Abstract
GET_HOMOLOGUES is an open-source software package that builds on popular orthology-calling approaches making highly customizable and detailed pangenome analyses of microorganisms accessible to nonbioinformaticians. It can cluster homologous gene families using the bidirectional best-hit, COGtriangles, or OrthoMCL clustering algorithms. Clustering stringency can be adjusted by scanning the domain composition of proteins using the HMMER3 package, by imposing desired pairwise alignment coverage cutoffs, or by selecting only syntenic genes. The resulting homologous gene families can be made even more robust by computing consensus clusters from those generated by any combination of the clustering algorithms and filtering criteria. Auxiliary scripts make the construction, interrogation, and graphical display of core genome and pangenome sets easy to perform. Exponential and binomial mixture models can be fitted to the data to estimate theoretical core genome and pangenome sizes, and high-quality graphics can be generated. Furthermore, pangenome trees can be easily computed and basic comparative genomics performed to identify lineage-specific genes or gene family expansions. The software is designed to take advantage of modern multiprocessor personal computers as well as computer clusters to parallelize time-consuming tasks. To demonstrate some of these capabilities, we survey a set of 50 Streptococcus genomes annotated in the Orthologous Matrix (OMA) browser as a benchmark case. The package can be downloaded at http://www.eead.csic.es/compbio/soft/gethoms.php and http://maya.ccg.unam.mx/soft/gethoms.php.
INTRODUCTION
The ever-growing number of sequenced genomes in public databases such as GenBank has prompted the development of tools aimed at comparing the gene repertoires of species. Such comparisons include the identification of orthologous genes, assumed to diverge from a common ancestor after a speciation event and more likely to conserve their functions across organisms than paralogues (1). For this reason, orthologues are key elements in genome annotation and evolutionary studies (2, 3). Among bacteria, which are being sequenced faster than any other domain of life (4), a popular heuristic recipe for detecting orthologous sequences is simply looking for reciprocal BLAST hits (5, 6), and different software choices are available for this task (7). By combining these tools with a growing number of genomic sequences, several recent studies have provided evidence suggesting that bacterial genomes are actually mosaics that include genes shared by all isolates of a group of interest (core genome) as well as strain-specific/partially shared genes (8). The sum of the core genome and the remaining genes within the group is defined as the pangenome (9).
Here we present GET_HOMOLOGUES, an open-source software package released under the GNU General Public License, specifically designed and tested for the pangenomic and comparative-genomic analysis of bacterial strains at different phylogenetic distances on Linux/Mac OS X computer systems. The software is unique in several respects. It implements a fully automatic and highly customizable analysis pipeline, including genome data download, extraction of user-selected sequence features, running of BLAST and HMMER jobs, and indexing, clustering, and parsing of results. It can take advantage of modern multiprocessor architectures, as well as computer clusters, to parallelize time-consuming BLAST and HMMER jobs. It can handle large data sets (for instance, we have analyzed 101 Escherichia coli genomes) on reasonably modest machines (<8 GB RAM) by using Berkeley DB to write temporary data to a disk and/or by calling a heuristic version of our bidirectional best-hit (BDBH) algorithm. Auxiliary scripts are integrated to facilitate the parsing and generation of gene families, including the computation of consensus clusters recovered by combinations of the sequence-clustering algorithms supported. Other scripts are provided for the statistical analysis and graphical display of results, including core and pangenome plots, by calling R functions. Diverse comparative-genomics analyses can be also performed. Finally, an installation script is provided to simplify the installation process, and a very detailed manual with hands-on tutorials is also provided to make this software package reasonably user-friendly.
Here we show some of these capabilities by analyzing a set of 50 Streptococcus genomes downloaded from the most recent version of OMA (Orthologous Matrix), a database that identifies orthologues among publicly available, complete genomes (10). We chose this genus for several reasons. It exhibits very high levels of genome plasticity (11). The first pangenomic analyses were conducted on Streptococcus agalactiae in the pioneering work of Tettelin and colleagues (12), and very detailed comparative-genomics studies have followed for diverse species in the genus, including the major human pathogens S. pyogenes (13) and S. pneumoniae (14), making Streptococcus an excellent test case for the GET_HOMOLOGUES software.
MATERIALS AND METHODS
Input data and output formats.
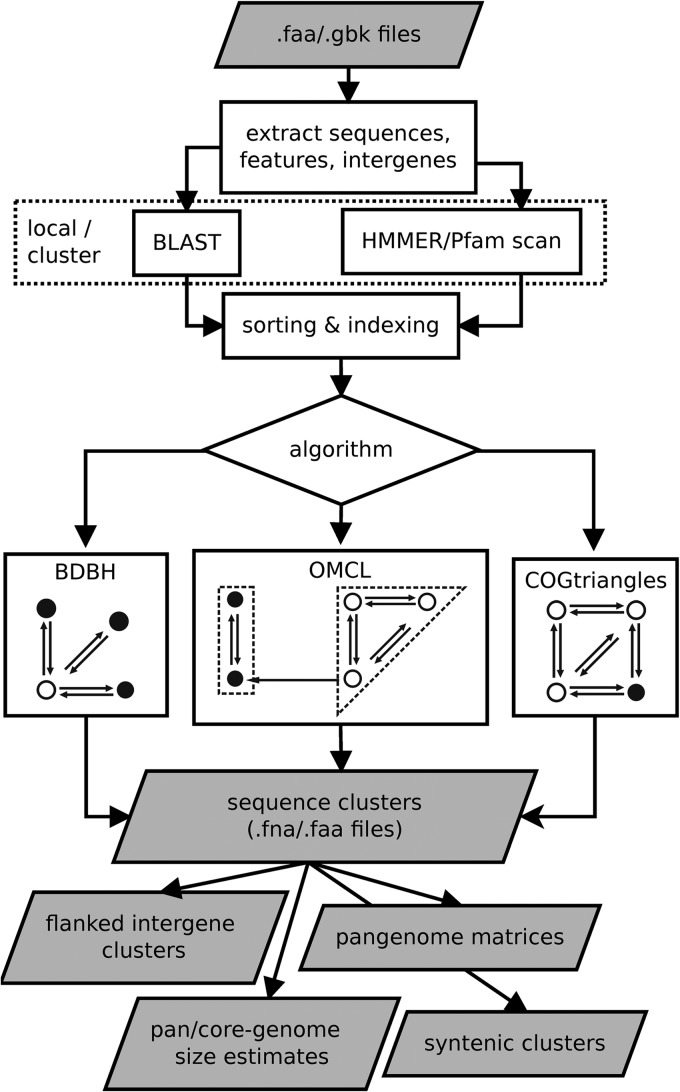
GET_HOMOLOGUES takes GenBank or FASTA input files and can produce different outputs, as summarized in Fig. 1, including orthologous gene families in FASTA and OrthoXML formats (15), at both the DNA and amino acid levels.
Fig 1.
GET_HOMOLOGUES flow chart and its outcomes. BLAST and optional Pfam searches are optimized for local (multicore) and cluster computer environments. While the BDBH algorithm uses one sequence from the reference genome to grow clusters, the COG algorithm requires a triangle of reciprocal hits. Instead, the OMCL algorithm groups nodes in a BLAST graph to build clusters. Note that these clustering algorithms can be fine-tuned by customizing parameters such as -C (minimum percentage of coverage in pairwise BLAST alignments), -E (maximum E value for a hit to be considered), -D (require equal Pfam domain composition when defining similarity-based orthology composition), -S (minimum percentage of sequence identity in BLAST query/subject pairs [BDBH|OMCL]) and -N (minimum BLAST neighborhood correlation [BDBH|OMCL]). In addition, the user can choose which genome should be used as the reference using option -r.
Third-party software dependencies, data processing, and sequence clustering.
The software is built on top of BLAST+ (16) and the code base of OrthoMCL, version 1.4 (17), and supports three popular sequence-clustering algorithms: OrthoMCL (OMCL), COGtriangles (18), and our own implementation of the bidirectional best hit (BDBH) algorithm (see Fig. S1 in the supplemental material). Despite their distinct strategies, these approaches call orthologous sequences using BLAST reciprocal best hits as evidence (19), and distinguish inparalogues (20) as genes with best hits in the same genome, i.e., recent paralogues. Moreover, HMMER (http://hmmer.org) is integrated to facilitate Pfam annotation of protein domains (21), so that clusters containing sequences with different domain architectures, which can confound orthology assignment, can be filtered out. If input files are in GenBank format, both nucleotide and amino acid sequence clusters are produced, and orthologous intergenic regions, flanked by orthologous genes, can also be extracted if required. In addition, genome coordinates in GenBank files can be used for selecting syntenic clusters, those with at least one conserved neighbor (see Fig. S2 in the supplemental material). Finally, since GenBank files contain a variety of DNA features, the software can also be asked to focus on, for instance, tRNA genes. In this case, BLASTN searches are performed instead of default BLASTP jobs.
Auxiliary scripts for cluster parsing and analysis.
In addition to the main Perl script get_homologues.pl, this software bundles a few auxiliary scripts to help with subsequent analyses. For instance, intersection clusters produced by several algorithms (such as COGtriangles and OMCL) can be easily selected with compare_clusters.pl, allowing the user to work only with consensus clusters. In addition, the script plot_pancore_matrix.pl is provided for plotting pangenomes and core genomes and for fitting the exponential models of Tettelin et al. (9) and Willenbrock et al. (22) to estimate core and pangenome sizes. Pangenomic matrices are conveniently provided in tabular and PHYLIP format for the automatic generation of pangenomic trees under the parsimony criterion, using the PARS program from the PHYLIP package (23), as illustrated here. The parse_pangenome_matrix.pl script is useful for comparative genomics, focusing on the identification of lineage-specific gene families or expansions, as well as for computing and graphing core, cloud, and shell genome compartments (24). GET_HOMOLOGUES defines these compartments empirically, as follows: core, genes contained in all genomes/taxa considered; soft core, genes contained in 95% of the genomes/taxa considered, as in the work of Kaas and collaborators (25); cloud, genes present only in a few genomes/taxa (the cutoff was defined as the most populated noncore cluster class and its immediately neighboring classes); shell, the remaining genes, present in several genomes/taxa. This script will also compute estimates of core and pangenome sizes under the binomial mixture model of Snipen and colleagues (26). If the source genomic data are provided in GenBank format, parse_pangenome_matrix.pl can be asked to plot the clade-specific genes on a linearized genetic map of a reference genome selected from that lineage. The pangenomic tree computed by compare_clusters.pl can be useful for selecting the members of the groups to be compared by parse_pangenome_matrix.pl.
Benchmark data sets.
In order to test our software pipeline and demonstrate its capabilities, 50 Streptococcus proteomes from 14 species were downloaded in FASTA format from the December 2012 release of the OMA browser (27), together with their orthologous groups (http://omabrowser.org) (see Table ST1 in the supplemental material). We chose OMA for this benchmark because it is a validated and updated repository of orthologous genes across genomes spanning all domains of life (27). OMA is based on an algorithm that compares genes on the basis of pairwise evolutionary distances instead of BLAST-scores, considering distance estimation uncertainty, and accounts for differential gene losses (10, 28). In this work we also reanalyze the 26 Streptococcus genomes analyzed by Lefébure and Stanhope (11) (see Table ST2 in the supplemental material).
Proteome annotation.
It was necessary to annotate the OMA clusters, because their sequence headers contain only OMA identifiers. To do so, we used a modified version of AutoFACT (29), which uses recent, curated, comprehensive sequence databases, such as NCBI's Conserved Domains Database (30) and Protein Clusters Database (31), in addition to the COG (Clusters of Orthologous Groups) database (32), the Kyoto Encyclopedia of Genes and Genomes (KEGG) PATHWAY Database (33), the Enzyme Nomenclature Database (http://www.expasy.org/enzyme/), and the Pfam (21) and UniRef90 (34) databases.
Supported platforms and availability.
This software is written in Perl and R (http://www.R-project.org) and is best run on a multicore Linux/Mac OS X box or on a Sun Grid Engine (SGE) computer cluster (tested with Rocks, versions 4.3 and 5.4 [http://www.rocksclusters.org]). The compressed software package is available for 32- and 64-bit processors and includes a user manual with detailed hands-on tutorials, as well as an installation script that checks for optional software dependencies and guides the user on how to proceed to install them. This script also supports downloading and formatting of the current version of Pfam. The package can be downloaded at http://www.eead.csic.es/compbio/soft/gethoms.php and http://maya.ccg.unam.mx/soft/gethoms.php.
RESULTS AND DISCUSSION
Comparison of GET_HOMOLOGUES clusters with those provided by the OMA browser for completely sequenced genomes.
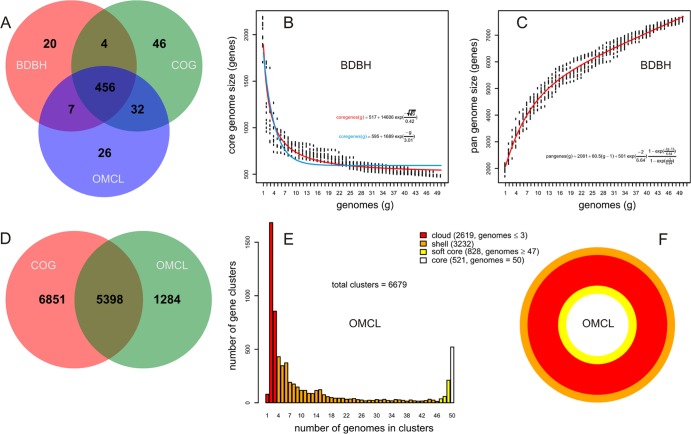
The core genome for the 50 Streptococcus proteomes from 14 species available in the last release of OMA was calculated with all three clustering strategies (BDBH, OMCL, and COG), imposing a minimum pairwise alignment coverage of 75%. Using these parameters, we obtained 487 BDBH, 521 OMCL, and 538 COG clusters; 456 consensus clusters were detected by all three algorithms (Fig. 2A; see also Table ST3 in the supplemental material). The robustness of GET_HOMOLOGUES is demonstrated by the fact that these core sets contain all of the 177 core genes reported by the OMA project of orthologous protein families (OMAc), as well as many more genes, including 391 BDBH, 413 OMCL, and 428 COG clusters with the same Pfam domain architecture (see Table ST4 in the supplemental material). Among these genes are those encoding essential proteins, such as seven 50S and six 30S ribosomal subunit proteins, translation initiation factor IF-2 (InfB), elongation factor G (FusA), a transcription termination factor (NusA), a transcription antitermination protein (NusG), a DNA topoisomerase (TopA), an ATP-dependent zinc metalloprotease (FtsH), and a recombinase (RecA), to mention but a few. These are bona fide core genes, most of them present in the “universal or extended universal” core computed from 12 bacterial and 2 archaeal phyla (35) (see Table ST5 in the supplemental material). The size of the strict core computed by GET_HOMOLOGUES is actually within the range (446 to 491 genes) of the strict core computed by Charlebois and Doolittle for 23 complete Bacillus/Streptococcus genomes (35), further highlighting the robustness of this calculation. In addition, sampling experiments similar to those carried out by Tettelin et al. (12), in which Streptococcus genomes are randomly added to the pangenome pool in order to track the fractions of unique and common genes contributed, were performed with the BDBH strategy to estimate the theoretical core genome size (Fig. 2B). The fitted functions converged to sizes of 517 and 595 genes when the Willenbrock (22) and Tettelin (12) fits, respectively, were used, clearly much larger than the core genome reported by OMAc. The pangenome samples (Fig. 2C) seem to converge to linear growth, as already observed by Tettelin for 8 S. agalactiae genomes (12). Note, however, that as expected, the slope of the pangenome curve fitted to the larger and taxonomically more diverse OMA data set (a slope of 60.5) is almost twice the slope of 33 reported by Tettelin. The pangenomes obtained by OMCL and COG agree for 5,398 clusters but disagree mainly for clusters of <3 genomes, because COGtriangles does not resolve them (Fig. 2D). Figure 2E and F show the partition of OMCL pangenome clusters. Note that for 50 Streptococcus strains, the soft-core compartment includes sequences found in at least 47 genomes.
Fig 2.
Pangenome analysis of 50 Streptococcus genomes from 14 species. (A) Venn diagram of core genomes generated by the BDBH, COG, and OMCL strategies. (B) Estimate of core genome size with the Tettelin (blue) and Willenbrock (red) fits (12, 22). (C) Estimate of pangenome size with the Tettelin fit. (D) Venn analysis of pangenomes generated by COG and OMCL. (E and F) Partition of the OMCL pangenomic matrix into shell, cloud, soft-core, and core compartments. These plots can be easily created with GET_HOMOLOGUES auxiliary scripts, as explained in the manual.
Computing soft-core genomes with GET_HOMOLOGUES on data sets containing draft genomes.
The performance of GET_HOMOLOGUES for core genomes was further validated with 26 genomes, from 6 Streptococcus species, previously analyzed by Lefébure and Stanhope (11), a data set that (currently) contains 6 draft genomes. Of the 611 core genes reported by these authors, the consensus (OMCL and COGtriangles) soft-core genome recovered 529, as illustrated in Fig. S3 in the supplemental material. Soft-core genes are by definition required to be present in at least 95% of the input genomes, allowing for missing or fragmented genes, which are expected when one is comparing data sets that include draft genomes (25). A strict core genome for this data set, based on the consensus between the BDBH, COG, and OMCL algorithms and on default 75% pairwise alignment coverage, contains only 412 gene families. This figure is clearly an underestimation of the core genome size, as judged from the estimates obtained with 50 proteomes from fully sequenced genomes presented in Fig. 2B and Table ST4 in the supplemental material. This misleading result is caused by missing or incomplete genes in the 6 draft genomes included in the data set, demonstrating the value of defining a “soft” core genome (25) and the flexibility of the GET_HOMOLOGUES package in adapting to different types of data sets and analysis requirements.
Use of pangenome trees to aid in group selection for comparative genomics.
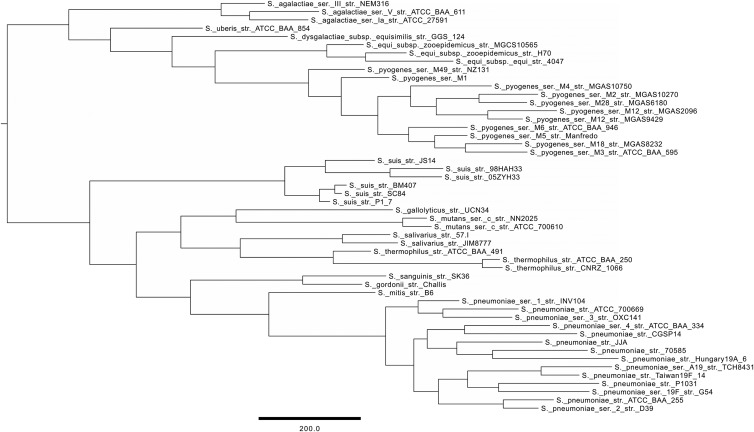
The auxiliary script compare_clusters.pl can be asked to generate a pangenomic matrix of the presence or absence of genes, which is inferred from the clusters generated by the main script get_homologues.pl when run with the option -t 0 (which retrieves clusters of all sizes, as opposed to default core clusters). Such a matrix was calculated for the 50 Streptococcus proteomes extracted from OMA and was then used to calculate the parsimony pangenomic tree shown in Fig. 3. This kind of tree shows the phylogenetic relationship of proteomes based on their gene family contents. Such phylogenies arguably reflect better the genetic affinities of the proteomes based on their composition (presence or absence of homologous genes), and therefore their phenotypic potential, than conventional species phylogenies estimated from concatenated alignments of core genome genes or gene products (36). For comparison purposes, we also computed a maximum likelihood tree from 364 concatenated alignments of the corresponding “strict” single-copy consensus core genes (see Fig. S4 in the supplemental material), reported by all BDBH, COGtriangles, and OMCL algorithms with the Pfam domain-scanning option enabled (-D) (the corresponding core genome estimates are shown in Table ST4 in the supplemental material). In both phylogenies, the species appear as monophyletic entities. However, comparison of the relationships between species in the two phylogenies reveals some differences. For example, in the pangenome tree, S. equi strains appear as the sister group to S. pyogenes, while on the core genome phylogenetic hypothesis, S. dysgalactiae would be the closest relative to S. pyogenes, as was found by Lefébure et al. (13) in a similar analysis. Yet in another study (37), the S. dysgalactiae strains analyzed grouped as the sister clade of S. pyogenes by use of hierarchical clustering (unweighted-pair group method using average linkages [UPGMA]) based on dissimilarities in gene content. GET_HOMOLOGUES uses the parsimony optimality criterion to compute pangenomic trees because the accuracy of the UPGMA clustering algorithm is well known to degrade with increasing deviation of the underlying distance data from ultrametricity (38). Given the important differences in the rates of gene gain or loss frequently observed between closely related bacterial lineages (13), deviations from ultrametricity should be observed frequently in pangenomic matrices. However, the tab-delimited pangenome matrix can be used to compute distance matrices in R or other packages. The differences between the groupings revealed by the two types of core and pangenome trees can be exploited as alternative evolutionary hypotheses to guide the selection of species or proteome groups for comparative-genomics analysis of clade pairs.
Fig 3.
Parsimony pangenome tree for 50 Streptococcus proteomes derived from presence/absence data in a consensus (OMCL and COGtriangles) pangenome matrix computed from the OMA data set, as detailed in the text. This phylogeny was the most parsimonious tree found in a tree search performed with PARS from the PHYLIP suite, using 50 data jumbles. The tree has a total length of 11,473 steps.
Identifying clade-specific genes and gene family expansions.
The auxiliary script parse_pangenome_matrix.pl can be used to report cloud, shell, soft-core, and core clusters (24) and to display them graphically (if R is installed), as shown in Fig. 2E and F, which indicate that the shell genome component is the largest one (3,232 genes present in 4 to 46 strains) for the 50 streptococci analyzed, followed by the cloud genome (2,619 genes present in ≤3 genomes). These data clearly illustrate the complex structure of the Streptococcus pangenome. The same script is also useful for performing basic comparative-genomics studies, specifically to identify lineage-specific genes or gene expansions in a target group “A” with regard to a reference group “B.” Option -P can be used to define the percentage of genomes that must comply with the presence or absence of a particular cluster (the default is 100%) in order to define genes specifically found or expanded in the focal lineage “A.” As an example, we performed an analysis to identify such gene families among the 11 S. pyogenes proteomes from the OMA data set, using as a close reference group the S. equi strains. Group selection was based on the pangenomic tree shown in Fig. 3. The analysis was first performed using the default option -P 100 to identify those gene clusters present or amplified in all 11 S. pyogenes strains. We found 42 S. pyogenes-specific gene clusters and 3 that were selectively expanded in this lineage in comparison to S. equi. Among the first class of genes were those encoding well-established streptococcal virulence factors, such as pyrogenic exotoxins, which are associated with streptococcal toxic shock syndrome and scarlet fever (39), and a hyaluronic acid hydrolase, involved in the degradation of connective tissue and in pathogen spread (40). Other annotated proteins include salivaricin A precursor, a lantibiotic produced by >90% of oral S. pyogenes strains found in human saliva (41), and a putative ATP-binding cassette (ABC) transporter previously shown to be involved in the virulence of S. pneumoniae in a mouse model of infection (42). All these genes were also found as part of the S. pyogenes gene repertoire in a recent study by Lefébure and colleagues (13). An additional 13 S. pyogenes-specific gene clusters were found when -P was relaxed to 90 (i.e., a cluster was considered lineage specific when at least 90% of the genomes in the target group A contained it). Among them were genes encoding bacteriocin UviB, a plasmid stabilization system toxin protein, and the sensor kinase DpiB from the dpiA-dpiB two-component regulatory system. The possibility of relaxing this value is particularly useful in dealing with draft genomes, because it makes this type of comparative-genomics analysis robust even when missing genes are expected due to incomplete genome sequencing. Table ST6 in the supplemental material shows the complete list of the genes found in these analyses along with their annotations.
Software design and performance benchmarks.
Unlike similar tools (43–45), which require the user to provide precomputed sequence similarity results, GET_HOMOLOGUES explicitly takes care of BLAST and optional Pfam searches. The software can take advantage of modern multiprocessor architectures to parallelize expensive BLAST+ and HMMScan analyses or to submit jobs to a computer cluster (see the manual and Fig. S5 in the supplemental material). For instance, 20 Escherichia coli genomes (93,612 genes) can be analyzed in <6 h on a commodity laptop (4 GB RAM, 4 cores). Figure S6 in the supplemental material plots the memory footprint and run time of increasingly larger sequence sets, clearly indicating that the software is more scalable when one is using Berkeley DB, a high-performance embedded database. For instance, 101 Escherichia coli genomes can be successfully processed with modest RAM requirements by invoking the -s option in the command line: 880 MB, 932 MB, and 902 MB for BDBH, OMCL, and COGtriangles, respectively. For extremely large data sets, GET_HOMOLOGUES also includes a heuristic option to minimize the number of BLAST searches required for a core genome BDBH job, which grows linearly instead of quadratically (see Fig. S7 and Table ST7 in the supplemental material).
In conclusion, we have shown that GET_HOMOLOGUES is a powerful, highly customizable, and fully automatic analysis pipeline that makes robust and rigorous pangenomic and comparative-genomics analyses much easier to perform for microbiologists without strong bioinformatics skills or dedicated hardware. In addition, GET_HOMOLOGUES is scalable, can deal with dozens of genomes on relatively modest computer systems, and will handle hundreds of genomes on more-powerful servers or computer clusters, making it suitable for large-scale pangenomic and comparative-genomics studies. It is open-source software, freely available for academic use.
Supplementary Material
ACKNOWLEDGMENTS
We thank Romualdo Zayas and Víctor del Moral at CCG-UNAM for technical support. We also thank David M. Kristensen and the development team of OrthoMCL for permission to use their code in our project.
Funding for this work was provided by the Fundación ARAID, Consejo Superior de Investigaciones Científicas (grant 200720I038), DGAPA-PAPIIT UNAM-México (grant IN212010), and CONACyT-México (grants P1-60071 and 179133).
Footnotes
Published ahead of print 4 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02411-13.
REFERENCES
- 1.Altenhoff AM, Studer RA, Robinson-Rechavi M, Dessimoz C. 2012. Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS Comput. Biol. 8:e1002514. 10.1371/journal.pcbi.1002514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altenhoff AM, Dessimoz C. 2012. Inferring orthology and paralogy. Methods Mol. Biol. 855:259–279 [DOI] [PubMed] [Google Scholar]
- 3.Koonin EV. 2005. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 39:309–338 [DOI] [PubMed] [Google Scholar]
- 4.Pagani I, Liolios K, Jansson J, Chen IM, Smirnova T, Nosrat B, Markowitz VM, Kyrpides NC. 2012. The Genomes OnLine Database (GOLD) v. 4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 40:D571–D579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rivera MC, Jain R, Moore JE, Lake JA. 1998. Genomic evidence for two functionally distinct gene classes. Proc. Natl. Acad. Sci. U. S. A. 95:6239–6244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kristensen DM, Wolf YI, Mushegian AR, Koonin EV. 2011. Computational methods for Gene Orthology inference. Brief. Bioinform. 12:379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch RA, Burland V, Plunkett G, III, Redford P, Roesch P, Rasko D, Buckles EL, Liou SR, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HL, Donnenberg MS, Blattner FR. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:17020–17024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tettelin H, Riley D, Cattuto C, Medini D. 2008. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 11:472–477 [DOI] [PubMed] [Google Scholar]
- 10.Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. 2011. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 39:D289–D294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lefébure T, Stanhope MJ. 2007. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 8:R71. 10.1186/gb-2007-8-5-r71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lefébure T, Richards VP, Lang P, Pavinski-Bitar P, Stanhope MJ. 2012. Gene repertoire evolution of Streptococcus pyogenes inferred from phylogenomic analysis with Streptococcus canis and Streptococcus dysgalactiae. PLoS One 7:e37607. 10.1371/journal.pone.0037607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Dunning Hotopp JC, Hu FZ, Riley DR, Covacci A, Mitchell TJ, Bentley SD, Kilian M, Ehrlich GD, Rappuoli R, Moxon ER, Masignani V. 2010. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 11:R107. 10.1186/gb-2010-11-10-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmitt T, Messina DN, Schreiber F, Sonnhammer EL. 2011. SeqXML and OrthoXML: standards for sequence and orthology information. Brief. Bioinform. 12:485–488 (Letter.) [DOI] [PubMed] [Google Scholar]
- 16.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kristensen DM, Kannan L, Coleman MK, Wolf YI, Sorokin A, Koonin EV, Mushegian A. 2010. A low-polynomial algorithm for assembling clusters of orthologous groups from intergenomic symmetric best matches. Bioinformatics 26:1481–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolf YI, Koonin EV. 2012. A tight link between orthologs and bidirectional best hits in bacterial and archaeal genomes. Genome Biol. Evol. 4:1286–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonnhammer EL, Koonin EV. 2002. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 18:619–620 [DOI] [PubMed] [Google Scholar]
- 21.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willenbrock H, Hallin PF, Wassenaar TM, Ussery DW. 2007. Characterization of probiotic Escherichia coli isolates with a novel pan-genome microarray. Genome Biol. 8:R267. 10.1186/gb-2007-8-12-r267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package), version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle, WA [Google Scholar]
- 24.Koonin EV, Wolf YI. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 36:6688–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaas RS, Friis C, Ussery DW, Aarestrup FM. 2012. Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics 13:577. 10.1186/1471-2164-13-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snipen L, Almoy T, Ussery DW. 2009. Microbial comparative pan-genomics using binomial mixture models. BMC Genomics 10:385. 10.1186/1471-2164-10-385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider A, Dessimoz C, Gonnet GH. 2007. OMA Browser—exploring orthologous relations across 352 complete genomes. Bioinformatics 23:2180–2182 [DOI] [PubMed] [Google Scholar]
- 28.Roth AC, Gonnet GH, Dessimoz C. 2008. Algorithm of OMA for large-scale orthology inference. BMC Bioinformatics 9:518. 10.1186/1471-2105-9-518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koski LB, Gray MW, Lang BF, Burger G. 2005. AutoFACT: an automatic functional annotation and classification tool. BMC Bioinformatics 6:151. 10.1186/1471-2105-6-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. 2013. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 41:D348–D352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, Kiryutin B, O'Neill K, Resch W, Resenchuk S, Schafer S, Tolstoy I, Tatusova T. 2009. The National Center for Biotechnology Information's Protein Clusters Database. Nucleic Acids Res. 37:D216–D223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. 2004. The KEGG resource for deciphering the genome. Nucleic Acids Res. 32:D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. 2007. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23:1282–1288 [DOI] [PubMed] [Google Scholar]
- 35.Charlebois RL, Doolittle WF. 2004. Computing prokaryotic gene ubiquity: rescuing the core from extinction. Genome Res. 14:2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snipen L, Ussery DW. 2010. Standard operating procedure for computing pangenome trees. Stand. Genomic Sci. 2:135–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki H, Lefébure T, Hubisz MJ, Pavinski Bitar P, Lang P, Siepel A, Stanhope MJ. 2011. Comparative genomic analysis of the Streptococcus dysgalactiae species group: gene content, molecular adaptation, and promoter evolution. Genome Biol. Evol. 3:168–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Felsenstein J. 2004. Inferring phylogenies, 1st ed. Sinauer Associates, Inc., Sunderland, MA [Google Scholar]
- 39.Lappin E, Ferguson AJ. 2009. Gram-positive toxic shock syndromes. Lancet Infect. Dis. 9:281–290 [DOI] [PubMed] [Google Scholar]
- 40.Starr CR, Engleberg NC. 2006. Role of hyaluronidase in subcutaneous spread and growth of group A Streptococcus. Infect. Immun. 74:40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wescombe PA, Upton M, Dierksen KP, Ragland NL, Sivabalan S, Wirawan RE, Inglis MA, Moore CJ, Walker GV, Chilcott CN, Jenkinson HF, Tagg JR. 2006. Production of the lantibiotic salivaricin A and its variants by oral streptococci and use of a specific induction assay to detect their presence in human saliva. Appl. Environ. Microbiol. 72:1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Basavanna S, Khandavilli S, Yuste J, Cohen JM, Hosie AH, Webb AJ, Thomas GH, Brown JS. 2009. Screening of Streptococcus pneumoniae ABC transporter mutants demonstrates that LivJHMGF, a branched-chain amino acid ABC transporter, is necessary for disease pathogenesis. Infect. Immun. 77:3412–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fouts DE, Brinkac L, Beck E, Inman J, Sutton G. 2012. PanOCT: automated clustering of orthologs using conserved gene neighborhood for pan-genomic analysis of bacterial strains and closely related species. Nucleic Acids Res. 40:e172. 10.1093/nar/gks757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laing C, Buchanan C, Taboada EN, Zhang Y, Kropinski A, Villegas A, Thomas JE, Gannon VP. 2010. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics 11:461. 10.1186/1471-2105-11-461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.