Abstract
Human norovirus (NoV) outbreak investigations suggest that the hands of infected individuals play an important role in NoV transmission. However, there is no experimental evidence documenting the likelihood and degree of NoV contamination on hands. As part of a clinical trial designed to evaluate the efficacy of high-pressure processing for Norwalk virus (NV) inactivation in oysters, 159 hand rinse samples were collected from 6 infected and 6 uninfected subjects. NV was concentrated from the samples by polyethylene glycol precipitation, followed by RNA extraction using an automated guanidinium isothiocyanate-silica method. NV RNA was detected and quantified using multiple NV-specific reverse transcription-quantitative PCR (RT-qPCR) assays. A total of 25.4% (18/71) of the hand rinse samples collected from 6 infected volunteers were presumptively positive for NV, with an average of 3.86 log10 genomic equivalent copies (GEC) per hand. Dot blot hybridization of PCR products obtained using a different primer set, and DNA sequencing of selected amplicons, provided further confirmation of the presence of NV in the hand rinses. NV contamination was also detected in two hand rinse samples obtained from one uninfected subject. These findings provide definitive evidence of NV contamination on the hands of infected subjects observed under controlled clinical research conditions. Such data support the need for better hand hygiene strategies to prevent NoV transmission.
INTRODUCTION
Human noroviruses (NoVs) are the most common cause of acute viral gastroenteritis worldwide (1) and a leading cause of food-borne disease (2, 3). They are spread primarily by the fecal-oral route but are also shed in vomitus. As such, NoV can be transmitted via consumption of fecally contaminated food or water or by contact with contaminated fomites and hands. The relative importance of each of these transmission routes is not well studied, but the potential for human hands to facilitate NoV transmission is widely recognized.
A recent epidemiological study by the CDC (4) identified NoV as the predominant etiology of food-borne disease outbreaks, and the largest proportion of these outbreaks were associated with food handlers implicated as the source of contamination. Food handlers are of particular concern (5) because they may shed NoV at extremely high titers for days or weeks during a symptomatic or asymptomatic NoV infection and subsequently transfer viruses from their hands to food. Furthermore, both laboratory and epidemiological data (6–8) provide evidence that NoV may persist on hands and fomites for extended periods of time. For example, Malek et al. (9) described an outbreak of NoV infection in which the index case was an infected food handler who worked for a delicatessen meat supplier company and handled sliced delicatessen meat with bare hands 1 day after recovering from gastroenteritis symptoms. This investigation documented a clear association between the contaminated hands of the food handler and the subsequent NoV outbreak. Because of NoV-contaminated hands, ready-to-eat foods and other products that are subject to extensive human handling immediately preceding consumption are a common cause of NoV outbreaks (10–13). Poor personal hygiene practices of infected food handlers provide the source of NoV contamination to these foods, and risk-modeling efforts have sought to model the importance of hands in the transmission of NoV in the food preparation environment (14). Unfortunately, no studies have been performed to quantitatively detect human NoV on contaminated hands. In this report, we provide direct laboratory evidence of NoV contamination on the hands of human subjects challenged with Norwalk virus (NV, the prototype, genogroup I [GI] human NoV).
MATERIALS AND METHODS
Volunteer study and sample pool.
The samples in this study were collected in conjunction with a clinical trial that evaluated the effect of high-hydrostatic-pressure processing (HPP) on NV inactivation in seeded oysters. Forty-four healthy adult subjects with positive secretor status [defined as individuals carrying at least one functional FUT2 allele and thus expressing alpha-(1,2)-fucosyltransferase-2 (15)] were enrolled, and each received a total of 1.0 × 104 genomic equivalent copies (GEC) of NV (8FIIb) in artificially seeded oysters with or without HPP treatment. The study was conducted at the Emory University Hospital, and the results have been previously reported (16).
Before challenge (day 0) and during the first 4 days postchallenge, when infection and symptoms typically occur, hand rinse samples were collected from all subjects at the time of vital sign measurement (3 times/day) and immediately after defecation. After determining subjects' infection statuses, all hand rinse samples (a total of 71 from six infected volunteers and 88 from six uninfected volunteers) were selected for inclusion in this study. Table 1 shows details about sample types and collection times.
Table 1.
Volunteer infection status and number/type of hand rinse samples collected post-NV challengea
| Volunteer status | Subject ID | No. of hand rinse samples |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day 0 | Day 2 |
Day 3 |
Day 4 |
Total | ||||||||
| Bath | Vital | N/A | Bath | Vital | N/A | Bath | Vital | N/A | ||||
| Infected | 34 | 2 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 |
| 36 | 2 | 4 | 0 | 0 | 6 | 6 | 0 | 2 | 2 | 0 | 22 | |
| 37 | 2 | 2 | 0 | 2 | 3 | 0 | 2 | 1 | 0 | 0 | 12 | |
| 40 | 0 | 2 | 0 | 0 | 5 | 0 | 0 | 2 | 0 | 0 | 9 | |
| 46 | 0 | 3 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 8 | |
| 54 | 2 | 2 | 0 | 2 | 0 | 6 | 0 | 0 | 0 | 0 | 12 | |
| Total | 71 | |||||||||||
| Uninfected | 38 | 2 | 4 | 0 | 0 | 6 | 2 | 0 | 2 | 0 | 0 | 16 |
| 41 | 2 | 3 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 9 | |
| 42 | 2 | 2 | 4 | 2 | 0 | 0 | 6 | 1 | 0 | 0 | 17 | |
| 44 | 2 | 6 | 0 | 0 | 6 | 0 | 0 | 2 | 0 | 0 | 16 | |
| 47 | 2 | 6 | 0 | 0 | 4 | 2 | 0 | 2 | 0 | 0 | 14 | |
| 55 | 2 | 6 | 0 | 0 | 4 | 2 | 0 | 0 | 0 | 2 | 16 | |
| Total | 88 | |||||||||||
ID, identification number. Bath samples were collected after defecation. Vital sign samples were collected during vital sign measurement. N/A, sample collection time not clear.
Hand rinse sample collection.
Hand rinse samples were collected using a glove juice method (17) after informed consent (approved by the institutional review board at Emory University) was obtained from each participant. Briefly, both hands of each subject were rinsed in a sterile, powder-free nitrile surgical glove filled with 50 ml of sterile glove juice solution (pH 7.8) composed of 0.4 g potassium phosphate monobasic (Fisher, Hampton, NH), 10.1 g sodium phosphate dibasic (Sigma-Aldrich Co., St. Louis, MO), and 1.0 ml Triton X-100 (Alfa Aesar, A Johnson Matthey Company, Ward Hill, MA) dissolved in 1 liter of distilled water. During the sample collection, hands were continuously massaged around the fingers, thumb, nails, and palm by the nurses for 60 s. Each rinse volume was introduced into a 50-ml conical tube and immediately frozen until being assayed for NV.
Determination of volunteer infection status.
The infection status of volunteers was defined as (i) reverse transcription-PCR (RT-PCR) detection of NV RNA in any postchallenge stool (16) or vomitus sample(s) and (ii) seroconversion as determined using an enzyme-linked immunosorbent assay (ELISA) with Norwalk virus-like particles (18).
Viral concentration and RNA extraction.
The viruses in 25-ml volumes of each hand rinse sample were precipitated by 12% polyethylene glycol (PEG; molecular weight of 7,000 to 9,000) after samples were adjusted to 0.9 M NaCl and supplemented with 1% bovine serum albumin (Sigma-Aldrich). After incubation for 2 h at 4°C, samples were spun at 10,000 × g for 20 min, and the precipitated pellet was suspended in 1 ml of 1× phosphate-buffered saline (PBS; MP Biomedicals, Solon, OH). Extraction of NV RNA was performed on the entire 1-ml concentrate using a NucliSENS easyMAG guanidinium isothiocyanate-silica automated system (bioMérieux SA, Marcy l'Etoile, France) according to the manufacturer's instructions, with a final elution volume of 50 μl.
Detection of NV in hand rinse samples by RT-qPCR.
To monitor PCR inhibition, a homologous internal amplification control (IAC) was generated in accordance with the method of Abdulmawjood et al. (19). This IAC could be amplified using the NV diagnostic primers but had an internal sequence corresponding to a region of the pUC19 vector (New England BioLabs, Ipswich, MA). To account for the possibility of outcompeting the IAC when target template levels were low, NV RNA extracted from hand rinse samples was amplified by RT-qPCR both with and without inclusion of the IAC. Sequences of primers and probes (20) used in these RT-qPCRs, including the IAC probe, are detailed in Table 2. The RT-quantitative PCR (RT-qPCR) protocol used was as previously reported (20). Briefly, RT-qPCRs (Qiagen, Valencia, CA) consisted of 5 μl of 5× RT-PCR buffer, 0.48 mM deoxynucleoside triphosphate (dNTP) mix, 600 nM each primer, 200 nM each probe, 0.25 μl of RNase inhibitor (Promega, Madison, WI) (40 U/μl), 1.4 pg IAC, 1.3 μl enzyme mix (HotStart Taq DNA polymerase and reverse transcriptase; Qiagen), and 10 μl of the RNA template. The final volume of the reaction was 25 μl. Amplification was performed under the following conditions: 50°C for 30 min and 95°C for 15 min followed by 45 cycles of 94°C for 15 s, 55°C for 15 s, and 60°C for 30 s.
Table 2.
RT-PCR primers and probes used for detection and confirmation of NV in hand rinse samples
| Assay | Primer sequence | Probe sequencea | Reference or source |
|---|---|---|---|
| Diagnostic RT-qPCR | NVKS1-Forward: 5′-ACAGCATGGGACTCAACACA-3′ | NVKS3 probe: 5′-(FAM)TCACCAGAATTGGCCGAGGTTGT(BHQ-1)-3′ | Liu et al. (20) |
| NVKS2-Reverse: 5′-GGGAAGTACATGGGAATCCA-3′ | IAC probe: 5′-(TET)ATCTCAGTTCGGTGTAGGTCGTTCGCTCC(BHQ-1)-3′ | This study | |
| Confirmation RT-PCR + hybridization | COG1F: 5′-CGYTGGATGCGNTTYCATGA-3′ | Ring a: 5′-(FAM)AGATYGCGATCYCCTGTCCA(TAMRA)-3′ | Kageyama et al. (21) |
| COG1R: 5′-CTTAGACGCCATCATCATTYAC-3′ | Ring b: 5′-(FAM)AGATCGCGGTCTCCTGTCCA(TAMRA)-3′ |
FAM, 6-carboxyfluorescein; TET, tetrachlorofluorescein; BHQ-1, black hole quencher 1; TAMRA, 6-carboxytetramethylrhodamine.
Determination of NV contamination status.
The presence of NV RNA in hand rinse samples was determined by two separate trials of RNA extraction and RT-qPCR. In each trial, RT-qPCR was performed in duplicate for each sample, and each reaction was performed with and without the IAC. In the first trial, samples were designated NV negative if they had threshold cycle (CT) values of 45 (no amplification) in two RT-qPCR duplicate amplifications in which the IAC was included. Samples were also considered NV negative if CT values were between 38 and 45 with coamplification of the IAC (total of 17 samples). When the IAC was not amplified, samples were diluted 1:4 and/or 1:10 and reamplified. Those samples that consistently produced CT values ≤ 38 in two duplicate amplifications with IAC coamplification were considered presumptively positive.
The remaining hand rinse volumes (ranging from 15 to 20 ml) of the presumptively positive samples and a selected subset of negative samples from the first trial were reprocessed by the sequential steps of PEG precipitation and RNA extraction and then retested by RT-qPCR amplification exactly as in the first trial. Sample positive/negative status was determined as described above for this second round of testing. Samples that were presumptively positive for both testing rounds were subjected to further confirmation by hybridization and/or sequencing.
Confirmation of positive samples by RT-PCR, hybridization, and sequencing.
Positive samples detected by the aforementioned RT-qPCR method were further confirmed by a conventional RT-PCR amplification using a COG1F/COG1R primer set (21) (Table 2), DNA hybridization, and, in some instances, cloning and sequencing. For confirmation, dot blot hybridization of amplicons was performed using a digoxigenin (DIG) nucleic acid detection kit (Roche Diagnostics Corp., Indianapolis, IN) according to the manufacturer's instructions. After hybridization, the membrane was washed and then processed using a DIG wash block buffer set (Roche) and DIG nucleic acid detection kit (Roche). A positive result was identified by a blue precipitate.
For further confirmation, a subset of the conventional RT-PCR amplicons corresponding to hybridization-confirmed NV-positive hand rinse samples with relatively low CT values (<30) were purified using a QIAquick PCR purification kit (Qiagen Inc., Valencia, CA) and cloned into TOPO vector using a TOPO cloning kit (Invitrogen, Carlsbad, CA). The DNA insertion in each clone was sequenced by Genewiz Inc. (Research Triangle Park, NC) using the universal M13R and M13F primers.
Estimation of viral load in hand rinse samples.
An RT-qPCR standard curve served as the basis for quantification of the viral load. The standard curve (corresponding to 49 to 4.9 × 105 GEC) was constructed using serially diluted full-length NV genomic RNA that was transcribed from a full-length Norwalk virus cDNA clone. The concentration of the NV RNA transcript was determined as previously described (22). The approximate virus concentration in the hand rinse samples was calculated directly from the standard curve with corrections made to account for total sample volume.
Statistical analysis.
NV concentrations in stool samples and hand rinse samples were expressed as log10 GEC per gram of feces and total log10 GEC per hand, respectively. The unpaired t test was applied to compare NV log10 concentrations obtained from positive hand rinse samples with respect to volunteer infection status (infected versus uninfected) and sample collection time (i.e., during routine vital sign measurement, after defecation, or without clear sample collection information). Data were analyzed using SAS 9.2 (SAS, Cary, NC), and P values < 0.05 were considered statistically significant.
RESULTS
Limits of detection for the EasyMAG/RT-qPCR assay.
The limit of detection of the RT-qPCR method used in the screening assay was 4.9 GEC per reaction. The limit of detection for the combined virus concentration-RNA extraction method was determined by seeding approximately 7.1 × 101 to 7.1 × 105 GEC of serially diluted (20%) NV-positive stool suspensions into 25 ml of glove juice solution and processing through RT-qPCR in triplicate trials. The lowest detectable concentration was considered the limit of detection for the combined assay and was equivalent to approximately 1.4 × 102 (2.15 log10) GEC per 50 ml of hand rinse sample.
Summary of NV screening results.
Based on the criteria of NV positivity and negativity, 125/159 (78.6%) of samples were negative and 34/159 (21.4%) were presumptively positive (data not shown) in the first trial. For all 34 presumptively positive samples and a subset of NV-negative samples (n = 36), the remaining rinse volumes were reprocessed by PEG precipitation and RNA extraction and then tested again by RT-qPCR amplification (with and without IAC) with duplicate amplifications. All negative samples remained negative. The final number of positive samples was reduced from 34 to 20 (Table 3).
Table 3.
Clinical symptoms, seroconversion, and RT-PCR detection of NV in stool samples and hand rinse samples collected from 12 subjectsc
| Subject status | Subject ID | Illnessa | Seroconversionb | Stool sample result |
Hand rinse sample result |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| RT-PCR | RT-qPCR | Max titer (log10/g) | Total no. | No. of Pos RT-qPCRs | No. of Pos Hyb | Mean log10 (no. of GEC/hand) | ||||
| Infected | 34 | Yes | Yes | Pos | Pos | 8.3 | 8 | 2 | 2 | 3.94 |
| 36 | Yes | Yes | Pos | Pos | 8.1 | 22 | 12d | 18 | 3.74 | |
| 37 | Yes | Yes | Pos | Pos | 7.4 | 12 | 0 | 1 | ||
| 40 | Yes | Yes | Pos | Pos | 7.5 | 9 | 0 | 0 | ||
| 46 | Yes | Yes | Pos | Pos | 7.4 | 8 | 2 | 2 | 3.30 | |
| 54 | Yes | Yes | Pos | Pos | 8.2 | 12 | 2 | 2 | 4.45 | |
| 71 | 18 | 25 | 3.86 | |||||||
| Uninfected | 38 | No | No | Neg | Neg | 16 | 0 | 0 | ||
| 41 | Yes | No | Neg | Neg | 9 | 0 | 0 | |||
| 42 | Yes | No | Neg | Neg | 17 | 2 | 3 | 2.81 | ||
| 44 | No | No | Neg | Neg | 16 | 0 | 0 | |||
| 47 | No | No | Neg | Neg | 14 | 0 | 0 | |||
| 55 | No | No | Neg | Neg | 16 | 0 | 0 | |||
| 88 | 2 | 3 | 2.81 | |||||||
Illness, ≥3 loose or watery stool samples and/or any vomiting within 24 h.
≥4-fold difference between the prechallenge and convalescent-phase serum NV-specific IgG titers.
Pos, positive; Neg, negative; Hyb, hybridizations.
At least one sample was confirmed positive by sequencing.
Confirmation of NV-positive samples.
All 20 NV-positive and 14 representative NV-negative hand rinse samples in the second round of testing were tested by conventional RT-PCR followed by dot blot hybridization with genogroup I (GI)-specific probes. All of the samples that were positive by RT-qPCR were also positive by conventional RT-PCR followed by hybridization. In general, there was a reverse correlation between the RT-qPCR CT value and the intensity of the hybridization signal (Fig. 1). An additional 8 samples that were negative by RT-qPCR in round 2 of testing were weakly positive by RT-PCR and hybridization. Two hand rinse samples that tested positive for NV by RT-qPCR (CT values were 26 and 28, respectively), along with the corresponding (same-day) stool samples, were reamplified by conventional RT-PCR, and the PCR products were cloned and sequenced. The NV sequences of the amplified PCR products in these hand rinse samples were confirmed to be Norwalk virus sequences and were identical to those sequences isolated from the same subjects' stool samples (data not shown).
Fig 1.
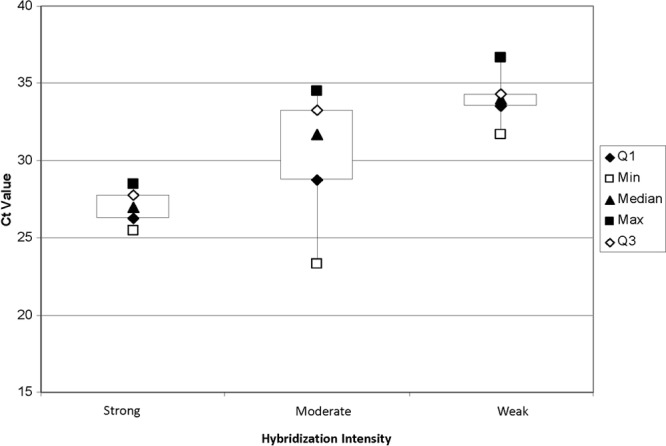
Relationship between average CT value for presumptively positive samples and intensity of confirmatory DNA hybridization as demonstrated in a box plot format. Hybridization intensity was classified into 3 categories: strong (n = 4), moderate (n = 5), and weak (n = 19). Box plot keys are as follows: Median, the median CT; Min, minimum CT; Max, maximum CT; Q3, 75th percentile; Q1, 25th percentile.
Interpretation of NV test results.
Based on RT-qPCR results, there was evidence of NV contamination in 25.4% (18/71) of hand rinse samples obtained from infected subjects, with an average titer of 3.86 (range of 3.30 to 4.45) log10 GEC per hand. Interestingly, subject 36 (the infected subject from whom the largest number of hand rinse samples were collected) had a high frequency (12/22 [54.5%]) of NV hand contamination, while subjects 34 and 46 had relatively lower positive rates (2/8 [25%]). Subject 36 was particularly interesting because of the high proportion of NV-positive hand samples (six samples) that were negative by RT-qPCR but positive by DNA hybridization. No evidence of NV contamination was identified on the hands of two infected subjects (subjects 37 and 40), although there was trace evidence of hand contamination in one of the rinses from subject 37 when the samples were screened by conventional RT-PCR followed by DNA hybridization (Table 3).
With respect to sample type, NV was detected in 37.5% (6/16) of samples collected during routine vital sign measurements, with an average of 3.32 log10 GEQ per hand. That proportion of NV-positive samples and that mean titer were significantly higher (P < 0.05) than the proportion and mean titer of the NV-positive samples (12.4% [11/89]; 2.30 log10) collected immediately after bathroom use. In contrast, the proportion of NV-positive samples without clear collection information was 25% (10/40), with an average titer of 2.46 log10 GEQ, and there were no statistically significant differences (P > 0.05) from the proportions and mean titers of positive samples collected during routine vital sign measurements and after bathroom use.
Surprisingly, NV was also identified in 2/88 (2.3%) hand rinse samples (and in one additional sample by RT-PCR and hybridization) obtained from uninfected subject 42 (Table 3). The mean log10 NV GEC of the positive samples derived from this uninfected subject was 2.81, which was significantly lower (P < 0.05) than the viral load estimated from the positive samples obtained from NV-infected subjects.
Relationship between clinical symptoms, viral shedding in stool, and hand contamination.
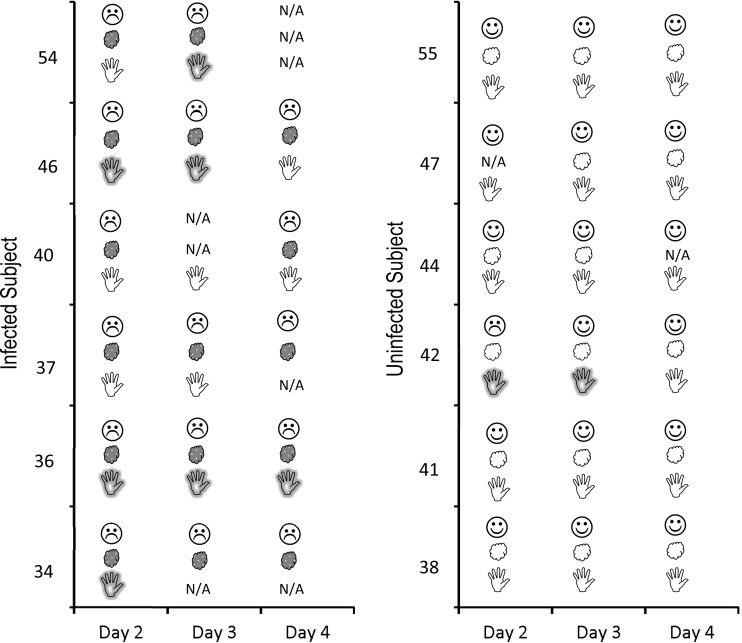
Fig. 2 shows the temporal relationship between NV contamination on hands, occurrence of clinical symptoms, and virus shedding in stool. NV RNA was detected in 18 of 71 (25.4%) hand rinse samples collected from 4 of 6 (66.7%) infected individuals who were experiencing diarrhea and/or vomiting and who were also excreting NV in their stool on the day of hand sample collection. Surprisingly, NV was also identified in hand rinse samples from subject 42, who demonstrated diarrhea/and or fever during the time of hand rinse sample collection but for whom we were unable to confirm NV shedding or seroconversion despite repeated testing and multiple RT-PCR attempts.
Fig 2.
Temporal relationship between clinical symptoms (diarrhea and/or vomiting), virus shedding in stools, and virus contamination on hands as determined by RT-qPCR in infected (left) and uninfected (right) subjects.  , absence of clinical symptoms;
, absence of clinical symptoms;  , presence of clinical (diarrhea or vomit and fever) symptoms;
, presence of clinical (diarrhea or vomit and fever) symptoms;  , no NV in stool;
, no NV in stool;  , NV shedding in stool;
, NV shedding in stool;  , NV not detected on hands;
, NV not detected on hands;  , NV contamination on hands; N/A, not applicable (sample not available for analysis).
, NV contamination on hands; N/A, not applicable (sample not available for analysis).
DISCUSSION
To our knowledge, this is the first direct experimental evidence of NoV contamination on the hands of individuals with known NV exposure in a controlled setting. A total of 25.4% of hand rinse samples obtained from 4 of 6 infected subjects showed evidence of NV contamination. The infected subjects were all secretor positive, experienced clinical symptoms, and exhibited NV shedding in fecal specimens and NV-specific IgG seroconversion. Low levels of NV contamination were found in hand rinses from 1 of 6 uninfected volunteers.
Not unexpectedly, interpretation of RT-qPCR results from the hand rinse samples was not always straightforward. Specifically, even for highly contaminated samples, CT values were rarely below 32. For this reason, presumptively positive samples were tested twice and subjected to multiple amplification reactions and confirmatory tests. Based on the criterion that both replicate samples must yield RT-qPCR values of CT ≤ 38, 18 of these samples were confirmed positive. As previously reported by others, it is possible that very high CT values in some samples actually corresponded to very low levels of NV contamination (23). The value of the confirmatory steps is illustrated by the fact that, for a select group of negative samples further tested by hybridization, some samples with average RT-qPCR CT values > 38 produced weak hybridization signals and a subset of 6 RT-qPCR negative samples were also weakly positive by hybridization.
NV contamination was also detected in two hand rinse samples from one uninfected subject (subject 42) who had no detectable NV shedding in his stool and no NV-specific IgG seroconversion but who did have the clinical symptoms of diarrhea and fever. These results were confirmed by replicate testing of hand rinse samples, two different RT-PCR protocols, and DNA hybridization. A previous study conducted in a diarrheal disease treatment center reported evidence of rotavirus contamination in the hand washings of attendants caring for patients without rotavirus, although other patients in the facility did indeed have rotavirus infection (24). While we do not know exactly how the hands of subject 42 became contaminated, contact with a contaminated fomite is a possibility. The potential for NoV surface contamination is well documented in outbreak settings (25, 26), and these viruses are readily transferred between surfaces and fingers (27). Hand rinse samples from subject 42 were collected in the same room 1 week after an episode occurred of virus shedding from a previous NV-infected subject in the same study. Although routine decontamination efforts had been performed in the ward environment between the two NV challenge events, NV environmental contamination can persist for long periods (6), and the viruses show resistance to many disinfectants (28).
This report has significance for both the health care and food service sectors. In health care, there is mounting evidence of the importance of hands (29) and surfaces (30) in the transmission of nosocomial infections. Likewise, the importance of hands in NoV transmission via food has been demonstrated through epidemiological investigation of outbreaks (10, 13), with more evidence provided recently (31). Efforts have also been undertaken to mathematically model the likelihood of NoV transmission in the food service environment (14). However, there are many unanswered questions that this study can help answer. For example, what degree of efficacy might be required for a hand sanitizer manufacturer to make specific antiviral claims? We do not yet have such standards in the United States, but in Canada and Europe, the standard is a 4 log10 reduction (in vitro) and a 2 log10 reduction (in vivo) (32) of the level of murine NoV, which may be more easily inactivated than human NoVs (33, 34). If, indeed, a dose of 1 to 2 log10 NV GEC is enough to cause infection in susceptible individuals (22), and the hands of NV-infected individuals with symptoms harbor around 4 log10 GEC, then a 2 log10 reduction in vivo efficacy might not be sufficiently stringent to ensure adequate public health protection. A combination of hand washing and hand sanitizer may be more stringent, and a previous study reported that a regimen of hand washing with antibacterial soap followed by a 70% ethanol advanced-formula gel sanitizer achieved a 3.19 to 4.04 log10 reduction of the level of cultivable surrogate murine NoV (35). If these findings could be translated into the removal of human NoV, the combined use of hand washing and hand sanitizer would provide added protection against transmission of human NoV.
The U.S. Food and Drug Administration (FDA) Food Code (36) for retail and food service settings recommends hand washing as a method to reduce transmission of food-borne pathogens from hands to food and other objects. The recommended hand-washing procedure includes washing hands using running warm water for at least 20 s, rubbing one hand against the other for 10 to 20 s, and finally rinsing and drying with towels or hot air. The FDA also suggests using gloves as a barrier to further prevent the transmission of pathogens. If these hand hygiene practices were properly used, opportunities for food contamination would be considerably reduced. However, factors such as improper use of gloves, loss of glove integrity, and, particularly, variable worker compliance can limit the effectiveness of Food Code hand hygiene interventions.
In conclusion, these laboratory results strongly support previous epidemiological evidence suggesting the importance of human hands in the transmission of NoV infection. The evidence of NV RNA on the hands of an uninfected individual suggests that hands can become contaminated from NoV-contaminated surfaces. These results further support the need for more-effective hand hygiene agents and for continued emphasis on dedicated compliance with recommended hand hygiene practices.
ACKNOWLEDGMENTS
This work was supported by Gojo Industries, Inc. Partial funding was also provided by a Food Virology Collaborative grant (no. 1111-2011-0494) from the National Institute of Food and Agriculture (NIFA) at the U.S. Department of Agriculture (USDA).
We thank scientists David Macinga and Carrie Zapka from Gojo for their contributions to develop the hand rinse sample collection method and the nurses and Marshall Lyon at Emory Hospital for their help with hand rinse sample collection. We are grateful to Juan Leon, who led the clinical trial evaluating NV inactivation in shellfish by high pressure at Emory Hospital. We also thank Songli Xu, who helped with the statistical analysis in this study.
Footnotes
Published ahead of print 11 October 2013
REFERENCES
- 1.Glass RI, Parashar UD, Estes MK. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. 2011. Foodborne illness acquired in the United States—unspecified agents. Emerg. Infect. Dis. 17:16–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dreyfuss MS. 2009. Is norovirus a foodborne or pandemic pathogen? An analysis of the transmission of norovirus-associated gastroenteritis and the roles of food and food handlers. Foodborne Pathog. Dis. 6:1219–1228 [DOI] [PubMed] [Google Scholar]
- 4.Hall AJ, Eisenbart VG, Etingue AL, Gould LH, Lopman BA, Parashar UD. 2012. Epidemiology of foodborne norovirus outbreaks, United States, 2001–2008. Emerg. Infect. Dis. 18:1566–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moe CL. 2009. Preventing norovirus transmission: how should we handle food handlers? Clin. Infect. Dis. 48:38–40 [DOI] [PubMed] [Google Scholar]
- 6.Liu P, Chien YW, Papafragkou E, Hsiao HM, Jaykus LA, Moe C. 2010. Persistence of human noroviruses on food preparation surfaces and human hands. Food Environ. Virol. 1:141–147 [Google Scholar]
- 7.Liu P, Jaykus LA, Wong E, Moe C. 2012. Persistence of Norwalk virus, male-specific coliphage, and Escherichia coli on stainless steel coupons and in phosphate-buffered saline. J. Food Prot. 75:2151–2157 [DOI] [PubMed] [Google Scholar]
- 8.Isakbaeva ET, Widdowson MA, Beard RS, Bulens SN, Mullins J, Monroe SS, Bresee J, Sassano P, Cramer EH, Glass RI. 2005. Norovirus transmission on cruise ship. Emerg. Infect. Dis. 11:154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malek M, Barzilay E, Kramer A, Camp B, Jaykus LA, Escudero-Abarca B, Derrick G, White P, Gerba C, Higgins C, Vinje J, Glass R, Lynch M, Widdowson MA. 2009. Outbreak of norovirus infection among river rafters associated with packaged delicatessen meat, Grand Canyon, 2005. Clin. Infect. Dis. 48:31–37 [DOI] [PubMed] [Google Scholar]
- 10.Barrabeig I, Rovira A, Buesa J, Bartolome R, Pinto R, Prellezo H, Dominguez A. 2010. Foodborne norovirus outbreak: the role of an asymptomatic food handler. BMC Infect. Dis. 10:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozawa K, Oka T, Takeda N, Hansman GS. 2007. Norovirus infections in symptomatic and asymptomatic food handlers in Japan. J. Clin. Microbiol. 45:3996–4005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniels NA, Bergmire-Sweat DA, Schwab KJ, Hendricks KA, Reddy S, Rowe SM, Fankhauser RL, Monroe SS, Atmar RL, Glass RI, Mead P. 2000. A foodborne outbreak of gastroenteritis associated with Norwalk-like viruses: first molecular traceback to deli sandwiches contaminated during preparation. J. Infect. Dis. 181:1467–1470 [DOI] [PubMed] [Google Scholar]
- 13.Parashar UD, Dow L, Fankhauser RL, Humphrey CD, Miller J, Ando T, Williams KS, Eddy CR, Noel JS, Ingram T, Bresee JS, Monroe SS, Glass RI. 1998. An outbreak of viral gastroenteritis associated with consumption of sandwiches: implications for the control of transmission by food handlers. Epidemiol. Infect. 121:615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mokhtari A, Jaykus LA. 2009. Quantitative exposure model for the transmission of norovirus in retail food preparation. Int. J. Food Microbiol. 133:38–47 [DOI] [PubMed] [Google Scholar]
- 15.Chang JG, Yang TY, Liu TC, Lin TP, Hu CJ, Kao MC, Wang NM, Tsai FJ, Peng CT, Tsai CH. 1999. Molecular analysis of secretor type alpha(1,2)-fucosyltransferase gene mutations in the Chinese and Thai populations. Transfusion 39:1013–1017 [DOI] [PubMed] [Google Scholar]
- 16.Leon JS, Kingsley DH, Montes JS, Richards GP, Lyon GM, Abdulhafid GM, Seitz SR, Fernandez ML, Teunis PF, Flick GJ, Moe CL. 2011. Randomized, double-blinded clinical trial for human norovirus inactivation in oysters by high hydrostatic pressure processing. Appl. Environ. Microbiol. 77:5476–5482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larson EL, Strom MS, Evans CA. 1980. Analysis of three variables in sampling solutions used to assay bacteria of hands: type of solution, use of antiseptic neutralizers, and solution temperature. J. Clin. Microbiol. 12:355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moe CL, Sair A, Lindesmith L, Estes MK, Jaykus LA. 2004. Diagnosis of norwalk virus infection by indirect enzyme immunoassay detection of salivary antibodies to recombinant norwalk virus antigen. Clin. Diagn. Lab. Immunol. 11:1028–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdulmawjood A, Roth S, Bulte M. 2002. Two methods for construction of internal amplification controls for the detection of Escherichia coli O157 by polymerase chain reaction. Mol. Cell. Probes 16:335–339 [DOI] [PubMed] [Google Scholar]
- 20.Liu P, Hsiao HM, Jaykus LA, Moe C. 2010. Quantification of Norwalk virus inocula: comparison of endpoint titration and real-time reverse transcription-PCR methods. J. Med. Virol. 82:1612–1616 [DOI] [PubMed] [Google Scholar]
- 21.Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Takeda N, Katayama K. 2003. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 41:1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teunis PF, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, Le Pendu J, Calderon RL. 2008. Norwalk virus: how infectious is it? J. Med. Virol. 80:1468–1476 [DOI] [PubMed] [Google Scholar]
- 23.Phillips G, Lopman B, Tam CC, Iturriza-Gomara M, Brown D, Gray J. 2009. Diagnosing norovirus-associated infectious intestinal disease using viral load. BMC Infect. Dis. 9:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Samadi AR, Huq MI, Ahmed QS. 1983. Detection of rotavirus in handwashings of attendants of children with diarrhoea. Br. Med. J. (Clin. Res. ed) 286:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu HM, Fornek M, Schwab KJ, Chapin AR, Gibson K, Schwab E, Spencer C, Henning K. 2005. A norovirus outbreak at a long-term-care facility: the role of environmental surface contamination. Infect. Control Hosp. Epidemiol. 26:802–810 [DOI] [PubMed] [Google Scholar]
- 26.Jones EL, Kramer A, Gaither M, Gerba CP. 2007. Role of fomite contamination during an outbreak of norovirus on houseboats. Int. J. Environ. Health Res. 17:123–131 [DOI] [PubMed] [Google Scholar]
- 27.Sharps CP, Kotwal G, Cannon JL. 2012. Human norovirus transfer to stainless steel and small fruits during handling. J. Food Prot. 75:1437–1446 [DOI] [PubMed] [Google Scholar]
- 28.Nowak P, Topping JR, Fotheringham V, Gallimore CI, Gray JJ, Iturriza-Gomara M, Knight AI. 2011. Measurement of the virolysis of human GII.4 norovirus in response to disinfectants and sanitisers. J. Virol. Methods 174:7–11 [DOI] [PubMed] [Google Scholar]
- 29.Tschudin-Sutter S, Pargger H, Widmer AF. 2010. Hand hygiene in the intensive care unit. Crit. Care Med. 38:S299–S305 [DOI] [PubMed] [Google Scholar]
- 30.Otter JA, Yezli S, French GL. 2011. The role played by contaminated surfaces in the transmission of nosocomial pathogens. Infect. Control Hosp. Epidemiol. 32:687–699 [DOI] [PubMed] [Google Scholar]
- 31.Boxman I, Dijkman R, Verhoef L, Maat A, van Dijk G, Vennema H, Koopmans M. 2009. Norovirus on swabs taken from hands illustrate route of transmission: a case study. J. Food Prot. 72:1753–1755 [DOI] [PubMed] [Google Scholar]
- 32.Health Canada 2009. Guidance document—human-use antiseptic drugs. http://www.hc-sc.gc.ca/dhp-mps/prodpharma/applic-demande/guide-ld/antiseptic_guide_ld-eng.php
- 33.Richards GP. 2012. Critical review of norovirus surrogates in food safety research: rationale for considering volunteer studies. Food Environ. Virol. 4:6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park GW, Barclay L, Macinga D, Charbonneau D, Pettigrew CA, Vinje J. 2010. Comparative efficacy of seven hand sanitizers against murine norovirus, feline calicivirus and GII.4 norovirus. J. Food Prot. 73:2232–2238 [DOI] [PubMed] [Google Scholar]
- 35.Edmonds SL, McCormack RR, Zhou SS, Macinga DR, Fricker CM. 2012. Hand hygiene regimens for the reduction of risk in food service environments. J. Food Prot. 75:1303–1309 [DOI] [PubMed] [Google Scholar]
- 36.FDA 2009. U.S. FDA food code. http://www.fda.gov/Food/GuidanceRegulation/RetailFoodProtection/FoodCode/default.htm U.S. Food and Drug Administration, Silver Spring, MD [Google Scholar]