Abstract
We assessed Escherichia coli ST131 and its H30 and H30-Rx subclones for virulence genes, antimicrobial resistance, and extended-spectrum beta-lactamase (ESBL) type. Although both subclones were associated with ESBL production, H30-Rx isolates had higher resistance scores and were associated specifically with CTX-M-15. Three virulence genes (iha, sat, and iutA) were more prevalent among H30 than non-H30 ST131 isolates. Thus, the H30 and H30-Rx subclones are more antimicrobial resistant and have virulence profiles that are distinct from those of non-H30 ST131 isolates.
TEXT
The H30 and H30-Rx subclones of Escherichia coli sequence type 131 (ST131) have expanded more extensively than other ST131 variants, for as-yet-unexplained reasons. The H30 subclone, so named because it contains allele 30 of fimH (type 1 fimbrial adhesin gene), comprises almost all current fluoroquinolone-resistant ST131 isolates (1). Within the H30 subclone, the H30-Rx subset often carries blaCTX-M-15 and may constitute its main repository within ST131 (2). Here we determined the prevalence of ST131 and its H30 and H30-Rx subclones among isolates from a case-control study of infections caused by extended-spectrum-β-lactamase (ESBL)-producing E. coli strains (3) and compared these groups for virulence genotypes, antimicrobial resistance, and ESBL type.
A total of 267 (100 ESBL-positive and 167 ESBL-negative) E. coli isolates were collected prospectively between 2007 and 2010 for a case-control study, as described in detail elsewhere (3), with approval by the NorthShore and VA Medical Center institutional review boards. Fifteen health care-associated isolates, collected during the prospective study >2 days after hospital admission, were excluded from the study reported in reference 3 but were evaluated here. Established PCR-based methods were used to define E. coli phylogenetic group (A, B1, B2, and D) (4), ST131 and its H30 subclone (1, 2, 5), major CTX-M groups (6), the presence of blaCTX-M-15 (7), and extended virulence genotypes (8–10). The H30-Rx subclone was identified by PCR detection of a specific single-nucleotide polymorphism (SNP) (G723A) within the allantoin-encoding gene, ybbW (2). Primers APfor63 (5′-GGTTGCGGTCTGGGCA-3′) and APrev66 (5′-CAATATCCAGCACGTTCCAGGTG-3′), with a cycling routine of 95°C for 8 min, 31 cycles of 94°C for 20 s and 72°C for 40 s, and a final extension at 72°C (for 5′), yielded a 194-bp amplicon. The resistance score was the total number of agents (among 7) to which an isolate was resistant. The virulence score was as described previously (11). Fisher's exact test and the Mann-Whitney U test were used for comparisons involving dichotomous and continuous variables, respectively; a P value of <0.05 was considered statistically significant. Principal coordinate analysis (PCoA) was used to reduce the dimensionality of the molecular data set for simplified comparisons (12).
The 100 ESBL-positive (49% ST131) and 167 ESBL-negative (13% ST131; P < 0.001) study isolates were predominantly from urine (92%) and community-associated infections (95%). The most prevalent ESBL type overall was CTX-M (84%), predominantly CTX-M-15 (73%), followed by group 9 CTX-M variants (11%). CTX-M-15 was not significantly associated with ST131 (data not shown).
Among the 49 ESBL-positive ST131 isolates, 48 (98%) represented the H30 subclone, and 44 (92%) of these represented the H30-Rx subclone. In contrast, among the 22 ESBL-negative ST131 isolates, only 14 (64%) represented the H30 subclone (P < 0.001), and only 3 (14%) of these represented the H30-Rx subclone (P < 0.001).
H30 ST131 subclone isolates were overwhelmingly fluoroquinolone resistant (98%) and ESBL positive (77%), whereas few non-H30 ST131 isolates were (Fig. 1A). Among H30 isolates, 94% of H30-Rx isolates, but only 27% of other H30 isolates, were ESBL positive (Fig. 1B). Within ST131, ESBL subtype varied significantly by subclone, with CTX-M-15 being associated with H30-Rx and CTX-M-9 being associated with other H30 isolates (Fig. 1A).
Fig 1.
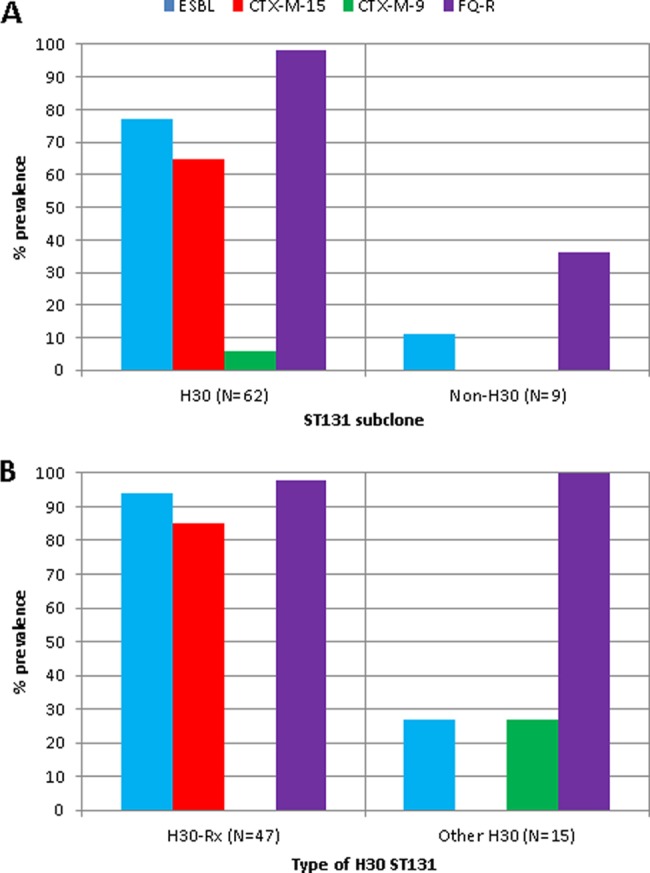
Resistance phenotypes and ESBL types among 71 ST131 isolates. (A) All 71 ST131 isolates, stratified by H30 subclone status. (B) The 62 H30 subclone isolates, stratified by H30-Rx status. ESBL, extended-spectrum β-lactamase; FQ-R, fluoroquinolone resistant. Prevalence values reflect percentages of isolates within each indicated subgroup.
Antimicrobial resistance also varied by subclone, with H30 subclone isolates having a higher prevalence of beta-lactam and ciprofloxacin resistance (Table 1) and higher resistance scores (median, 5 versus 3; P < 0.001). Similarly, among H30 isolates, H30-Rx isolates had a higher prevalence of resistance to cephalosporins (Table 1) and higher resistance scores (median, 6 versus 3; P = 0.01).
Table 1.
Antimicrobial resistance by ST131 subclone among 71 Escherichia coli ST131 isolates
| Antimicrobial agent | Within ST131 |
Within H30 ST131 subclone |
||||
|---|---|---|---|---|---|---|
| No. (%) of resistant strains |
Pa | No. (%) of resistant strains |
Pa | |||
| H30 (n = 62) | Non-H30 (n = 9) | H30-Rx (n = 47) | Other H30 (n = 15) | |||
| Ampicillin | 61 (98) | 7 (78) | 0.04 | 46 (98) | 15 (100) | |
| Cefazolin | 48 (77) | 1 (11) | <0.001 | 43 (91) | 5 (33) | <0.001 |
| Ceftriaxone | 46 (74) | 1 (11) | <0.001 | 42 (89) | 4 (27) | <0.001 |
| Ceftazidime | 44 (71) | 1 (11) | 0.001 | 40 (85) | 4 (27) | <0.001 |
| Gentamicin | 23 (37) | 5 (56) | 15 (32) | 8 (53) | ||
| Ciprofloxacin | 61 (98) | 2 (22) | <0.001 | 46 (98) | 15 (100) | |
| TMP-SMZb | 31 (50) | 6 (67) | 21 (45) | 10 (67) | ||
Determined by Fisher's exact test. Values are shown where P is <0.05; for all other comparisons, P was >0.10.
TMP-SMZ, trimethoprim-sulfamethoxazole.
Virulence scores were significantly lower overall among ESBL-positive than ESBL-negative isolates (median, 9 versus 11; P < 0.001) and among non-ST131 ESBL-positive than non-ST131 ESBL-negative isolates (median, 6 versus 12; P < 0.001). In contrast, ST131 isolates had similar virulence scores (median, 10) regardless of ESBL status. Among ESBL-negative isolates, virulence scores were slightly lower among ST131 than non-ST131 isolates (median, 10 versus 12; P = 0.03). In contrast, among ESBL-positive isolates, virulence scores were much greater among ST131 than non-ST131 isolates (median, 10 versus 6; P < 0.001).
Within ST131, several virulence factor (VF) genes were subclone specific, with iha, sat, and iutA occurring predominantly among H30 isolates but iroN, K1, and ibeA occurring predominantly among non-H30 isolates (Table 2). In contrast, among H30 isolates, only kpsMII and K5 differed in prevalence between H30-Rx and non-H30-Rx isolates (Table 2). Virulence scores were similar across the ST131 subclones (median for each, 10).
Table 2.
Virulence gene distribution by ST131 subclone among 71 ST131 Escherichia coli isolates
| Functional category | Genea | Within ST131 |
Within H30 ST131 subclone |
||||
|---|---|---|---|---|---|---|---|
| No. (%) of strains carrying gene |
Pb | No. (%) of strains carrying gene |
Pb | ||||
| H30 (n = 62) | Non-H30 (n = 9) | H30-Rx (n = 47) | Other H30 (n = 15) | ||||
| Adhesin | iha | 60 (97) | 5 (56) | 0.002 | 45 (96) | 15 (100) | |
| Toxin | sat | 61 (98) | 3 (33) | <0.001 | 46 (98) | 15 (100) | |
| Siderophores | iroN | 1 (2) | 2 (22) | 0.04 | 1 (2) | 0 | |
| iutA | 56 (90) | 5 (56) | 0.02 | 42 (89) | 14 (93) | ||
| Protectins | kpsMII | 53 (85) | 8 (89) | 44 (94) | 9 (60) | 0.004 | |
| K1 | 2 (3) | 3 (33) | 0.01 | 1 (2) | 1 (7) | ||
| K5 | 17 (27) | 1 (11) | 9 (19) | 8 (53) | 0.02 | ||
| Other | ibeA | 0 | 6 (67) | <0.001 | 0 | 0 | |
Virulence genes listed are those that yielded P values of <0.05 in at least one comparison.
Determined by Fisher's exact test. Values are shown where P is <0.05; for all other comparisons, the differences were not significant.
According to PCoA, ST131 isolates had distinctive virulence profiles relative to non-ST131 isolates, and each subclone group within ST131 (H30-Rx, other H30, and non-H30) had a characteristic profile (Fig. 2). Among the ST131 isolates, H30 (non-H30-Rx) profiles were most homogeneous, whereas non-H30 profiles were most diverse and overlapped most with those of non-ST131 isolates (Fig. 2).
Fig 2.
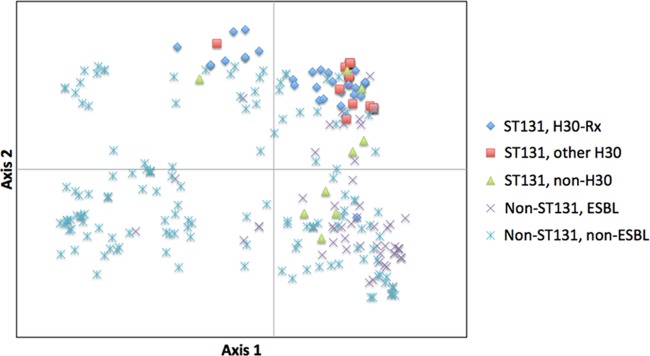
Principal coordinate analysis (PCoA) of virulence gene profiles among 267 Escherichia coli isolates. The PCoA was based on results for all 50 virulence genes studied. Each isolate is plotted based on its values for PCoA coordinates 1 (x axis) and 2 (y axis), which collectively capture 64.5% of total variance in the data set. For ST131, 47 H30-Rx, 15 other H30, and 9 non-H30 isolates were evaluated; for non-ST131, 51 ESBL-producing and 145 non-ESBL-producing isolates were evaluated.
Here we determined the prevalence of ST131 and its H30 and H30-Rx subclones and explored their associations with resistance phenotypes, ESBL types, and virulence profiles, among prospectively collected E. coli clinical isolates from the Chicago area (2007 to 2010). We confirmed the well-established association between ST131 and ESBL production (13–15), and found that the recently identified H30 ST131 subclone (1, 2, 5, 16) has expanded in the study region more than non-H30 ST131 subclones. Furthermore, we found that within ST131, CTX-M-15 was confined almost exclusively to the H30-Rx subset.
Our findings confirm, in a geographically distinct population, recent whole-genome-based evidence that within ST131, blaCTX-M-15 is transmitted mainly vertically within the H30-Rx lineage, after what was probably a single ancestral acquisition event (2). Our findings also uniquely document a continuum of increasing antimicrobial resistance within ST131, from the non-H30 (most susceptible) isolates to the H30-Rx isolates (most resistant).
Furthermore, we document that, among ESBL-positive isolates, ST131 isolates—mostly representing the H30-Rx subclone—have higher virulence scores than non-ST131 isolates, implying greater virulence potential and thereby possibly explaining their high prevalence. We also identified three VF genes (iha, sat, and iutA) that are more prevalent among H30 than non-H30 ST131 isolates. The mechanisms whereby specific accessory traits may facilitate the epidemiologic success of ST131 and its principal subclones deserve further study.
Study limitations include the fact that most isolates were from urine and community onset infections and hence may not be representative of isolates causing invasive infections. We also lacked clinical data, so we could not correlate bacterial traits with infection severity. Study strengths include the fact that that we evaluated a large number of ESBL-positive and ESBL-negative clinical isolates and used novel SNP-based PCR assays to identify the recently recognized H30 and H30-Rx subclones within ST131.
In conclusion, among ESBL-positive E. coli strains in the study region, the H30 ST131 subclone now accounts for almost half of ESBL-positive E. coli strains causing infections, with CTX-M-15, the most common ESBL type, being carried almost exclusively by the H30-Rx subset within H30. Elucidation of the molecular and ecologic basis for the epidemiologic success of ST131, especially its H30 and H30-Rx components, could inform the development of interventions against further spread of these highly antimicrobial-resistant lineages.
ACKNOWLEDGMENTS
This material is based upon work supported by Office of Research and Development, Medical Research Service, Department of Veterans Affairs, grant 1 I01 CX000192 01 (J.R.J.); NIH grants RC4-AI092828 (E.V.S. and J.R.J.) and 2KL2RR024151-07 (R.B.); and a research grant from Merck, Inc.
J.R.J. has received grants, contracts, or consultancies from ICET, Rochester Medical, and Syntiron. J.R.J., L.B.P., and E.V.S. have patent applications related to diagnostic tests for E. coli clonal groups.
Footnotes
Published ahead of print 30 September 2013
REFERENCES
- 1. Johnson J, Tchesnokova V, Johnston B, Clabots C, Roberts P, Billig M, Riddell K, Rogers P, Qin X, Butler-Wu S, Price L, Aziz M, Nicolas-Chanoine M, Debroy C, Robicsek A, Hansen G, Urban C, Platell J, Trott D, Zhanel G, Weissman S, Cookson B, Fang F, Limaye A, Scholes D, Chattopadhyay S, Hooper D, Sokurenko E. 2013. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J. Infect. Dis. 207:919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Price L, Johnson J, Aziz M, Clabots C, Johnston B, Tchesnokova V, Nordstrom L, Pearson T, Keim P, Sokurenko E. Epidemic clonal expansion of CTX-M-15-producing Escherichia coli ST131. mBio, in press. [Google Scholar]
- 3. Banerjee R, Strahilevitz J, Johnson J, Nagwekar P, Schora D, Shevrin I, Du H, Peterson L, Robicsek A. 2013. Predictors and molecular epidemiology of community-onset extended-spectrum beta-lactamase (ESBL) Escherichia coli infection in a Midwestern community Infect. Control Hosp. Epidemiol. 34:947–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Colpan A, Porter S, Johnston B, Clabots C, Anway R, Thao L, Kuskowski MA, Tchesnokova V, Sokurenko EV, Johnson J. Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among U.S. veterans. Clin. Infect. Dis. 57:1256–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu L, Ensor V, Gossain S, Nye K, Hawkey P. 2005. Rapid and simple detection of bla CTX-M genes by multiplex PCR assay. J. Med. Microbiol. 54:1183–1187 [DOI] [PubMed] [Google Scholar]
- 7. Johnson JR, Urban C, Weissman SJ, Jorgensen JH, Lewis JS, II, Hansen G, Edelstein PH, Robicsek A, Cleary T, Adachi J, Paterson D, Quinn J, Hanson ND, Johnston BD, Clabots C, Kuskowski MA. 2012. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and blaCTX-M-15 among extended-spectrum-β-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob. Agents Chemother. 56:2364–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnson J, Johnston B, Clabots C, Kuskowski M, Castanheira M. 2010. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin. Infect. Dis. 51:286–294 [DOI] [PubMed] [Google Scholar]
- 9. Johnson J, Menard M, Johnston B, Kuskowski M, Nichol K, Zhanel G. 2009. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002–2004. Antimicrob. Agents Chemother. 53:2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson J, Stell A. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261–272 [DOI] [PubMed] [Google Scholar]
- 11. Johnson J, Murray A, Gajewski A, Sullivan M, Snippes P, Kuskowski M, Smith K. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47:2161–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peakall R, Smouse P. 2006. GENALEX 6: genetic analysis in Excel: population genetic software for teaching and research. Mol. Ecol. Notes 6:288–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olesen B, Hansen D, Nilsson F, Frimodt-Møller J, Leihof R, Struve C, Scheutz F, Johnston B, Krogfelt K, Johnson J. 2013. Prevalence and characteristics of the epidemic multiresistant Escherichia coli ST131 clonal group among extended-spectrum beta-lactamase-producing E. coli isolates in Copenhagen, Denmark. J. Clin. Microbiol. 51:1779–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pitout JDD, Campbell L, Church DL, Gregson DB, Laupland KB. 2009. Molecular characteristics of travel-related extended-spectrum beta-lactamase-producing Escherichia coli isolates from the Calgary health region. Antimicrob. Agents Chemother. 53:2539–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blanco J, Mora A, Mamani R, López C, Blanco M, Dahbi G, Herrera A, Marzoa J, Fernández V, de la Cruz F, Martínez-Martínez L, Alonso MP, Nicolas-Chanoine M-H, Johnson JR, Johnston B, López-Cerero L, Pascual A, Rodríguez-Baño J, the Spanish Group for Nosocomial Infections (GEIH) 2013. Four main virotypes among extended-spectrum β-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J. Clin. Microbiol. 51:3358–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tchesnokova V, Billig M, Chattapadhyay S, Linardopoulou E, Aprikian P, Roberts PL, Skrivankova PV, Johnston B, Gileva A, Igusheva I, Toland A, Riddell K, Rogers P, Qin X, Bulter-Wu S, Cookson BT, Fang FC, Kahl B, Price LB, Weissman SJ, Limaye A, Scholes D, Johnson JR, Sokurenko EV. 2013. Predictive diagnostics for Escherichia coli infections based on the clonal association of antimicrobial resistance and clinical outcome. J. Clin. Microbiol. 51:2991–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]


