Abstract
It was found in our previous study that berberine (BBR) and fluconazole (FLC) used concomitantly exhibited a synergism against FLC-resistant Candida albicans in vitro. The aim of the present study was to clarify how BBR and FLC worked synergistically and the underlying mechanism. Antifungal time-kill curves indicated that the synergistic effect of the two drugs was BBR dose dependent rather than FLC dose dependent. In addition, we found that BBR accumulated in C. albicans cells, especially in the nucleus, and resulted in cell cycle arrest and significant change in the transcription of cell cycle-related genes. Besides BBR, other DNA intercalators, including methylene blue, sanguinarine, and acridine orange, were all found to synergize with FLC against FLC-resistant C. albicans. Detection of intracellular BBR accumulation by fluorescence measurement showed that FLC played a role in increasing intracellular BBR concentration, probably due to its effect in disrupting the fungal cell membrane. Similar to the case with FLC, other antifungal agents acting on the cell membrane were able to synergize with BBR. Interestingly, we found that the efflux of intracellular BBR was FLC independent but strongly glucose dependent and associated with the drug efflux pump Cdr2p. These results suggest that BBR plays a major antifungal role in the synergism of FLC and BBR, while FLC plays a role in increasing the intracellular BBR concentration.
INTRODUCTION
Candida albicans, one of the most prevalent human fungal pathogens, causes superficial mycoses, invasive mucosal infections, and disseminated systemic disease (1–4). Although the need for effective antifungal therapy is increasing, the available antifungal agents are still limited. Fluconazole (FLC) is most widely used due to its high bioavailability and low toxicity (5, 6). However, with the increasing clinical use of FLC, drug-resistant isolates are emerging rapidly (7–11). The high mortality rate of invasive Candida infections and limited availability of highly effective antifungal agents make it necessary to develop new antifungal therapeutics.
The combinations of antimicrobial agents against drug-resistant Candida have been studied (12–15). Members of our group have shown that concomitant use of berberine (BBR; an alkaloid with a long history of medicinal use in traditional Chinese medicine [16]) and FLC is highly efficacious in killing FLC-resistant C. albicans in vitro, and the fractional inhibitory concentration (FIC) index ranges from 0.017 to 0.127 (17, 18). Our comparative proteomic study and further investigations indicated that FLC and BBR treatment affected the expression of proteins functioning in energy metabolism, increased mitochondrial membrane potential, decreased intracellular ATP level, inhibited ATP-synthase activity, and increased generation of endogenous reactive oxygen species (ROS) in FLC-resistant C. albicans strains (19). Nevertheless, the mechanism for how the combination of BBR and FLC augments the efficacy of each agent alone remains unclear.
In this study, we found that BBR played a major antifungal role in the synergism of FLC and BBR. BBR accumulated in C. albicans cells, especially in the nucleus, where it probably binds to DNA, causing cell cycle arrest and DNA damage. Interestingly, FLC increased the intracellular BBR concentration.
MATERIALS AND METHODS
Strains, culture, and agents.
All strains used in this study are listed in Table 1. Strains were routinely grown in YPD (1% yeast extract, 2% peptone, and 2% dextrose) liquid medium at 30°C in a shaking incubator. FLC (Pfizer-Roerig Pharmaceuticals, New York, NY) was prepared in sterile H2O, and BBR (Sigma-Aldrich, St. Louis, MO) was prepared in dimethyl sulfoxide (DMSO).
Table 1.
Strains used in this study
| Strain(s) | Characteristics | Fluconazole MIC80 (μg/ml) | Reference |
|---|---|---|---|
| 0304103 | Clinical isolate, fluconazole resistant | >64 | |
| 01010 | Clinical isolate, fluconazole resistant | >64 | 17 |
| 632 | Clinical isolate, fluconazole resistant | >64 | This study |
| 15 fluconazole-resistant strainsa | Clinical isolate, fluconazole resistant | >64 | This study |
| 3 caspofungin-resistant strainsa | FKS hot spot mutants, caspofungin resistant | 64 | 28 |
| CAF2-1 | ura3Δ::imm434/URA3 | 0.5 | 30 |
| DSY448 | cdrlΔ::hisG-URA3-hisG/cdrlΔ::hisG | 0.125 | 30 |
| DSY465 | mdrlΔ::hisG-URA3-hisG/mdrlΔ::hisG | 0.5 | 30 |
| DSY653 | cdr2Δ::hisG-URA3-hisG/cdr2Δ::hisG | 0.25 | 30 |
| DSY294 | TAC1-3/TAC1-4 | 0.25 | 31 |
| DSY296 | TAC1-5/TAC1-5 | 64 | 31 |
| DSY3606 | tac1-5Δ::hisG/tac1-5Δ::hisG, RPS1::TAC1-5 | 4 | 31 |
| DSY3706 | tac1-5Δ::hisG/tac1-5Δ::hisG, RPS1::CIp10 | 0.25 | 31 |
Names and MIC results for these strains are shown in Tables S1 and S2 in the supplemental material.
Time-kill curve assay.
Time-kill curve assay was performed according to a previous protocol (17), with a few modifications. Briefly, exponentially growing C. albicans cells were harvested and resuspended in fresh YPD medium to 5 × 104 cells/ml. Various concentrations of FLC and BBR were added. The cells were cultured at 30°C with constant shaking (200 rpm) and counted at designated time points after culture (0, 3, 6, 9, and 12 h). No FLC or BBR was added in the control group. Three independent experiments were performed.
Subcellular localization of BBR.
Exponentially growing C. albicans cells were treated with 16 μg/ml of FLC and 8 μg/ml of BBR for 8 h, harvested by centrifugation, washed thrice with phosphate-buffered saline (PBS), and treated with 1 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI) at 30°C for 10 min in the dark. Living cells were immobilized on 0.1% poly-l-lysine-coated slides and observed under a Leica TCS SP2 confocal microscope equipped with argon and helium-neon lasers (Leica, Germany).
Cell cycle analysis by flow cytometry.
Exponentially growing C. albicans cells were harvested, washed twice with PBS, and resuspended in RPMI 1640 medium at 5 × 107 cells/ml. Different concentrations of FLC and BBR were added. After incubation at 30°C with constant shaking (200 rpm) for 12 and 24 h, 75 ml of each sample was centrifuged, washed, and fixed with 70% ethanol overnight. The cells were then stained with 50 μg/ml of propidium iodide at 4°C for 30 min. The samples were sonicated to obtain separate cells. Data were collected using a FACSCalibur cytometer (Becton, Dickinson, San Jose, CA) and analyzed with Cell Quest 3.0 software.
Gene expression analysis.
Exponentially growing C. albicans cells were harvested and resuspended in fresh YPD medium at 1.5 × 106 cells/ml. Either 64 μg/ml of FLC or 64 μg/ml of FLC plus 16 μg/ml of BBR was added. The samples were incubated at 30°C for 6 h. Cells were collected by centrifugation and stored in liquid nitrogen. Total RNA preparation and Candida genome microarray (CapitalBio Corp., Beijing, China) were performed as described previously (20). Three different clinical FLC-resistant strains (0304103, 01010, and 632) were selected to carry out the expression profile microarray, and only genes showing a consistent pattern were considered. The genes were named according to the Candida Genome Database (CGD) (http://www.candidagenome.org/) and divided according to their biological functions defined by the CGD.
Antifungal susceptibility test.
Antifungal susceptibility testing was carried out in 96-well microtiter plates (Greiner, Germany) using a broth microdilution protocol of the Clinical and Laboratory Standards Institute M27-A3 method, with a few modifications (17, 19, 21). Briefly, the initial concentration of fungal suspension in RPMI 1640 medium was 103 CFU/ml, and the final concentrations ranged from 0.125 to 64 μg/ml for FLC and 0.5 to 16 μg/ml for BBR. Plates were incubated at 35°C for 24 h. MIC80s were determined as the lowest concentration of the drugs (alone or in combination) that inhibited growth by 80%.
Intracellular BBR accumulation assay.
Intracellular BBR concentration was detected according to a previously described protocol (22, 23), with a few modifications. Briefly, exponentially growing C. albicans cells were harvested, washed twice with PBS, and resuspended in RPMI 1640 medium at 5 × 107 cells/ml. Different concentrations of FLC and BBR were added. One milliliter of each sample was cultured at 30°C with constant shaking (200 rpm) for 0, 2, 4, 6, 8, and 10 h, centrifuged, washed twice, and resuspended in 1 ml of PBS. One hundred microliters of each disposed sample was transferred into a black 96-well microplate with a clear bottom (Greiner, Germany). Fluorescence measurement of BBR was performed with an Infinite 200 Universal microplate reader (Tecan Group Ltd., Switzerland) and FACSCalibur cytometer (Becton Dickinson, San Jose, CA) at 405-nm excitation and 520-nm emission wavelengths, respectively.
Transmission electron microscopy.
Exponentially growing C. albicans cells which had been treated with 10 μg/ml of FLC and/or 16 μg/ml of BBR for 24 h were washed twice with PBS and fixed in 500 μl of fixative solution (sodium cacodylate buffer, pH 7.2, containing 4% polyoxymethylene) at 4°C for 24 h. The samples were then washed with saline and postfixed with 1% phosphotungstic acid for 90 min. The fixed cells were dehydrated through a graded series of ethanol and embedded with EPON-812. Ultrathin sections were prepared, double stained with uranium and lead, and observed under a transmission electron microscope (Hitachi H-800; Japan) at a magnification of ×10,000.
Microarray data accession number.
All of the transcription data were deposited at the NCBI Gene Expression Omnibus (GEO) database with accession number GSE43662 and platform identification GPL9728.
RESULTS
BBR plays a major antifungal role in the synergism of FLC and BBR.
In this study, we investigated the synergistic mechanism of FLC and BBR against FLC-resistant C. albicans. The relationship between the synergistic effect and the dose of the two drugs was first studied by orthogonal experiments using the clinical FLC-resistant C. albicans strain 0304103. Different concentrations of FLC (16 μg/ml to 64 μg/ml) and BBR (2 μg/ml to 8 μg/ml) were used in the time-kill curve assay, and our results indicated that the synergistic effect of the two drugs depended on the concentration of BBR but not FLC (Fig. 1). More specifically, 8 μg/ml of BBR alone had no antifungal effect and 64 μg/ml of FLC alone had a weak antifungal effect, but the antifungal effect was improved significantly when the two drugs were used together (Fig. 1). Interestingly, the antifungal effect of FLC plus BBR was improved when the dose of BBR was increased even though the dose of FLC remained unchanged. In contrast, increasing the dose of FLC did not enhance the synergistic antifungal effect when the dose of BBR remained unchanged (Fig. 1). These results suggested that BBR had a major antifungal effect in the synergism of FLC and BBR, while FLC played an auxiliary role.
Fig 1.
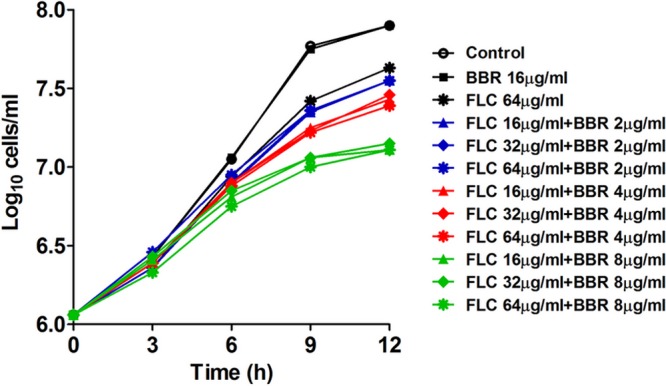
Time-kill curves of C. albicans strain 0304103 at different concentrations of FLC and BBR.
BBR accumulates in C. albicans cells, especially in the nucleus.
Since BBR is a compound with fluorescence, we examined the localization of BBR in C. albicans by confocal microscopy. Our result indicated that although weak fluorescence of BBR could be observed throughout the cytoplasm, the BBR fluorescence was particularly strong in the nucleus (Fig. 2A) and overlapped with the nucleus dye DAPI (Fig. 2C), suggesting that BBR accumulated in C. albicans cells, especially in the nucleus.
Fig 2.
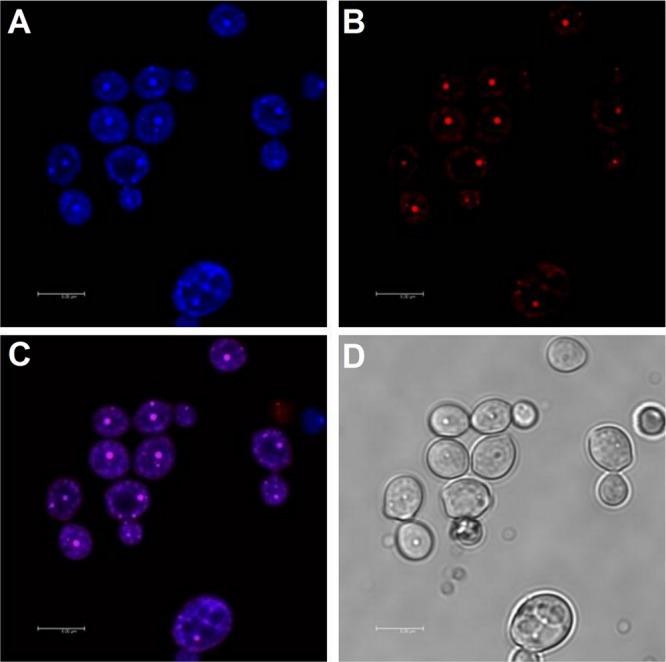
Subcellular localization of BBR. (A) The fluorescence of intracellular BBR was observed with excitation at 405 nm and emission at 520 nm. (B) The fluorescence of DAPI was observed with excitation at 364 nm and emission at 454 nm. (C) Fluorescence of intracellular BBR and DAPI. (D) Bright field. Scale bars = 5 μm.
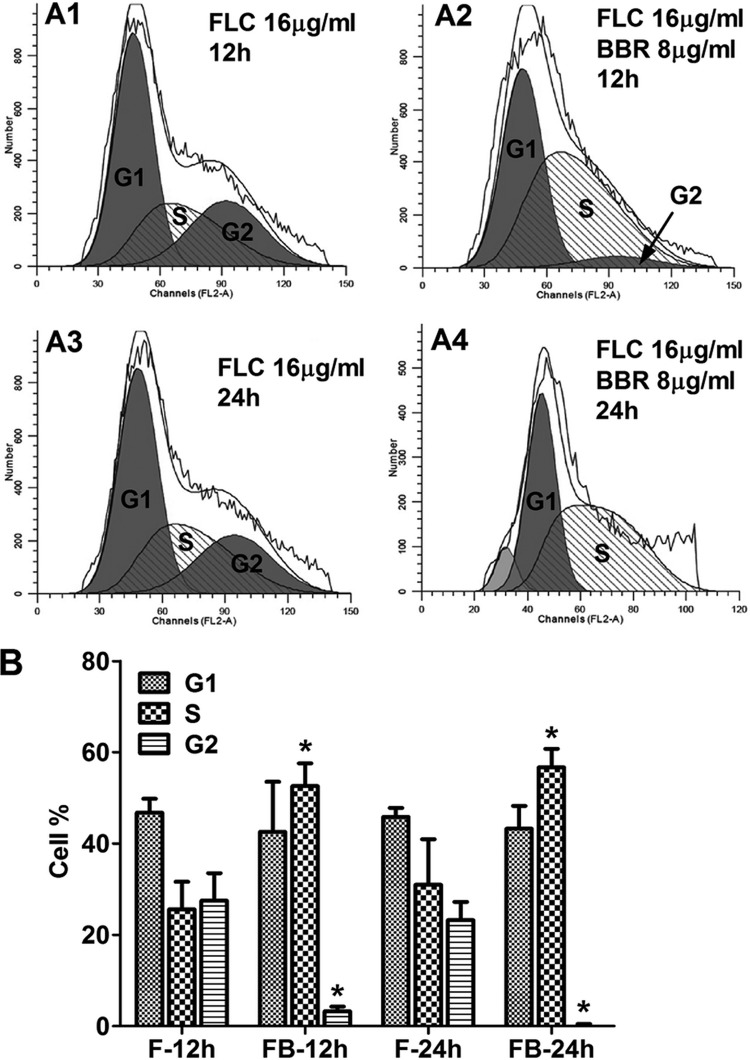
Combination of FLC and BBR induces S/G2 phase arrest and DNA damage.
After showing that intracellular BBR accumulated especially in the nucleus, we studied the impact of the two drugs on cell cycle using flow cytometric analysis. Interestingly, we found that after 12 h of exposure, 25.6% of cells were in S phase and 27.5% of cells were in G2 phase in the group treated with 10 μg/ml of FLC alone, compared to 52.6% and 3.28% in the group treated with 10 μg/ml of FLC and 16 μg/ml of BBR (Fig. 3A1, A2, and B). Similarly, after 24 h of treatment, 30.9% of cells were in S phase and 23.2% cells were in G2 phase in the group treated with 10 μg/ml of FLC alone, compared to 56.7% and 0% in the group treated with 10 μg/ml of FLC and 16 μg/ml of BBR (Fig. 3A3, A4, and B). These results indicated that the combination of FLC and BBR significantly increased the subpopulation of cells in S phase (P < 0.05) (Fig. 3B) and reduced the number of cells in G2 phase (P < 0.05) (Fig. 3B), resulting in cell cycle arrest at S/G2 phase.
Fig 3.
(A) Cell cycle analysis by flow cytometry. C. albicans 0304103 cells were treated for 12 h or 24 h with 10 μg/ml of FLC (A1 and A3) or with 10 μg/ml of FLC plus 16 μg/ml of BBR (A2 and A4). Determination was performed in triplicate, and one representative experiment is shown. (B) Mean percentages of cells in G1, S, and G2. F, 10 μg/ml of FLC; FB, 10 μg/ml of FLC plus 16 μg/ml of BBR. The percentages in different phases were calculated by flow cytometry software. Statistical significance among groups was determined by analyses of variance. Comparison between the FLC monotreatment group and the FLC-BBR combination treatment group was performed by Student t test. The data are shown as means ± SDs from three independent experiments.
We performed expression profile microarrays to study the gene expression changes in response to combination treatment with 64 μg/ml of FLC and 16 μg/ml of BBR compared to monotreatment with 64 μg/ml of FLC. Three different clinical FLC-resistant strains (0304103, 01010, and 632) were used to carry out the expression profile microarray, and only genes showing a consistent pattern were considered. We found that 76 genes were altered at the transcription level, including 28 downregulated genes and 48 upregulated genes (cutoff value = 1.5). Of the 76 genes, the expression of cell cycle- and DNA metabolic process-related genes changed significantly. More specifically, DNA metabolic process-related genes, including orf19.7234 and orf19.5569, were overexpressed after FLC-BBR combination treatment (Table 2), while the expression of cell cycle-related genes, including SAC7, orf19.1185, orf19.4906, and orf19.5457, was reduced (Table 3). Besides these genes, the expression of energy metabolism-related genes, including COX9, orf19.4727, GPM2, QCR7, orf19.2067, GIT2, LSC1, and HXT5, changed significantly (Tables 2 and 3). Moreover, there was a significant change in the expression of genes involved in transcription regulation (MET28, MBF1, SWI1, SRR1, and EFG1), RNA and protein binding (TIM9, YTH1, SMD3, and CTA3), hydrolase activity (UBP6, PRE10, YPT1, SSU72, PRE1, and PHO15), vacuole function (VPS4, VMA8, VMA2, and NCR1), and structural molecule activity (RPL17B and RPL35) (Tables 2 and 3).
Table 2.
Genes upregulated in response to combination treatment with 64 μg/ml of FLC and 16 μg/ml of BBR versus monotreatment with 64 μg/ml of FLCa
| Gene name (CGD) | Description of gene or product (CGD) | Ratio (FB treated vs F treated) |
||
|---|---|---|---|---|
| 0304103 | 01010 | 632 | ||
| DNA metabolic process | ||||
| orf19.7234 | Putative RSC chromatin remodeling complex component | 1.82 | 1.55 | 2.00 |
| orf19.5569 | Ortholog(s) have role in maintenance of rDNA | 1.70 | 1.75 | 1.55 |
| Energy metabolism related | ||||
| COX9 | Putative subunit VIIa of cytochrome c oxidase; flucytosine induced | 4.52 | 1.95 | 1.93 |
| orf19.4727 | Ortholog(s) have succinate dehydrogenase (ubiquinone) activity | 4.62 | 1.68 | 1.63 |
| GPM2 | Putative phosphoglycerate mutase; induced by high levels of peroxide stress | 2.53 | 1.79 | 1.69 |
| QCR7 | Putative ubiquinol-cytochrome c reductase, subunit 7; Hap43p-repressed gene | 2.13 | 2.09 | 1.77 |
| orf19.2067 | Putative protein with a predicted role in mitochondrial iron metabolism | 1.87 | 2.27 | 1.69 |
| Transcription regulator activity | ||||
| MET28 | Protein induced during the mating process | 4.12 | 2.22 | 1.61 |
| MBF1 | Putative transcriptional coactivator; caspofungin repressed | 3.27 | 2.29 | 1.94 |
| RNA and protein binding | ||||
| TIM9 | Predicted protein of the mitochondrial intermembrane space | 4.93 | 1.88 | 2.00 |
| YTH1 | Putative mRNA cleavage and polyadenylation specificity factor | 2.53 | 1.82 | 1.52 |
| SMD3 | Protein described as a core snRNP protein; induced upon adherence to polystyrene | 1.85 | 1.73 | 1.69 |
| Hydrolase activity | ||||
| UBP6 | Putative ubiquitin-specific protease of the 26S proteasome; oxidative stress induced via Cap1p | 4.56 | 1.98 | 1.98 |
| PRE10 | Alpha7 (C8) subunit of the 20S proteasome; multiple phosphorylated residues | 2.73 | 1.69 | 1.83 |
| YPT1 | An essential small Ras-type GTPase involved in protein secretion at ER-to-Golgi transport | 2.65 | 1.55 | 1.58 |
| SSU72 | Orthologs have CTD phosphatase activity and protein tyrosine phosphatase activity | 2.34 | 1.57 | 1.67 |
| PRE1 | Upregulated by Rim101p at acid pH; flucytosine induced; amphotericin B repressed | 2.73 | 1.69 | 1.83 |
| PHO15 | Protein described as 4-nitrophenyl phosphatase; hypha downregulated | 2.22 | 1.55 | 1.53 |
| Vacuole related | ||||
| VPS4 | Protein involved in transport from multivesicular body to the vacuole | 1.49 | 1.69 | 1.55 |
| VMA8 | Protein similar to the S. cerevisiae Vma8p subunit of vacuolar H+-ATPase | 1.51 | 1.55 | 1.66 |
| Structural molecule activity | ||||
| RPL17B | Ribosomal protein L17; mutation confers hypersensitivity to flucytosine and tubercidin; genes encoding cytoplasmic ribosomal subunits, translation factors, and tRNA synthetases are downregulated on phagocytosis by macrophages; Hap43p induced | 3.94 | 2.17 | 2.35 |
| RPL35 | Ribosomal protein; downregulation correlates with clinical development of fluconazole resistance; shows colony morphology-related gene regulation by Ssn6p; Hap43p induced | 3.45 | 2.51 | 1.86 |
| Not classified and not yet annotated | ||||
| PRN4 | Protein with similarity to pirins; increased transcription is observed upon benomyl treatment | 3.47 | 2.36 | 1.81 |
| PHB2 | Prohibitin 2; plasma membrane localized | 3.87 | 1.69 | 1.65 |
| BMT8 | Putative beta-mannosyltransferase, member of a 9-gene family including characterized BMT genes with roles in beta-1,2-mannosylation of cell wall phosphopeptidomannan | 2.54 | 1.97 | 1.55 |
| LAB5 | Orthologs have roles in protein lipoylation and mitochondrion localization | 2.04 | 1.56 | 1.90 |
| IFG3 | Putative d-amino acid oxidase | 1.83 | 1.50 | 1.54 |
| orf19.1306 | Orthologs have roles in cytosol and nucleus localization | 3.70 | 1.57 | 2.79 |
| orf19.2397.3 | Putative aminotransferase; Hap43p-repressed gene; homozygous transposon insertion causes decreased colony wrinkling under filamentous growth-inducing conditions but does not block true hyphal formation in liquid media | 2.99 | 1.85 | 2.03 |
| orf19.5985 | Orthologs have tubulin binding activity, roles in tubulin complex assembly and nucleus, polysome, and prefoldin complex localization | 2.56 | 2.50 | 1.61 |
| orf19.3537 | Putative sulfiredoxin; biofilm-induced gene; regulated by Tsa1p and Tsa1Bp in minimal media at 37°C | 1.62 | 3.16 | 1.61 |
| orf19.2755 | Orthologs have roles in proteasomal ubiquitin-dependent protein catabolic process, proteasomal ubiquitin-independent protein catabolic process and cytosol, nucleus, proteasome core complex, and beta-subunit complex localization | 3.28 | 1.55 | 1.51 |
| orf19.2196 | Orthologs have ubiquitin binding activity and cytoplasm localization | 1.79 | 1.52 | 2.60 |
| orf19.2621 | Orthologs have roles in mRNA splicing, via spliceosome and U1 snRNP, U2-type prespliceosome, U4/U6 × U5 tri-snRNP complex, U5 snRNP, and cytosol localization | 2.02 | 1.86 | 2.01 |
| orf19.4503 | Orthologs have roles in cytosol and nucleus localization | 2.06 | 1.98 | 1.57 |
| orf19.3914 | Has domain(s) with predicted translation initiation factor activity, role in translational initiation and cytoplasm localization | 1.54 | 2.24 | 1.60 |
| orf19.2794 | Putative nonspecific single-domain racemase; repressed in response to amino acid starvation (3-aminotriazole treatment) | 1.70 | 1.82 | 1.71 |
| orf19.4449 | Orthologs have superoxide dismutase copper chaperone activity, roles in cellular copper ion homeostasis, cellular response to metal ion, intracellular copper ion transport and cytosol, mitochondrial inner membrane, and nucleus localization | 1.94 | 1.61 | 1.62 |
| orf19.1632 | Orthologs have roles in cytosol and nucleus localization | 1.71 | 1.50 | 1.80 |
| orf19.3482 | Orthologs have NAD+ diphosphatase activity, role in NADH metabolic process and cytosol, nucleus, and peroxisome localization | 1.86 | 1.57 | 1.57 |
| orf19.5817 | Orthologs have Rab guanyl-nucleotide exchange factor activity, role in ER-to-Golgi vesicle-mediated transport | 1.68 | 1.61 | 1.65 |
| orf19.715 | Predicted ORF from assembly 19 | 8.22 | 1.66 | 2.03 |
| orf19.4830 | Predicted ORF from assembly 19 | 1.56 | 2.96 | 2.19 |
| orf19.2853 | Predicted ORF from assembly 19 | 2.45 | 1.87 | 2.09 |
| orf19.6132 | Predicted ORF from assembly 19 | 2.23 | 1.81 | 1.97 |
| orf19.649 | Predicted ORF from assembly 19 | 1.67 | 1.81 | 1.85 |
| orf19.1574 | Predicted ORF from assembly 19 | 1.61 | 2.04 | 1.53 |
F, monotreatment with 64 μg/ml of FLC; FB, 64 μg/ml of FLC and 16 μg/ml of BBR; ER, endoplasmic reticulum; ORF, open reading frame; RSC, chromatin structure remodeling; CTD, carboxyl-terminal domain; BMT, beta-mannosyltransferase.
Table 3.
Genes downregulated in response to combination treatment with 64 μg/ml of FLC and 16 μg/ml of BBR versus monotreatment with 64 μg/ml of FLCa
| Gene name (CGD) | Description of gene or product (CGD) | Ratio (FB treated vs F treated) |
||
|---|---|---|---|---|
| 0304103 | 01010 | 632 | ||
| Cell cycle | ||||
| SAC7 (orf19.7115) | Putative GTPase-activating protein for RHO1; supposed to be related with actin filament reorganization involved in cell cycle | 0.66 | 0.59 | 0.63 |
| orf19.1185 | Orthologs have ubiquitin-protein ligase activity and roles in establishment of mitotic spindle orientation, mitotic cell cycle spindle assembly checkpoint, and protein autoubiquitination | 0.67 | 0.48 | 0.44 |
| orf19.4906 | Putative adhesin-like protein; transcription is positively regulated by Tbf1p | 0.38 | 0.52 | 0.46 |
| orf19.5457 | Orthologs have role in meiosis | 0.31 | 0.52 | 0.48 |
| Energy metabolism related | ||||
| GIT2 | Putative glycerophosphoinositol permease; fungus specific | 0.62 | 0.47 | 0.52 |
| LSC1 | Protein described as succinate-CoA ligase subunit; transcriptionally regulated by iron | 0.48 | 0.45 | 0.56 |
| HXT5 | Putative sugar transporter; transcription upregulated in response to treatment with ciclopirox olamine | 0.14 | 0.43 | 0.48 |
| Transcription regulator activity | ||||
| SWI1 | Protein involved in transcription regulation; SWI/SNF (switch/sucrose nonfermentable) complex is essential for hyphal growth and virulence | 0.43 | 0.64 | 0.61 |
| SRR1 | Response regulator of a two-component system involved in stress response | 0.39 | 0.57 | 0.67 |
| EFG1 | Transcriptional repressor; required for white-phase cell type | 0.42 | 0.55 | 0.47 |
| RNA and protein binding | ||||
| CTA3 | Protein similar to S. cerevisiae Ede1p, which is involved in endocytosis | 0.36 | 0.59 | 0.51 |
| Vacuole related | ||||
| VMA2 | Predicted ORF from assembly 19; amphotericin B repressed, caspofungin repressed | 0.57 | 0.64 | 0.66 |
| NCR1 | Putative vacuolar membrane protein with a predicted role in sphingolipid metabolism | 0.34 | 0.66 | 0.59 |
| Not classified and not yet annotated | ||||
| PRP8 | Protein similar to S. cerevisiae Prp8p; transcription is repressed in response to alpha pheromone in SpiderM medium | 0.49 | 0.66 | 0.57 |
| YTA6 | Protein similar to S. cerevisiae Yta6p ATPase; transposon mutation affects filamentous growth | 0.56 | 0.61 | 0.53 |
| MVD | Mevalonate diphosphate decarboxylase; functional homolog of S. cerevisiae Erg19p; possible drug target; transcriptionally regulated by carbon source, yeast-hyphal switch, growth phase, and antifungals; gene has intron | 0.57 | 0.58 | 0.53 |
| LEU5 | Putative mitochondrial carrier protein; Hap43p-repressed gene; transcription is upregulated in both intermediate and mature biofilms | 0.58 | 0.43 | 0.60 |
| orf19.2315 | Putative transcription factor with bZIP DNA-binding motif | 0.59 | 0.64 | 0.63 |
| orf19.684 | Putative transcription factor with zinc finger DNA-binding motif | 0.60 | 0.62 | 0.58 |
| orf19.2515 | Has domain(s) with predicted zinc ion binding activity | 0.48 | 0.61 | 0.62 |
| orf19.6443 | Has domain(s) with predicted catalytic activity and role in metabolic process | 0.50 | 0.50 | 0.64 |
| orf19.7554 | Putative transporter; slightly similar to the Sit1p siderophore transporter; induced by nitric oxide independently of Yhb1p | 0.34 | 0.64 | 0.56 |
| orf19.6654 | Predicted membrane transporter, member of the l-amino acid transporter-3 family | 0.29 | 0.56 | 0.63 |
| orf19.7261 | Putative Rab GDP-dissociation inhibitor; GlcNAc-induced protein | 0.53 | 0.33 | 0.47 |
| orf19.5461 | Predicted ORF from assembly 19 | 0.65 | 0.47 | 0.57 |
| orf19.2791 | Predicted ORF from assembly 19 | 0.44 | 0.57 | 0.60 |
| orf19.6181 | Predicted ORF from assembly 19 | 0.35 | 0.47 | 0.64 |
| orf19.2777 | Predicted ORF from assembly 19 | 0.26 | 0.55 | 0.62 |
| tT(UGU)1 | tRNA-Thr, predicted by tRNAscan-SE; UGU anticodon | 0.12 | 0.46 | 0.63 |
F, monotreatment with 64 μg/ml of FLC; FB, 64 μg/ml of FLC and 16 μg/ml of BBR; ORF, open reading frame.
DNA intercalators synergize with FLC against FLC-resistant C. albicans.
To validate that the synergism of FLC and BBR against FLC-resistant C. albicans was mainly due to the cell cycle arrest and DNA damage caused by intracellular BBR, we used DNA intercalators. Three DNA intercalators, including methylene blue (MB), sanguinarine (SN), and acridine orange (AO), were used (Fig. 4), all of which were found to have the antifungal effect due to their ability to bind DNA (24–27). Meanwhile, five clinical FLC-resistant C. albicans isolates (MIC80 > 64 μg/ml) were used in this study (Table 4). MB, SN, and AO all showed a weak antifungal effect when they were used alone. Their MIC80s were 16 μg/ml, 5 to 10 μg/ml, and 8 to 16 μg/ml, respectively (Table 4). Compared with these agents used alone, their combination with FLC showed a significant synergistic effect by reducing the dose of FLC to 0.25 to 2 μg/ml, the doses of MB, SN, and AO to 1 μg/ml, 0.63 to 1.25 μg/ml, and 1 to 2 μg/ml, respectively, and the FIC indices to 0.07, 0.13 to 0.27, and 0.08 to 0.26, respectively (Table 4).
Fig 4.
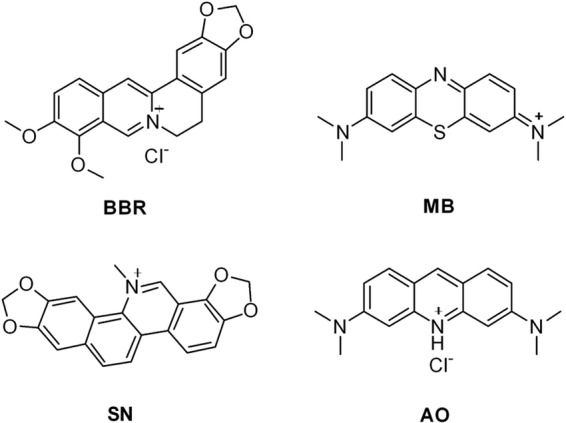
Chemical structures of selected DNA intercalators.
Table 4.
Interaction of FLC and DNA intercalators against FLC-resistant C. albicans by microdilution assay
| Clinical isolate | MIC80 (μg/ml) of intercalator alone |
MIC80 (μg/ml) of intercalators in combination |
FIC index for combinationa |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FLC | MB | SN | AO | FLC-MB | FLC-SN | FLC-AO | FLC-MB | FLC-SN | FLC-AO | |
| 101117 | >64 | 16 | 10 | 16 | 1/1 | 0.63/1.25 | 2/2 | 0.07 | 0.13 | 0.14 |
| 101118 | >64 | 16 | 5 | 8 | 1/1 | 1/0.63 | 0.25/2 | 0.07 | 0.13 | 0.25 |
| 1103379 | >64 | 16 | 10 | 16 | 0.5/1 | 2/1.25 | 0.5/2 | 0.07 | 0.14 | 0.13 |
| 1011655 | >64 | 16 | 5 | 16 | 1/1 | 1/1.25 | 2/1 | 0.07 | 0.26 | 0.08 |
| 1012782 | >64 | 16 | 5 | 8 | 1/1 | 2/1.25 | 1/2 | 0.07 | 0.27 | 0.26 |
Synergy and antagonism were defined by FIC indices of ≤0.5 and >4, respectively. An FIC index result of >0.5 but ≤4 was considered indifferent (18).
FLC plays a role in increasing the intracellular BBR concentration.
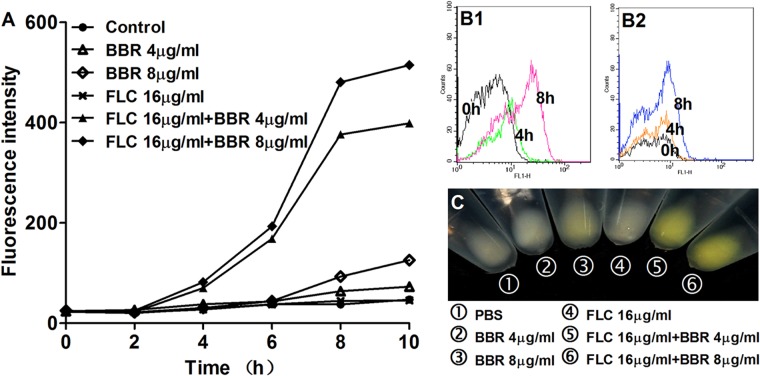
On the basis of the above findings, we postulated that FLC could increase intracellular BBR concentration. To validate this hypothesis, we evaluated the intracellular concentration of BBR with or without FLC treatment. We found that there was minimal BBR fluorescence detectable in the cells after 6 h of treatment with 4 μg/ml or 8 μg/ml of BBR alone (Fig. 5A), and the fluorescence was insignificantly higher than that in the control group even after 8 h of treatment (Fig. 5A). Interestingly, the addition of 16 μg/ml of FLC markedly increased the intracellular BBR fluorescence, as presented by intracellular BBR accumulation assay (Fig. 5A). A similar result was also detected by flow cytometry, showing that the addition of 16 μg/ml of FLC significantly increased the subpopulation of cells with high intracellular BBR fluorescence over time (Fig. 5B1), while the intracellular BBR fluorescence remained weak even after 8 h of incubation in the 4-μg/ml BBR monotreatment group (Fig. 5B2). Notably, the bright yellow color of BBR was visually observed in the FLC-BBR combination treatment groups (16 μg/ml of FLC plus 8 μg/ml of BBR and 16 μg/ml of FLC plus 4 μg/ml BBR), while only light yellow color was observed in the 8-μg/ml BBR monotreatment group, and no yellow color was observed in the 4-μg/ml BBR monotreatment group as well as the 16-μg/ml FLC group and the PBS control group (Fig. 5C). Subsequently, intracellular BBR concentration in media treated with different concentrations of FLC was further detected. It was interesting to find that under the same exogenous BBR concentration, different concentrations of FLC (16 μg/ml, 32 μg/ml, and 64 μg/ml) did not cause different intracellular BBR concentrations (Fig. 6). In contrast, with certain FLC doses, a higher concentration of exogenous BBR caused higher intracellular BBR fluorescence intensity. More specifically, when 16 μg/ml of FLC was added, the intracellular BBR fluorescence intensities caused by 2 μg/ml, 4 μg/ml, and 8 μg/ml of exogenous BBR were 35, 50, and 70, respectively (P < 0.05 and P < 0.01) (Fig. 6).
Fig 5.
FLC increases the intracellular BBR concentration in C. albicans. C. albicans strain 0304103 was treated with different concentrations of FLC and BBR. (A) Fluorescence intensity was measured with an Infinite 200 Universal microplate reader at 405-nm excitation and 520-nm emission wavelengths over time. (B) Fluorescence intensity of the group treated with 16 μg/ml of FLC plus 4 μg/ml of BBR (B1) and the group treated with 4 μg/ml of BBR (B2) was measured at 0, 4, and 8 h (shown in the graph) by FACSCalibur cytometry. (C) Color of samples from different groups. Strains were photographed at 8 h.
Fig 6.
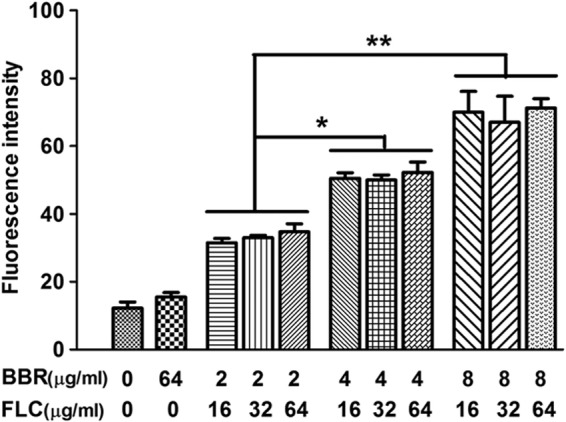
Fluorescence intensity of samples treated with different concentrations of FLC and BBR for 8 h. Statistical significance among groups was determined by analyses of variance. Comparison between two groups was performed by Student t test. The data are shown as means ± SDs from three independent experiments. *, P < 0.05; **, P < 0.01.
Cell membrane disruption effect of FLC contributes to its synergism with BBR against FLC-resistant C. albicans.
Because the major antifungal effect of FLC is attributed to its damage to the cell membrane, we postulated that FLC could promote BBR to enter cells by disrupting the cell membrane of the FLC-resistant strain 0304103. To validate this hypothesis, we performed electron microscopy of cells treated with FLC and BBR alone or in combination. As shown in Fig. 7A and B, electron microscopy results indicated that cells in the untreated control group and the 16-μg/ml BBR group were well preserved, with an intact cell wall, a plasma membrane with normal shape, and a homogeneous cytoplasm. It was also found that although strain 0304103 was highly resistant to FLC (MIC80 > 64 μg/ml), 10 μg/ml of FLC was able to cause modest damage to the cell membrane. As shown in Fig. 7C, the cell membrane detached from the cell wall in the strain treated with 10 μg/ml of FLC (white arrowhead). Notably, FLC and BBR combination treatment caused severe damage to the cells, as represented by detachment of the cell membrane from the cell wall (Fig. 7D, white arrowhead) and extensive solubilization in the cytoplasm (Fig. 7D, white arrow).
Fig 7.
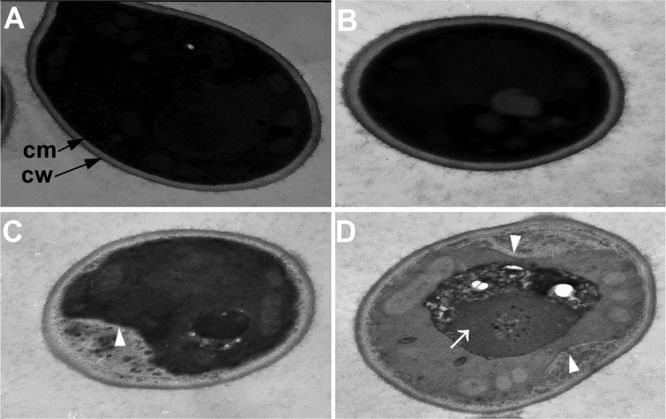
Ultrastructure of C. albicans strain 0304103. (A) Untreated control. (B) Cells treated with 16 μg/ml of BBR for 24 h. (C) Cells treated with 10 μg/ml of FLC for 24 h. (D) Cells treated with 10 μg/ml of FLC and 16 μg/ml of BBR for 24 h. White arrowhead, detachment of the cell membrane from the cell wall; white arrow, extensive solubilization in the cytoplasm.
To further evaluate our hypothesis that FLC increased the intracellular BBR concentration by causing cell membrane damage, other antifungal agents were used in combination with BBR to treat drug-resistant C. albicans. Five other antifungal agents and 15 clinical FLC-resistant C. albicans isolates (MIC80 > 64 μg/ml) were used (Table 5). The results indicated that all the antifungal agents acting on the cell membrane, including ketoconazole (KCZ), itraconazole (ICZ), and amphotericin B (AMB), demonstrated synergy with BBR against the drug-resistant strains of C. albicans, with the FIC index ranging from 0.02 to 0.38 (Table 5), while the antifungal agents not acting on the cell membrane, including flucytosine (5-FC) and caspofungin (CAS) (28), failed to synergize with BBR, with the FIC index ranging from 0.52 to 1.50 (Table 5). Meanwhile, three CAS-resistant strains were also used to investigate the effect of FLC-CAS combination treatment, and no obvious synergistic effect was observed (Table 5). These results supported our postulation, on the other hand.
Table 5.
Interaction of antifungal agents and BBR against drug-resistant C. albicans by microdilution assaya
| Parameter | MIC80 (μg/ml) of 15 FLC-resistant strains |
MIC80 (μg/ml) of 3 CAS-resistant strains |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| FLC | KCZ | ICZ | AMB | 5-FC | CAS | BBR | CAS | BBR | |
| Antifungal agent alone | >64 | 0.13–>16 | 0.25–>16 | 0.5–1 | 0.06–0.25 | 0.13–1 | >16 | >16 | >16 |
| Combination with BBRb | ≤0.13–1/1–4 | ≤0.03–0.25/0.5–4 | ≤0.03–0.25/0.5–8 | ≤0.03–0.13/1–8 | 0.06–0.25/0.5–16 | 0.13–1/0.5–16 | >16/0.5 | ||
| FIC indexc | 0.04–0.13 | 0.02–0.25 | 0.02–0.31 | 0.19–0.38 | 0.52–1.50 | 0.52–1.03 | 1.02 | ||
Detailed MIC results of these strains are shown in Tables S1, S2, and S3 in the supplemental material.
MIC80s for combinations are expressed as MIC80 of drug/MIC80 of BBR. High off-scale MIC80 was converted to the next highest concentration.
Synergy and antagonism were defined by FIC indices of ≤0.5 and >4, respectively. An FIC index result of >0.5 but ≤4 was considered indifferent (18).
BBR efflux is FLC independent but associates with the drug efflux pump Cdr2p.
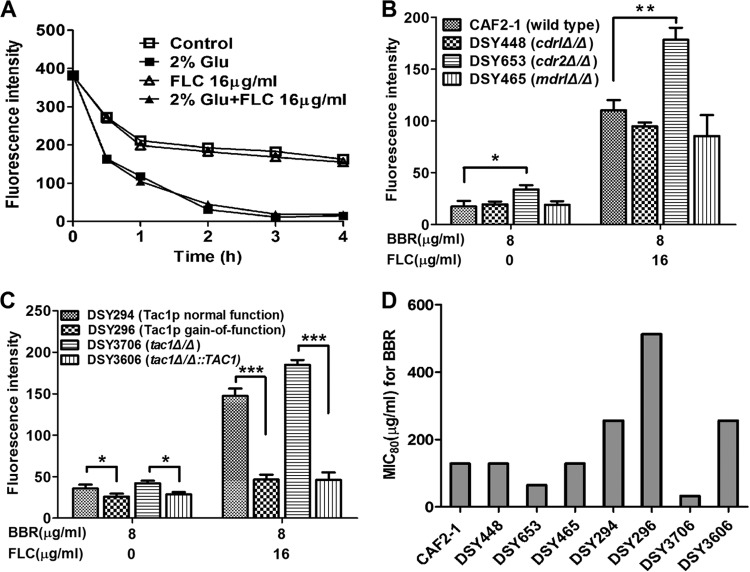
To evaluate the hypothesis that FLC may inhibit the efflux of intracellular BBR, increasing the intracellular BBR concentration (29), we studied the efflux of intracellular BBR. Our results indicated that FLC had no effect on the efflux of intracellular BBR (Fig. 8A). More specifically, we treated C. albicans strains with 16 μg/ml of FLC and 8 μg/ml of BBR for 8 h to obtain high intracellular BBR accumulation. Subsequently, the C. albicans cells were resuspended with RPMI 1640 medium containing 16 μg/ml of FLC or control medium without FLC. The efflux of intracellular BBR was monitored over time. The results showed that the addition of FLC did not influence the efflux of BBR, as the decreases of the fluorescence intensity were almost the same in groups with or without FLC (Fig. 8A). However, we were interested to find that the efflux of BBR in C. albicans was strongly glucose dependent. As shown in Fig. 8A, the fluorescence intensity of intracellular BBR decreased about 50% from the initial value in 2 h without glucose and remained almost unchanged thereafter. However, it continued to decrease to almost none with the addition of 2% glucose (Fig. 8A).
Fig 8.
(A) Efflux of intracellular BBR. Strains were treated with 16 μg/ml of FLC and 8 μg/ml of BBR for 8 h in RPMI 1640 medium, washed twice with PBS, and then resuspended with RPMI 1640 medium at 5 × 107 cells/ml. With the addition of 2% glucose (Glu) or 16 μg/ml of FLC, BBR fluorescence measurement was performed with an Infinite 200 Universal microplate reader at 405-nm excitation and 520-nm emission wavelengths over time. (B and C) Intracellular BBR accumulation of different strains. Strains were treated with different concentrations of FLC and BBR, and fluorescence intensity was measured at 8 h. Statistical significance among groups was determined by analyses of variance. Comparison between two groups was performed by Student t test. The data are shown as means ± SDs from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) Susceptibilities of different C. albicans strains to BBR. The strains included CAF2-1 (wild type), DSY448 (cdr1Δ/Δ), DSY653 (cdr2Δ/Δ), DSY465 (mdr1Δ/Δ), DAY294 (Tac1p normal-function strain), DAY296 (Tac1p gain-of-function mutant), DAY3706 (tac1Δ/Δ), and DAY3606 (tac1Δ/Δ::TAC1).
Based on the findings above, we developed the hypothesis that the efflux of BBR might be related to the ABC (ATP binding cassette) family of transporters, which play efflux roles in an energy-dependent manner. We used mutant stains with deletions involving the ABC family transporter genes, including DSY448 (cdrlΔ/Δ) and DSY653 (cdr2Δ/Δ) (30). Also, we used strain DSY465 (mdrlΔ/Δ), with a deletion involving the MFS (major facilitator superfamily) transporter gene MDR1. The results showed that after treatment with 8 μg/ml of BBR, the fluorescence intensity of intracellular BBR was almost the same within the cdrlΔ/Δ and mdrlΔ/Δ strains and the wild-type strain CAF2-1, while the fluorescence intensity within the cdr2Δ/Δ strain was significantly higher than that in the wild-type strain (P < 0.05) (Fig. 8B), and findings were similar when 16 μg/ml of FLC was added (P < 0.01) (Fig. 8B). To verify the role of Cdr2p in BBR efflux, we used another CDR2-overexpressing strain, DAY296, in which the CDR2 overexpression is caused by the gain of function of its transcription factor Tac1p (31). The BBR fluorescence intensity within DAY296 (Tac1p gain-of-function mutant) was significantly lower than that in the control strain DAY294 (Tac1p normal-function strain) (Fig. 8C). Consistently, the BBR fluorescence intensity within DAY3606 (tac1Δ/Δ::TAC1) was significantly lower than that in control strain DAY3706 (tac1Δ/Δ) (Fig. 8C). Similar results were obtained in both the BBR monotreatment group and the BBR-plus-FLC treatment group (Fig. 8C). Moreover, checkerboard microdilution assays were performed to investigate the susceptibility of these mutants to BBR. In accordance with our prediction, DSY653 (cdr2Δ/Δ) was more susceptible to BBR than the wild-type control strain CAF2-1. Consistently, DAY296 was more resistant than DAY294, and DAY3606 was more resistant than DAY3706 upon BBR treatment (Fig. 8D). In contrast, the susceptibility of DSY448 and DSY465 to BBR was the same as that of the control wild-type strain CAF2-1 (Fig. 8D). These results indicated that Cdr2p, but neither Cdr1p nor Mdr1p, played a role in the efflux of BBR.
DISCUSSION
Previously we have carried out some studies comparing the antifungal effect of FLC and BBR in combination relative to their stand-alone activity and revealed that the concomitant use of BBR and FLC exhibited synergism in killing FLC-resistant C. albicans in vitro (17), and the production of ROS contributed to the synergism of FLC and BBR (19). We also observed that BBR and FLC did not exhibit synergism against FLC-sensitive strains (see Table S4 in the supplemental material), and it seems that FLC-BBR synergism is relevant to FLC resistance. To determine if the synergistic mechanism was relevant to the resistance mechanisms of C. albicans, we screened quite a lot of FLC-resistant strains with different resistance mechanisms. FLC and BBR exhibited synergism against all the strains tested (17). Thus, it seemed that the synergism is relevant to FLC resistance, rather than a specific resistance mechanism. Based on these findings, we assumed that with the FLC-sensitive strains, FLC monotreatment at a low dose was effective enough and the FLC-BBR synergism was not exhibited. In contrast, with FLC-resistant strains, FLC monotreatment at a low dose was not effective, and the FLC-BBR synergism was revealed easily.
In this study, we further investigated the synergistic mechanism of FLC and BBR against FLC-resistant C. albicans. Our results demonstrated that BBR had a major antifungal effect by causing cell cycle arrest and DNA damage. The time-kill curves showed that the synergistic effect of the two drugs was dependent on the concentration of BBR rather than FLC. Confocal scanning microscopy showed that intracellular BBR accumulated especially in the nucleus. Flow cytometric analysis and expression profile microarray showed that the cell cycle was arrested and the expression of cell cycle- and DNA metabolic process-related genes changed significantly, indicating that cell cycle arrest and DNA damage contribute to the synergism of FLC and BBR. In the subsequent study, we found that FLC could increase the intracellular BBR concentration. Transmission electron microscopy showed that a low concentration of FLC was able to cause modest damage to the cell membrane, which probably promoted the ingression of BBR. In addition, we found that the efflux of BBR was strongly glucose dependent and was related to the drug efflux pump Cdr2p.
In this study, our results indicated that BBR had a major antifungal effect on the synergism of FLC and BBR by causing cell cycle arrest and DNA damage. BBR exhibits multiple biological and pharmacological effects, including antimicrobial and antitumor activities (16, 32), and the cytotoxic effect of BBR has been studied extensively (16, 33–35). It was reported that BBR could bind DNA to affect DNA replication, transcription, and the cell cycle (36, 37). In the present study, we found that intracellular BBR accumulated especially in the nucleus and caused cell cycle arrest of C. albicans, which made us form the hypothesis that cell cycle arrest and DNA damage may contribute to the synergism of FLC and BBR against FLC-resistant C. albicans. Meanwhile, the expression profile microarray showed that the expression of cell cycle-related genes, including SAC7, orf19.1185, orf19.4906, and orf19.5457, was reduced, while DNA metabolic process-related genes, including orf19.7234 and orf19.5569, were overexpressed after FLC-BBR combination treatment. SAC7 is a cell cycle-related gene, and its homologous gene in Saccharomyces cerevisiae is related to actin filament reorganization (38). Orthologs of orf19.1185 have ubiquitin-protein ligase activity and play a role in establishment of mitotic spindle orientation (CGD; http://www.candidagenome.org/). orf19.7234 encodes a putative chromatin remodeling complex component and is related to G1/S transition of the mitotic cell cycle (39). The overexpression of orf19.7234 indicated DNA damage and mitosis inhibition (40). And orf19.5569 is involved in the maintenance of ribosomal DNA (rDNA) (CGD). The microarray results were in accordance with the results of flow cytometric analysis and confirmed that cell cycle arrest and DNA damage contribute to the synergism of FLC and BBR. In addition, all DNA intercalators used in this study could synergize with FLC against FLC-resistant C. albicans, which further supported our conclusion that cell cycle arrest and DNA damage contribute to FLC-BBR synergism.
Interestingly, the microarray results showed that the expression of energy metabolism-related genes, including COX9, orf19.4727, GPM2, QCR7, orf19.2067, GIT2, LSC1, and HXT5, changed significantly. COX9, orf19.4727, QCR7, and LSC1 encode a putative subunit VIIa of cytochrome c oxidase, succinate dehydrogenase, ubiquinol-cytochrome c reductase, and succinate coenzyme A (succinate-CoA) ligase subunit, respectively, and all of them play roles in electron transfer in the mitochondrial respiratory chain (41). GPM2 encodes putative phosphoglycerate mutase, which is involved in glycolysis (42). The changed expression of these genes was consistent with our previous findings that mitochondrial aerobic respiration shift and endogenous ROS augmentation contributed to the synergistic action of FLC and BBR (19).
Of note, we also performed another expression profile microarray to find out gene expression changes in response to 16-μg/ml BBR monotreatment. The results showed that only two genes (PRX1 and orf19.3451) were in a consistent pattern for all the three strains tested (0304103, 01010, and 632), indicating that BBR monotreatment caused rather limited and unspecific gene expression changes (GEO series accession number GSE43662).
In this study, we also found that FLC could cause modest damage to the cell membrane of drug-resistant C. albicans and increase the intracellular BBR concentration. BBR has been reported as an isoquinoline alkaloid with poor bioavailability (43). Some studies have demonstrated that the weak antibacterial effect of BBR was mainly due to the low concentration of intracellular BBR, and agents which could promote the ingression of BBR were able to enhance the antibacterial effect of BBR (29, 44–46). In this study, we postulated that FLC could promote the ingression of BBR. FLC targets the fungal cell membrane (47). In agreement with the study by Iwazaki et al. (48), our findings indicated that FLC could cause a modest damage to the cell membrane of some FLC-resistant C. albicans strains. It can be inferred that by causing cell membrane damage, FLC increased the intracellular concentration of BBR, resulting in an enhanced effect against FLC-resistant C. albicans. In addition, we found that other cell membrane-targeted antifungal drugs, including KCZ, ICZ, and AMB, could synergize with BBR against drug-resistant C. albicans, while the antifungal agents not acting on the cell membrane, including 5-FC and CAS, did not synergize with BBR, which further supported our postulation that FLC promotes the ingression of BBR.
Interestingly, we found that the efflux of BBR was related to the drug efflux pump Cdr2p. Some previous studies demonstrated that BBR exhibited relatively weak antibiotic properties due to some drug efflux pumps (29, 49, 50). In Staphylococcus aureus, for example, the MFS (major facilitator superfamily) pump NorA could efflux structurally diverse antibacterial agents like BBR (51), and the NorA inhibitor 5′-methoxyhydnocarpin (5′-MHC) could strongly potentiate the action of BBR (29). In this study, we found that the efflux of BBR was not related to the major MFS pump MDR1 but was strongly glucose dependent and was related to the ABC family efflux pump Cdr2p in C. albicans.
Since the combination of FLC and BBR exhibited synergism in vitro, we would like to investigate their in vivo antifungal effect. Previous studies have shown serious toxicity of BBR when it was given intravenously (52). For drug discovery, we have been investigating the use of BBR and FLC together in the treatment of superficial fungal infections. Our preliminary results indicate that the azole-BBR combination treats superficial fungal infection more effectively than azole monotherapy and has advantages in preventing relapses (unpublished data). We hope that this ongoing project will prove helpful in clinical practice.
In conclusion, we revealed that BBR plays a major antifungal role in the synergism of FLC and BBR. BBR accumulated in C. albicans cells, especially in the nucleus, where it binds with DNA, resulting in cell cycle arrest and DNA damage. Meanwhile, FLC is able to increase the intracellular BBR concentration by causing modest damage to the cell membrane. The results in this study provide new insights into the synergistic mechanisms of antifungal combinations.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81072678, 21072226, 81273558, and 90913008), the National Key Basic Research Program of China (2013CB531602), the National Science and Technology Major Project of the Ministry of Science and Technology of China (2011ZX09102-002-01), the Shanghai Educational Development Foundation (2007CG51), and the Shanghai Science and Technology Major Project (10431902200).
We thank Dominique Sanglard (Institute of Microbiology, University of Lausanne and University Hospital Center, Lausanne, Switzerland) for providing C. albicans strains CAF2-1, DSY448, DSY465, DSY653, DSY294, DSY296, DSY3606, and DSY3706. We thank David Perlin (Public Health Research Institute, New Jersey Medical School, University of Medicine and Dentistry of New Jersey, Newark, NJ) for providing the caspofungin-resistant C. albicans strains 1105S20, 1105S33, and 1105S36. We also thank Jun Gu from Changhai Hospital (Shanghai, China) for providing the clinical C. albicans strains.
Footnotes
Published ahead of print 23 September 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00499-13.
REFERENCES
- 1. Gudlaugsson O, Gillespie S, Lee K, Vande Berg J, Hu J, Messer S, Herwaldt L, Pfaller M, Diekema D. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37:1172–1177 [DOI] [PubMed] [Google Scholar]
- 2. Wilson LS, Reyes CM, Stolpman M, Speckman J, Allen K, Beney J. 2002. The direct cost and incidence of systemic fungal infections. Value Health 5:26–34 [DOI] [PubMed] [Google Scholar]
- 3. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 4. Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kohli A, Smriti Mukhopadhyay K, Rattan A, Prasad R. 2002. In vitro low-level resistance to azoles in Candida albicans is associated with changes in membrane lipid fluidity and asymmetry. Antimicrob. Agents Chemother. 46:1046–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. White TC, Holleman S, Dy F, Mirels LF, Stevens DA. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 46:1704–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. White TC, Marr KA, Bowden RA. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Angiolella L, Stringaro AR, De Bernardis F, Posteraro B, Bonito M, Toccacieli L, Torosantucci A, Colone M, Sanguinetti M, Cassone A, Palamara AT. 2008. Increase of virulence and its phenotypic traits in drug-resistant strains of Candida albicans. Antimicrob. Agents Chemother. 52:927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wilkerson J, McPherson C, Donze A. 2010. Fluconazole to prevent systemic fungal infections in infants: reviewing the evidence. Neonatal Netw. 29:323–333 [DOI] [PubMed] [Google Scholar]
- 10. Niimi M, Firth NA, Cannon RD. 2010. Antifungal drug resistance of oral fungi. Odontology 98:15–25 [DOI] [PubMed] [Google Scholar]
- 11. Gullo A. 2009. Invasive fungal infections: the challenge continues. Drugs 69(Suppl 1):65–73 [DOI] [PubMed] [Google Scholar]
- 12. Wei GX, Xu X, Wu CD. 2011. In vitro synergism between berberine and miconazole against planktonic and biofilm Candida cultures. Arch. Oral Biol. 56:565–572 [DOI] [PubMed] [Google Scholar]
- 13. Kamysz E, Simonetti O, Cirioni O, Arzeni D, Ganzetti G, Campanati A, Giacometti A, Gabrielli E, Silvestri C, Kamysz W, Offidani A, Barchiesi F. 2011. In vitro activity of the lipopeptide PAL-Lys-Lys-NH2, alone and in combination with antifungal agents, against clinical isolates of Candida spp. Peptides 32:99–103 [DOI] [PubMed] [Google Scholar]
- 14. Kiraz N, Dag I, Yamac M, Kiremitci A, Kasifoglu N, Oz Y. 2010. Synergistic activities of three triazoles with caspofungin against Candida glabrata isolates determined by time-kill, Etest, and disk diffusion methods. Antimicrob. Agents Chemother. 54:2244–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karwa R, Wargo KA. 2009. Efungumab: a novel agent in the treatment of invasive candidiasis. Ann. Pharmacother. 43:1818–1823 [DOI] [PubMed] [Google Scholar]
- 16. Tang J, Feng Y, Tsao S, Wang N, Curtain R, Wang Y. 2009. Berberine and Coptidis rhizoma as novel antineoplastic agents: a review of traditional use and biomedical investigations. J. Ethnopharmacol. 126:5–17 [DOI] [PubMed] [Google Scholar]
- 17. Quan H, Cao YY, Xu Z, Zhao JX, Gao PH, Qin XF, Jiang YY. 2006. Potent in vitro synergism of fluconazole and berberine chloride against clinical isolates of Candida albicans resistant to fluconazole. Antimicrob. Agents Chemother. 50:1096–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Odds FC. 2003. Synergy, antagonism, and what the chequerboard puts between them. J. Antimicrob. Chemother. 52:1. 10.1093/jac/dkg301 [DOI] [PubMed] [Google Scholar]
- 19. Xu Y, Wang Y, Yan L, Liang RM, Dai BD, Tang RJ, Gao PH, Jiang YY. 2009. Proteomic analysis reveals a synergistic mechanism of fluconazole and berberine against fluconazole-resistant Candida albicans: endogenous ROS augmentation. J. Proteome Res. 8:5296–5304 [DOI] [PubMed] [Google Scholar]
- 20. Liang RM, Yong XL, Jiang YP, Tan YH, Dai BD, Wang SH, Hu TT, Chen X, Li N, Dong ZH, Huang XC, Chen J, Cao YB, Jiang YY. 2011. 2-amino-nonyl-6-methoxyl-tetralin muriate activity against Candida albicans augments endogenous reactive oxygen species production—a microarray analysis study. FEBS J. 278:1075–1085 [DOI] [PubMed] [Google Scholar]
- 21. CLSI 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts, vol 28, no. 14 Approved standard—third edition; CLSI document M27-A3. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 22. de la Fuente-Salcido N, Salcedo-Hernandez R, Alanis-Guzman MG, Bideshi DK, Barboza-Corona JE. 2007. A new rapid fluorogenic method for measuring bacteriocin activity. J. Microbiol. Methods 70:196–199 [DOI] [PubMed] [Google Scholar]
- 23. Pereira GC, Branco AF, Matos JA, Pereira SL, Parke D, Perkins EL, Serafim TL, Sardao VA, Santos MS, Moreno AJ, Holy J, Oliveira PJ. 2007. Mitochondrially targeted effects of berberine [Natural Yellow 18, 5,6-dihydro-9,10-dimethoxybenzo (g)-1,3-benzodioxolo(5,6-a) quinolizinium] on K1735-M2 mouse melanoma cells: comparison with direct effects on isolated mitochondrial fractions. J. Pharmacol. Exp. Ther. 323:636–649 [DOI] [PubMed] [Google Scholar]
- 24. Sinha R, Islam MM, Bhadra K, Kumar GS, Banerjee A, Maiti M. 2006. The binding of DNA intercalating and non-intercalating compounds to A-form and protonated form of poly(rC)·poly(rG): spectroscopic and viscometric study. Bioorg. Med. Chem. 14:800–814 [DOI] [PubMed] [Google Scholar]
- 25. Teichert MC, Jones JW, Usacheva MN, Biel MA. 2002. Treatment of oral candidiasis with methylene blue-mediated photodynamic therapy in an immunodeficient murine model. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 93:155–160 [DOI] [PubMed] [Google Scholar]
- 26. Giuliana G, Pizzo G, Milici ME, Musotto GC, Giangreco R. 1997. In vitro antifungal properties of mouthrinses containing antimicrobial agents. J. Periodontol. 68:729–733 [DOI] [PubMed] [Google Scholar]
- 27. Oliver G, de Ruiz Holgado AP, Salim R. 1982. Dimorphism in Candida albicans, effect of cycloheximide and acridine orange on germ-tube formation. Mycopathologia 79:43–47 [DOI] [PubMed] [Google Scholar]
- 28. Garcia-Effron G, Park S, Perlin DS. 2011. Improved detection of Candida sp. fks hot spot mutants by using the method of the CLSI M27-A3 document with the addition of bovine serum albumin. Antimicrob. Agents Chemother. 55:2245–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stermitz FR, Lorenz P, Tawara JN, Zenewicz LA, Lewis K. 2000. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. U. S. A. 97:1433–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sanglard D, Ischer F, Monod M, Bille J. 1997. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology 143(Part 2):405–416 [DOI] [PubMed] [Google Scholar]
- 31. MacCallum DM, Coste A, Ischer F, Jacobsen MD, Odds FC, Sanglard D. 2010. Genetic dissection of azole resistance mechanisms in Candida albicans and their validation in a mouse model of disseminated infection. Antimicrob. Agents Chemother. 54:1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vuddanda PR, Chakraborty S, Singh S. 2010. Berberine: a potential phytochemical with multispectrum therapeutic activities. Expert Opin. Investig. Drugs 19:1297–1307 [DOI] [PubMed] [Google Scholar]
- 33. Wu M, Wang J, Liu LT. 2010. Advance of studies on anti-atherosclerosis mechanism of berberine. Chin. J. Integr. Med. 16:188–192 [DOI] [PubMed] [Google Scholar]
- 34. Chou HC, Lu YC, Cheng CS, Chen YW, Lyu PC, Lin CW, Timms JF, Chan HL. 2012. Proteomic and redox-proteomic analysis of berberine-induced cytotoxicity in breast cancer cells. J. Proteomics 75:3158–3176 [DOI] [PubMed] [Google Scholar]
- 35. Boberek JM, Stach J, Good L. 2010. Genetic evidence for inhibition of bacterial division protein FtsZ by berberine. PLoS One 5:e13745. 10.1371/journal.pone.0013745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bhadra K, Kumar GS. 2011. Therapeutic potential of nucleic acid-binding isoquinoline alkaloids: binding aspects and implications for drug design. Med. Res. Rev. 31:821–862 [DOI] [PubMed] [Google Scholar]
- 37. Yadav RC, Kumar GS, Bhadra K, Giri P, Sinha R, Pal S, Maiti M. 2005. Berberine, a strong polyriboadenylic acid binding plant alkaloid: spectroscopic, viscometric, and thermodynamic study. Bioorg. Med. Chem. 13:165–174 [DOI] [PubMed] [Google Scholar]
- 38. Novick P, Osmond BC, Botstein D. 1989. Suppressors of yeast actin mutations. Genetics 121:659–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249–1260 [DOI] [PubMed] [Google Scholar]
- 40. Neely KE, Workman JL. 2002. The complexity of chromatin remodeling and its links to cancer. Biochim. Biophys. Acta 1603:19–29 [DOI] [PubMed] [Google Scholar]
- 41. Lenaz G, Genova ML. 2007. Kinetics of integrated electron transfer in the mitochondrial respiratory chain: random collisions vs. solid state electron channeling. Am. J. Physiol. Cell Physiol. 292:C1221–C1239. 10.1152/ajpcell.00263.2006 [DOI] [PubMed] [Google Scholar]
- 42. Swoboda RK, Bertram G, Delbruck S, Ernst JF, Gow NA, Gooday GW, Brown AJ. 1994. Fluctuations in glycolytic mRNA levels during morphogenesis in Candida albicans reflect underlying changes in growth and are not a response to cellular dimorphism. Mol. Microbiol. 13:663–672 [DOI] [PubMed] [Google Scholar]
- 43. Pan GY, Wang GJ, Liu XD, Fawcett JP, Xie YY. 2002. The involvement of P-glycoprotein in berberine absorption. Pharmacol. Toxicol. 91:193–197 [DOI] [PubMed] [Google Scholar]
- 44. Ettefagh KA, Burns JT, Junio HA, Kaatz GW, Cech NB. 2011. Goldenseal (Hydrastis canadensis L.) extracts synergistically enhance the antibacterial activity of berberine via efflux pump inhibition. Planta Med. 77:835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ball AR, Casadei G, Samosorn S, Bremner JB, Ausubel FM, Moy TI, Lewis K. 2006. Conjugating berberine to a multidrug efflux pump inhibitor creates an effective antimicrobial. ACS Chem. Biol. 1:594–600 [DOI] [PubMed] [Google Scholar]
- 46. Samosorn S, Tanwirat B, Muhamad N, Casadei G, Tomkiewicz D, Lewis K, Suksamrarn A, Prammananan T, Gornall KC, Beck JL, Bremner JB. 2009. Antibacterial activity of berberine-NorA pump inhibitor hybrids with a methylene ether linking group. Bioorg. Med. Chem. 17:3866–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Carrillo-Muñoz AJ, Giusiano G, Ezkurra PA, Quindos G. 2006. Antifungal agents: mode of action in yeast cells. Rev. Esp. Quimioter. 19:130–139 [PubMed] [Google Scholar]
- 48. Iwazaki RS, Endo EH, Ueda-Nakamura T, Nakamura CV, Garcia LB, Filho BP. 2010. In vitro antifungal activity of the berberine and its synergism with fluconazole. Antonie Van Leeuwenhoek 97:201–205 [DOI] [PubMed] [Google Scholar]
- 49. Amin AH, Subbaiah TV, Abbasi KM. 1969. Berberine sulfate: antimicrobial activity, bioassay, and mode of action. Can. J. Microbiol. 15:1067–1076 [DOI] [PubMed] [Google Scholar]
- 50. Hsieh PC, Siegel SA, Rogers B, Davis D, Lewis K. 1998. Bacteria lacking a multidrug pump: a sensitive tool for drug discovery. Proc. Natl. Acad. Sci. U. S. A. 95:6602–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Borges-Walmsley MI, Walmsley AR. 2001. The structure and function of drug pumps. Trends Microbiol. 9:71–79 [DOI] [PubMed] [Google Scholar]
- 52. Kheir MM, Wang Y, Hua L, Hu J, Li L, Lei F, Du L. 2010. Acute toxicity of berberine and its correlation with the blood concentration in mice. Food Chem. Toxicol. 48:1105–1110 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.