Abstract
Many antibiotics inhibit the growth of sensitive bacteria by interfering with ribosome function. However, discovery of new protein synthesis inhibitors is curbed by the lack of facile techniques capable of readily identifying antibiotic target sites and modes of action. Furthermore, the frequent rediscovery of known antibiotic scaffolds, especially in natural product extracts, is time-consuming and expensive and diverts resources that could be used toward the isolation of novel lead molecules. In order to avoid these pitfalls and improve the process of dereplication of chemically complex extracts, we designed a two-pronged approach for the characterization of inhibitors of protein synthesis (ChIPS) that is suitable for the rapid identification of the site and mode of action on the bacterial ribosome. First, we engineered antibiotic-hypersensitive Escherichia coli strains that contain only one rRNA operon. These strains are used for the rapid isolation of resistance mutants in which rRNA mutations identify the site of the antibiotic action. Second, we show that patterns of drug-induced ribosome stalling on mRNA, monitored by primer extension, can be used to elucidate the mode of antibiotic action. These analyses can be performed within a few days and provide a rapid and efficient approach for identifying the site and mode of action of translation inhibitors targeting the bacterial ribosome. Both techniques were validated using a bacterial strain whose culture extract, composed of unknown metabolites, exhibited protein synthesis inhibitory activity; we were able to rapidly detect the presence of the antibiotic chloramphenicol.
INTRODUCTION
Pathogenic bacteria readily develop resistance to antibiotics. This generates a perpetual need for new antimicrobial agents to combat infections caused by resistant pathogens (1). Although the majority of clinical antibiotics are either natural products or their semisynthetic derivatives (2), the discovery of novel scaffolds has faded over the years (3) due in part to the continuous reisolation of known compounds. As a result, many antibiotic discovery programs have been abandoned, all while the spread of drug-resistant bacteria forges ahead of the development of lead compounds capable of treating such infections. In order to close this gap, new technologies are being developed that allow researchers to access natural and synthetic products that have been previously overlooked by traditional screening programs. In particular, developing new antibiotic assays has been one strategy to quickly screen for small molecules that act through novel mechanisms of action (MOA) (4–6).
A significant number of clinically used drugs inhibit the growth of pathogenic bacteria by binding to the ribosome and interfering with protein synthesis (7). The ribosome is a 2.5-MDa multimolecular ribonucleoprotein complex composed of two subunits, small and large. Functional complexity of the ribosome offers many opportunities to inhibit its activity. Antibiotic binding at specific functional centers interferes with various facets of protein synthesis including but not limited to tRNA binding, mRNA translocation, peptide bond formation, and egress of the nascent peptide. This complexity makes it difficult to pinpoint the precise site and mode of action of a new inhibitor. The analysis usually involves an extended series of sophisticated assays, requiring individually purified components of translation machinery, in which inhibition of specific ribosomal activities is tested or the site of the drug action is painstakingly characterized. Thus, the availability of more general biochemical assays capable of rapidly pinpointing the MOA of novel inhibitors could significantly streamline the drug lead discovery process.
Genetic approaches that provide important clues about protein synthesis inhibitors often supplement biochemical MOA investigation. Since most ribosome-targeting inhibitors interact directly with rRNA (reviewed in reference 8), isolation of resistance mutants possessing nucleotide changes in rRNA is the most straightforward way for understanding where antibiotics bind to the ribosome, as well as the manner in which they interfere with translation. However, due to the redundancy of rRNA genes in bacterial genomes, it is difficult to isolate a mutant that developed resistance by acquiring a mutation in rRNA. This is greatly beneficial from a clinical perspective because resistance is rarely associated with rRNA mutations but severely hampers experimental identification of the sites of drug action. Selection of resistant mutants with alterations in rRNA has been traditionally carried out using specialized model organisms which either naturally possess a single rRNA allele (e.g., Halobacterium halobium [9, 10]) or have been engineered to carry a single copy of rRNA genes in their chromosomes (e.g., Thermus thermophilus [11, 12] or Mycobacterium smegmatis [13]). Although these experimental organisms were useful in characterizing the sites of action of many antibiotics, most of these models have drawbacks. The archaeon H. halobium is slow growing and is insensitive to many antibacterials due to significant evolutionary divergence of bacterial and archaeal ribosomes. The engineered strain of T. thermophilus with a single rrn allele (11, 12) requires an inconveniently high growth temperature (ca. 65°C) at which stability of many antibiotics may be compromised. The single rRNA operon variant of Mycobacterium smegmatis (13), although instrumental in elucidating modes of action of important antibiotics (14, 15), grows fairly slowly and possesses peculiar lipids in the outer cell wall layer that impair the uptake of some drugs (16). Other experimental systems, which use plasmid-borne rRNA genes, do not allow for easy isolation of a homogenous population of the resistant ribosomes for subsequent biochemical characterization (17, 18). Thus, the availability of an efficient and biochemically tractable model for isolation of rRNA resistance mutations would significantly speed up the investigation of new antibiotic leads.
Here, we describe a two-pronged approach that we called ChIPS (Characterization of Inhibitors of Protein Synthesis) for the rapid identification of the site of binding and MOA of ribosome-targeting antibiotics. The combination of two assays that comprise ChIPS allows the rapid characterization of unknown protein synthesis inhibitors and prioritizing hits identified via high-throughput screening of natural product extracts or chemical libraries. First, we engineered two antibiotic-hypersensitive Escherichia coli strains that possess a single rrn allele and can be used for rapid isolation of resistance mutations and identification of the ribosomal site of antibiotic action. Then, we show that in a cell-free translation system, antibiotic-induced ribosome stalling on mRNA exhibits inhibitor-specific, idiosyncratic patterns. These patterns can be used as fingerprints for the rapid identification of the MOA of known and novel protein synthesis inhibitors.
MATERIALS AND METHODS
Construction of the SQ110, SQ110DTC, and SQ110LPTD strains.
E. coli strain SQ110 [Δ(rrsH-aspU)794(::FRT) Δ(rrfG-rrsG)791(::FRT) Δ(rrfF-rrsD)793(::FRT) Δ(rrsC-trpT)795(::FRT) Δ(rrsA-rrfA)792(::FRT) Δ(rrsB-rrfB)790(::FRT) rph-1 λ−; ptRNA67] was prepared from the K-12 strain MG1655 by individually inactivating six rrn alleles in different strains by allelic replacement with the kanamycin resistance marker (19) and combining them together in one strain by P1 phage transduction and removing the markers using FRT recombinase. The SQ110 strain represents an intermediate product in the preparation of the Δ7′ strain SQ171 described previously (20). The experimental details of engineering the SQ110 strain, which will be published elsewhere, are currently available at the Coli Genetic Stock Center (CGSC) site (http://cgsc.biology.yale.edu/CGSCDocs/rrn7v2.pdf).
The ΔtolC mutation was incorporated into the SQ110 strain via P1 phage transduction of the tolC::kan allele from the JW3003 strain from the Keio collection (21). Replacement of the tolC gene with the kan marker in the SQ110DTC strain was confirmed by PCR using the primers TCTATCGCCTTCTTGACGAGT and CTGGATTGCTGGGCCTGCGC.
For preparing the SQ110LPTD strain, first a BW25113 mutant strain containing a chloramphenicol resistance marker (cat) next to the truncated lptD gene was prepared. For that, a hybrid DNA product was generated by overlapping PCR that contained a mutant lptD gene with the 69 bases deletion and a cat gene inserted 3′ from lptD. The lptD gene with the deletion of 23 codons (Asp330-Asp352) was PCR amplified from the imp strain (22, 23) using the primers GAGTTCTACCTGCCATATTACTGG and GGTAAATCAACAAATCACAAAGTGTTTTG. The cat gene was PCR amplified from the pKD3 plasmid (19) using the primers CAAAACACTTTGTGATTTGTTGATTTACCACATATGAATATCCTCCTTA and TCCAGTTCTTCATACTTTTTCCATTTCAATTAACCGCACTGCGGATTACGGTGTAGGCTGGAGCTGCTTCG. The overlapping PCR was carried out using the primers GAGTTCTACCTGCCATATTACTGG and TCCAGTTCTTCATACTTTTTCCATTTC. The recombinant mutant lptD/cat product was used to replace the wild-type lptD allele in the BW25113 strain by recombineering (19). The mutant allele was then transferred from BW25113 cells to the SQ110 strain by P1 phage transduction. The presence of the 69-bp deletion and the lack of six rrn alleles in the resulting SQ110LPTD strain were verified by PCR.
Selection of resistant mutants.
The SQ110DTC strain was grown overnight at 37°C in Luria-Bertani (LB) medium containing 50 μg of spectinomycin/ml and 50 μg of kanamycin/ml. The cells were then diluted to an optical density at 600 nm (OD600) of ca. 0.05 and grown at 37°C in the presence of spectinomycin and kanamycin until they reached mid-log phase (OD600 ∼ 0.4 to 0.7). At an A600 of 1.25, ca. 109 cells of the cell culture were plated onto the LB agar plate supplemented with a 20-μg/ml fraction G006-24 prepared from the Micromonospora sp. extract (see Fig. S1 in the supplemental material). Plates were incubated overnight at 37°C.
Sequencing of the 16S and 23S genes in the selected mutants.
16S and 23S rRNA genes were amplified either directly from the resistant colonies or using genomic DNA prepared from liquid cultures using a GenElute bacterial genomic DNA kit (Sigma). The 16S rRNA gene (rrsE) was amplified using the primers GTCTCAAGAGTGAACACGTAATTC and CGCAAGACGCCTTGCTTTTCA. The 23S rRNA gene (rrlE) was PCR amplified in three fragments using the primer pairs CAAATTTTCGCAACACGATGATG and CGCCTTAGGGGTCGACTC, GAGGGAAACAACCCAGACCG and GGGTGGTATTTCAAGGTCGG, and CCCGAGACTCAGTGAAATTGAACTC and GCGTTCTGATTTAATCTGTATCAG. The PCR products were purified and sequenced at the UIC DNA sequencing facility.
Toe-printing analysis.
The in vitro translation in the PURExpress cell-free transcription-translation system, and toe-printing was carried out essentially as described previously (24, 25). The DNA templates contained the T7 promoter and optimized ribosome-binding sites that were introduced on the forward PCR primers.
The osmC template was PCR amplified from E. coli genomic DNA using the primers ATTAATACGACTCACTATAGGGATATAAGGAGGAAAACATATGACAATCCATAAGAAAGG and TTACGATTTCAACTGGTAATCC. The ermBL and RST1 templates were prepared by overlapping PCR as previously described (24, 26). The primer AAAACGCGTGTTAAATCCAT was used for toe-printing analysis with the osmC template, and the primer NV1 GGTTATAATGAATTTTGCTTATTAAC was used for toe-printing with the ermBL and RST1 templates (24, 25).
Isolation and characterization of the antibiotic-producing marine actinomycete strain.
Actinomycete strain G006 was isolated from sediment collected using PONAR from Lan Ha Bay, Cát B̂, Vietnam (20°43′41.00″N, 107°3′39.00″E; 5.5 m) on 8 July 2011. The strain was identified using 16S rRNA gene sequence analysis. It displayed 98.7% identity by 16S to Micromonospora matsumotoense reference strain S000388244 (RDP ID). The partial 16S rRNA gene sequence (1,476 bp) has been deposited in GenBank under accession number KC121341.1.
Preparation of natural product extract, fractionation and isolation of chloramphenicol.
Strain G006 was cultured in high-nutrient medium (30 g of instant ocean, 10 g of starch, 4 g of yeast, 2 g of peptone, 1 g of calcium carbonate, 40 mg of iron sulfate, and 100 mg of potassium bromate) for 7 days at 25°C while shaking at 200 rpm. Amberlie XAD-16 resin (20 g/liter) was added to each flask to absorb the extracellular metabolites. The culture medium was shaken with resin for 6 h and filtered using cheesecloth to collect the resin. After washing the resin with deionized water, the resin and cell mass were extracted with acetone overnight. The acetone extract was dried under vacuum, and partitioned between water and ethyl acetate. The compounds in the organic phase were fractionated using silica gel column chromatography into four fractions. Fraction 2, eluted with 80:20 ethyl acetate-hexane, exhibited protein synthesis inhibitory activity and was selected for further study. Fraction 2 was then further fractionated using reversed-phase C18 flash column chromatography with a step gradient of increasing aqueous methanol from 10 to 100%. The 40% aqueous methanol fraction (fraction 24) inhibited the in vitro translation and growth of E. coli strain SQ110DTC. This fraction was then further separated by using semipreparative phenyl-hexyl high-pressure liquid chromatography at an isocratic flow of 20% aqueous acetonitrile. The fraction eluting at tR = 71.5 min (fraction 249) exhibited the highest inhibition of in vitro translation. Chloramphenicol was identified in this fraction using high-resolution liquid chromatography-mass spectrometry (LC-MS). Fraction 249 was separated over C18 using an isocratic flow of 50% aqueous acetonitrile, tR = 19.0 min (fraction 2492) yielded 0.1 mg of chloramphenicol. High-resolution electrospray ionization-mass spectra were obtained using a Shimadzu IT-TOF spectrometer at the University of Illinois at Chicago Mass Spectrometry, Metabolomics, and Proteomics Facility. A selection of fractions was also analyzed using a Shimadzu-LTII evaporative light-scattering detector with the following parameters: N2 pressure, 52 lb/in2; temperature, 45°C; gain, 6; and flow rate, 0.5 ml min−1.
RESULTS
Engineered E. coli strains for the rapid identification of antibiotic target sites.
Compared to other model bacterial systems, E. coli provides a tremendous advantage since its biochemistry, molecular biology, and genetics are well understood. Indeed, the majority of key findings in the mechanisms of translation and the mode of its inhibition have been obtained using E. coli ribosomes. However, E. coli presents two serious problems as an experimental model for studying ribosome-targeting antibiotics. This Gram-negative bacterium carries seven copies of rRNA operon in the chromosome and in addition, it is naturally resistant to a number of inhibitors due to the relative impermeability of its outer membrane, in addition to the presence of multidrug efflux pumps (27, 28). In order to overcome these problems, we engineered two E. coli strains (SQ110DTC and SQ110LPTD) that are particularly suitable for the selection of mutants resistant to ribosome-targeting inhibitors because they have only one chromosomal rrn allele and are hypersensitive to many antibiotics.
The initial SQ110 strain was prepared by deleting six out of seven rrn alleles from the chromosome of the E. coli strain MG1655, leaving intact only one rRNA operon (rrnE). The general strategy used for the preparation of the SQ110 strain (20) was similar to the one used previously for generation of the TA series of the rrn deletion E. coli strains (29, 30), but an alternative gene replacement strategy based on the recombineering approach has been used (19) (see Materials and Methods).
Although the doubling time of the SQ110 strain was reduced compared to the parental MG1655 strain (41 ± 5 min versus 26 ± 1 min, respectively; Table 1), it nevertheless showed robust growth under laboratory conditions. Similar to the parental MG1655 strain, the SQ110 strain was intrinsically resistant to many protein synthesis inhibitors (Table 2), and two independent strategies have been used to boost its sensitivity to antibiotics. First, the tolC gene, which encodes an outer membrane component of several multidrug transporters (31), was deleted by transducing the tolC::kan allele from the JW3003 clone from the Keio collection (21). The resulting SQ110DTC strain showed increased sensitivity to a wide range of antibiotics (Table 2). Independently, we mutated the lptD gene in the SQ110 strain by deleting its codons 330 to 352. LptD is an essential protein functioning in the final stages of assembly of lipopolysaccharides into the outer membrane (32); the 23-amino-acid deletion interferes with its functions rendering the outer membrane more permeable to antibiotics (23). In order to introduce a mutant lptD allele in the SQ110 strain, the 23 codon deletion mutation in lptD, which is associated with the downstream chloramphenicol resistance gene cat, was first engineered by recombineering in the E. coli strain BW25113 and then transduced into the SQ110 strain (see Materials and Methods). The deletion in the lptD gene significantly increased the sensitivity of the resulting SQ110LPTD strain to a number of inhibitors, including large molecular weight compounds such as the 1,665-Da thiostrepton (Table 2). Introduction of the tolC deletion and lptD mutations in the SQ110 strain had a modest negative effect on cell growth, increasing doubling time to 47 ± 2 min for SQ110DTC and 81 ± 4 min for SQ110LPTD. The growth rate was sufficient for both strains to form large colonies on the LB agar plates after overnight (SQ110DTC) or 24-h (SQ110LPTD) incubation periods. We also succeeded in combining the tolC deletion and the lptD mutation in one strain, but the resulting cells showed an unstable phenotype and were not used in further experiments.
Table 1.
Growth rates of the engineered E. coli strains
| Strain | Mean doubling timea (min) ± SD |
|---|---|
| MG1655 | 26 ± 1 |
| SQ110 | 41 ± 5 |
| SQ110DTC | 47 ± 2 |
| SQ110LPTD | 81 ± 4 |
In liquid cultures.
Table 2.
MICs of antibiotics for the engineered E. coli strains
| Antibiotic | MIC (μg/ml) |
||
|---|---|---|---|
| SQ110 | SQ110DTC | SQ110LPTD | |
| Antibiotics acting upon the large ribosomal subunit | |||
| Clindamycin | > 256 | 8 | 4 |
| Linezolid | > 256 | 4 | 32 |
| Erythromycin | 256 | 2 | 0.25 |
| Telithromycin | 64 | 2 | ≤0.25 |
| Tylosin | >256 | 16 | 1 |
| Evernimicin | >256 | >256 | ≤0.25 |
| Thiostrepton | >128 | >128 | 0.25 |
| Pristinamycin IB | >256 | 64 | 4 |
| Antibiotics acting upon the small ribosomal subunit | |||
| Tetracycline | 2 | 0.5 | 0.5 |
| Pactamycin | 8 | 1 | ≤0.06 |
| Kasugamycin | 512 | 128 | 64 |
| Gentamicin | 1 | 1 | 0.25 |
The engineered SQ110DTC and SQ110LPTD strains, which have a single rrn allele in the chromosome and exhibit high sensitivity to antibiotics, have been successfully tested for the selection of resistant mutants using a number of commercially available drugs. By simply plating ca 109 cells on an LB agar plate containing antibiotic at concentrations exceeding an MIC of 2- to 25-fold, we routinely obtained several resistant colonies after an overnight incubation at 37°C. The presence and location of the resistance mutations could be easily assessed by PCR amplification of 16S and 23S rRNA genes or their fragments directly from the resistant colonies. Using this strategy, we were able to isolate many known as well as some novel rRNA mutations conferring resistance to such antibiotics as kanamycin, gentamicin, neomycin, clindamycin, erythromycin, pactamycin, and kasugamycin (Table 3). With the proper DNA sequencing support, rRNA mutations conferring resistance to the new protein synthesis inhibitors can be isolated and identified in less than 4 days.
Table 3.
Resistant mutants selected in the presence of various protein synthesis inhibitors
| Strain and antibiotic | Drug concn (μg/ml) used during selection | Mutanta | Reference |
|---|---|---|---|
| SQ110DTC | |||
| Clindamycin | 50 | 23S rRNA:A2058G | 33 |
| 23S rRNA:A2059G | 34 | ||
| Gentamicin | 32 | 16S rRNA:A1408G | 35 |
| Pactamycin | 5–10 | 16S rRNA:G693A | |
| 16S rRNA:A695G | |||
| 16S rRNA:C795U | 36 | ||
| Kasugamycin | 256 | 16S rRNA:A794G | 37 |
| SQ110LPTD | |||
| Gentamicin | 16 | 16S rRNA:A1408G | 35 |
| Kanamycin | 16 | 16S rRNA:G1491U | 38 |
| 16S rRNA:G1491C | 38 | ||
| Erythromycin | 50 | 23S rRNA:A2058G | 39 |
| 23S rRNA:A2059G | 40 |
Most of the mutations shown in the table have been previously isolated and characterized either in E. coli or in other organisms (see the references column) and are listed in the Ribosomal Mutation Database (41).
Toe-printing pattern as a tool for identifying the mode of action of ribosome-targeting antibiotics.
Because ribosome-targeting antibiotics inhibit specific functions of the ribosome, their action usually results in halted progression of the ribosome along mRNA. The site of translation arrest, however, should critically depend on the antibiotic MOA. The inhibitors of translation initiation are expected to either arrest the ribosome at the start codon of the gene or prevent binding of the ribosomes to mRNA (7). The inhibitors of peptide bond formation, tRNA binding, or translocation (if present at high concentrations at the onset of the gene translation) are anticipated to arrest the translation at the initiator codon of the gene, whereas at intermediate concentrations or when added at the elongation phase of protein synthesis, such drugs should cause ribosome pausing at random codons. On the other hand, antibiotics that bind to the exit tunnel and interfere with the egress of the nascent peptide should inhibit translation after a few amino acids have been polymerized and thus are predicted to arrest the ribosome at a specific mRNA codon close to the start site (25, 42). With these considerations in mind, we aimed to explore whether the pattern of translation arrest could be used as an approach for identifying the mode of antibiotic action.
The sites of ribosome stalling on mRNA could be determined by primer extension inhibition analysis (commonly known as “toe-printing”) where the product of reverse transcriptase-catalyzed extension of the DNA primer defines the position of the arrested ribosome on mRNA (43) (Fig. 1A). Primer extension on any RNA template usually generates some spurious bands (resulting from the template secondary structure or nuclease cuts), which hypothetically can complicate the analysis. However, the inclusion of the “no-antibiotic” control sample enables the easy identification of toe-printing bands resulting specifically from antibiotic action upon the translating ribosome.
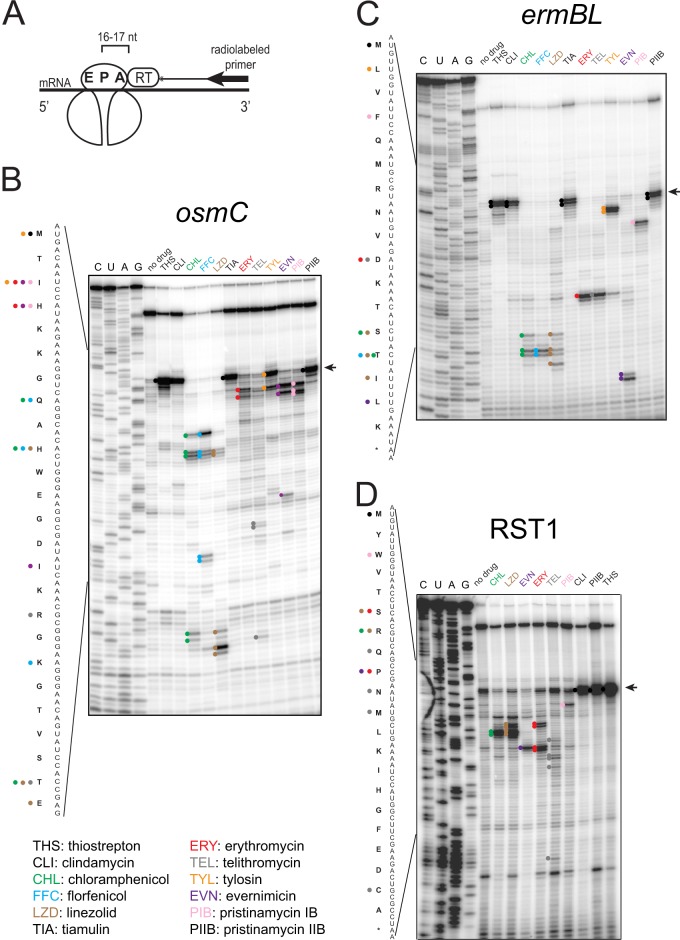
Fig 1.
Toe-printing analysis of protein synthesis inhibitors acting upon the large ribosomal subunit. (A) Principle of toe-printing analysis. The antibiotic arrests the ribosome at a specific mRNA codon. A radioactive primer, annealed to the 3′ end of the transcript, is extended with reverse transcriptase. The enzyme stops polymerization of cDNA when it encounters the ribosome. The 3′ end of the cDNA product is separated by 16 to 17 nucleotides from the first nucleotide of the mRNA codon in the ribosomal P site. (B to D) Toe-printing patterns generated by different antibiotics with osmC (B), ermBL (C), or RST1 (D) templates. The toe-print bands are marked with colored dots on the gel, and the codons occupied by the arrested ribosomes are indicated in the gene sequence by the dots with the corresponding colors. The panel below the gels provides the legend with antibiotic names and abbreviations. All antibiotics but linezolid were tested at 50 μM. Because linezolid exhibited poor activity in cell-free translation assay (IC50 ∼100 μM), its concentration in panels B and C was raised to 800 μM. The bands corresponding to the ribosome occupying the initiator codons are indicated by the arrows.
The toe-printing experiments were performed using the commercially available cell-free transcription-translation system composed of purified components (PURExpress; New England BioLabs) (44). We tested the approach with several different mRNA templates. One template is the osmC gene specifying an osmotically inducible envelope protein whose translation is known to be highly sensitive to inhibition by macrolide antibiotics (Fig. 1B) (45). Another template is a 17-codon long open reading frame (ORF) ermBL encoding leader peptide that controls expression of macrolide-inducible resistance gene ermB (26) (Fig. 1C). In addition, we also used a synthetic 21-codon synthetic gene RST1, which besides the initiator codon, has one codon for each amino acid (24) (Fig. 1D).
In spite of our expectation to observe a fairly simple and uniform distribution of translation arrest sites, where many different antibiotics would cause the ribosomal pausing at the same codons, many tested drugs exhibited highly specific individual patterns (Fig. 1 and 2). Strikingly, several antibiotics with various modes of action arrested the ribosome at a number of defined codons. This trend was especially pronounced with the antibiotics acting upon the large ribosomal subunit. As anticipated, the tunnel-binding macrolides (ERY and TEL) and streptogramin B type inhibitor pristinamycin IB (PIB) arrested the ribosome close to the 5′ end of the coding sequence. However, the sites of arrest differed between the three tested tunnel-binding compounds (Fig. 1A to C). Unexpectedly, several classic peptidyl transferase inhibitors (chloramphenicol, florfenicol, and linezolid) did not arrest the ribosome at the initiator codons of the tested genes but instead at one of the downstream codons in the vicinity of the 5′ end of the ORF. A 16-member ring macrolide tylosin, which is also known to inhibit peptide bond formation, caused translation arrest at the first codon of the osmC gene, but on the ermBL ORF the drug stalled the ribosome only at the second codon (Fig. 1, lane TYL). Evernimicin, which was proposed to inhibit IF2-dependent initiation complex formation (46), instead caused translation arrest at various, but specific codons within the tested genes. The other analyzed large subunit-binding antibiotics, including peptide bond formation inhibitors clindamycin, streptogramin A-type pristinamycin IIB and tiamulin, as well as the inhibitor of the ribosome-associated GTPase center, thiostrepton, arrested translation at the start codon (Fig. 1B to D).
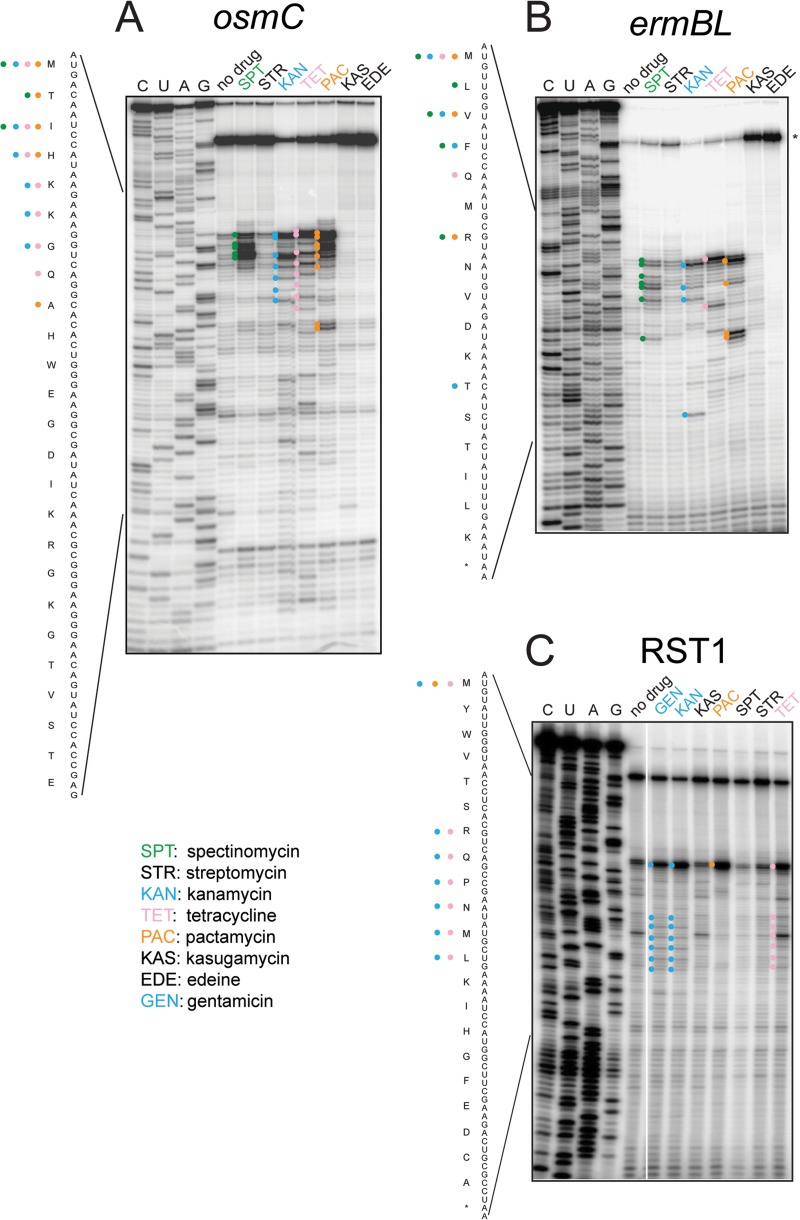
Fig 2.
Toe-printing analysis of protein synthesis inhibitors acting upon the small ribosomal subunit. (A) osmC template; (B) ermBL; (C) RST1. Antibiotics and their abbreviations are listed at the bottom of the figure. In panels A and B, antibiotics were tested at 100 μM (except for kasugamycin, 1 mM). In panel C, antibiotics were tested at 50 μM (except for kasugamycin, 5 mM).
Among the small subunit-targeting antibiotics (Fig. 2), spectinomycin and pactamycin (commonly viewed as inhibitors of translocation [7]) caused translation arrest at specific mRNA codons. The aminoglycoside kanamycin, as well as tetracycline, produced periodic patterns of translation arrest at several consecutive codons. Characteristically, the bands in the kanamycin sample were shifted by one nucleotide relative to the tetracycline sample, an effect known to reflect the binding of aminoacyl-tRNA in the ribosomal A-site (47). Indeed, the kanamycin-stalled ribosomes are expected to carry aminoacyl-tRNA in the A-site, whereas tetracycline-bound ribosomes should have an empty A-site because the antibiotic prevents aminoacyl-tRNA binding (7). Edeine and kasugamycin, which are classic translation initiation inhibitors known to interfere with mRNA placement in the small ribosomal subunit, appeared to prevent binding of ribosomes to mRNA so that a full-length transcript was reversed transcribed during primer extension (marked with an asterisk on top of the gels on Fig. 2B).
The toe-printing patterns observed with different mRNA templates were highly reproducible and could be reliably obtained with the drugs present at concentrations greater than or equal to the 50% inhibitory concentration (IC50) of the inhibitor in the conventional in vitro translation assay. However, the higher concentrations of antibiotics tend to generate stronger toe-print bands near the 5′ end of mRNA and thus are beneficial for the analysis. Although the molecular bases for the specific toe-printing patterns observed with a variety of antibiotics are not entirely clear, the fact that different antibiotics arrest the ribosome at different codons allows for the opportunity to use this technique as a rapid tool for assessing the general MOA of select protein synthesis inhibitors. Comparison of the toe-printing pattern generated by an unknown inhibitor present in natural product extracts or in other small molecule libraries to those afforded by a panel of known drugs shows whether the unknown antibiotic inhibits the ribosome in a way similar to one of the known compounds or, alternatively, represents a lead with a novel MOA.
Use of ChIPS for testing a natural products extract exhibiting protein synthesis inhibitory activity.
Using our dual-pronged ChIPS approach described above, which includes the use of the engineered E. coli strain for rapid selection of resistant mutations that define the site of antibiotic binding and toe-printing as a method to define the MOA, we tested extracts of the marine-derived actinomycete bacterium Micromonospora sp. (strain G006, isolated from Halong Bay, Vietnam). In preliminary experiments, an extract of G006 exhibited antibiotic activity against a range of Gram-positive and Gram-negative bacterial strains. Furthermore, macromolecular synthesis inhibition, as well as preliminary in vitro translation testing suggested that the active compound targets protein synthesis (data not shown). However, the nature of the inhibitor, its target site, and its MOA were all unknown.
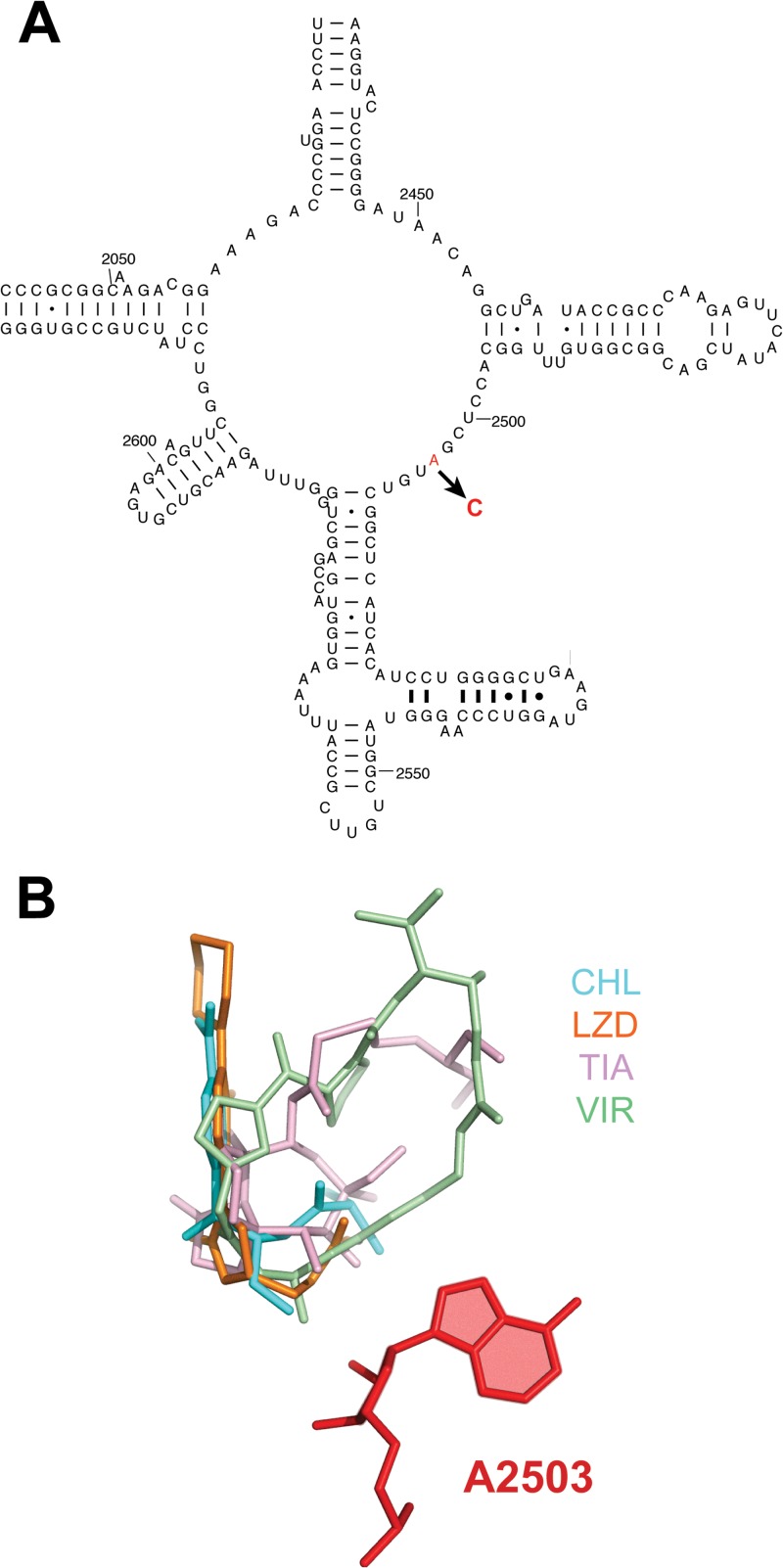
In order to define the site of action of the antibiotic, we used one of the subfractions of the extract (G006-24; see Fig. S1 in the supplemental material), which exhibited antibacterial activity, to select resistant mutants. This fraction inhibited growth of the SQ110DTC strain at 10 μg/ml. When ca. 109 (A600 = 1.25) SQ110DTC cells were plated onto LB-agar supplemented with 20 μg of G006-24/ml, one resistant colony appeared after <24 h of incubation. The 16S and 23S rRNA genes from the resistant mutant were PCR amplified and sequenced, and a single mutation, A2503C, was identified in the 23S rRNA (Fig. 3A). A2503 is located in the peptidyl transferase center (PTC), thereby revealing that the active compound in the G006 extract targets the PTC. Indeed, mutations of A2503 or its C-8 methylation are known to render cells resistant to various PTC-targeting inhibitors (48–55) (Fig. 3B).
Fig 3.
Mutation, selected in the SQ110DTC strain, conferring resistance to the inhibitor present in extract of strain G006. (A) Position of the mutation within the central loop of domain V of 23S rRNA that participates in formation of the active site of the PTC. (B) Placement of A2503 (red color) in the PTC relative to the binding sites of antibiotics that inhibit peptide bond formation. The aligned structures of the ribosome antibiotic complexes were taken from the DARC server (http://darcsite.genzentrum.lmu.de/darc/) (56). The following antibiotics are shown: virginiamycin M1 (streptogramin A) (VIR), PDB 1YIT; linezolid (LZD), PDB 3CPW; tiamulin (TIA), PDB 3G4S; and chloramphenicol (CHL), PDB 3OFC.
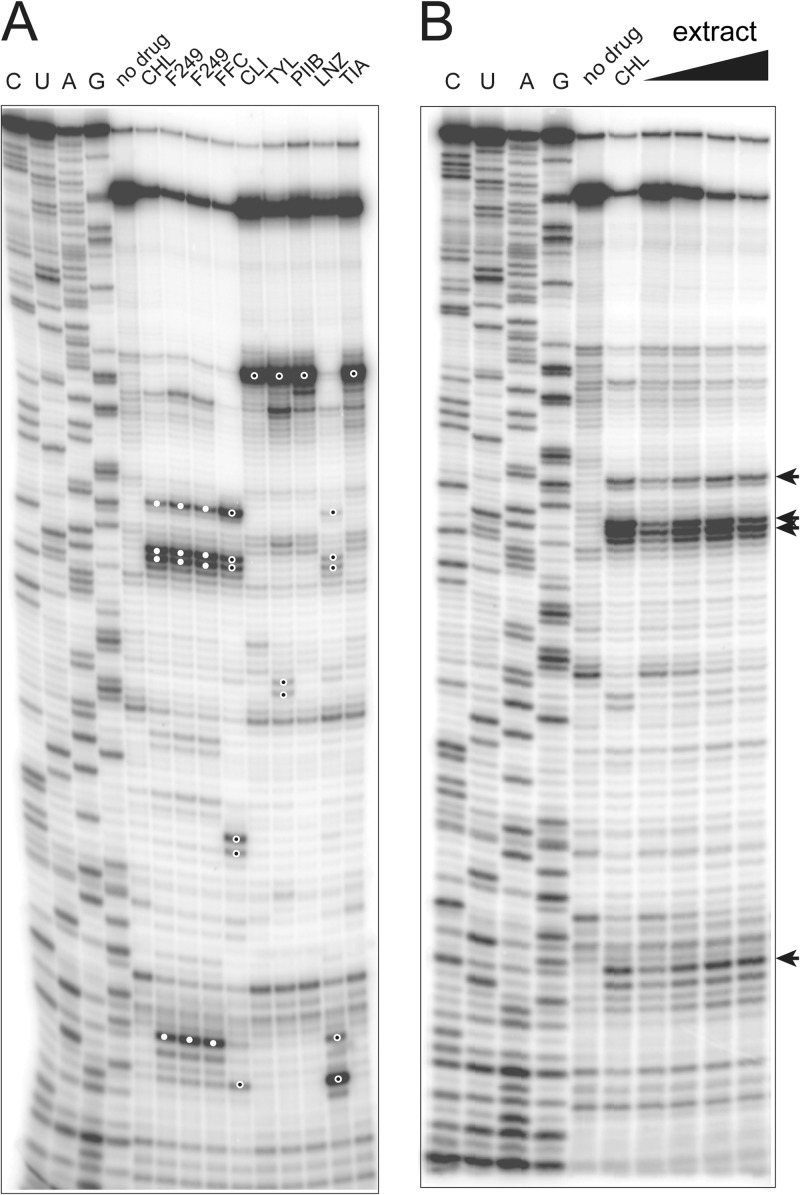
In order to narrow down the possible mode of action of the G006 inhibitor, we then used the toe-printing assay (Fig. 4A). This experiment, which was completed in less than a day, revealed that the main stalling site observed with the OsmC template in the presence of the G006 fraction exactly matched that of chloramphenicol (white-filled dots in Fig. 4A) and differed from the toe-prints generated by other tested PTC inhibitors (florfenicol, clindamycin, tylosin, streptogramin A, linezolid, or tiamulin) (black-filled dots in Fig. 4A). Thus, the results of the ChIPS approach rapidly identified the active substance in the G006 extract as a compound with properties similar to chloramphenicol. In order to verify the accuracy of our prediction, additional spectroscopic characterization of the extract was carried out and we confirmed the presence of chloramphenicol via LC-MS (Fig. 5).
Fig 4.
Toe-printing analysis of the MOA of the translation inhibitor present in the G006 extract. (A) Comparative toe-printing was performed using the osmC template. For the better reproducibility, two independent samples with fraction G006-249 were independently processed. Characteristic toe-print bands obtained with the extract fractions and with chloramphenicol are indicated by filled white circles on the gel. The toe-printing bands in the other control samples, which do not match those observed with the extract, are marked with open circles. (B) Toe-printing in the presence of the crude material present in fraction 2 of the extract (0.25 to 2 mg/ml final concentration) (see Fig. S1 in the supplemental material).
Fig 5.
Mass spectrometry (IT-TOF) analysis of the compounds present in the fraction G006-249. The chloramphenicol control (A) and fraction G006-249 (B) were analyzed under positive ionization modes. The molecular ion (m/z 323.02) corresponding to intact chloramphenicol is indicated by the arrows.
In the process of isolating the active compound, a number of intermediate fractions were generated, each of which contained a considerable metabolic complexity (ca. 5 to 15 compounds per fraction). We used this opportunity to assess whether more complex fractions (see Fig. S2 and S3 in the supplemental material) could be used directly in the toe-printing experiments. As shown in Fig. 4B, even the most complex fraction (F2) produced a distinct and characteristic “chloramphenicol-like” toe-print. These results demonstrate that complex extracts, if used at concentrations sufficient to inhibit protein synthesis, may be directly tested in toe-printing experiments. The antibiotic MOA of the active compound could be identified without further purification and could greatly streamline the potential discovery of novel antibiotics and/or serve to triage uninteresting hits.
DISCUSSION
We describe here a two-pronged research approach, ChIPS, which could be used as a convenient tool during the discovery of new protein synthesis inhibitors. ChIPS affords rapid information about the site and MOA of an unidentified ribosome inhibitor in a natural product extract or a chemical library and, as a consequence, provides essential information for prioritizing antibiotic leads.
The SQ110DTC and SQ110LPTD strains allow for the isolation and characterization of mutants resistant to the ribosome-targeting antibiotics in as little as 4 days. Because the engineered strains are highly sensitive to many antibiotics, the mutants can be selected using not only the purified compounds, but even crude extracts or other complex mixtures of molecules. Since the strains possess a single rrn allele, characterization of the mutants is extremely straightforward: the mutant rRNA genes can be PCR amplified directly from the resistant colony or a culture using few universal primer combinations without the need for allele-specific PCR. Our strains provide considerable advantage compared to the other “popular” single rRNA operon models (e.g., H. halobium, T. thermophilus, or M. smegmatis [10–13]). The engineered E. coli strains are sensitive to a broad range of antibacterials, do not require elevated temperature for proliferation, and exhibit a robust and fairly rapid growth under the laboratory conditions. Furthermore, if subsequent biochemical or structural characterization of the hits is required, the strains can be directly used as a source of both wild-type and mutant ribosomes for the in vitro experiments or crystallographic studies, whose results could be directly correlated with the effects observed in vivo. The SQ110DTC and SQ110LPTD strains possess different antibiotic sensitivity profiles (Table 2). SQ110DTC is 8-fold more sensitive to linezolid than SQ110LPTD, whereas it is highly resistant to the larger-molecular-weight inhibitors such as evernimicin or thiostrepton that readily inhibit the SQ110LPTD growth. The parallel use of both strains in selection of the resistance marker could be a preferred strategy if the chemical nature of the inhibitor is unknown. In the future, extended collections of resistant mutants could be prepared on the basis of the SQ110DTC and SQ110LPTD strains and used in screens of new extracts or compounds, thereby further accelerating the identification of compounds acting upon the known sites. However, even with the availability of a panel of resistant mutants, de novo selection of rRNA mutants will remain a useful universal strategy that will facilitate analysis of antibiotics binding to new sites in the ribosome.
Like any other approach, selection of resistant mutants using the SQ110DTC and SQ110LPTD strains has its limitations. These strains do not provide any additional advantage for the selection of mutants resistant to protein synthesis inhibitors that act upon translation factors or aminoacyl-tRNA synthetases. Similarly, if a new antibiotic interacts exclusively with a ribosomal protein rather than rRNA, our strains will not facilitate the isolation and mapping of resistance mutations. Nevertheless, given the prevalence of rRNA-binding antibiotics among protein synthesis inhibitors, our strains remain useful for the majority of new leads.
We discovered that many antibiotics arrest translation at specific mRNA codons and generate a specific pattern of cDNA bands in toe-printing experiments. This finding helped us to develop the toe-printing assay as a tool for rapidly assessing the mechanism of translation inhibition by the unknown compounds targeting the ribosome. This observation expands the previously proposed use of toe-printing for characterizing protein synthesis inhibitors acting upon aminoacyl-tRNA synthetases (24). Similar to the mutant selection, toe-printing can be carried out not only with purified compounds but also with crude extracts. In the current format, the use of radiolabeled toe-printing primers and slab gel electrophoresis limits the number of samples that could be analyzed in a single experiment to approximately 20. However, the throughput could be easily increased by using fluorescently labeled DNA primers and capillary electrophoresis of standard DNA sequencers (57).
We noted that in toe-printing experiments, different mRNA templates yield somewhat different stalling patterns. For example, tylosin arrested the ribosome at the first codon of osmC and with the use of this template could not be distinguished from several other PTC inhibitors, e.g., clindamycin or tiamulin (Fig. 1B). In contrast, when ermBL was used as a template, tylosin generated an idiosyncratic toe-print at the second codon (Fig. 1C) that differed from the clindamycin or tiamulin toe-prints. As a result, the use of two or more templates is recommended to differentiate a largest number of drugs. More extensive testing of a range of natural or synthetic templates with different antibiotics will help to identify the single most “discriminating” mRNA template that could be superior to the genes we have tested in our experiments.
The two components of the ChIPS approach are equally important because their combined use provides critical information that can be missed if only one of its elements is used. Thus, finding out that an unknown inhibitor acts upon the “old” site should not be sufficient for discarding it. For example, oxazolidinones (e.g., linezolid), which represent one of the newest clinically successful antibiotic scaffolds, act upon exactly the same PTC site that is targeted by one of the oldest antibiotics, chloramphenicol. Conversely, in certain cases the sole use of the toe-printing patterns cannot distinguish between compounds acting upon principally different ribosomal sites. Thus, arrest of the ribosome at the initiator codon would not discriminate between thiostrepton and clindamycin, which bind at different functional centers of the large ribosomal subunit (Fig. 1). Regardless, the combination of both techniques comprising ChIPS provides more reliable information regarding whether the unknown inhibitor is principally new and whether it is worth further exploration.
We illustrated the triaging utility of the ChIPS approach by rapidly demonstrating that a Micromonospora sp. (strain G006) produced chloramphenicol, which is responsible for the antibacterial activity of the culture extract. The resistant mutant, selected using the active G006-24 fraction, suggested the PTC as the antibiotic target. Subsequently, the toe-printing pattern, which was indistinguishable from that generated by chloramphenicol, helped to narrow the identity of our small molecule to a specific class among many PTC-targeting antibiotics. This example shows that with the use of our approach, the triaging process is significantly streamlined. On the other hand, if the resistance mutation was located in a new ribosomal site or the toe-printing pattern significantly differed from those generated by our panel of known antibiotics, these results would be a strong incentive to prioritize the producing strain for large-scale fermentation studies aimed at isolating and characterizing the putative new antibiotic. The identification of chloramphenicol in the G006 extract was facilitated by the fact that this was the only inhibitor of bacterial growth produced by this organism. In the event that the producing organism secreted a combination of inhibitors, the mixture of compounds would have to be simplified through one or more fractionation steps.
In summary, our ChIPS approach offers a fast and simple way to characterize unknown protein synthesis inhibitors and among other advantages helps to address a significant impediment to antibiotic discovery—the rediscovery of known compounds. The tools we developed can be used in a stand-alone mode or in combination with other techniques aimed at streamlining the small molecule triaging process (6). Finally, although the method is particularly valuable for natural product extracts, it is fully applicable to synthetic protein synthesis inhibitors identified in generalized translation inhibition screens for which the ribosomal target and mode of action are unknown.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karen J. Shaw (Trius Therapeutics, Inc) for providing the imp (lptD) mutant E. coli strain, Krishna Kannan (University of Illinois) for the suggestion of using the osmC template, Teresa Szal, James Marks, and Trupti Patel (all at the University of Illinois) for help with some experiments. We also thank K. J. Shaw and K. Kannan for helpful discussions. We thank the Ministry of Science and Technology of Vietnam and the Vietnam Academy of Science and Technology (grant VAST.TD.DAB.13-15) for their collaboration and work collecting marine sediment samples.
This study was supported by the grants R01 GM106386 and R03 DA035191 from the National Institutes of Health.
Footnotes
Published ahead of print 16 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01673-13.
REFERENCES
- 1. Levy SB, Marshall B. 2004. Antibacterial resistance worldwide: causes, challenges, and responses. Nat. Med. 10:S122–S129 [DOI] [PubMed] [Google Scholar]
- 2. Newman DJ, Cragg GM. 2012. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75:311–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li J, Vederas J. 2009. Drug discovery and natural products: end of an era or an endless frontier? Science 325:161–165 [DOI] [PubMed] [Google Scholar]
- 4. Peach KC, Bray WM, Winslow D, Linington PF, Linington RG. 2013. Mechanism of action-based classification of antibiotics using high-content bacterial image analysis. Mol. Biosyst. 9:1837–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schulze CJ, Bray WM, Woerhmann MH, Stuart J, Lokey RS, Linington RG. 2013. “Function-first” lead discovery: mode of action profiling of natural product libraries using image-based screening. Chem. Biol. 20:285–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wong WR, Oliver AG, Linington RG. 2012. Development of antibiotic activity profile screening for the classification and discovery of natural product antibiotics. Chem. Biol. 19:1483–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilson DN. 2009. The A-Z of bacterial translation inhibitors. Crit. Rev. Biochem. Mol. Biol. 44:393–433 [DOI] [PubMed] [Google Scholar]
- 8. Poehlsgaard J, Douthwaite S. 2005. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 3:870–881 [DOI] [PubMed] [Google Scholar]
- 9. Hummel H, Bock A. 1987. Thiostrepton resistance mutations in the gene for 23S rRNA of halobacteria. Biochimie 69:857–861 [DOI] [PubMed] [Google Scholar]
- 10. Hummel H, Bock A. 1987. 23S rRNA mutations in halobacteria conferring resistance to the anti-80S ribosome targeted antibiotic anisomycin. Nucleic Acids Res. 15:2431–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gregory ST, Dahlberg AE. 2009. Genetic and structural analysis of base substitutions in the central pseudoknot of Thermus thermophilus 16S rRNA. RNA 15:215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Monshupanee T, Gregory ST, Douthwaite S, Chungjatupornchai W, Dahlberg AE. 2008. Mutations in conserved helix 69 of 23S rRNA of Thermus thermophilus that affect capreomycin resistance but not posttranscriptional modifications. J. Bacteriol. 190:7754–7761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sander P, Prammananan T, Bottger EC. 1996. Introducing mutations into a chromosomal rRNA gene using a genetically modified eubacterial host with a single rRNA operon. Mol. Microbiol. 22:841–848 [DOI] [PubMed] [Google Scholar]
- 14. Sander P, Belova L, Kidan YG, Pfister P, Mankin AS, Bottger EC. 2002. Ribosomal and non-ribosomal resistance to oxazolidinones: species-specific idiosyncrasy of ribosomal alterations. Mol. Microbiol. 46:1295–1304 [DOI] [PubMed] [Google Scholar]
- 15. Springer B, Kidan YG, Prammananan T, Ellrott K, Bottger EC, Sander P. 2001. Mechanisms of streptomycin resistance: selection of mutations in the 16S rRNA gene conferring resistance. Antimicrob. Agents Chemother. 45:2877–2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lambert PA. 2002. Cellular impermeability and uptake of biocides and antibiotics in Gram-positive bacteria and mycobacteria. J. Appl. Microbiol. 92(Suppl):46S–54S [PubMed] [Google Scholar]
- 17. Cochella L, Green R. 2004. Isolation of antibiotic resistance mutations in the rRNA by using an in vitro selection system. Proc. Natl. Acad. Sci. U. S. A. 101:3786–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ettayebi M, Prasad SM, Morgan EA. 1985. Chloramphenicol-erythromycin resistance mutations in a 23S rRNA gene of Escherichia coli. J. Bacteriol. 162:551–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bollenbach T, Quan S, Chait R, Kishony R. 2009. Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell 139:707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Braun M, Silhavy TJ. 2002. Imp/OstA is required for cell envelope biogenesis in Escherichia coli. Mol. Microbiol. 45:1289–1302 [DOI] [PubMed] [Google Scholar]
- 23. Sampson BA, Misra R, Benson SA. 1989. Identification and characterization of a new gene of Escherichia coli K-12 involved in outer membrane permeability. Genetics 122:491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Orelle C, Szal T, Klepacki D, Shaw KJ, Vazquez-Laslop N, Mankin AS. 2013. Identifying the targets of aminoacyl-tRNA synthetase inhibitors by primer extension inhibition. Nucleic Acids Res. 41:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vazquez-Laslop N, Thum C, Mankin AS. 2008. Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell 30:190–202 [DOI] [PubMed] [Google Scholar]
- 26. Vazquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. 2010. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 29:3108–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nikaido H. 2009. Multidrug resistance in bacteria. Annu. Rev. Biochem. 78:119–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nikaido H, Pages JM. 2012. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 36:340–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Asai T, Condon C, Voulgaris J, Zaporojets D, Shen B, Al-Omar M, Squires C, Squires CL. 1999. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J. Bacteriol. 181:3803–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Asai T, Zaporojets D, Squires C, Squires CL. 1999. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl. Acad. Sci. U. S. A. 96:1971–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sulavik MC, Houseweart C, Cramer C, Jiwani N, Murgolo N, Greene J, DiDomenico B, Shaw KJ, Miller GH, Hare R, Shimer G. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45:1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu T, McCandlish AC, Gronenberg LS, Chng SS, Silhavy TJ, Kahne D. 2006. Identification of a protein complex that assembles lipopolysaccharide in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 103:11754–11759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Douthwaite S. 1992. Functional interactions within 23S rRNA involving the peptidyltransferase center. J. Bacteriol. 174:1333–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poehlsgaard J, Pfister P, Bottger EC, Douthwaite S. 2005. Molecular mechanisms by which rRNA mutations confer resistance to clindamycin. Antimicrob. Agents Chemother. 49:1553–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Recht MI, Douthwaite S, Puglisi JD. 1999. Basis for prokaryotic specificity of action of aminoglycoside antibiotics. EMBO J. 18:3133–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mankin AS. 1997. Pactamycin resistance mutations in functional sites of 16 S rRNA. J. Mol. Biol. 274:8–15 [DOI] [PubMed] [Google Scholar]
- 37. Vila-Sanjurjo A, Squires CL, Dahlberg AE. 1999. Isolation of kasugamycin-resistant mutants in the 16 S rRNA of Escherichia coli. J. Mol. Biol. 293:1–8 [DOI] [PubMed] [Google Scholar]
- 38. De Stasio EA, Moazed D, Noller HF, Dahlberg AE. 1989. Mutations in 16S rRNA disrupt antibiotic-RNA interactions. EMBO J. 8:1213–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vester B, Garrett RA. 1987. A plasmid-coded and site-directed mutation in Escherichia coli 23S RNA that confers resistance to erythromycin: implications for the mechanism of action of erythromycin. Biochimie 69:891–900 [DOI] [PubMed] [Google Scholar]
- 40. Prunier AL, Malbruny B, Tande D, Picard B, Leclercq R. 2002. Clinical isolates of Staphylococcus aureus with ribosomal mutations conferring resistance to macrolides. Antimicrob. Agents Chemother. 46:3054–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Triman KL. 2007. Mutational analysis of the ribosome. Adv. Genet. 58:89–119 [DOI] [PubMed] [Google Scholar]
- 42. Tenson T, Lovmar M, Ehrenberg M. 2003. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330:1005–1014 [DOI] [PubMed] [Google Scholar]
- 43. Hartz D, McPheeters DS, Traut R, Gold L. 1988. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 164:419–425 [DOI] [PubMed] [Google Scholar]
- 44. Shimizu Y, Inoue A, Tomari Y, Suzuki T, Yokogawa T, Nishikawa K, Ueda T. 2001. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19:751–755 [DOI] [PubMed] [Google Scholar]
- 45. Kannan K, Vazquez-Laslop N, Mankin AS. 2012. Selective protein synthesis by ribosomes with a drug-obstructed exit tunnel. Cell 151:508–520 [DOI] [PubMed] [Google Scholar]
- 46. Belova L, Tenson T, Xiong L, McNicholas PM, Mankin AS. 2001. A novel site of antibiotic action in the ribosome: interaction of evernimicin with the large ribosomal subunit. Proc. Natl. Acad. Sci. U. S. A. 98:3726–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jerinic O, Joseph S. 2000. Conformational changes in the ribosome induced by translational miscoding agents. J. Mol. Biol. 304:707–713 [DOI] [PubMed] [Google Scholar]
- 48. Dujon B. 1980. Sequence of the intron and flanking exons of the mitochondrial 21S rRNA gene of yeast strains having different alleles at the omega and rib-1 loci. Cell 20:185–197 [DOI] [PubMed] [Google Scholar]
- 49. Gregory ST, Carr JF, Rodriguez-Correa D, Dahlberg AE. 2005. Mutational analysis of 16S and 23S rRNA genes of Thermus thermophilus. J. Bacteriol. 187:4804–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li BB, Shen JZ, Cao XY, Wang Y, Dai L, Huang SY, Wu CM. 2010. Mutations in 23S rRNA gene associated with decreased susceptibility to tiamulin and valnemulin in Mycoplasma gallisepticum. FEMS Microbiol. Lett. 308:144–149 [DOI] [PubMed] [Google Scholar]
- 51. Li BB, Wu CM, Wang Y, Shen JZ. 2011. Single and dual mutations at positions 2058, 2503, and 2504 of 23S rRNA and their relationship to resistance to antibiotics that target the large ribosomal subunit. J. Antimicrob. Chemother. 66:1983–1986 [DOI] [PubMed] [Google Scholar]
- 52. Long KS, Munck C, Andersen TM, Schaub MA, Hobbie SN, Bottger EC, Vester B. 2010. Mutations in 23S rRNA at the peptidyl transferase center and their relationship to linezolid binding and cross-resistance. Antimicrob. Agents Chemother. 54:4705–4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vester B, Garrett RA. 1988. The importance of highly conserved nucleotides in the binding region of chloramphenicol at the peptidyl transfer centre of Escherichia coli 23S rRNA. EMBO J. 7:3577–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Long KS, Poehlsgaard J, Kehrenberg C, Schwarz S, Vester B. 2006. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob. Agents Chemother. 50:2500–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smith LK, Mankin AS. 2008. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 52:1703–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jarasch A, Dziuk P, Becker T, Armache JP, Hauser A, Wilson DN, Beckmann R. 2012. The DARC site: a database of aligned ribosomal complexes. Nucleic Acids Res. 40:D495–D500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gould PS, Bird H, Easton AJ. 2005. Translation toe-printing assays using fluorescently labeled primers and capillary electrophoresis. Biotechniques 38:397–400 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.