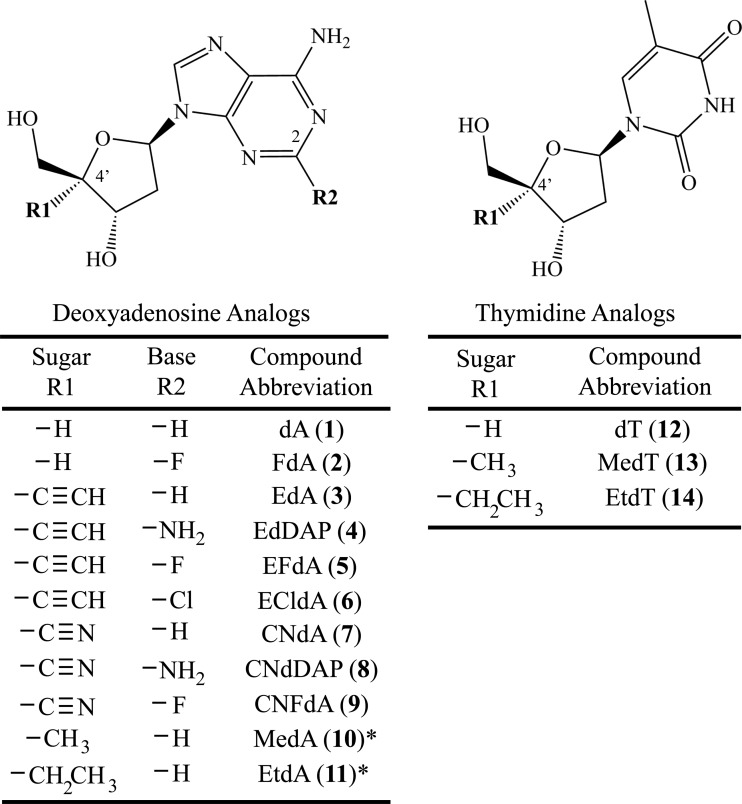
Abstract
Nucleos(t)ide reverse transcriptase inhibitors (NRTIs) form the backbone of most anti-HIV therapies. We have shown that 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) is a highly effective NRTI; however, the reasons for the potent antiviral activity of EFdA are not well understood. Here, we use a combination of structural, computational, and biochemical approaches to examine how substitutions in the sugar or adenine rings affect the incorporation of dA-based NRTIs like EFdA into DNA by HIV RT and their susceptibility to deamination by adenosine deaminase (ADA). Nuclear magnetic resonance (NMR) spectroscopy studies of 4′-substituted NRTIs show that ethynyl or cyano groups stabilize the sugar ring in the C-2′-exo/C-3′-endo (north) conformation. Steady-state kinetic analysis of the incorporation of 4′-substituted NRTIs by RT reveals a correlation between the north conformation of the NRTI sugar ring and efficiency of incorporation into the nascent DNA strand. Structural analysis and the kinetics of deamination by ADA demonstrate that 4′-ethynyl and cyano substitutions decrease the susceptibility of adenosine-based compounds to ADA through steric interactions at the active site. However, the major determinant for decreased susceptibility to ADA is the 2-halo substitution, which alters the pKa of N1 on the adenine base. These results provide insight into how NRTI structural attributes affect their antiviral activities through their interactions with the RT and ADA active sites.
INTRODUCTION
There are 10 nucleos(t)ide reverse transcriptase inhibitors (NRTIs) that are currently approved for the treatment of human immunodeficiency virus type 1 (HIV-1) infections (1–5). Several more nucleoside analog drugs are approved or being studied for the treatment of viruses, such as herpes simplex virus (HSV), hepatitis C virus (HCV), and hepatitis B virus (HBV), or as anticancer agents (6–10). NRTIs are among the most effective anti-HIV drugs. All approved anti-HIV NRTIs lack a 3′-hydroxyl moiety and, thus, act as chain terminators following their incorporation by the viral reverse transcriptase (RT) into the nascent DNA chain. However, the absence of a 3′-OH, while essential for the inhibition of DNA synthesis, also imparts detrimental properties to these inhibitors, including reduced intracellular phosphorylation to the active triphosphate form and reduced RT binding affinity (11). Prolonged exposure to NRTI-based treatments causes mitochondrial toxicity (12–14) and leads to the development of NRTI resistance mutations (15–18), giving rise to complications in the treatment of HIV-infected patients.
Ideally, an NRTI should have a strong binding affinity for the RT target, a high barrier for the development of resistance, and low toxicity. We have reported that a series of 4′-substituted nucleosides in which the 3′-OH is retained has exceptional inhibitory activity against HIV-1 RT (19). Among these compounds, 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA) is a highly active RT inhibitor that prevents translocation of the nucleic acid from the nucleotide-binding or pretranslocation site to the primer-binding or posttranslocation site on RT following its incorporation at the 3′ primer terminus (20). Depending on the DNA template sequence, EFdA can also act (albeit less frequently) as a delayed chain terminator, incorporating one incoming deoxynucleoside triphosphate (dNTP) before DNA synthesis is blocked (20). We previously showed that EFdA inhibits HIV replication in peripheral blood mononuclear cells (PBMCs) with a 50% effective concentration (EC50) of 0.05 nM (20). Additionally, EFdA effectively inhibits the replication of many common drug-resistant strains of HIV, including strains that carry a substitution of R for K at position 65 (K65R) (a 0.2-fold change in EC50) (21, 22), N348I (a 0.9-fold change in 50% inhibitory concentration [IC50]) (23), the excision-enhancing mutations M41L and T215Y (a 1.5-fold change in EC50) (21), and multidrug resistance Q151M complex mutations (A62V/V75I/F77L/F116Y/Q151M) (a 0.7-fold change in EC50) (21). EFdA is also able to inhibit HIV containing the M184V drug resistance mutation (with an EC50 of 8.3 nM, or a 7.5-fold change) (21, 24). EFdA is generally a poor substrate for human DNA polymerases in in vitro experiments (IC50s of >100 μM for polymerase α and β and 10 μM for polymerase γ; EFdA-triphosphate [EFdA-TP] is incorporated by polymerase γ 4,300-fold less efficiently than dATP) and, thus, has a low potential for cytotoxicity (25, 26). Because the 50% cytotoxic concentration (CC50) for EFdA is more than 10 μM in MT4 cells or PBMCs, the selectivity of the compound is high. Moreover, in studies with simian immunodeficiency virus (SIV)-infected macaques, no signs of clinical or pathological drug toxicity were observed after 6 months of continuous EFdA monotherapy (24). These data suggest that EFdA is a strong candidate for further development as a therapeutic agent.
The EC50s of NRTIs are also affected by cellular uptake, activation to the active metabolite by host kinases, and catabolism by cellular enzymes. Previous studies have shown that EFdA has a favorable cellular uptake profile and is efficiently phosphorylated to EFdA-TP (25). Additional reported data strongly suggest that, in cells, EFdA is phosphorylated to EFdA-monophosphate (EFdA-MP) by deoxycytidine kinase (dCK) (21). The molecular details of the efficiency and specificity of this process are being investigated in separate studies. The enzyme adenosine deaminase (ADA), which is present at high concentrations in human serum, is involved in the catabolism of dA and its analogs (27, 28). ADA is responsible for the deamination of adenosine or deoxyadenosine analogs to inosine- or deoxyinosine-based products (27, 29–33) and can therefore affect the activation pathway and intracellular concentration of dA-based compounds. For example, ADA converts dideoxyadenosine (ddA) to ddI, leading to a different and indirect activation pathway for ddA (28, 34, 35). Thus, it is important to study the interaction of dA-based compounds, such as EFdA, with ADA to gain insights into their catabolism and subsequent activation pathways.
We have also previously reported that small changes in the chemical composition of dA analogs can have a pronounced effect on their antiviral potency. These changes can be complex, because they can have different and at times opposing effects on the recognition of NRTIs by the RT target and the various enzymes involved in activation and catabolism. For example, the presence of fluorine or another halogen at the 2 position on the adenine base of 4′-substituted dA analogs significantly enhanced their antiviral activity (21). Similarly, the nature of the 4′ substitutions on the deoxyribose ring also affected the EC50s of dA-based compounds against both wild-type (WT) and NRTI-resistant HIV-1 variants (19).
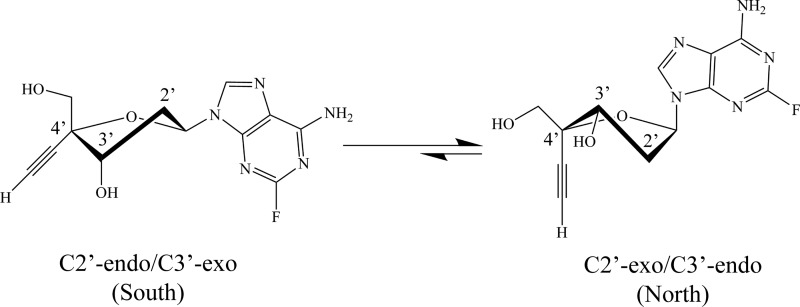
Additionally, changes in the conformation of the sugar ring and in the nature of the nucleobase can affect the biological activity of NRTIs. In solution, the structure of the deoxyribose ring of nucleosides exists in a dynamic equilibrium between the C-2′-exo/C-3′-endo (north) and C-2′-endo/C-3′-exo (south) conformations. We have previously shown that EFdA exists primarily in the north conformation (36) and that HIV-1 RT prefers the NRTI or incoming dNTP sugar ring in the north conformation for optimal binding (37–40). In contrast, host cell kinases appear to prefer the sugar ring in the south conformation; compounds that favor the north conformation are generally phosphorylated inefficiently (41, 42). Sugar ring conformation has also been reported to play an important role in ADA recognition; the enzyme has been reported to deaminate nucleosides biased toward the north conformation up to 65-fold faster than those biased toward the south conformation (43–46). Additionally, 2-fluoro-2′-deoxyadenosine (FdA) analogs are somewhat resistant to ADA-mediated catabolism due to the electron withdrawing properties of the 2-fluoro group (47, 48), and we have previously reported that EFdA is poorly deaminated by ADA because of the 2-fluoro group (21). However, the effects of specific 4′ substitutions on sugar ring conformation and how these substitutions would affect the recognition of the compounds by HIV RT and ADA have not been determined.
In the present work, we carried out a series of structural, computational, and biochemical studies using a variety of 2- and 4′-substituted dA and dT analogs (Fig. 1) to understand the specific effects of these different substitutions on sugar ring conformation, recognition by HIV RT, and deamination by ADA. Our data show how the 2-fluoro and 4′-ethynyl moieties affect the antiviral potential of NRTIs and help determine the basis for the favorable inhibitory profile of EFdA.
Fig 1.
Deoxyadenosine and thymidine analogs used in the study. *, only the triphosphate forms of these compounds were used in this study.
MATERIALS AND METHODS
Chemicals.
Compounds dA, FdA, and dT (compound abbreviations are defined and their numbers specified in Results) were purchased from Sigma-Aldrich (St. Louis, MO). Triphosphate forms of dA and dT were purchased from Fermentas (Glen Burnie, MD). Compounds 13 to 14 and the triphosphate forms of compounds 10, 11, 13, and 14 were synthesized by Marquez and colleagues as previously described (49, 50). ADA (bovine intestine, EC 3.5.4.4) was purchased from Calbiochem (San Diego, CA). Deuterated dimethyl sulfoxide (DMSO-D6) was purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA).
Nucleic acid substrates.
DNA oligomers were synthesized by Integrated DNA Technologies (Coralville, IA). In steady-state kinetics assays of dATP and dATP analogs, a 26-nucleotide-long DNA template (Td26A; CCA TAG ATA GCA TTG GTG CTC GAA CA) was used. In steady-state kinetics assays of dTTP and dTTP analogs, a 26-nucleotide-long template (Td26T; CCA TAG TAA GCA TTG GTG CTC GAA CA) was used. An 18-nucleotide-long 5′-fluorescently labeled DNA primer (Pd18; TGT TCG AGC ACC AAT GCT) was annealed to both Td26A and Td26T.
NMR spectroscopy.
One-dimensional 1H nuclear magnetic resonance (NMR) spectra were collected in 10°C increments from 20 to 50°C on a Bruker Avance DRX500 operating at 500 MHz and equipped with a 5-mm HCN cryoprobe, as previously described (36). Samples were dissolved in DMSO-D6 to give concentrations of 2 to 4 mM. Spectra were processed with resolution enhancement using negative line broadening (lb = −0.3). Coupling constants were analyzed by spectral simulation using SpinWorks 3.0 (Kirk Marat, University of Manitoba, Winnipeg, Canada). Simulations were performed for the spin systems on the deoxyribose ring only. The root mean square (RMS) deviations of calculated and experimental coupling constants for all compounds were <0.15 Hz.
Pseudorotational analysis.
The sugar ring conformations of the nucleoside analogs were determined using the program PSEUROT 6.3 (DOS version, purchased from Cornelis Altona, University of Leiden, Leiden, The Netherlands) as previously described (36). PSEUROT assumes a two-state approximation (equilibrium between north and south conformations) and utilizes a generalized Karplus equation combined with a nonlinear Newton-Raphson minimization process to examine the conformational flexibility of five-membered rings (51). An iterative methodology was used to determine the optimal pseudorotational parameters PN and PS (pseudorotational phase angle [P] for the north [N]or south [S] conformation) and ϕm(N) and ϕm(S) (degree of maximum ring pucker [ϕm] for the north or south conformation), in which some of these parameters were fixed during refinement in order to constrain the values within the range commonly observed for nucleosides (average observed, ϕm = 39°).
Molecular orbital calculations.
In order to determine the role of molecular orbitals in the stability of the north and south sugar ring conformations of EFdA, we carried out molecular orbital calculations using semiempirical quantum chemical methods. All quantum chemical calculations were performed by the density functional theory (DFT) method using the B3LYP approach with 6-31G** basis sets. The quantum chemistry program Jaguar (Schrödinger, Inc., NY) was used for all computations. The structures of north and south conformations of EFdA were completely optimized by restricting the position of the 3′-C either in the north or south conformation. To obtain insight into the interaction of the lone pairs of the O4 and 3′-O atoms with the antibonding (π*) orbitals of the 4′-ethynyl group of EFdA, the natural bond orbitals (NBOs), natural (semi-)localized molecular orbitals (NLMOs), and canonical molecular orbitals (CMOs) were computed using the program NBO (version 5.0) (52, 53).
HIV-1 RT steady-state kinetic assays.
The steady-state kinetic parameters, Km and kcat, for incorporation of dNTPs and dNTP analogs were determined using single-nucleotide incorporation in gel-based assays under saturating substrate conditions. The reactions were carried out in 50 mM Tris-HCl, pH 7.8, 50 mM NaCl, 6 mM MgCl2, 100 nM Td26A/Pd18 for dATP analogs, 100 nM Td26T/Pd18 for dTTP analogs, and 10 nM RT in a final volume of 20 μl and stopped at the reaction times of 1.5 to 3 min. The products were resolved on 15% polyacrylamide 7 M urea gels, and the gels were scanned with a Typhoon FLA 9000 phosphorimager (GE Healthcare). The bands were quantified using Multi Gauge software (FujiFilm). Km and kcat values were determined graphically using the Michaelis-Menten equation in GraphPad Prism 4 (GraphPad Software, Inc.).
ADA kinetic assays.
The deamination rates of dA (compound 1), EdA (compound 3), and CNdA (compound 7) were determined by following the disappearance of the substrate on a BioTek Synergy HT spectrophotometer at 265 nm in 100-μl solutions at 25°C with 0.1 M phosphate, pH 7.4. A change in molar extinction coefficients for all of the deaminated products was assumed to be Δε = −7,900 for all dA analogs (45). For each assay, the substrate concentrations varied between 15 μM and 240 μM. Absorbance was measured every 20 s. Three determinations were made for each analog. Km and kcat for each compound were determined using Michaelis-Menten plots (GraphPad Prism 4).
Because the reactions of ADA with FdA (compound 2), EFdA (compound 5), ECldA (compound 6), and CNFdA (compound 9) were very slow under these conditions, a high-pressure liquid chromatography (HPLC) method was used to observe the hydrolytic deamination of these compounds (54). In order to separate the substrate and product of the reaction, an elution gradient was developed using various combinations of 0.1 M phosphate, pH 7.4, and methanol. The following gradient was run on an Agilent 1100 series HPLC system: 7% methanol for 3 min, 14% methanol for 2 min, 21% methanol for 2 min, 28% methanol for 2 min, 35% methanol for 5 min, and 7% methanol for the final 2 min, returning the conditions to the original state. The gradient ran for a total of 16 min at a flow rate of 1 ml/min and an injection volume of 60 μl. The method ran at room temperature, and the absorbance was measured at a wavelength of 260 nm. The solvents used for the HPLC gradient were vacuum filtered prior to use.
Kinetic measurements were made using dA (compound 1), FdA (compound 2), EdA (compound 3), EFdA (compound 5), ECldA (compound 6), CNdA (compound 7), and CNFdA (compound 9) as ADA substrates. The reactions were carried out in 0.1 M phosphate buffer, pH 7.4. The FdA (compound 2) and EFdA (compound 5) analogs were tested using concentrations ranging from 10 μM to 9 mM. Compounds dA (compound 1), EdA (compound 3), ECldA (compound 6), CNdA (compound 7), and CNFdA (compound 9) were tested at 500 μM. Reactions were started by the addition of 2.4 mU ADA. The reactions were run for 45 min at 25°C before being terminated by the addition of perchlorate at 0.3 M final concentration. Denatured protein was removed from the reaction mixture by using centrifugation at 10,000 rpm for 5 min to pellet the protein. The clear supernatant was then removed, and the final pH was adjusted to 7.4 by adding Na2CO3 to reach a final concentration of 2.5 mM (54).
Structural analysis of predicted EFdA interactions at the active site of ADA.
Structural analysis of EFdA interactions at the ADA active site were carried out using the coordinates of human ADA in complex with dA obtained from the RCSB Protein Data Bank (PDB ID 3IAR). EFdA was drawn and minimized using the ChemBioDraw Ultra 13.0 and ChemBio3D Ultra 13.0 software from the ChemBioOffice Suite (PerkinElmer, Waltham, MA). Using the Coot model building software package (55), the structure of EFdA was superposed to the dA molecule, followed by the removal of dA.
RESULTS
Effect of 4′ substitutions on the structure of dA and dT analogs.
To evaluate the structural effects of the 4′ substitutions on the sugar ring conformations of the dA and dT analogs, we collected one-dimensional 1H NMR spectra over a range of physiologically relevant temperatures for the following compounds: 2′-deoxyadenosine (dA, compound 1), 2-fluoro-2′-deoxyadenosine (FdA, compound 2), 4′-ethynyl-2′-deoxyadenosine (EdA, compound 3), 4′-ethynyl-2′-deoxyribofuranosyl-2,6-diaminopurine (EdDAP, also known as ENdA, compound 4), 4′-ethynyl-2-fluoro-2′-deoxyadenosine (EFdA, compound 5), 4′-ethynyl-2-chloro-2′-deoxyadenosine (ECldA, compound 6), 4′-cyano-2′-deoxyadenosine (CNdA, compound 7), 4′-cyano-2′-deoxyribofuranosyl-2,6-diaminopurine (CNdDAP, compound 8), 4′-cyano-2-fluoro-2′-deoxyadenosine (CNFdA, compound 9), thymidine (dT, compound 12), 4′-methyl-thymidine (MedT, compound 13), and 4′-ethyl-thymidine (EtdT, compound 14) (Fig. 1) (36). Resolving the coupling constants between most of the hydrogen atoms on the deoxyribose ring required spectral simulation (see Table S1 in the supplemental material). The coupling constants determined from each spectrum were used to calculate the structural parameters of the deoxyribose ring of each compound. The changes in the value of each coupling constant with temperature were used to calculate the pseudorotational phase angle, P, and the degree of maximum ring pucker, ϕm, for the C-2′-exo/C-3′-endo (north or N) and C-2′-endo/C-3′-exo (south or S) sugar ring conformations (see Fig. S1 in the supplemental material) of the nucleoside analogs in this study.
The pseudorotational parameters listed in Table 1 were calculated using PSEUROT 6.3 as described in Materials and Methods and our previous report (36). The value of PN for each of the 4′-ethynyl- or cyano-substituted deoxyadenosine compounds 3 to 9 (PN = 37.3 to 47.8°) lies in the northern hemisphere of the pseudorotational pathway (PN = 0 to 90°), although these values are higher than what is typically observed for most nucleosides (PN = 0 to 36°) (see Fig. S1 in the supplemental material). These values correspond to a conformation between 4T3 (PN = 36°) and 4E or 4′-exo (PN = 54°) in the pseudorotational pathway (see Fig. S1) (56). A similar conformation was determined by Siddiqui et al. from the crystal structure of 4′-ethynyl-2′,3′-dideoxycytidine (PN = 43.5°) (57). The PN values of the 4′-cyano-substituted compounds 7 to 9 (PN = 41.6 to 47.8°) are slightly higher than those observed for the 4′-ethynyl-substituted compounds 3 to 6 (PN = 37.3 to 39.7°). The values of PN for the 4′-methyl- and 4′-ethyl-substituted thymidine compounds (PN = 24.3° and 23.3°, respectively) were also in the northern hemisphere but were much lower than those observed for the 4′-ethynyl and 4′-cyano analogs. These values correspond to a conformation between 3E or 3′-endo (PN = 18°) and 4T3 (PN = 36°) in the pseudorotational pathway (see Fig. S1) (56).
Table 1.
Summary of pseudorotational analysis of the sugar ring of deoxyadenosine and thymidine analogs
| Compound (no.) | PN (°) | PS (°) | ϕm(N) (°) | ϕm(S) (°) | % North | RMS error (Hz)a |
|---|---|---|---|---|---|---|
| Deoxyadenosine analogs | ||||||
| dA (1)b | 18.7 | 169.1 | 39.0c | 31.1 | 23 | 0.03 |
| FdA (2) | 3.4 | 169.1 | 38.7 | 29.2 | 24 | 0.01 |
| EdA (3) | 37.3 | 152.1 | 39.0c | 39.0c | 64 | 0.40 |
| EdDAP (4) | 39.7 | 152.1 | 39.0c | 39.0c | 63 | 0.49 |
| EFdA (5)b | 38.7 | 146.5 | 39.0c | 39.0c | 75 | 0.20 |
| ECldA (6) | 38.4 | 148.0 | 39.0c | 39.0c | 73 | 0.21 |
| CNdA (7) | 47.8 | 152.8 | 38.0c | 38.0c | 61 | 0.12 |
| CNdDAP (8) | 45.0 | 154.0c | 37.2 | 38.0c | 56 | 0.12 |
| CNFdA (9) | 41.6 | 154.2 | 38.0c | 38.0c | 59 | 0.23 |
| Thymidine analogs | ||||||
| MedT (13) | 24.3 | 132.5 | 38.0c | 31.5 | 43 | 0.05 |
| EtdT (14) | 23.3 | 150.3 | 38.0c | 30.2 | 39 | 0.18 |
Root mean square deviation between experimental and calculated coupling constants.
Values previously reported (36).
Value fixed during calculations.
The pseudorotational values determined for dA (compound 1) are in agreement with previous reports, demonstrating that dA strongly prefers the south conformation (36, 45). Comparison of the conformational parameters of dA (compound 1) versus EdA (compound 3) (23% versus 64% north) and FdA (compound 2) versus EFdA (compound 5) (24% versus 75% north) showed that the 4′-ethynyl strongly favors the north conformation of the sugar (Table 1 and Fig. 2). Comparison of dA (compound 1) versus CNdA (compound 7) (23% versus 61% north) and FdA (compound 2) versus CNFdA (compound 9) (24% versus 59% north) showed that the 4′-cyano also has a large effect on sugar conformation, albeit smaller than that of the 4′-ethynyl group (% north in EdA [compound 3] > % north in CNdA [compound 7] and % north in EFdA [compound 5] > % north in CNFdA [compound 9]) (Table 1).
Fig 2.
Dynamic equilibrium between the south and north sugar ring conformations of 4′-ethynyl-substituted compounds in solution (EFdA is shown). Natural deoxynucleoside substrates favor the south conformation.
Evaluation of the conformational parameters of the thymidine analogs dT (compound 12), MedT (compound 13), and EtdT (compound 14) demonstrated that 4′-methyl and ethyl substitutions have smaller effects on the sugar ring conformation than 4′-ethynyl and cyano groups. Although we were unable to accurately determine the coupling constants for dT (compound 12) due to overlap of one of the 2′-H signals with the DMSO solvent peak, it has previously been determined that the deoxyribose ring of dT favors the south conformation, which is similar to dA (56, 58). 4′-Methyl and 4′-ethyl substitutions shift the equilibrium toward the north conformation relative to the equilibrium of thymidine (43% and 39% north, respectively), but this effect is smaller than that of a 4′-ethynyl or 4′-cyano substitution in dA analogs (64% and 61% north compared to 23% in dA) (Table 1).
Hence, the 4′-ethynyl substitution imparts the largest effect on the conformation of the deoxyribose ring, followed by the 4′-cyano, the 4′-methyl, and the 4′-ethyl groups.
Effect of 2 position substitutions on the structure of dA analogs.
Using the same method, we also assessed the effect of substitutions at the 2 position of the adenine base on the structure of dA analogs. Evaluation of the conformation parameters of dA (compound 1) versus FdA (compound 2) showed that substitutions at the 2 position of the adenine base have little to no effect on sugar ring conformation. Similarly, comparisons of the 4′-ethynyl-substituted EdA (compound 3), EdDAP (compound 4), EFdA (compound 5), and ECldA (compound 6) showed that the 2-fluoro and 2-chloro substitutions have only a modest effect on the sugar ring conformation (Table 1). Finally, comparisons of CNdA (compound 7), CNdDAP (compound 8), and CNFdA (compound 9) also confirmed that 2-fluoro and 2-amino substitutions on the adenine ring have no significant impact on the sugar ring conformation (61%, 56%, and 59% north, respectively) (Table 1).
Molecular orbital calculations.
Potential stabilizing orbital interactions can occur between a lone pair of electrons in a p-like fully occupied orbital and an unoccupied antibonding orbital, allowing the delocalization of electrons and, thus, increasing molecular stability. To determine whether there are any favorable orbital interactions in the north or south sugar ring conformations of EFdA that could stabilize one conformation versus the other, we carried out quantum chemical computations to assess the orbital interactions of the lone pairs of the O4 and 3′-O atoms with the antibonding (π*) orbitals of the 4′-ethynyl moiety. We first determined the electronic composition of natural bond orbitals (NBOs) and their contribution in the formation of natural (semi-)localized molecular orbitals (NLMOs). The NBOs are localized orbitals that describe the Lewis-like molecular bonding pattern of electron pairs (59). The NLMOs have been described as semilocalized alternatives to canonical molecular orbitals (CMOs) for representing the electron pairs of MO-type wave functions. Each NLMO consists of a parent NBO (strictly localized) and associated delocalizations needed to describe the density of a full electron pair (59). The NBOs were used to compute CMOs. Both the north and south sugar ring conformations of EFdA have 375 NBOs (which include bonding orbitals, core orbitals, lone pairs, antibonding orbitals, and Rydberg orbitals), 76 NLMOs, and 375 CMOs. To assess the orbital interaction involving the lone pairs of O4 and 3′-O and the antibonding orbitals (π*) of the 4′-ethynyl group, we examined the second-order perturbative estimates of donor-acceptor (bond-antibond) NBO interactions for both north and south conformations. These estimates provide the measure of delocalization due to the interaction of the O4 or 3′-O lone pair with the π* antibonding orbital of the 4′-ethynyl group. Table 2 summarizes the energy of the donor-acceptor NBO interactions. These results suggest that the interaction of the lone pairs of the O4 and 3′-O atoms with the antibonding orbitals of the 4′-ethynyl moiety contributes to the greater stabilization energy (∼2-fold) of the north conformation. Further NBO analyses show the reduced occupancy of the O4 and 3′-O lone pairs (10% and 6%, respectively) and the remote interaction of both lone pairs with the antibonding NBOs of the 4′-C C in the north conformation. In the south conformation, the O4 and 3′-O lone pairs have a reduced occupancy of 7% and 4%, respectively, and only the O4 lone pair has a remote interaction with the antibonding NBOs of the 4′-C
C in the north conformation. In the south conformation, the O4 and 3′-O lone pairs have a reduced occupancy of 7% and 4%, respectively, and only the O4 lone pair has a remote interaction with the antibonding NBOs of the 4′-C C. The NLMO analyses of the north conformation showed that the lone pairs of both O4 and 3′-O atoms are delocalized into the vicinal region (C-5′ and 4′-C
C. The NLMO analyses of the north conformation showed that the lone pairs of both O4 and 3′-O atoms are delocalized into the vicinal region (C-5′ and 4′-C C, ∼5% and 2%, respectively). The delocalization of the O4 lone pair in the vicinal C-5′ and 4′-C
C, ∼5% and 2%, respectively). The delocalization of the O4 lone pair in the vicinal C-5′ and 4′-C C groups for the south conformation is significantly less than ∼1%, and there is almost no delocalization of the 3′-O lone pair in the C-5′ and 4′-C
C groups for the south conformation is significantly less than ∼1%, and there is almost no delocalization of the 3′-O lone pair in the C-5′ and 4′-C C vicinity. Taken together, these data suggest (i) that the north sugar ring conformation is more stable than the south and (ii) that the north conformation is stabilized by the interactions of the O4 and 3′-O lone pairs with the antibonding orbitals of the 4′-ethynyl group.
C vicinity. Taken together, these data suggest (i) that the north sugar ring conformation is more stable than the south and (ii) that the north conformation is stabilized by the interactions of the O4 and 3′-O lone pairs with the antibonding orbitals of the 4′-ethynyl group.
Table 2.
Summary of molecular orbital calculationsa
| NBO donor (LP) | NBO acceptor (π*) | Interaction energy (kcal/mol) for EFdA conformation |
|
|---|---|---|---|
| North | South | ||
| O4 | 4′-C C C |
1.63 | 1.25 |
| 3′-O | 4′-C C C |
0.71 | 0.0 |
| Total energy | 2.34 | 1.25 | |
LP, lone pair; π*, antibonding orbital.
Effect of 4′ substitutions on the ability of dA and dT analogs to block reverse transcription in vitro.
In order to assess the effect of various 4′ substitutions on the ability of dA and dT analogs to block reverse transcription in vitro, we tested the triphosphate forms of compounds 1, 4, 5, and 10 to 14 for their ability to block DNA synthesis by HIV-1 RT (Table 3). Additional previously published in vitro and cell-based inhibition data for dA and dT analogs are summarized in Table S2 in the supplemental material. Remarkably, almost all 4′-substituted analogs are excellent substrates of HIV-1 RT and are incorporated even more efficiently than the canonical substrate (higher kcat/Km). Specifically, the 4′-ethynyl substitution resulted in the highest efficiencies of inhibitor incorporation (EdDAP-TP and EFdA-TP have 60- and 45-fold higher kcat/Km than dATP). The higher efficiency appears to be the result of both increased catalytic turnover rate (kcat) and decreased Km (Table 3). Notably, the enhancement in incorporation efficiency in vitro appears to be more pronounced for the dA than for the dT analogs (Table 3). Moreover, 4′-ethynyl dA analogs are 3 to 4 times more efficiently incorporated by HIV-1 RT than 4′-methyl compounds. Inhibitors containing a 4′-ethyl modification are the least efficiently incorporated by HIV-1 RT.
Table 3.
Steady-state kinetic parameters for incorporation of the triphosphate forms of the 4′-substituted compounds by HIV-1 RT
| dNTP | Km (μM)a | kcat (min−1)a | kcat/Km (min−1 · μM−1) | Fold change in incorporation efficiencyb |
|---|---|---|---|---|
| dATP | 2.35 ± 0.06 | 0.66 ± 0.13 | 0.3 | 1 |
| EFdA-TP | 0.27 ± 0.01 | 3.47 ± 0.04 | 12.9 | 45.9 |
| EdDAP-TP | 0.15 ± 0.01 | 2.55 ± 0.10 | 17.0 | 60.7 |
| MedA-TP | 0.29 ± 0.01 | 1.24 ± 0.05 | 4.3 | 15.3 |
| EtdA-TP | 0.88 ± 0.19 | 3.41 ± 0.12 | 3.9 | 13.9 |
| dTTP | 0.60 ± 0.01 | 1.04 ± 0.12 | 1.7 | 1 |
| MedT-TP | 0.68 ± 0.22 | 1.58 ± 0.15 | 2.3 | 1.3 |
| EtdT-TP | 2.00 ± 0.22 | 1.44 ± 0.20 | 0.7 | 0.4 |
Values are means ± standard deviations of two independent experiments and were determined from the Michaelis-Menten equation using GraphPad Prism 4.
Fold change in incorporation efficiency is the ratio of the incorporation efficiency (kcat/Km) of NRTI-TP over that of dATP or dTTP [(kcat/Km)NRTI-TP/(kcat/Km)dATP or (kcat/Km)NRTI-TP/(kcat/Km)dTTP].
Effect of 4′ substitutions on susceptibility to ADA.
Bovine ADA was chosen to study the hydrolytic deamination of compounds 1 to 7 and 9 because it is readily available and because it is highly similar to human ADA (91% overall and 100% active-site sequence identity) (60, 61), allowing us to use the biochemical data to infer what would happen with the human enzyme. We found that 4′ substitutions decreased the ability of ADA to use these nucleosides as substrates, primarily due to an increase in Km (Table 4). The 4′-ethynyl and 4′-cyano groups have similar effects on ADA catabolism.
Table 4.
Summary of steady-state kinetic data for the deamination of dA, EdA, and CNdA by ADAa
| Compound (no.) | Mean Km ± SD (μM) | kcat (s−1) | kcat/Km (s−1 · μM−1) |
|---|---|---|---|
| dA (1) | 42.6 ± 15.0 | 13.5 | 0.32 |
| EdA (3) | 145.9 ± 37.6 | 11.3 | 0.08 |
| CNdA (7) | 156.9 ± 84.2 | 8.9 | 0.06 |
Data were determined spectrophotometrically.
Effect of 2 position substitutions on susceptibility to ADA.
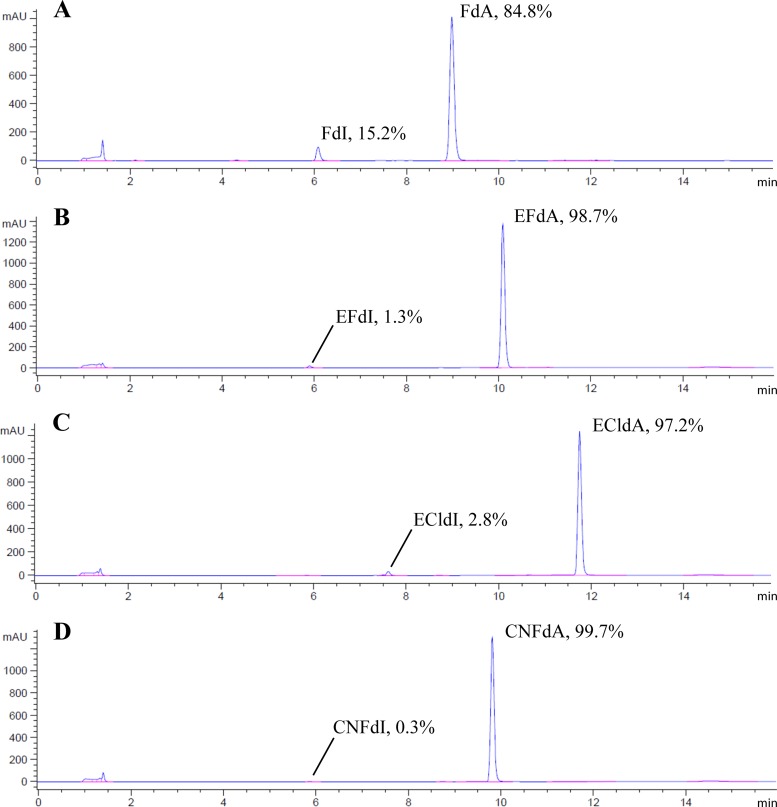
Because the 2 position-substituted compounds were refractory to hydrolytic deamination by ADA, we were unable to obtain reliable kinetic measurements using the spectrophotometric assay. In order to at least obtain a relative order of susceptibility to ADA, we used an HPLC-based assay (54), which allowed long incubation times and a determination of the amount of product from the slow hydrolytic deamination of compounds 2, 5, 6, and 9 by ADA. These experiments showed that incubation of 500 μM substrate for 45 min resulted in 15% conversion of FdA (compound 2), 1.3% of EFdA (compound 5), 2.8% of ECldA (compound 6), and <1% of CNFdA (compound 9) (Fig. 3). These data suggest that the 2-halo combined with 4′ substitutions resulted in significantly decreased susceptibility of the dA compounds to ADA and that the decreasing order of stability was CNFdA (compound 9) > EFdA (compound 5) > ECldA (compound 6) > FdA (compound 2). This is consistent with our previous report that EFdA is not efficiently deaminated by ADA (21).
Fig 3.
HPLC data taken after reaction of dA analogs (500 μM) with ADA for 45 min (260-nm signal is shown in blue). FdA (A), EFdA (B), ECldA (C), and CNFdA (D) were converted to the corresponding deoxyinosine (dI) forms to different extents. Percentages of dA-based reactant and dI-based product were calculated based on peak areas.
Structural analysis of predicted EFdA interactions at the ADA active site.
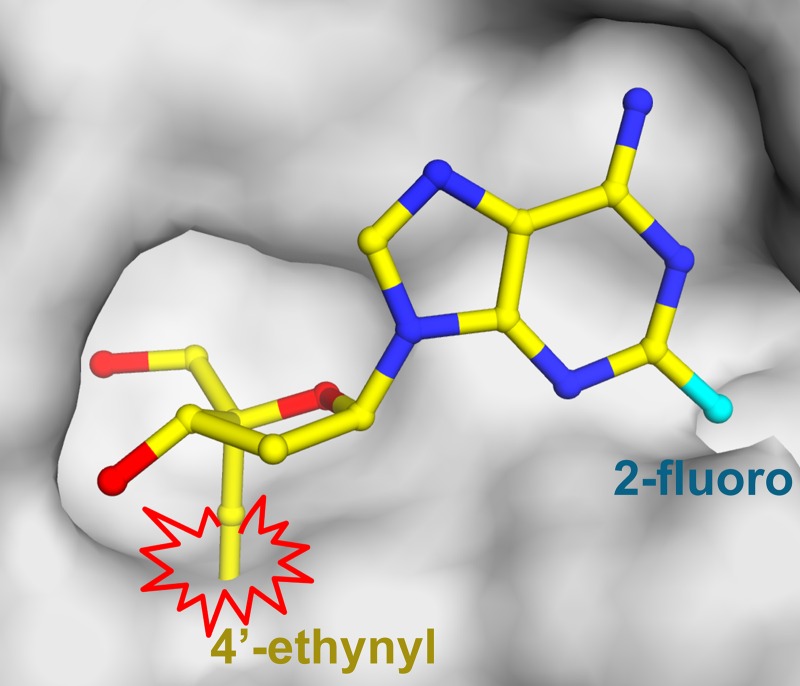
Using the crystal structure coordinates of human ADA, we looked for potential interactions between the active-site residues and the modeled EFdA that could lead to decreased susceptibility of this inhibitor to ADA. As shown in Fig. 4, the placement of EFdA at the expected position of a substrate would result in unfavorable steric interactions between the 4′-ethynyl group of EFdA and the sulfur and ε-carbon of Met155 (d = 2.2 and 2.0 Å, respectively). These potential steric effects could contribute to a lower binding affinity for EdA than for dA, consistent with an ∼3.5-fold difference in the corresponding Km values (Km-EdA = 146 μM versus Km-dA = 43 μM), which is the primary contributing factor in the observed decrease in the catalytic efficiency of ADA when EdA is the substrate.
Fig 4.
Structural analysis of the predicted effect of the 4′-ethynyl group of EFdA (yellow sticks) on interactions with the binding pocket of ADA (gray surface). When EFdA binds in a manner similar to that of other substrates, the 4′-ethynyl group may clash with the sulfur and ε-carbon atoms of Met155 in ADA. The figure was made using the PyMOL molecular graphics software package (http://www.pymol.org/).
DISCUSSION
We have previously demonstrated that EFdA is more potent against WT and drug-resistant HIV strains than other approved NRTIs (19–24) in both biochemical and virological assays and that it has low cytotoxicity (24–26). This suggests that EFdA not only has a high affinity toward HIV-1 RT but is also likely to be efficiently activated by host kinases (21, 25) and is relatively stable in cells (21). In fact, a favorable NRTI EC50 depends on multiple desirable attributes that include (i) a strong binding affinity of the active metabolite for the RT target, (ii) efficient cellular uptake and activation by host kinases, (iii) an ability to withstand deactivation by cellular enzymes, (iv) a high barrier to resistance, and (v) low cytotoxicity. The present study investigates how different substitutions on the sugar ring and adenine base contribute to the structural features of NRTIs and the effects of these features on the first three of the factors listed above.
Sugar puckering affects several aspects of the activity of EFdA by influencing its interactions with HIV RT, dCK, and ADA. We have previously proposed that the extent to which an NRTI can exist in the north conformation affects its potency. In that respect, we have shown (Marquez and colleagues) that HIV-1 RT prefers as a substrate a version of zidovudine-triphosphate (AZT-TP) that is locked in the north rather than in the south conformation (38), suggesting that the HIV-1 RT active site prefers nucleosides or their analogs in the north conformation. Our NMR data and molecular orbital calculations demonstrate that the 4′-ethynyl substitution causes the sugar ring conformation of EFdA to be primarily in the north conformation. This causes it (and presumably the structure of its active metabolite EFdA-TP) to reside in a favorable conformation for binding at the polymerase active site of HIV-1 RT (36). This hypothesis is further supported by our steady-state kinetic assays and a previous report of in vitro HIV RT inhibition by EFdA-TP (20).
For a nucleoside analog to be a potent inhibitor of HIV replication, it must be converted to the triphosphate form in cells. The nucleoside substrate type or sugar ring conformation can affect the ability of cellular kinases to activate nucleoside analogs. The type of nucleoside substrate can determine which kinase will carry out the initial phosphorylation step. For example, dCK preferentially phosphorylates dC, dA, and dG analogs (62, 63), whereas thymidine kinase 1 phosphorylates dT analogs (64). Hence, the differences in phosphorylation efficiency of various NRTIs likely depend, at least in part, on the fact that different kinases are used to convert the various analogs to the active metabolite (65). Moreover, the activation of NRTIs involves multiple enzymes whose specificity may also impact the efficiency of activation. For example, AZT-MP is not phosphorylated efficiently to AZT-diphosphate (AZT-DP) by thymidylate kinase, and this is the rate-limiting step in AZT-TP formation (25, 66).
The sugar ring conformation can also affect the ability of host kinases to activate nucleoside analogs. For example, dCK is known to bind its natural nucleoside substrates in the south conformation (41, 42). Interestingly, although the EFdA sugar ring favors the north conformation, it appears to be phosphorylated efficiently by dCK in cells (25). This suggests that other structural features of EFdA may contribute to its efficient phosphorylation, such as the 4′-ethynyl, the 2-fluoro, or both. In fact, it has been previously reported that a 2-fluoro substitution on dA decreases the Km value 33-fold compared to that of dA, making FdA a very efficient substrate for dCK (63). Ongoing structural studies of EFdA in complex with dCK should reveal the basis of efficient activation of EFdA by dCK.
The EC50 of NRTIs can also be affected by catabolizing enzymes. For example, ADA can deaminate dA-based nucleosides and change the pathway and efficiency of their activation. The activity of ADA is affected by the structural determinants of its substrates. While we have previously demonstrated (Marquez and colleagues) that this enzyme efficiently deaminates nucleoside analogs in the north conformation (43–46), the kinetic analysis of ADA that we report here shows that the presence of a substituent at the 2 position of the adenine base makes these derivatives poor substrates for ADA. These results agree with previous studies reporting that 2-halo-substituted compounds are not efficiently deaminated by ADA (29, 31–33).
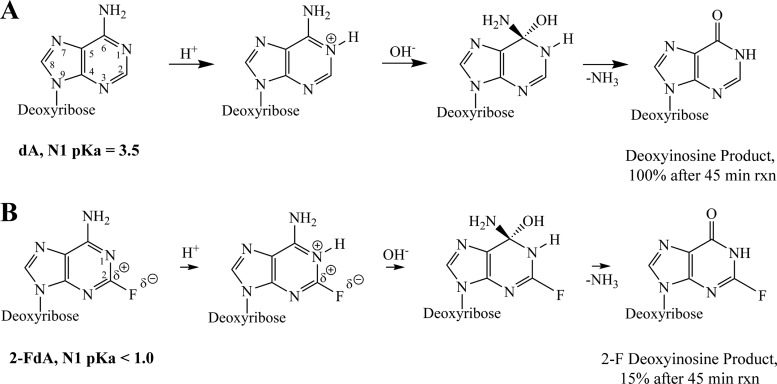
The mechanism by which 2-halo substitutions affect ADA activity does not appear to involve steric interactions in the ADA binding pocket (Fig. 4). This suggests that they decrease susceptibility through electronic rather than steric effects. The electronic effect of 2-fluoro can be explained in terms of changes in pKa and charge distribution on the adenine base. The deamination mechanism by ADA involves an addition-elimination reaction on the substrate to form an inosine-based product (Fig. 5A). The first step involves electrophilic attack to protonate the N1 position of the purine ring, followed by nucleophilic attack at the C-6 position of the ring to form a tetrahedral intermediate (67–70). The introduction of an electronegative fluorine atom at the 2 position has been reported to dramatically lower the pKa of the N1 atom of adenosine-based compounds (more than 100-fold) (Fig. 5B) (71). This is the result of the highly electronegative fluorine atom increasing the positive charge at the C-2 position (C-2 ∂+, F ∂−), which would raise the energy barrier of N1 protonation. Hence, the N1 protonation step becomes the rate-limiting step of the hydrolytic deamination reaction, thus reducing the yield of the 2-fluoro deoxyinosine product (Fig. 3). The presence of a 2-amino instead of a 2-halo substitution has a different effect on the mechanism and efficiency of the deamination reaction. In the case of EdDAP, a resonance effect caused by the 2-amino substitution would distribute the positive charge of the protonated N1 to all neighboring nitrogen atoms. Hence, differences in the deamination efficiency of EdDAP and EFdA can be accounted for in terms of different charge distribution effects on the adenine base. This explanation is consistent with experimental results that demonstrated that EdDAP was catabolized more efficiently than EFdA but less efficiently than either dA or EdA, suggesting that a 2-amino substitution has a smaller impact on deamination by ADA than a 2-fluoro substitution (E. N. Kodama, personal communication).
Fig 5.
Reaction mechanism of hydrolytic deamination of dA (A) and FdA (B) by adenosine deaminase (67–70). The electronegative 2-fluoro group causes a dramatic decrease in pKa at the N1 position (71), resulting in a high-energy barrier for the N1 protonation step of the reaction. rxn, reaction.
In addition to 2 position substitutions, the presence of a 4′-ethynyl or cyano group also reduces deamination by ADA. For example, ADA deaminates EdA or CNdA less efficiently than dA (Table 4) and EFdA or CNFdA less efficiently than FdA (Fig. 3). Our structural analysis of the predicted interactions of EFdA at the ADA active site suggests that the 4′-ethynyl group may sterically interact with the flexible Met155 side chain (Fig. 4), resulting in a moderate decrease in the binding and reactivity of EFdA compared to the binding and reactivity of dA (Table 4). These data suggest that the effect of 4′-ethynyl on ADA activity is caused primarily by steric interactions. Hence, the substrate specificity of ADA is affected by both electronic and steric interactions from different structural determinants of dA-based NRTIs (2-fluoro and 4′-ethynyl, respectively).
In conclusion, strategic substitutions have a pronounced and complex effect on the ability of dA-based inhibitors to block HIV-1 replication. In particular, substitutions at the 4′ position of the deoxyribose ring and the 2 position of the adenine base of EFdA have separate effects that enhance its antiviral activity by favoring structural conformations that improve its interactions with the RT target and by decreasing its susceptibility to ADA. Because EFdA is a poor substrate for ADA, it is likely to remain intact for longer periods of time, thus reducing the dosage required to achieve an antiviral effect.
Supplementary Material
ACKNOWLEDGMENTS
We thank Wei Wycoff for assistance with variable-temperature NMR data collection and processing.
The 500-MHz NMR spectrometer is supported by NSF grant CHE-89-08304 and NIH/NCRR grant S10 RR022341-01 (cold probe). This work was supported, in whole or in part, by National Institutes of Health grants AI076119, AI099284, AI100890, and GM103368 (S.G.S.) and AI079801 (M.A.P.). This study was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, Center for Cancer Research. We also acknowledge support from Ministry of Knowledge and Economy, Bilateral International Collaborative R&D Program, Republic of Korea. B.M. is a recipient of the amfAR Mathilde Krim Fellowship and a Canadian Institutes of Health Research (CIHR) Fellowship. E.N.K. and H.M. are coinventors of EFdA.
Footnotes
Published ahead of print 7 October 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01703-13.
REFERENCES
- 1. Parniak MA, Sluis-Cremer N. 2000. Inhibitors of HIV-1 reverse transcriptase. Adv. Pharmacol. 49:67–109 [DOI] [PubMed] [Google Scholar]
- 2. Painter GR, Almond MR, Mao S, Liotta DC. 2004. Biochemical and mechanistic basis for the activity of nucleoside analogue inhibitors of HIV reverse transcriptase. Curr. Top. Med. Chem. 4:1035–1044 [DOI] [PubMed] [Google Scholar]
- 3. Hammer SM, Saag MS, Schechter M, Montaner JS, Schooley RT, Jacobsen DM, Thompson MA, Carpenter CC, Fischl MA, Gazzard BG, Gatell JM, Hirsch MS, Katzenstein DA, Richman DD, Vella S, Yeni PG, Volberding PA. 2006. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society—U. S. A. panel. Top. HIV Med. 14:827–843 [PubMed] [Google Scholar]
- 4. De Clercq E. 2007. Anti-HIV drugs. Verh. K. Acad. Geneeskd. Belg. 69:81–104 [PubMed] [Google Scholar]
- 5. Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. 2009. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 385:693–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schinazi RF, Bassit L, Gavegnano C. 2010. HCV drug discovery aimed at viral eradication. J. Viral Hepat. 17:77–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wauchope OR, Tomney MJ, Pepper JL, Korba BE, Seley-Radtke KL. 2010. Tricyclic 2′-C-modified nucleosides as potential anti-HCV therapeutics. Org. Lett. 12:4466–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doong SL, Tsai CH, Schinazi RF, Liotta DC, Cheng YC. 1991. Inhibition of the replication of hepatitis B virus in vitro by 2′,3′-dideoxy-3′-thiacytidine and related analogues. Proc. Natl. Acad. Sci. U. S. A. 88:8495–8499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Michailidis E, Kirby KA, Hachiya A, Yoo W, Hong SP, Kim SO, Folk WR, Sarafianos SG. 2012. Antiviral therapies: focus on hepatitis B reverse transcriptase. Int. J. Biochem. Cell Biol. 44:1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wauchope OR, Johnson C, Krishnamoorthy P, Andrei G, Snoeck R, Balzarini J, Seley-Radtke KL. 2012. Synthesis and biological evaluation of a series of thieno-expanded tricyclic purine 2′-deoxy nucleoside analogues. Bioorg. Med. Chem. 20:3009–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gallois-Montbrun S, Schneider B, Chen Y, Giacomoni-Fernandes V, Mulard L, Morera S, Janin J, Deville-Bonne D, Veron M. 2002. Improving nucleoside diphosphate kinase for antiviral nucleotide analogs activation. J. Biol. Chem. 277:39953–39959 [DOI] [PubMed] [Google Scholar]
- 12. Brinkman K, ter Hofstede HJ, Burger DM, Smeitink JA, Koopmans PP. 1998. Adverse effects of reverse transcriptase inhibitors: mitochondrial toxicity as common pathway. AIDS 12:1735–1744 [DOI] [PubMed] [Google Scholar]
- 13. Kakuda TN. 2000. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin. Ther. 22:685–708 [DOI] [PubMed] [Google Scholar]
- 14. Lewis W, Day BJ, Copeland WC. 2003. Mitochondrial toxicity of NRTI antiviral drugs: an integrated cellular perspective. Nat. Rev. Drug Discov. 2:812–822 [DOI] [PubMed] [Google Scholar]
- 15. Sluis-Cremer N, Arion D, Parniak MA. 2000. Molecular mechanisms of HIV-1 resistance to nucleoside reverse transcriptase inhibitors (NRTIs). Cell. Mol. Life Sci. 57:1408–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Menendez-Arias L. 2008. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res. 134:124–146 [DOI] [PubMed] [Google Scholar]
- 17. Singh K, Marchand B, Kirby KA, Michailidis E, Sarafianos SG. 2010. Structural aspects of drug resistance and inhibition of HIV-1 reverse transcriptase. Viruses 2:606–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Arts EJ, Hazuda DJ. 2012. HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2:a007161. 10.1101/cshperspect.a007161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kodama EI, Kohgo S, Kitano K, Machida H, Gatanaga H, Shigeta S, Matsuoka M, Ohrui H, Mitsuya H. 2001. 4′-Ethynyl nucleoside analogs: potent inhibitors of multidrug-resistant human immunodeficiency virus variants in vitro. Antimicrob. Agents Chemother. 45:1539–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Michailidis E, Marchand B, Kodama EN, Singh K, Matsuoka M, Kirby KA, Ryan EM, Sawani AM, Nagy E, Ashida N, Mitsuya H, Parniak MA, Sarafianos SG. 2009. Mechanism of inhibition of HIV-1 reverse transcriptase by 4′-ethynyl-2-fluoro-2′-deoxyadenosine triphosphate, a translocation-defective reverse transcriptase inhibitor. J. Biol. Chem. 284:35681–35691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kawamoto A, Kodama E, Sarafianos SG, Sakagami Y, Kohgo S, Kitano K, Ashida N, Iwai Y, Hayakawa H, Nakata H, Mitsuya H, Arnold E, Matsuoka M. 2008. 2′-Deoxy-4′-C-ethynyl-2-halo-adenosines active against drug-resistant human immunodeficiency virus type 1 variants. Int. J. Biochem. Cell Biol. 40:2410–2420 [DOI] [PubMed] [Google Scholar]
- 22. Michailidis E, Ryan EM, Hachiya A, Kirby KA, Marchand B, Leslie MD, Huber AD, Ong YT, Jackson JC, Singh K, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. 2013. Hypersusceptibility mechanism of tenofovir-resistant HIV to EFdA. Retrovirology 10:65. 10.1186/1742-4690-10-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michailidis E, Singh K, Ryan EM, Hachiya A, Ong YT, Kirby KA, Marchand B, Kodama EN, Mitsuya H, Parniak MA, Sarafianos SG. 2012. Effect of translocation defective reverse transcriptase inhibitors on the activity of N348I, a connection subdomain drug resistant HIV-1 reverse transcriptase mutant. Cell. Mol. Biol. 58:187–195 [PMC free article] [PubMed] [Google Scholar]
- 24. Murphey-Corb M, Rajakumar P, Michael H, Nyaundi J, Didier PJ, Reeve AB, Mitsuya H, Sarafianos SG, Parniak MA. 2012. Response of simian immunodeficiency virus to the novel nucleoside reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine in vitro and in vivo. Antimicrob. Agents Chemother. 56:4707–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nakata H, Amano M, Koh Y, Kodama E, Yang G, Bailey CM, Kohgo S, Hayakawa H, Matsuoka M, Anderson KS, Cheng YC, Mitsuya H. 2007. Activity against human immunodeficiency virus type 1, intracellular metabolism, and effects on human DNA polymerases of 4′-ethynyl-2-fluoro-2′-deoxyadenosine. Antimicrob. Agents Chemother. 51:2701–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sohl CD, Singh K, Kasiviswanathan R, Copeland WC, Mitsuya H, Sarafianos SG, Anderson KS. 2012. Mechanism of interaction of human mitochondrial DNA polymerase gamma with the novel nucleoside reverse transcriptase inhibitor 4′-ethynyl-2-fluoro-2′-deoxyadenosine indicates a low potential for host toxicity. Antimicrob. Agents Chemother. 56:1630–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Adams A, Harkness RA. 1976. Adenosine deaminase activity in thymus and other human tissues. Clin. Exp. Immunol. 26:647–649 [PMC free article] [PubMed] [Google Scholar]
- 28. Johnson MA, Ahluwalia G, Connelly MC, Cooney DA, Broder S, Johns DG, Fridland A. 1988. Metabolic pathways for the activation of the antiretroviral agent 2′,3′-dideoxyadenosine in human lymphoid cells. J. Biol. Chem. 263:15354–15357 [PubMed] [Google Scholar]
- 29. Chilson OP, Fisher JR. 1963. Some comparative studies of calf and chicken adenosine deaminase. Arch. Biochem. Biophys. 102:77–85 [DOI] [PubMed] [Google Scholar]
- 30. Baer HP, Drummond GI, Gillis J. 1968. Studies on the specificity and mechanism of action of adenosine deaminase. Arch. Biochem. Biophys. 123:172–178 [DOI] [PubMed] [Google Scholar]
- 31. Simon LN, Bauer RJ, Tolman RL, Robins RK. 1970. Calf intestine adenosine deaminase. Substrate specificity. Biochemistry 9:573–577 [DOI] [PubMed] [Google Scholar]
- 32. Maguire MH, Sim MK. 1971. Studies on adenosine deaminase. 2. Specificity and mechanism of action of bovine placental adenosine deaminase. Eur. J. Biochem. 23:22–29 [DOI] [PubMed] [Google Scholar]
- 33. Agarwal RP, Sagar SM, Parks RE., Jr 1975. Adenosine deaminase from human erythrocytes: purification and effects of adenosine analogs. Biochem. Pharmacol. 24:693–701 [DOI] [PubMed] [Google Scholar]
- 34. Cooney DA, Ahluwalia G, Mitsuya H, Fridland A, Johnson M, Hao Z, Dalal M, Balzarini J, Broder S, Johns DG. 1987. Initial studies on the cellular pharmacology of 2′,3′-dideoxyadenosine, an inhibitor of HTLV-III infectivity. Biochem. Pharmacol. 36:1765–1768 [DOI] [PubMed] [Google Scholar]
- 35. Yarchoan R, Mitsuya H, Thomas RV, Pluda JM, Hartman NR, Perno CF, Marczyk KS, Allain JP, Johns DG, Broder S. 1989. In vivo activity against HIV and favorable toxicity profile of 2′,3′-dideoxyinosine. Science 245:412–415 [DOI] [PubMed] [Google Scholar]
- 36. Kirby KA, Singh K, Michailidis E, Marchand B, Kodama EN, Ashida N, Mitsuya H, Parniak MA, Sarafianos SG. 2011. The sugar ring conformation of 4′-ethynyl-2-fluoro-2′-deoxyadenosine and its recognition by the polymerase active site of HIV reverse transcriptase. Cell Mol. Biol. 57:40–46 [PMC free article] [PubMed] [Google Scholar]
- 37. Marquez VE, Ezzitouni A, Russ P, Siddiqui MA, Ford H, Jr, Feldman RJ, Mitsuya H, George C, Barchi JJ., Jr 1998. Lessons from the pseudorotational cycle: conformationally rigid AZT carbocyclic nucleosides and their interaction with reverse transcriptase. Nucleosides Nucleotides 17:1881–1884 [Google Scholar]
- 38. Marquez VE, Ezzitouni A, Russ P, Siddiqui MA, Ford H, Jr, Feldman RJ, Mitsuya H, George C, Barchi JJ., Jr 1998. HIV-1 reverse transcriptase can discriminate between two conformationally locked carbocyclic AZT triphosphate analogues. J. Am. Chem. Soc. 120:2780–2789 [Google Scholar]
- 39. Mu L, Sarafianos SG, Nicklaus MC, Russ P, Siddiqui MA, Ford H, Jr, Mitsuya H, Le R, Kodama E, Meier C, Knispel T, Anderson L, Barchi JJ, Jr, Marquez VE. 2000. Interactions of conformationally biased north and south 2′-fluoro-2′, 3′-dideoxynucleoside 5′-triphosphates with the active site of HIV-1 reverse transcriptase. Biochemistry 39:11205–11215 [DOI] [PubMed] [Google Scholar]
- 40. Boyer PL, Julias JG, Marquez VE, Hughes SH. 2005. Fixed conformation nucleoside analogs effectively inhibit excision-proficient HIV-1 reverse transcriptases. J. Mol. Biol. 345:441–450 [DOI] [PubMed] [Google Scholar]
- 41. Van Roey P, Taylor EW, Chu CK, Schinazi RF. 1990. Correlation of molecular conformation and activity of reverse transcriptase inhibitors. Ann. N. Y. Acad. Sci. 616:29–40 [DOI] [PubMed] [Google Scholar]
- 42. Wang J, Choudhury D, Chattopadhyaya J, Eriksson S. 1999. Stereoisomeric selectivity of human deoxyribonucleoside kinases. Biochemistry 38:16993–16999 [DOI] [PubMed] [Google Scholar]
- 43. Marquez VE, Russ P, Alonso R, Siddiqui MA, Shin KJ, George C, Nicklaus MC, Dai F, Ford H., Jr 1999. Conformationally restricted nucleosides. The reaction of adenosine deaminase with substrates built on a bicyclo[3.1.0]hexane template. Nucleosides Nucleotides 18:521–530 [DOI] [PubMed] [Google Scholar]
- 44. Marquez VE, Russ P, Alonso R, Siddiqui MA, Hernandez S, George C, Nicklaus MC, Dai F, Ford H., Jr 1999. Synthesis of conformationally restricted carbocyclic nucleosides: the role of the O(4′)-atom in the key hydration step of adenosine deaminase. Helv. Chim. Acta 82:2119–2129 [Google Scholar]
- 45. Ford H, Jr, Dai F, Mu L, Siddiqui MA, Nicklaus MC, Anderson L, Marquez VE, Barchi JJ., Jr 2000. Adenosine deaminase prefers a distinct sugar ring conformation for binding and catalysis: kinetic and structural studies. Biochemistry 39:2581–2592 [DOI] [PubMed] [Google Scholar]
- 46. Marquez VE, Schroeder GK, Ludek OR, Siddiqui MA, Ezzitouni A, Wolfenden R. 2009. Contrasting behavior of conformationally locked carbocyclic nucleosides of adenosine and cytidine as substrates for deaminases. Nucleosides Nucleotides Nucleic Acids 28:614–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shuto S, Obara T, Itoh H, Kosugi Y, Saito Y, Toriya M, Yaginuma S, Shigeta S, Matsuda A. 1994. New neplanocin analogues. IV. 2-Fluoroneplanocin A: an adenosine deaminase-resistant equivalent of neplanocin A. Chem. Pharm. Bull. (Tokyo) 42:1688–1690 [DOI] [PubMed] [Google Scholar]
- 48. Obara T, Shuto S, Saito Y, Snoeck R, Andrei G, Balzarini J, De Clercq E, Matsuda A. 1996. New neplanocin analogues. 7. Synthesis and antiviral activity of 2-halo derivatives of neplanocin A. J. Med. Chem. 39:3847–3852 [DOI] [PubMed] [Google Scholar]
- 49. Boyer PL, Julias JG, Ambrose Z, Siddiqui MA, Marquez VE, Hughes SH. 2007. The nucleoside analogs 4′C-methyl thymidine and 4′C-ethyl thymidine block DNA synthesis by wild-type HIV-1 RT and excision proficient NRTI resistant RT variants. J. Mol. Biol. 371:873–882 [DOI] [PubMed] [Google Scholar]
- 50. Vu BC, Boyer PL, Siddiqui MA, Marquez VE, Hughes SH. 2011. 4′-C-methyl-2′-deoxyadenosine and 4′-C-ethyl-2′-deoxyadenosine inhibit HIV-1 replication. Antimicrob. Agents Chemother. 55:2379–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de Leeuw FAAM, Altona C. 1983. Computer assisted pseudorotational analysis of 5-membered rings by means of 3J.HH coupling constants: program PSEUROT. J. Comp. Chem. 4:428–437 [Google Scholar]
- 52. Glendening ED, Badenhoop JK, Reed AE, Carpenter JE, Bohmann JA, Morales CM, Weinhold F. 2001. NBO 5.0 program. Theoretical Chemistry Institute, University of Wisconsin, Madison, WI [Google Scholar]
- 53. Weinhold F, Landis CR. 2001. Introduction to Natural Bond Orbitals, NBO 5.0 program, and recent extensions of localized bonding concepts. Chem. Educ. Res. Pract. 2:91–104 [Google Scholar]
- 54. Paul MK, Grover V, Mukhopadhyay AK. 2005. Merits of HPLC-based method over spectrophotometric method for assessing the kinetics and inhibition of mammalian adenosine deaminase. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 822:146–153 [DOI] [PubMed] [Google Scholar]
- 55. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 56. Altona C, Sundaralingam M. 1972. Conformational analysis of the sugar ring in nucleosides and nucleotides: a new description using the concept of pseudorotation. J. Am. Chem. Soc. 94:8205–8212 [DOI] [PubMed] [Google Scholar]
- 57. Siddiqui MA, Hughes SH, Boyer PL, Mitsuya H, Van QN, George C, Sarafinanos SG, Marquez VE. 2004. A 4′-C-ethynyl-2′,3′-dideoxynucleoside analogue highlights the role of the 3′-OH in anti-HIV active 4′-C-ethynyl-2′-deoxy nucleosides. J. Med. Chem. 47:5041–5048 [DOI] [PubMed] [Google Scholar]
- 58. Young DW, Tollin P, Wilson HR. 1969. The crystal and molecular structure of thymidine. Acta Crystallogr. B 25:1423–1432 [Google Scholar]
- 59. Weinhold F, Landis CR. 2012. Discovering chemistry with natural bond orbitals. John Wiley and Sons, Inc, Hoboken, NJ [Google Scholar]
- 60. Kelly MA, Vestling MM, Murphy CM, Hua S, Sumpter T, Fenselau C. 1996. Primary structure of bovine adenosine deaminase. J. Pharm. Biomed. Anal. 14:1513–1519 [DOI] [PubMed] [Google Scholar]
- 61. Wiginton DA, Adrian GS, Hutton JJ. 1984. Sequence of human adenosine deaminase cDNA including the coding region and a small intron. Nucleic Acids Res. 12:2439–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Durham JP, Ives DH. 1970. Deoxycytidine kinase. II. Purification and general properties of the calf thymus enzyme. J. Biol. Chem. 245:2276–2284 [PubMed] [Google Scholar]
- 63. Krenitsky TA, Tuttle JV, Koszalka GW, Chen IS, Beacham LM, III, Rideout JL, Elion GB. 1976. Deoxycytidine kinase from calf thymus. Substrate and inhibitor specificity. J. Biol. Chem. 251:4055–4061 [PubMed] [Google Scholar]
- 64. Weissman SM, Smellie RM, Paul J. 1960. Studies on the biosynthesis of deoxyribonucleic acid by extracts of mammalian cells. IV. The phosphorylation of thymidine. Biochim. Biophys. Acta 45:101–110 [DOI] [PubMed] [Google Scholar]
- 65. Bazzoli C, Jullien V, Le Tiec C, Rey E, Mentre F, Taburet AM. 2010. Intracellular pharmacokinetics of antiretroviral drugs in HIV-infected patients, and their correlation with drug action. Clin. Pharmacokinet. 49:17–45 [DOI] [PubMed] [Google Scholar]
- 66. Furman PA, Fyfe JA, St Clair MH, Weinhold K, Rideout JL, Freeman GA, Lehrman SN, Bolognesi DP, Broder S, Mitsuya H. 1986. Phosphorylation of 3′-azido-3′-deoxythymidine and selective interaction of the 5′-triphosphate with human immunodeficiency virus reverse transcriptase. Proc. Natl. Acad. Sci. U. S. A. 83:8333–8337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wolfenden R, Frick L. 1986. Mechanisms of enzyme action and inhibition: transition state analogues for acid-base catalysis. J. Protein Chem. 5:147–155 [Google Scholar]
- 68. Frick L, Wolfenden R, Smal E, Baker DC. 1986. Transition-state stabilization by adenosine deaminase: structural studies of its inhibitory complex with deoxycoformycin. Biochemistry 25:1616–1621 [DOI] [PubMed] [Google Scholar]
- 69. Orozco M, Lluis C, Mallol J, Canela EI, Franco R. 1989. Theoretical approximation to the reaction mechanism of adenosine deaminase. Quant. Struct. Act. Relat. 8:109–114 [Google Scholar]
- 70. Orozco M, Canela EI, Franco R. 1990. Theoretical study of the hydroxyl nucleophilic attack on the 6-aminopyrimidine molecule: functional implications in the reaction mechanism of nucleoside deaminative enzymes. J. Org. Chem. 55:2630–2637 [Google Scholar]
- 71. Suydam IT, Strobel SA. 2008. Fluorine substituted adenosines as probes of nucleobase protonation in functional RNAs. J. Am. Chem. Soc. 130:13639–13648 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.