miles downstream from the source in the forests of the Palouse Range. A total of 500 ml of water was collected in sterile bottles for each mating experiment. To remove suspended solid particles from water, the samples were filtered through autoclaved, grade 1 Whatman filter paper (11 μm) and then through sterile Fisher P5 filter paper (5 μm) into sterile 1-liter bottles, using a sterile vacuum filtration system.
Triparental matings.
To capture a wide range of self-transmissible mobilizing plasmids, two types of mobilizable plasmids were used: (i) pSU4814 (30), which has a CloDF13 mob region and has been shown to be mobilized efficiently by IncF, IncN, IncI, and IncW plasmids (31), and (ii) pBBR1MCS (32), derived from plasmid pBBR1 from Bordetella bronchiseptica, known to be mobilized by IncP-1 plasmid RK2 (33), and its derivative pBBR1MCS-5 (34). Triparental matings were carried out as shown in Fig. 1 and described here. First, E. coli DH5α harboring either of the mobilizable plasmids and a recipient E. coli strain CV601gfp were grown overnight. Then, 500 ml of filtered water was mixed with 500 μl of 100-fold diluted overnight cultures of the two E. coli strains and filtered through a sterile Nalgene cellulose nitrate analytical filter (0.22-μm pore size) (Nalge Nunc International, Rochester, NY). The filters were transferred onto an R2A agar plate supplemented with 300 μg/ml of cycloheximide to prevent fungal growth and incubated at 30°C overnight. The mating mixtures from the filters were then resuspended in 2 ml of sterile saline (0.85% NaCl), and appropriate dilutions were plated onto LBA containing Rif and Cm (for pBBR1MCS and pSU4814) or on LBA containing Rif and Gm (for pBBR1MCS-5) to select transconjugants. The presence of captured mobilizing plasmids was verified as described below. At various stages, some of the plasmids that were initially captured were subsequently eliminated from the list of candidate plasmids, for example, because they were not self-transmissible or not successfully marked with the transposon. As far as we know, our procedure was not biased toward plasmids without known accessory genes. Because of this lack of quantitation, we do not draw any conclusions on the relative abundance of the captured plasmids in their natural habitats.
In a second set of triparental matings to compare the proportion of antibiotic resistance plasmids from creek water and WWTP effluent, E. coli HY842 was used as recipient instead of CV601gfp.
General DNA manipulations.
Plasmid presence in bacterial strains was confirmed by alkaline lysis extraction and agarose gel electrophoresis (35). Restriction fragment length polymorphisms of captured plasmids were determined by restriction digestion with enzymes from New England BioLabs (Ipswich, MA, USA), followed by agarose gel electrophoresis. Plasmid DNA to be used for sequence determination was obtained using a Plasmid Midi kit (Qiagen, Valencia, CA) according to the manufacturer's instructions for low-copy-number plasmids. PCR was carried out using Phusion DNA polymerase (New England BioLabs, Ipswich, MA, USA).
Plasmid marking with tetracycline resistance.
Since none of the newly captured plasmids showed resistance to antibiotics or mercuric chloride, they were marked with tetracycline resistance transposons. To do this, mini-Tn21 donor plasmids pMS0507 (kind gift from Masahiro Sota) and pMT1749Tc were introduced into E. coli CV601gfp carrying a captured plasmid (Table 1). The resulting strains were mated with E. coli K12Nal, and the transconjugants were selected for Tc and Nal resistance. This typically yielded K12Nal derivatives carrying a self-transmissible plasmid tagged with mini-Tn21OTc or mini-Tn21Tc, respectively. Mini-Tn21OTc on pMS0507 contains pBR322 oriV, which was used to increase the copy number of the tagged plasmids in E. coli and therefore improved the quantity and quality of purified plasmid DNA required for sequencing. Only plasmids that showed unique restriction fragment length patterns were sequenced. The tagged plasmids were designated pMBUI1T, pMBUI2T, pMBUI3T, pMBUI4T, pMBUI6T, pMBUI7T, pMBUI8T, pDS1T, pDS2T, and pDS3T.
Host range assays.
The host range of the marked plasmids was evaluated by conjugative transfer to three bacterial strains: Pseudomonas putida UCW1gfp, Cupriavidus pinatubonensis JMP228gfp, and Sinorhizobium meliloti RM1021, belonging to the Gamma-, Beta-, and Alphaproteobacteria, respectively (36). The donor strain, E. coli K12Nal bearing the plasmid to be tested, was grown at 37°C in LB broth supplemented with 10 μg/ml tetracycline. All recipients were grown overnight in LB broth at 30°C. Biparental matings between the donor strain and each of the recipients was carried out at 30°C as previously described (37) with selection for Rif and Tc resistance.
Incompatibility testing.
To determine incompatibility with a known IncP-1 plasmid, pDS1T was first transferred by conjugation to E. coli DH10B with prototype IncP-1γ plasmid pQKH54 (37). Three transconjugants were grown overnight in 5 ml of LB containing Tc to select for pDS1T. Every 24 h for 10 days, 50 μl of culture was transferred to 5 ml of fresh medium containing Tc. Dilutions were plated on LBA every other day, followed by replica plating of 52 colonies onto LBA, LBA with Tc, and LBA with HgCl2. The proportion of cells sensitive to HgCl2 is a measure of the proportion that lost pQKH54. In parallel, E. coli DH10B bearing only pQKH54 was propagated in the absence of Tc for 10 days to determine its stability. Dilutions were plated on LBA, and colonies were replica plated on LBA and LBA with Hg. The same experiment was repeated for the IncP-1γ plasmid pMBUI1T as the positive control and for IncN plasmid pDS2T as a negative control.
Sequencing and annotation.
The DNA sequences of the plasmids were determined by pyrosequencing at the Department of Energy (DOE) Joint Genome Institute (Walnut Creek, CA). The shotgun sequencing protocol using the Roche/454 platform has been described previously (38). The plasmids were sequenced with GS FLX Titanium Sequencing chemistry (Roche/454 Life Sciences, Branford, CT). Approximately 199.6 million bases of sequence information were generated from half of a sequencing run. The sequences were assembled using the Newbler software (Roche/454 Life Sciences, Branford, CT). Gaps between contigs were closed by PCR amplification and subsequent Sanger sequencing of the PCR product using BigDye Terminator, version 3.2, cycle sequencing (Applied Biosystems, Foster City, CA). The IGS Annotation Engine at the Institute for Genome Sciences, School of Medicine, University of Maryland (http://ae.igs.umaryland.edu/), provided automatic annotation of the plasmid sequences, which were further annotated manually by us. To reconstruct the genome sequences of the originally captured plasmids, the DNA sequences of the minitransposons used to tag the plasmids were removed from each plasmid sequence. The presence of repeated sequences at each side of the transposon was confirmed in each case, indicating that the transposition had not caused deletions of flanking regions.
Phylogenetic analysis of IncP-1 plasmids.
The part of the trfA gene that is conserved among the IncP-1 plasmids was used to infer the phylogenetic relationship of pDS1 with the other IncP-1 subgroups. The complete sequences of trfA genes from 45 IncP-1 plasmids (AB237782, GQ495894, GQ983559, HQ891317, JF274988, JF274990, JN106164, JN106165, JN106166, JN106167, JN106168, JN106169, JN106170, JN106171, JN106172, JN106173, JN106174, JN106175, NC_001621, NC_003430, NC_004840, NC_004956, NC_005088, NC_005793, NC_006352, NC_006388, NC_007337, NC_007502, NC_007680, NC_008055, NC_008272, NC_008357, NC_008385, NC_008766, NC_010935, JX847411, NC_014911, NC_016968, NC_016978, JQ432563, NC_006830, NC_001735, EF107516, JQ432564, and KC170283) were aligned by first translating the genes to protein sequences, aligning the protein sequences using ClustalW (39), and then aligning the nucleotide sequences to the protein sequences using the transalign function of EMBOSS (40). Only the homologous 957 nucleotides (nt) of the 3′ end of the alignment were used in the phylogenetic analysis. This fragment coincides with the last 915 nt of pDS1. Sites that had gaps were excluded from the analysis, resulting in 843 aligned sites. A DNA sequence evolution model was chosen using DT-ModSel (41), and a phylogenetic tree was inferred using the maximum-likelihood method implemented in PAUP* (42).
Bioinformatics analyses and software.
The IGS Annotation Engine (http://ae.igs.umaryland.edu/cgi/index.cgi) was used to automatically annotate plasmid sequences. Annotations were corrected manually using Manatee (http://ae.igs.umaryland.edu/cgi/manatee_intro.cgi). All unique hypothetical proteins were compared against the Interpro database using BLAST with a default E value of 10 (43). Interpro (http://www.ebi.ac.uk/interpro/) is a database of protein families, domains, regions, repeats, and sites in which identifiable features of known proteins can be used to infer functions for uncharacterized proteins based upon sequence similarity.
Comparison of antibiotic resistance profiles of plasmids captured from creek water and effluent.
To compare resistance profiles of plasmids from the Moscow, ID, wastewater treatment plant to a new set of plasmids from Paradise Creek, triparental matings and disk diffusion assays were used. Plasmids from both locations were captured as described above, except that E. coli HY842 was used as a recipient instead of CV601gfp. HY842 has several advantages that CV601gfp does not have. First, this strain completely lacks genes for restriction enzymes, thus allowing establishment of more diverse plasmids. Second, this strain is easily and clearly distinguishable from environmental strains by colony PCR targeting the mini-Tn7–chromosome junction as well as by detection of the yellow fluorescence it encodes. The presence of captured plasmids was confirmed by plasmid DNA extraction, followed by gel electrophoresis. The recipient was confirmed to be E. coli HY842 by observing the fluorescence of the transconjugants under blue light of a fluorescence microscope. Plasmid DNA was digested with EcoRI, and only plasmids with distinct patterns were chosen for disk diffusion assays.
Overnight cultures of transconjugants in LB broth were diluted 100 times in fresh LB broth and grown at 37°C until they reached an optical density at 600 nm (OD600) of 0.4, corresponding to approximately 108 CFU/ml. Then 100 and 260 μl of this culture were spread onto 100-mm and 150-mm plates of Mueller-Hinton agar, respectively, with sterile cotton swabs. Negative controls were E. coli HY842 carrying only the mobilizable plasmids. Antibiotics were chosen from each of the main classes, and disks containing the following antibiotics were used (concentrations per disk in μg): kanamycin, 30; gentamicin, 10; spectinomycin, 100; polymyxin B, 300 IU; aztreonam, 30; cefoxitin, 30; imipenem, 10; ceftazidime, 30; ciprofloxacin, 5; amoxicillin and clavulanic acid, 30; trimethoprim, 5; sulfamethoxazole/trimethoprim, 23.75/1.25; tetracycline, 30; and chloramphenicol, 30 (BD BBL, USA). Plates were incubated at 37°C for 48 h, and transconjugants were classified as resistant or susceptible in comparison to the controls and in accordance with the guidelines provided by the Clinical and Laboratory Standards Institute (44).
Nucleotide sequence accession numbers.
Complete nucleotide sequences of untagged plasmids were deposited in GenBank under accession numbers KC170278 to KC170285.
RESULTS
Capture of plasmids from freshwater sources.
To capture self-transmissible plasmids from three freshwater sources in and near Moscow, ID, without selecting for a particular plasmid-encoded trait, multiple triparental matings were carried out with E. coli as the recipient. The first site was a pond in the university arboretum, which receives water from ground water, precipitation, and irrigation runoff of hyperchlorinated effluent from the municipal wastewater treatment plant (WWTP) (see Materials and Methods for details). The second sample was taken from Paradise Creek, which receives agricultural and urban runoff. The third was from Idler's Rest Creek in a sparsely inhabited forested area outside town, taken a few miles downstream from the spring. The genomes of 10 captured plasmids were completely sequenced. The plasmids were chosen based on their unique restriction profiles and because either they were shown to have a broad host range (Table 2) or their BHR nature was inferred from partial sequence information (from pDS1 and pDS2). None conferred resistance to any of the antibiotics tested. To facilitate host range and incompatibility assays, the plasmids were marked with a mini-Tn21 transposon encoding tetracycline resistance (Table 1). Except for pDS1 and pDS2, all plasmids showed a broad host range as they transferred and replicated in alpha-, beta-, and gammaproteobacteria (Table 2).
Table 2.
General features of completely sequenced plasmids
| Plasmid | Size (bp) | Origin | Mobilizable plasmid | Host rangea | BHR group | Accessory gene region(s) | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| pMBUI1 | 44,304 | Arboretum | pBBR1MCS | A, B, C | IncP-1γ | None | JQ432563 |
| pMBUI2 | 37,976 | Arboretum | pBBR1MCS | A, B, C | New | 6.6 kb, 6 unique ORFs | KC170285 |
| pMBUI3 | 33,736 | Arboretum | pBBR1MCS | A, B, C | IncU | None | KC170281 |
| pMBUI4 | 37,247 | Arboretum | pSU4814 | A, B, C | IncW-like | 12 kb, 21 unique ORFs | KC170278 |
| pMBUI6 | 47,999 | Arboretum | pSU4814 | A, B, C | PromA-like | 14 kb, 10 unique ORFs | KC170282 |
| pMBUI7 | 34,006 | Paradise Creek | pBBR1MCS-5 | A, B, C | IncU | None | KC170284 |
| pMBUI8b | 53,313 | Paradise Creek | pBBR1MCS-5 | A, B, C | IncP-1β | 9.3 kb, 3 unique ORFsc | KC170279 |
| pDS1 | 40,596 | Idler's Rest | pBBR1MCS | Cd | IncP-1η | None | KC170283 |
| pDS2 | 56,683 | Idler's Rest | pBBR1MCS | Cd | IncN | 15.6 kbe | KC170280 |
| pDS3 | 40,806 | Idler's Rest | pBBR1MCS | A, B, C | IncP-1β | None | JX469834 |
The recipients used in matings to assess host range were Pseudomonas putida UCW1gfp (Gammaproteobacteria), Cupriavidus pinatubonensis JMP228gfp (Betaproteobacteria), and Sinorhizobium meliloti RM1021 (Alphaproteobacteria). A, Alphaproteobacteria; B, Betaproteobacteria; C, Gammaproteobacteria.
Marked with mini-Tn21Tc; all others marked with mini-Tn21OTc (Table 1).
Putative functions encoded by genes in accessory region of pMBUI8: resolvase, PepSY, phosphoesterase, histidine kinase, triphenylmethane reductase, dihydrolipoamide dehydrogenase, transcriptional regulator, transposases.
Plasmids pDS1 and pDS2 were transferable into E. coli but could not be transferred to P. putida, C. pinatubonensis, or S. meliloti.
Putative functions encoded by genes in the accessory region of pDS2: sodium-proton antiporter, multidrug ABC transporter, transposases, and insertion elements.
Plasmids from the arboretum pond.
Five plasmids were captured from pond water at the university's arboretum: pMBUI1, pMBUI2, pMBUI3, pMBUI4, and pMBUI6. Each plasmid belonged to a different incompatibility group (Table 2), indicating high plasmid diversity in our sample. Two plasmids do not seem to encode accessory functions.
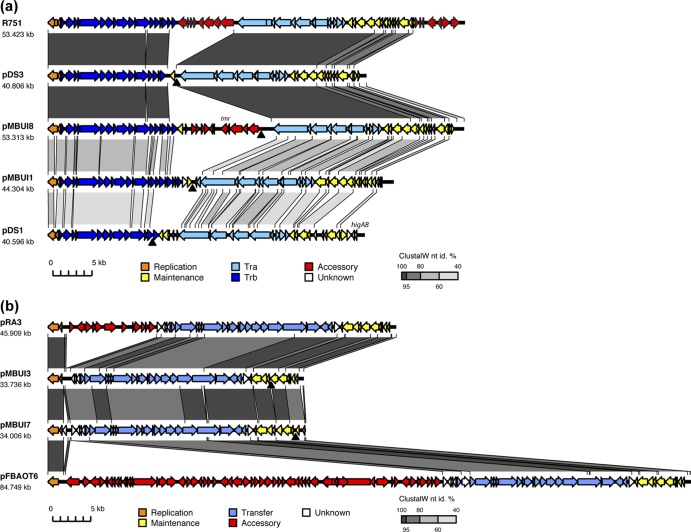
Only two of the five plasmid genomes showed high similarity to previously described BHR plasmids, i.e., IncP-1 plasmid pMBUI1 and IncU plasmid pMBUI3 (Fig. 2 and Table 3). They are also the only two plasmids from this habitat that seem to be devoid of known accessory genes. Plasmid pMBUI1 belongs to the IncP-1γ subgroup (45) and is closely related to the archetype pQKH54 (46). Its characteristics are described elsewhere (47). Plasmid pMBUI3 (Fig. 2b) is closely related to the completely sequenced IncU plasmids RA3 (48) and pFBAOT6 (6); therefore its annotation follows that of these two plasmids. Surprisingly, in contrast to these other two IncU plasmids, no known accessory genes were found on pMBUI3. Between the replication and transfer genes there are three ORFs of unknown function that are present on all IncU plasmids. Since expression of these and several plasmid backbone genes was recently shown to be regulated by the plasmid-encoded protein KorC, we infer that these ORFs are also backbone genes (49). We propose that pMBUI3 consists entirely of IncU plasmid backbone genes.
Fig 2.
Alignment of newly sequenced plasmids showing their relationship to each other and to well-studied members of their incompatibility group. (a) IncP-1 plasmids. (b) IncU plasmids. Coding regions are shown as colored arrows; putative functions are indicated by the color key. The triphenylmethane reductase gene is labeled as tmr on pMBUI8. The degree of similarity between plasmids (percent nucleotide identity [nt id] of alignments performed using ClustalW) is indicated by gray scale-shaded regions; the darker the shading between two segments, the higher their similarity as shown in the heat key. Sites of mini-Tn21 insertion in the tagged plasmids are marked with a filled black triangle.
Table 3.
Sequence identity of replication initiation proteins from each plasmid with most similar protein sequence in GenBank
| Plasmid | Most similar plasmid (Inc group) | Protein | GenBank accession no. | % Identity |
|---|---|---|---|---|
| pMBUI1 | pQKH54 (IncP-1 γ) | TrfA | YP_619825 | 98 |
| pMBUI2 | pMATVIM-7 (unknown) | Rep | YP_001427363 | 72 |
| pMBUI3 | RA3 (IncU) | RepB | YP_001966818 | 99 |
| pMBUI4 | R7K (IncW) | Rep | YP_001874895 | 62 |
| pMBUI6 | pMOL98 (PromA) | Rep | CAC93881 | 39 |
| pMBUI7 | RA3 (IncU) | RepB | YP_001966818 | 99 |
| pMBUI8 | pB10 (IncP-1β) | TrfA | NP_858039 | 100 |
| pDS1 | pBS228 (IncP-1α) | TrfA2 | YP_758655 | 72 |
| pDS2 | Klebsiella pneumoniae plasmid 9 (IncN) | RepA | YP_002286867 | 100 |
| pDS3 | pB10 (IncP-1β) | TrfA | NP_858039 | 100 |
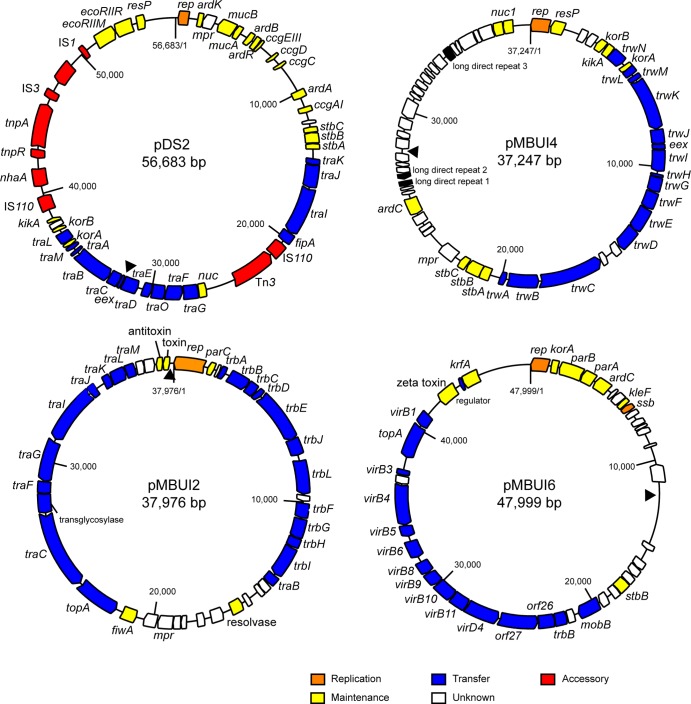
Plasmid pMBUI4 (Fig. 3) is an IncW-like plasmid that bears moderate gene synteny and sequence similarity in its replication (Table 3) and transfer regions to the IncW plasmid R388 (5). However, its establishment and stable inheritance and control regions are far less recognizable, and some of the genes in these regions on R388 are missing on pMBUI4 (5). The so-called establishment module of R388 is comprised of three genes, ardC (antirestriction activity), ssb (DNA metabolism), and ardK (transcriptional regulator of ardC). Of these, only ardC was found on pMBUI4 (Fig. 3), but its protein sequence was most similar to ArdC from pMOL98, a PromA plasmid (only 62% identity). The stable inheritance module of R388 is comprised of two operons. The first has kfrA (plasmid partitioning), nuc1 (thermonuclease; a homologue of parB from RK2), nuc2, and osa (oncogenicity suppression). Of these, only nuc1 could be identified on pMBUI4 (Fig. 3). The second operon of R388 is composed of genes stbA, stbB, and stbC, which are all present on pMBUI4. Their products have about 85% identity with the respective proteins in pIE321. Like R388, long direct repeats were found on pMBUI4, the functions of which are unknown. Of the six transcriptional regulators found on R388 (ardK, klcB, stbA, trwA, korA, and korB) (5), only the last four were identified on pMBUI4. Because of the many missing genes, we refer to pMBUI4 as an IncW-like plasmid (Table 2).
Fig 3.
Genetic maps of 4 of the 10 plasmids isolated from Moscow, ID: the IncN plasmid pDS2, the IncW-like plasmid pMBUI4, and two plasmids whose Inc groups are unknown, pMBUI2 and pMBUI6. The gene modules are shown as colored arrows representing different functions (as indicated by the color key). In pMBUI6, the genes named “orf” are most closely related to transfer genes of different BHR plasmids. Genes with the same names on different plasmids are not necessarily homologous due to gene naming conventions for plasmids. Sites of mini-Tn21 insertion in the tagged plasmids are marked with a filled black triangle.
The cluster of hypothetical genes on pMBUI4 into which our artificial Tn21 inserted does not have any homologues; even Interpro domains could not be detected. Of the genes with unknown functions, only mpr, which encodes a putative zinc metalloproteinase, has homologues in other plasmids.
Plasmid pMBUI2 is a member of a new group of BHR plasmids with similarities to pMATVIM-7 (50) from Pseudomonas aeruginosa (Fig. 3 and Table 3). The main difference between the backbone genes of these two plasmids is that pMBUI2 seems to have more transfer genes than pMATVIM-7. This new BHR group has several similarities to IncP-1 plasmids including the following: (i) the presence of transfer genes that show sequence similarity to those of IncP-1 plasmids and a similar gene arrangement to that of IncP-1 plasmid pNeutP1 from Nitrosomonas eutropha C91 (51, 52); (ii) a putative nick site, ATCTTG, between traJ and traK, with similarity to the nick site in the origin of transfer (oriT) on IncP-1 plasmids. Dissimilarities to IncP-1 plasmids include the following: (i) a replication initiation gene that bears no resemblance to trfA, (ii) the presence of a partitioning gene, and (iii) the absence of distinguishable iterons. In fact, most typical IncP-1-specific stable inheritance genes involved in partitioning, postsegregational killing, or multimer resolution were not identified on pMBUI2, with the exception of one encoding a putative fertility inhibition factor that has 37% identity to FiwA of IncP-1α plasmids. Finally, pMBUI2 has a cluster of hypothetical genes between traB and fiwA with no known homologues. Hidden Markov model (HMM)-based detection of Interpro domains (53) also did not help identify these gene products. Another putative mpr gene was found on this plasmid, whose protein product has 50% amino acid identity to its homologue on pMBUI4.
Plasmid pMBUI6 is a PromA-like plasmid (Fig. 3). Some of its backbone genes bear close similarity to pXF51 from Xylella fastidiosa (54) and more distantly to BHR plasmids of the PromA group (Table 3), which includes pMOL98, pSB102, pIPO2, pTER331, and pMRAD02 (8). Important PromA-like features identified on pMBUI6 include (i) the location of the topoisomerase gene (topA) among genes of the mating pair formation complex, (ii) reverse orientation of the relaxase gene (mobB) with respect to other transfer genes, and (iii) presence of long direct repeats in intergenic regions (at least 10 direct repeats of 168 bp downstream of rep and 4 more elsewhere) (Fig. 3). The primary difference between pMBUI6 and the PromA plasmids is the lack of synteny in the partitioning and regulatory region. Additionally, there is a total of 14 kb of sequence in which no known or conserved hypothetical genes are found; ORFs in these regions may encode host beneficial traits.
Plasmids from Paradise Creek.
The two plasmids isolated from Paradise Creek, pMBUI7 and pMBUI8, have typical IncU and IncP-1β backbones, respectively. Plasmid pMBUI7 is similar to the IncU plasmid pMBUI3 from the arboretum pond and also has no known accessory genes (Fig. 2b). Plasmid pMBUI8 is a typical IncP-1β plasmid with accessory genes, one of which putatively encodes triphenylmethane reductase (tmr) (Fig. 2a). This gene has 99% identity with tmr genes previously found on the IncP-1β plasmids pGNB1, pKV11, pKV29, and pKV36, where it was shown to code for decolorization of crystal violet (55, 56).
Plasmids from Idler's Rest.
Of the three plasmids isolated from Idler's Rest Creek, pDS1 and pDS3 are IncP-1 plasmids, and pDS2 belongs to the IncN group. Only pDS2 has identifiable accessory genes.
The genetic organization of pDS1 (Fig. 2a) is typical of IncP-1 plasmids, but the sequence identity to known IncP-1 plasmids is low. IncP-1 plasmids are typically classified into different subgroups based on sequence similarity of backbone genes. For a recent thorough phylogenetic analysis of the five major IncP-1 subgroups, we refer to Sen et al. (45). A phylogeny inferred from the trfA gene showed that pDS1 is distinct from other known subgroups including the new IncP-1ζ plasmids isolated from marine biofilms (57) (Fig. 4). Surprisingly, unlike most other IncP-1 plasmids, pDS1 could not be transferred to P. putida or C. pinatubonensis (Table 2). To verify the placement of pDS1 in the IncP-1 group, an incompatibility assay was performed. The marked plasmid pDS1T was introduced into E. coli DH10B(pQKH54) by conjugation, and loss of pQKH54 was monitored when we selected for the incoming plasmid only. Nearly 80% of the clones tested had lost pQKH54 by day 1, in contrast to only a 16% loss of pQKH54 in a separate stability assay. This indicates that pDS1T is incompatible with IncP-1 plasmids and can be classified as an IncP-1 plasmid. We therefore propose the new subgroup IncP-1η for pDS1.
Fig 4.
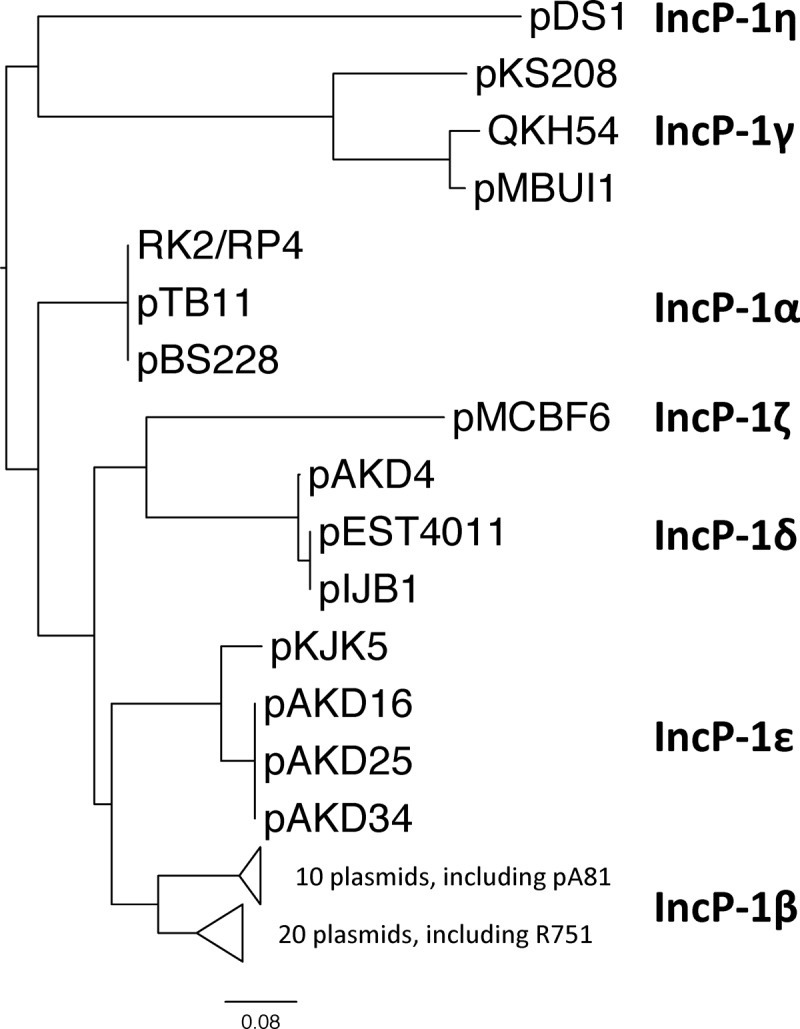
Phylogenetic relationship of pDS1 to other IncP-1 plasmid subgroups. The conserved 3′ end of the trfA gene was used to infer the phylogeny of 45 IncP-1 plasmids using the maximum-likelihood method (base frequencies, gamma shape parameter, proportion of invariant sites, and rate matrix [a b c c b a] were estimated from the data). The incompatibility subgroup of each clade is indicated on the right.
Although the gene content of pDS1 is conserved relative to other IncP-1 plasmids, some features stand out. Its putative oriT sequence is considerably divergent from that of other IncP-1 plasmids although a conserved nick site was found. The replication region consists of a replication initiation gene, trfA, and an origin of replication, oriV. The TrfA protein showed nearly 90% similarity to TrfA2 from IncP-1α plasmid pBS228 (Table 3) and is missing the N terminus found in many other TrfA proteins. The oriV has 10 direct repeats or iterons that differ in sequence from those of other IncP-1 plasmids. Like some other IncP-1 plasmids, pDS1 carries putative toxin-antitoxin genes (nt 39259 to 38951 and 39568 to 39269); they bear closest similarity to higBA, first identified on a Proteus vulgaris plasmid (58). No insertions of mobile elements or known accessory genes were detected on pDS1, suggesting that it is another “backbone-only” plasmid.
Plasmid pDS2 is a typical IncN plasmid (Fig. 3) with quite a narrow host range (Table 2), but it is the first IncN plasmid to be completely devoid of antibiotic resistance genes (59, 60). It has two accessory regions between fipA and nuc and between kikA and the EcoRII type II restriction modification gene complex. The first is composed of an IS110 family insertion sequence, and the second contains another IS110-like element, a Na+/H+ antiporter gene (61), a Tn3-family transposon with only transposition genes, and an IS3-like transposon.
Finally, like pMBUI8 from Paradise Creek, plasmid pDS3 is a typical IncP-1β plasmid (45). Unlike pMBUI8, it has no accessory genes and is thus the fifth plasmid of the 10 examined that seems to exist in nature as a bare plasmid backbone (Fig. 2a).
Comparison of antibiotic resistance profiles of plasmids captured from Paradise Creek and WWTP effluent.
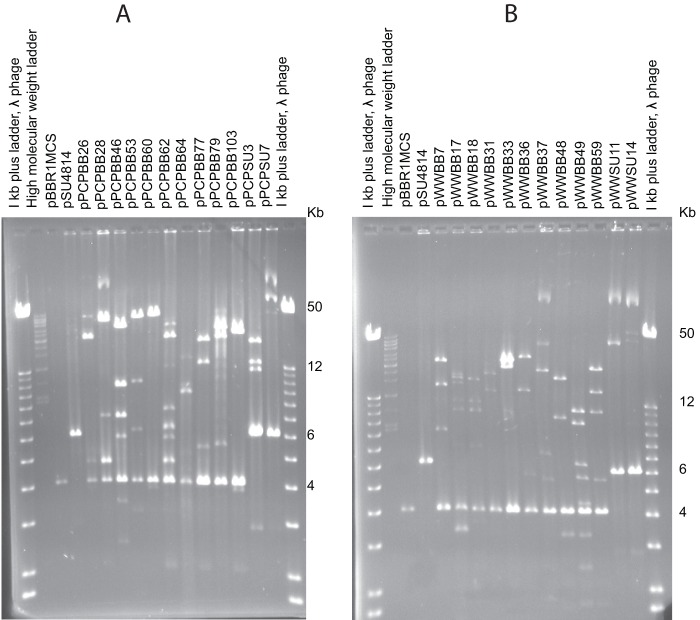
The striking lack of antibiotic resistance genes on all 10 plasmids prompted us to perform a comparative analysis of antibiotic resistance profiles. Using the same plasmid capture protocol, a new set of plasmids was captured from Paradise Creek, upstream of the WWTP in Moscow, ID, and from the WWTP effluent. Twelve unique plasmids were chosen from each site based on distinct restriction profiles (Fig. 5). Transconjugants harboring these plasmids were screened for resistance to 14 antibiotics using the disk diffusion method. Only one plasmid from the creek conferred resistance, namely, to trimethoprim and sulfamethoxazole, and two plasmids from the effluent each showed resistance to six antibiotics: aztreonam, tetracycline, kanamycin, polymyxin B, spectinomycin, and ceftazidime. Thus, while the number of resistance determinants was higher among plasmids from the effluent, there was no clear difference in the proportions of resistance plasmids captured from the two sites. Strikingly, all three resistance plasmids were captured in matings with mobilizable plasmid pSU4814, and none was from matings with pBBR1MCS. We tested 16 additional transconjugants from the pSU4814 mating with WWTP effluent, but none conferred resistance. Thus, among the plasmids in raw effluent that can be captured in E. coli based on their mobilization potential, only a relatively small proportion (<10%) encoded common antibiotic resistance determinants.
Fig 5.
Unique plasmid restriction fragment patterns of captured plasmids generated by EcoRI digestion. (A) Plasmids isolated from Paradise Creek (PCP). (B) Plasmids isolated from Moscow, ID, WWTP (WW). All plasmid DNA samples were completely digested, as indicated by the linearized mobilizable plasmid at 4 kb (pBBR1MCS) or 6.2 kb (pSU4814), and were run on a 0.5% agarose gel for 18 h at 30 V. Plasmids captured using pBBR1MCS contain BB in their names while those captured with pSU4814 contain SU.
DISCUSSION
One of the central questions in plasmid ecology and evolutionary biology is whether or not plasmids can be considered genetic parasites in their natural bacterial populations. Do they need to confer a benefit to their host to be successfully maintained in microbial communities, or can they survive by spreading horizontally by means of conjugation in spite of often imposing a cost to their hosts? As pointed out more than 35 years ago, for plasmids to persist in the absence of positive selection, high transfer rates must compensate for occasional plasmid loss and cost (14). One way to address this central question is to analyze the accessory gene content of extant pools of plasmids in various environments. Unfortunately, most studies on plasmid diversity have focused on plasmids that were isolated based on a trait of interest, such as virulence or resistance to or catabolism of antimicrobials or pollutants, and from habitats and bacteria that are under strong selection, such as polluted soils and water bodies, wastewater treatment plants, or known pathogens (18, 62–68). Not surprisingly, such plasmids contain accessory genes that encode the selected host-beneficial traits. Far fewer studies have explored plasmid diversity and gene content using plasmid isolation methods that are not based on a plasmid-encoded accessory phenotype and in environments not known to impose strong selection for accessory genes. While the relative abundance of plasmids or pollution-related accessory genes has been compared between polluted and clean control sites, no studies have so far compared gene contents of plasmids (64, 69–72). The recently developed metagenomics approaches to assess the “plasmidome” or “metamobilome” (73, 74) present one way to assess accessory gene content independent of what the plasmids encode, but obtaining reliable, closed genome sequences for large self-transmissible plasmids is still a challenge (75). Using the triparental mating approach, we showed the presence of diverse self-transmissible plasmids that do not carry known host-beneficial genes in natural environments. The few hypothetical ORFs detected in these plasmids may now become the focus of future research to determine whether they confer heretofore unknown host-beneficial traits.
In the first part of this study, two important observations were made. The first is the high diversity among the 10 plasmids captured from three sites: no less than six incompatibility groups were represented. Studies that selected for plasmids carrying specific traits appear to show noticeably lower plasmid diversity. For example, at least 14 of the 19 BHR plasmids captured from agricultural soils belonged to the IncP-1 group (76, 77). Of the 12 plasmids captured from activated sludge, 10 were members of the IncP-1 group (66). Older studies that characterized plasmids using probes only for known incompatibility groups were often not able to determine the diversity of plasmid groups since several plasmids would not hybridize to any of the probes (27, 46, 78). The mobilizable plasmids used in these studies may have selected for only certain Inc groups. Some plasmids are mobilized rarely by IncP-1 plasmids (31), while others like the IncQ plasmids used in previous studies (pD10 [26], pMOL155 [27], and pMOL187 [77]) may be preferentially mobilized by IncP-1 plasmids. By using different vectors known to be mobilized by different plasmids (30, 32), we captured 10 plasmids from no fewer than six divergent groups, as well as three different IncP-1 subgroups.
The second observation of interest is that in each of the three water bodies sampled around Moscow, ID, at least one self-transmissible plasmid was devoid of identifiable accessory gene regions (pDS1, pDS3, pMBUI1, pMBUI3, and pMBUI7). The two IncU plasmids captured in our study are the only plasmids of that group that lack known accessory genes (6, 48). Although the possibility exists that accessory genes were lost in the process of isolation, it seems unlikely that two such similar plasmids from two different locations would undergo the same loss of genetic material. Moreover, three of the four IncP-1 plasmids did not have any known accessory genes. This result is in stark contrast to the finding that only 2 of 65 previously sequenced IncP-1 plasmids encode no recognizable accessory functions (16, 17). These and other putative cryptic plasmids described in our study and other studies (8, 28) carry ORFs of unknown function that may, of course, provide unknown benefits to their hosts in their natural environment. The apparent success of these plasmids may also be due to known plasmid backbone genes that confer a host benefit, such as promotion of biofilm formation through the conjugation pilus (79), or to fluctuation selection through temporary benefits to the plasmid host provided by accessory genes that are frequently acquired and lost and not detected at the time of sampling. Whether these plasmids benefit their hosts in unknown ways or are simply persisting as parasitic elements due to efficient horizontal transfer is currently not known and should be investigated in future studies. A first step would be to determine if the ORFs of unknown functions are even expressed and what their role might be.
Five plasmids were identified as having accessory genes, but the functions encoded by these genes are currently mostly unknown. The large clusters of hypothetical genes found on pMBUI2, pMBUI4, and pMBUI6 (Fig. 3) may encode host-beneficial traits, and further work is needed to determine their function. The IncN plasmid, pDS2 has accessory genes but, surprisingly, none that encode antibiotic resistance. IncN plasmids are considered a health risk because they typically confer a variety of antibiotic resistances (59). As far as we know, pDS2 is the first IncN plasmid that is devoid of antibiotic resistance genes. The novel plasmid pMBUI2 and the IncW-like plasmid pMBUI4 encode homologous zinc metalloproteinases that are also encoded on pDS2, pMBUI3, and pMBUI7. These Mpr proteins share 50 to 60% sequence identity among the different Inc groups. They are unrelated to the Mpr proteins that are associated with bacterial pathogenesis (80) but are similar to those encoded by other plasmids such as IncU plasmid RA3 (48). Because the homologous mpr genes are consistently found near or within the control regions of diverse Inc groups, we hypothesize that they are backbone and not accessory genes.
The narrow host range of the IncP-1η plasmid pDS1 was unexpected, given that most IncP-1-like plasmids have a broad host range. A preliminary plasmid genomic signature analysis suggested a signature that is quite different from the other IncP-1 plasmids captured in this study (H. Suzuki, personal communication). As pDS1 transferred very well between E. coli strains, the plasmid's transfer system seems intact. Since the molecular basis of the apparent narrow host range of this IncP-1η plasmid is of great interest, a detailed host range investigation will be the subject of a future study, combining experimental host range testing with a plasmid genomic signature analysis to assess its potential host range (81).
Because of the predominance of antibiotic resistance plasmids shown in WWTPs around the world (23), we expected to find a large number of such plasmids in the Moscow, ID, WWTP. Indeed, using the biparental plasmid capture method and selection for antibiotic resistance, a plasmid with 11 genes encoding resistance to seven different antibiotics was isolated from this facility several years ago (data not shown). However, using the triparental plasmid capture method in the current study, less than 10% of the plasmids captured from both creek water and raw WWTP effluent conferred resistance to antibiotics. Such plasmids may, of course, occasionally act as vehicles for trafficking antibiotic resistance determinants and at other times persist as parasitic elements. Future studies should evaluate the potential role of these plasmids as in situ vectors for drug resistance spread.
Although triparental matings avoid the bias of selecting for specific accessory genes, they introduce other biases. For instance, this method selects for plasmids that efficiently transfer themselves under our specific mating conditions. Therefore, a bias might exist toward smaller conjugative plasmids with fewer accessory genes. However, there is no evidence for or against this conjecture. In previous studies, 50- to 80-kb plasmids with accessory genes were isolated (26, 27, 77). Given that matings were done overnight, it is unlikely that larger plasmids would be selected against because of the slightly longer time required for transfer or because of lower establishment efficiencies. Additionally, this study retrieved only plasmids that efficiently transferred the mobilizable plasmid pBBR1MCS or pSU4814 and stably replicated in the rapidly growing E. coli recipients. Strikingly, the only IncW-like plasmid isolated in this study was captured with pSU4814, a vector known to be mobilized well by the IncW transfer system (F. de la Cruz, personal communication). Thus, although the triparental mating approach cannot be used to infer relative abundance of cryptic plasmids or the plasmid richness in specific habitats because of these biases, our results clearly show the presence of diverse cryptic plasmids in natural freshwater communities.
Our study is a first attempt at assessing the occurrence of cryptic self-transmissible plasmids in various freshwater habitats. Future work will include whole-genome sequencing of much larger sets of plasmids captured in identical ways from different sites. Metagenomic approaches will also shed more light on the diversity of plasmids than triparental plasmid capture methods, but obtaining correctly closed genomes of large plasmids like the ones described in this study remains a challenge for complex communities. Thus, in the future a combination of different plasmid isolation and sequencing methods will be needed to improve our view of the diversity and accessory gene content of these important mobile genetic elements in the horizontal gene pool.
ACKNOWLEDGMENTS
This work was funded by National Science Foundation grant EF-0627988 and by National Institutes of Health (NIH) grant R01 AI084918 from the National Institute of Allergy and Infectious Diseases (NIAID). We are also thankful for support from the Idaho INBRE Program, NIH grants P20RR016454 and P20GM103408, and the COBRE program, NIH grants P20RR16448 and P20GM103397 through fellowships to D.S. via the BCB graduate program and through the IBEST Computational Resources Core. We are grateful to K. Barry, B. Foster, and A. Lapidus at the U.S. Department of Energy (DOE) Joint Genome Institute (JGI) for providing draft genome sequences, supported by the DOE Office of Science under contract no. DE-AC02-05CH11231.
We thank H. Schweizer at Colorado State University for providing the mini-Tn7 constructs and H. Suzuki for genomic signature analyses. We are also grateful for the services of the IGS Annotation Engine at the Institute for Genome Sciences, University of Maryland School of Medicine.
Footnotes
Published ahead of print 4 October 2013
REFERENCES
- 1.Thomas CM. 2004. Evolution and population genetics of bacterial plasmids p 509–528 In Funnell BE, Phillips GJ. (ed), Plasmid biology. ASM Press, Washington, DC [Google Scholar]
- 2.Szpirer C, Top E, Couturier M, Mergeay M. 1999. Retrotransfer or gene capture: a feature of conjugative plasmids, with ecological and evolutionary significance. Microbiology 145:3321–3329 [DOI] [PubMed] [Google Scholar]
- 3.Datta N, Hedges RW. 1972. Host ranges of R factors. J. Gen. Microbiol. 70:453–460 [DOI] [PubMed] [Google Scholar]
- 4.Thomas CM, Smith CA. 1987. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu. Rev. Microbiol. 41:77–101 [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Lopez R, Garcillan-Barcia M, Revilla C, Lazaro M, Vielva L, de la Cruz F. 2006. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol. Rev. 30:942–966 [DOI] [PubMed] [Google Scholar]
- 6.Rhodes G, Parkhill J, Bird C, Ambrose K, Jones M, Huys G, Swings J, Pickup R. 2004. Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity. Appl. Environ. Microbiol. 70:7497–7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rawlings D, Tietze E. 2001. Comparative biology of IncQ and IncQ-like plasmids. Microbiol. Mol. Biol. Rev. 65:481–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van der Auwera GA, Krol JE, Suzuki H, Foster B, Van Houdt R, Brown CJ, Mergeay M, Top EM. 2009. Plasmids captured in C. metallidurans CH34: defining the PromA family of broad-host-range plasmids. Antonie Van Leeuwenhoek 96:193–204 [DOI] [PubMed] [Google Scholar]
- 9.Guiney DG. 1993. Broad host range conjugative and mobilizable plasmids in Gram-negative bacteria, p 75–104 In Clewell DB. (ed), Bacterial conjugation. Plenum Press, New York, NY [Google Scholar]
- 10.Dahlberg C, Chao L. 2003. Amelioration of the cost of conjugative plasmid carriage in Escherichia coli K12. Genetics 165:1641–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Gelder L, Ponciano JM, Joyce P, Top EM. 2007. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology 153:452–463 [DOI] [PubMed] [Google Scholar]
- 12.Lenski R, Bouma J. 1987. Effects of segregation and selection on instability of plasmid pACYC184 in Escherichia coli B. J. Bacteriol. 169:5314–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lundquist PD, Levin BR. 1986. Transitory derepression and the maintenance of conjugative plasmids. Genetics 113:483–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart FM, Levin BR. 1977. The population biology of bacterial plasmids: a priori conditions for the existence of conjugationally transmitted factors. Genetics 87:209–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox RE, Zhong X, Krone SM, Top EM. 2008. Spatial structure and nutrients promote invasion of IncP-1 plasmids in bacterial populations. ISME J. 2:1024–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harada K, Aso Y, Hashimoto W, Mikami B, Murata K. 2006. Sequence and analysis of the 46.6-kb plasmid pA1 from Sphingomonas sp. A1 that corresponds to the typical IncP-1β plasmid backbone without any accessory gene. Plasmid 56:11–23 [DOI] [PubMed] [Google Scholar]
- 17.Kamachi K, Sota M, Tamai Y, Nagata N, Konda T, Inoue T, Top EM, Arakawa Y. 2006. Plasmid pBP136 from Bordetella pertussis represents an ancestral form of IncP-1β plasmids without accessory mobile elements. Microbiology 152:3477–3484 [DOI] [PubMed] [Google Scholar]
- 18.Anjum R, Grohmann E, Malik A. 2011. Molecular characterization of conjugative plasmids in pesticide tolerant and multi-resistant bacterial isolates from contaminated alluvial soil. Chemosphere 84:175–181 [DOI] [PubMed] [Google Scholar]
- 19.Halling-Sorensen B, Nors Nielsen S, Lanzky P, Ingerslev F, Holten Lutzhoft H, Jorgensen S. 1998. Occurrence, fate and effects of pharmaceutical substances in the environment—a review. Chemosphere 36:357–393 [DOI] [PubMed] [Google Scholar]
- 20.Karthikeyan K, Meyer M. 2006. Occurrence of antibiotics in wastewater treatment facilities in Wisconsin, USA. Sci. Total Environ. 361:196–207 [DOI] [PubMed] [Google Scholar]
- 21.Kümmerer K. 2001. Drugs in the environment: emission of drugs, diagnostic aids and disinfectants into wastewater by hospitals in relation to other sources—a review. Chemosphere 45:957–969 [DOI] [PubMed] [Google Scholar]
- 22.Kümmerer K. 2003. Significance of antibiotics in the environment. J. Antimicrob. Chemother. 52:5–7 [DOI] [PubMed] [Google Scholar]
- 23.Schlüter A, Szczepanowski R, Pühler A, Top EM. 2007. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol. Rev. 31:449–477 [DOI] [PubMed] [Google Scholar]
- 24.Szczepanowski R, Linke B, Krahn I, Gartemann KH, Gutzkow T, Eichler W, Pühler A, Schlüter A. 2009. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology 155:2306–2319 [DOI] [PubMed] [Google Scholar]
- 25.Smalla K, Sobecky PA. 2002. The prevalence and diversity of mobile genetic elements in bacterial communities of different environmental habitats: insights gained from different methodological approaches. FEMS Microbiol. Ecol. 42:165–175 [DOI] [PubMed] [Google Scholar]
- 26.Hill K, Weightman A, Fry J. 1992. Isolation and screening of plasmids from the epilithon which mobilize recombinant plasmid pD10. Appl. Environ. Microbiol. 58:1292–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Top E, De Smet I, Verstraete W, Dijkmans R, Mergeay M. 1994. Exogenous isolation of mobilizing plasmids from polluted soils and sludges. Appl. Environ. Microbiol. 60:831–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Elsas JD, Gardener BB, Wolters AC, Smit E. 1998. Isolation, characterization, and transfer of cryptic gene-mobilizing plasmids in the wheat rhizosphere. Appl. Environ. Microbiol. 64:880–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yano H, Deckert GE, Rogers LM, Top EM. 2012. Roles of long and short replication initiation proteins in the fate of IncP-1 plasmids. J. Bacteriol. 194:1533–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nunez B, De La Cruz F. 2001. Two atypical mobilization proteins are involved in plasmid CloDF13 relaxation. Mol. Microbiol. 39:1088–1099 [DOI] [PubMed] [Google Scholar]
- 31.Cabezon E, Sastre J, de la Cruz F. 1997. Genetic evidence of a coupling role for the TraG protein family in bacterial conjugation. Mol. Gen. Genet. 254:400–406 [DOI] [PubMed] [Google Scholar]
- 32.Kovach M, Phillips R, Elzer P, Roop RN, Peterson K. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802 [PubMed] [Google Scholar]
- 33.Antoine R, Locht C. 1992. Isolation and molecular characterization of a novel broad-host-range plasmid from Bordetella bronchiseptica with sequence similarities to plasmids from gram-positive organisms. Mol. Microbiol. 6:1785–1799 [DOI] [PubMed] [Google Scholar]
- 34.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36.Heuer H, Krögerrecklenfort E, Wellington E, Egan S, van Elsas J, van Overbeek L, Collard J, Guillaume G, Karagouni A, Nikolakopoulou T, Smalla K. 2002. Gentamicin resistance genes in environmental bacteria: prevalence and transfer. FEMS Microbiol. Ecol. 42:289–302 [DOI] [PubMed] [Google Scholar]
- 37.Sen D, Yano H, Suzuki H, Krol JE, Rogers L, Brown CJ, Top EM. 2010. Comparative genomics of pAKD4, the prototype IncP-1δ plasmid with a complete backbone. Plasmid 63:98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson J, Gibson T, Higgins D. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics Chapter 2:Unit 2.3. 10.1002/0471250953.bi0203s00 [DOI] [PubMed] [Google Scholar]
- 40.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 41.Minin V, Abdo Z, Joyce P, Sullivan J. 2003. Performance-based selection of likelihood models for phylogeny estimation. Syst. Biol. 52:674–683 [DOI] [PubMed] [Google Scholar]
- 42.Swofford D. 2003. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- 43.Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wikler MA, Cockerill FR, Craig WA, Dudley MN, Eliopoulos GM, Hecht DW, Hindler JF, Low WE, Sheehan DJ, Tenover FC, Turnidge JD, Weinstein MP, Zimmer BL, Ferraro MJ, Swenson JM. 2010. Performance standards for antimicrobial disk susceptibility tests. Approved standard, 9th ed. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 45.Sen D, Brown CJ, Top EM, Sullivan J. 2013. Inferring the evolutionary history of IncP-1 plasmids despite incongruence among backbone gene trees. Mol. Biol. Evol. 30:154–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haines A, Akhtar P, Stephens E, Jones K, Thomas C, Perkins C, Williams J, Day M, Fry J. 2006. Plasmids from freshwater environments capable of IncQ retrotransfer are diverse and include pQKH54, a new IncP-1 subgroup archetype. Microbiology 152:2689–2701 [DOI] [PubMed] [Google Scholar]
- 47.Yano H, Rogers LM, Knox M, Heuer H, Smalla K, Brown CJ, Top EM. 3 September 2013. Host range diversification within the IncP-1 plasmid group. Microbiology. [Epub ahead of print.] 10.1099/mic.0.068387-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulinska A, Czeredys M, Hayes F, Jagura-Burdzy G. 2008. Genomic and functional characterization of the modular broad-host-range RA3 plasmid, the archetype of the IncU group. Appl. Environ. Microbiol. 74:4119–4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ludwiczak M, Dolowy P, Markowska A, Szarlak J, Kulinska A, Jagura-Burdzy G. 2013. Global transcriptional regulator KorC coordinates expression of three backbone modules of the broad-host-range RA3 plasmid from IncU incompatibility group. Plasmid 70:131–145 [DOI] [PubMed] [Google Scholar]
- 50.Li H, Toleman MA, Bennett PM, Jones RN, Walsh TR. 2008. Complete Sequence of p07-406, a 24,179-base-pair plasmid harboring the blaVIM-7 metallo-β-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 52:3099–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcillan-Barcia M, Francia M, de la Cruz F. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33:657–687 [DOI] [PubMed] [Google Scholar]
- 52.Stein L, Arp D, Berube P, Chain P, Hauser L, Jetten M, Klotz M, Larimer F, Norton J, Op den Camp H, Shin M, Wei X. 2007. Whole-genome analysis of the ammonia-oxidizing bacterium, Nitrosomonas eutropha C91: implications for niche adaptation. Environ. Microbiol. 9:2993–3007 [DOI] [PubMed] [Google Scholar]
- 53.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33:W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marques MV, da Silva AM, Gomes SL. 2001. Genetic organization of plasmid pXF51 from the plant pathogen Xylella fastidiosa. Plasmid 45:184–199 [DOI] [PubMed] [Google Scholar]
- 55.Schlüter A, Krahn I, Kollin F, Bönemann G, Stiens M, Szczepanowski R, Schneiker S, Pühler A. 2007. IncP-1β plasmid pGNB1 isolated from a bacterial community from a wastewater treatment plant mediates decolorization of triphenylmethane dyes. Appl. Environ. Microbiol. 73:6345–6350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stolze Y, Eikmeyer F, Wibberg D, Brandis G, Karsten C, Krahn I, Schneiker-Bekel S, Viehover P, Barsch A, Keck M, Top EM, Niehaus K, Schluter A. 2012. IncP-1β plasmids of Comamonas sp. and Delftia sp. strains isolated from a wastewater treatment plant mediate resistance to and decolorization of the triphenylmethane dye crystal violet. Microbiology 158:2060–2072 [DOI] [PubMed] [Google Scholar]
- 57.Norberg P, Bergstrom M, Jethava V, Dubhashi D, Hermansson M. 2011. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat. Commun. 2:268. 10.1038/ncomms1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian Q, Ohnishi M, Tabuchi A, Terawaki Y. 1996. A new plasmid-encoded proteic killer gene system: cloning, sequencing, and analyzing hig locus of plasmid Rts1. Biochem. Biophys. Res. Commun. 220:280–284 [DOI] [PubMed] [Google Scholar]
- 59.Eikmeyer F, Hadiati A, Szczepanowski R, Wibberg D, Schneiker-Bekel S, Rogers LM, Brown CJ, Top EM, Pühler A, Schlüter A. 2012. The complete genome sequences of four new IncN plasmids from wastewater treatment plant effluent provide new insights into IncN plasmid diversity and evolution. Plasmid 68:13–24 [DOI] [PubMed] [Google Scholar]
- 60.Humphrey B, Thomson NR, Thomas CM, Brooks K, Sanders M, Delsol AA, Roe JM, Bennett PM, Enne VI. 2012. Fitness of Escherichia coli strains carrying expressed and partially silent IncN and IncP1 plasmids. BMC Microbiol. 12:53. 10.1186/1471-2180-12-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Padan E, Maisler N, Taglicht D, Karpel R, Schuldiner S. 1989. Deletion of ant in Escherichia coli reveals its function in adaptation to high salinity and an alternative Na+/H+ antiporter system(s). J. Biol. Chem. 264:20297–20302 [PubMed] [Google Scholar]
- 62.Top E, Holben W, Forney L. 1995. Characterization of diverse 2,4-dichlorophenoxyacetic acid-degradative plasmids isolated from soil by complementation. Appl. Environ. Microbiol. 61:1691–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heuer H, Smalla K. 2012. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 36:1083–1104 [DOI] [PubMed] [Google Scholar]
- 64.Akiyama T, Asfahl K, Savin M. 2010. Broad-host-range plasmids in treated wastewater effluent and receiving streams. J. Environ. Qual. 39:2211–2215 [DOI] [PubMed] [Google Scholar]
- 65.Inoue D, Murashige K, Sei K, Soda S, Ike M, Fujita M. 2007. Distribution and characteristics of plasmid mobilizers in aquatic and soil environments and activated sludge. J. Gen. Appl. Microbiol. 53:67–70 [DOI] [PubMed] [Google Scholar]
- 66.Dröge M, Pühler A, Selbitschka W. 2000. Phenotypic and molecular characterization of conjugative antibiotic resistance plasmids isolated from bacterial communities of activated sludge. Mol. Gen. Genet. 263:471–482 [DOI] [PubMed] [Google Scholar]
- 67.Moura A, Henriques I, Smalla K, Correia A. 2010. Wastewater bacterial communities bring together broad-host range plasmids, integrons and a wide diversity of uncharacterized gene cassettes. Res. Microbiol. 161:58–66 [DOI] [PubMed] [Google Scholar]
- 68.Fernandez-Alarcon C, Singer R, Johnson T. 2011. Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS One 6:e23415. 10.1371/journal.pone.0023415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burton NF, Day MJ, Bull AT. 1982. Distribution of bacterial plasmids in clean and polluted sites in a South Wales river. Appl. Environ. Microbiol. 44:1026–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Glassman DL, McNicol LA. 1981. Plasmid frequency in natural populations of estuarine microorganisms. Plasmid 5:224–231 [Google Scholar]
- 71.Hada H, Sizemore R. 1981. Incidence of plasmids in marine Vibrio spp. isolated from an oil field in the northwestern Gulf of Mexico. Appl. Environ. Microbiol. 41:199–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kristiansson E, Fick J, Janzon A, Grabic R, Rutgersson C, Weijdegard B, Soderstrom H, Larsson DG. 2011. Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS One 6:e17038. 10.1371/journal.pone.0017038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brown Kav A, Sasson G, Jami E, Doron-Faigenboim A, Benhar I, Mizrahi I. 2012. Insights into the bovine rumen plasmidome. Proc. Natl. Acad. Sci. U. S. A. 109:5452–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li LL, Norman A, Hansen LH, Sorensen SJ. 2012. Metamobilomics—expanding our knowledge on the pool of plasmid encoded traits in natural environments using high-throughput sequencing. Clin. Microbiol. Infect. 18(Suppl 4):5–7 [DOI] [PubMed] [Google Scholar]
- 75.Jones B, Sun F, Marchesi J. 2010. Comparative metagenomic analysis of plasmid encoded functions in the human gut microbiome. BMC Genomics 11:46. 10.1186/1471-2164-11-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sen D, Van der Auwera GA, Rogers LM, Thomas CM, Brown CJ, Top EM. 2011. Broad-host-range plasmids from agricultural soils have IncP-1 backbones with diverse accessory genes. Appl. Environ. Microbiol. 77:7975–7983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Drønen AK, Torsvik V, Goksøyr J, Top EM. 1998. Effect of mercury addition on plasmid incidence and gene mobilizing capacity in bulk soil. FEMS Microbiol. Ecol. 27:381–394 [Google Scholar]
- 78.Kobayashi N, Bailey MJ. 1994. Plasmids isolated from the sugar beet phyllosphere show little or no homology to molecular probes currently available for plasmid typing. Microbiology 140:289–296 [DOI] [PubMed] [Google Scholar]
- 79.Ghigo J. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445 [DOI] [PubMed] [Google Scholar]
- 80.Hase C, Finkelstein R. 1993. Bacterial extracellular zinc-containing metalloproteases. Microbiol. Rev. 57:823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Suzuki H, Yano H, Brown C, Top E. 2010. Predicting plasmid promiscuity based on genomic signature. J. Bacteriol. 192:6045–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
- 83.Choi KH, Schweizer HP. 2006. Mini-Tn7 insertion in bacteria with secondary, non-glmS-linked attTn7 sites: example Proteus mirabilis HI4320. Nat. Protoc. 1:170–178 [DOI] [PubMed] [Google Scholar]
- 84.Choi KH, Gaynor JB, White KG, Lopez C, Bosio CM, Karkhoff-Schweizer RR, Schweizer HP. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2:443–448 [DOI] [PubMed] [Google Scholar]
- 85.Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW, Crosa JH, Falkow S. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95–113 [PubMed] [Google Scholar]